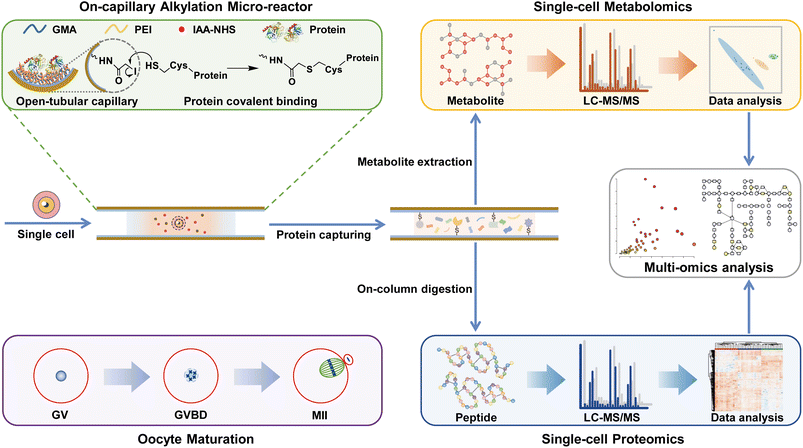
Open Access Article
Open Access ArticleOn-capillary alkylation micro-reactor: a facile strategy for proteo-metabolome profiling in the same single cells†
Yingyun
He
ab,
Huiming
Yuan
*a,
Yu
Liang
a,
Xinxin
Liu
a,
Xiaozhe
Zhang
a,
Yahui
Ji
a,
Baofeng
Zhao
 a,
Kaiguang
Yang
a,
Kaiguang
Yang
 a,
Jue
Zhang
c,
Shen
Zhang
c,
Yukui
Zhang
a and
Lihua
Zhang
a,
Jue
Zhang
c,
Shen
Zhang
c,
Yukui
Zhang
a and
Lihua
Zhang
 *a
*a
aCAS Key Laboratory of Separation Science for Analytical Chemistry, National Chromatographic Research and Analysis Center, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China. E-mail: huimingyuan@dicp.ac.cn; lihuazhang@dicp.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cClinical Research Center for Reproduction and Genetics in Hunan Province, Reproductive and Genetic Hospital of CITIC-XIANGYA, Changsha 410013, China
First published on 14th November 2023
Abstract
Single-cell multi-omics analysis can provide comprehensive insights to study cell-to-cell heterogeneity in normal and disease physiology. However, due to the lack of amplification technique, the measurement of proteome and metabolome in the same cell is challenging. Herein, a novel on-capillary alkylation micro-reactor (OCAM) was developed to achieve proteo-metabolome profiling in the same single cells, by which proteins were first covalently bound to an iodoacetic acid functionalized open-tubular capillary micro-reactor via sulfhydryl alkylation reaction, and metabolites were rapidly eluted, followed by on-column digestion of captured proteins. Compared with existing methods for low-input proteome sample preparation, OCAM exhibited improved efficiency, anti-interference ability and recovery, enabling the identification of an average of 1509 protein groups in single HeLa cells. This strategy was applied to single-cell proteo-metabolome analysis of mouse oocytes at different stages, 3457 protein groups and 171 metabolites were identified in single oocytes, which is the deepest coverage of proteome and metabolome from single mouse oocytes to date, achieving complementary characterization of metabolic patterns during oocyte maturation.
Introduction
Single-cell analysis has the unique ability to characterize cell-to-cell heterogeneity caused by stochastic expression of genes, proteins, and metabolites.1 This is particularly important for analyzing cells with highly dynamic and heterogeneous subpopulations, such as early embryonic development.2 Single-cell RNA sequencing (scRNAseq) has emerged as a powerful method for the study of cellular heterogeneity by determining genome-wide regulatory networks.3 However, the abundance of mRNA transcript is only partially correlated with protein abundance,4 and the complexity and dynamics of proteome and metabolome cannot be fully inferred from transcriptomic data.5 To comprehensively understand the link between genotype and phenotype in single cells, it is necessary to characterize cell heterogeneity at all molecular levels,6 especially for proteins and metabolites that are significant for intracellular function and intercellular communication. However, the contents of proteins and metabolites in individual mammalian cells are fairly low and cannot be amplified.7 Therefore, new techniques for achieving such proteo-metabolome analysis in the same single cells are urgently required.In the last few years, the advances in liquid chromatography-mass spectrometry (LC-MS) technology have greatly improved the identification coverage of proteins and metabolites in bulk cells in an unbiased manner.8,9 The main challenge for MS-based single-cell omics analysis lies significantly in sample preparation.10 Therefore, considerable efforts have been made to develop low-input sample preparation protocols.11–13 For single-cell proteomics, Zhu et al. for the first time developed nanodroplet processing platform (nanoPOTs),14 by which all sample preparation steps could be performed in a nano-well with a total volume of only 200 nL, and achieved an average of over 1000 protein identifications from single HeLa cells.15 Subsequently, various capillary column or chip-based micro-reactors16–18 have been developed as promising alternatives to improve the efficiency and recovery of sample preparation through minimizing reaction volume and eliminating sample transfer, by which 50–1200 protein identifications could be achieved from single HeLa cells. However, most of these methods are not compatible with strong extraction reagents such as sodium dodecyl sulfate (SDS), which might affect the efficiency of protein extraction, especially for hydrophobic proteins. For single-cell metabolomics, capillary probe micro-sampling has been typically used to extract metabolites from cells of interest.19 Patch-clamp technique enabled simultaneous monitoring of metabolic and electrophysiological state of single neurons in the mouse brain.20 Recently, double capillary micro-sampling coupled with ultrasensitive capillary electrophoresis (CE)-MS was developed for performing proteo-metabolome analysis of Xenopus laevis embryos at the cleavage stage.21 It enabled the detection of 150 metabolites (57 identified) and 738 proteins from aspirated <5% of cell volume (10 nL). However, considering the picoliter (pL) volume of a single mammalian cell, which contains trace analytes with a wide dynamic range, it is a great challenge to separate proteins and metabolites from the same cell. More recently, an automated picoliter extraction system was developed for tandem multi-omics sampling from the same single cells, enabling the detection of 651 proteins, 524 phospholipids, and 216 metabolites in a single cell.22 However, considering the overlapping polarity distribution of different components, it would be actually hard to achieve complete separation of metabolites, phospholipids, and proteins in single cells by tandem solvent extraction technology due to the lack of effective sorbents. Furthermore, the direct detection of protein digests by MS without chromatographic separation might affect the coverage and accuracy of protein identification due to the complexity of proteome.
Alternatively, the Oligo-EG modified microwell-chip was prepared for hydrophilic interaction liquid chromatography (HILIC)-based tandem extraction of metabolites and proteins, achieving the identification of 132 compounds and more than 1200 proteins in single cells.18 However, due to the weak interaction between proteins and the hydrophilic surface of the microwell-chip, it might still result in protein loss when extracting metabolites by organic solvent, especially for hydrophobic proteins. Therefore, chemically covalent immobilization of proteins onto functionalized materials may be a better solution.
In this work, we developed an on-capillary alkylation micro-reactor (OCAM) for “one-pot” sample preparation to achieve proteo-metabolome profiling in the same single cells. By the OCAM strategy illustrated in Scheme 1, the sample processing including cell lysis, protein reduction and covalent binding was firstly performed in a single step, and then high concentration organic solvent was introduced to elute metabolites, followed by on-column digestion of captured proteins. This method was used for parallel measurement of proteome and metabolome in the same single mouse oocytes during oocyte maturation (Scheme 1), enabling the identification of 3457 protein groups and 171 metabolites. To the best of our knowledge, this is the deepest coverage of proteome and metabolome from single mouse oocytes. By quantitative proteo-metabolome analysis of single oocytes at different stages, comprehensive characteristics of distinct metabolic patterns during oocyte maturation were revealed.
Results and discussion
Design and preparation of OCAM
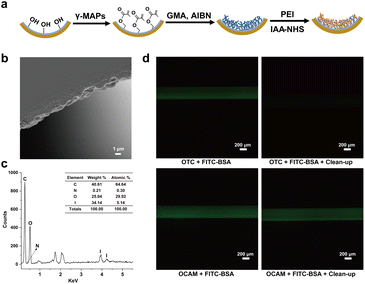
In contrast to conventional bulk sample processing, it is difficult to isolate proteins and metabolites in single cells by traditional organic solvent precipitation or centrifugal filtering.23,24 Herein, we develop an on-capillary alkylation micro-reactor (OCAM), by which proteins are first covalently bound onto the inner surface of the capillary via chemical reaction, then the unreacted metabolites are eluted by high concentration organic solvent, and finally on-column protein digestion is performed for bottom-up proteome analysis. As shown in Fig. 1a, in situ polymerization of glycidyl methacrylate (GMA) was carried out in a capillary with 2,2′-azobisisobutyronitrile (AIBN) as the initiator. The effect of reaction temperature on the morphology of the open tubular capillary column (OTC) was firstly investigated. It could be seen from Fig. 1b and S1a† that a homogeneous and wrinkled polymer layer tightly coated on the inner surface of the capillary could be obtained at 60 °C. However, with the reaction temperature increased to 70 °C, an inhomogeneous multilayer structure was observed due to excessive polymerization (Fig. S1b†). The batch-to-batch preparation reproducibility of OTC was also evaluated, and the thickness of the polymer layer could achieve 2.2 ± 0.6 μm (n = 3), demonstrating good reproducibility of OTC preparation (Fig. S2†).With OTC as the support, OCAM was developed by grafting polyetherimide (PEI) on the inner surface via epoxy ring opening reaction, followed by modification with iodoacetic acid N-succinimidyl iodoacetate (IAA-NHS). In this synthesis protocol, the branched chains of PEI can be used as spacer-arm to reduce steric hindrance for protein immobilization. In addition, the abundant primary, secondary, and tertiary amino groups on PEI were introduced onto the inner surface of OTC, not only to increase the amount of reaction sites with IAA-NHS, but also to decrease the nonspecific adsorption of hydrophobic interfering substances. More importantly, such flexible polymer chains with three-dimensional (3D) nanostructure can increase the interaction between proteins, thus improving the binding capacity of proteins. Energy dispersive spectrometer (EDS) was used to evaluate the elemental composition of the inner surface of OCAM. As shown in Fig. 1c, the atomic content of I could reach 5 mol%, demonstrating the successful modification of IAA-NHS. To further verify the binding mechanism of protein on OCAM, FITC-labeled BSA dissolved in 50 mM tris(2-carboxyethyl) phosphine (TCEP) solution was injected into bare OTC and OCAM, respectively. After incubation at 95 °C for 5 min, both of them were flushed with 50% (v/v) methanol to remove unreacted proteins, followed by characterization with fluorescence microscope. Compared to bare OTC, FITC-labeled BSA was still strongly retained on the inner surface of OCAM after extensive washing (Fig. 1d), which was attributed to the covalent binding of proteins via sulfhydryl reaction.
Furthermore, to determine the protein binding capacity, 1.0 mg mL−1 BSA dissolved in 5 mM TCEP solution was denatured and reduced, and then pumped into OCAM (200 μm i.d. × 110 cm). The concentration of residual BSA in the flow-through (30 μL) was analyzed by the Bradford Protein Assay Kit. It was calculated that the amount of BSA bound onto OCAM was 16 ng cm−1. Given that a single mammalian cell contains ∼0.15 ng of proteins,25 it is sufficient for 1 cm capillary micro-reactor to process 100 or fewer cells in a limited volume of 300 nL.
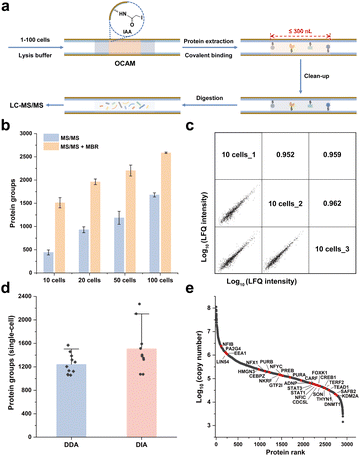
OCAM for proteome analysis of 1–100 mammalian cells
The workflow of OCAM coupled with nanoLC-MS/MS for proteome analysis of 1–100 cells was shown in Fig. 2a. Briefly, the lysis solution composed of SDS and TCEP, and the diluted cell solution containing different number of cells were aspirated sequentially into OCAM to form a “sandwich” structure, followed by ultra-sonication to accelerate cell lysis (Fig. S3†). Protein reduction and covalent binding were performed in a single step by incubation at 95 °C. After extensive washing with 50% (v/v) methanol to remove SDS, the captured proteins were on-column digested, followed by nanoLC-MS/MS analysis. To maintain low sample loss, the above-mentioned sample pretreatment processes were carried out in a single micro-reactor without any sample transfer.To achieve sufficient protein reduction and covalent binding in a single step, we firstly optimized the reaction time with 100 cells as the sample. As shown in Fig. S4a,† the number of identified proteins after 2 h incubation was comparable to that achieved with 5 min, and no bias towards the abundance range of identified proteins was observed (Fig. S4b†). These results indicated that the sample preparation procedures including protein denaturation, reduction and binding could be completed within 5 min, which might be mainly attributed to the greatly enhanced collision possibility of reactive molecules in the micro-scale confined space, thereby improving the efficiency of chemical reactions. Furthermore, to improve the digestion efficiency by eliminating most of missed cleavages in typical trypsin digests,26 we applied the combination of trypsin and Lys-C proteases with a fixed ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (m m−1), and evaluated a series of enzyme-to-protein ratios (2
1 (m m−1), and evaluated a series of enzyme-to-protein ratios (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, m m−1). For 20 ng HeLa cell lysate, although the number of protein identification was comparable at different enzyme-to-protein ratios, the lowest missed cleavage rate (0.38% ± 0.1%) and the highest peptide identification (5737 ± 201) could be achieved at the enzyme-to-protein ratio of 2
10, m m−1). For 20 ng HeLa cell lysate, although the number of protein identification was comparable at different enzyme-to-protein ratios, the lowest missed cleavage rate (0.38% ± 0.1%) and the highest peptide identification (5737 ± 201) could be achieved at the enzyme-to-protein ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. S5†), which was chosen in the following experiments, beneficial for improving the accuracy and precision of label-free proteome quantification.
1 (Fig. S5†), which was chosen in the following experiments, beneficial for improving the accuracy and precision of label-free proteome quantification.
Under the optimal conditions for sample preparation by OCAM, the proteome analysis for 10–100 HeLa cells was performed by nanoLC-MS/MS analysis using Q-Exactive mass spectrometer. As shown in Fig. 2b, 442 ± 54, 931 ± 64, 1187 ± 141 and 1680 ± 44 protein groups (n = 3) were identified by MS/MS from 10, 20, 50 and 100 HeLa cells, respectively. As the quality control, only 43 proteins were identified in the blank samples, demonstrating the negligible carry-over produced from OCAM-based sample preparation and nanoLC-MS/MS system. Using the match between runs (MBR) function, the identified protein number could be boosted by 242%, 111%, 86%, and 54% for 10, 20, 50 and 100 cells, respectively. Furthermore, the label-free quantitative (LFQ) reproducibility of this workflow was also evaluated. Pairwise analysis of any two 10-cell samples showed a Pearson correlation coefficient of 0.95–0.96 (Fig. 2c), and a higher correlation coefficient of 0.97–0.98 was observed for 100 cells (Fig. S6†), indicating that highly reproducible proteome analysis of small population cells was achieved.
Importantly, the OCAM method offers several advantages over other proteomic processing strategies for mass-limited samples. First, this method is compatible with detergents and chaotropic reagents such as SDS, which can be easily removed by washing with methanol. Second, due to the strong covalent interaction between proteins and OCAM, this method can efficiently minimize sample loss during extensive washing. Compared to recently reported Nano3 method,27 we noticed a significant increase of protein groups from 10 HeLa cells (1511 ± 110 vs. 983 ± 292) and 100 HeLa cells (2587 ± 15 vs. 1473 ± 169) identified by our method (Fig. S7a†). We further performed Gene Ontology cellular component (GOCC) analysis on the identified proteins. Notably, more membrane proteins (1.5 folds) could be identified by our method compared to the Nano3 method (Fig. S7b†), including some low-abundance membrane proteins such as GTF3C1.
To further investigate the capacity for processing single-cell samples, the front end of OCAM was pulled into a tip (20–40 μm i.d.) to aspirate single cells under the microscope. After processed by OCAM, the single-cell samples were further analyzed by Orbitrap Exploris 480 mass spectrometer coupled with the high-field asymmetric ion mobility spectrometry (FAIMS) Pro interface. As shown in Fig. 2d, 1243 ± 175 (n = 11) protein groups were identified from single HeLa cells using data-dependent acquisition (DDA) with MBR (Table S1†). Notably, the proteome coverage of single HeLa cells could be further improved using data-independent acquisition (DIA) strategy, and an average identification of 1509 ± 395 (n = 10) protein groups was achieved, which was 1.6 times more than those identified by the Oligo-EG modified microwell chip18 (Fig. S8a†). Furthermore, it was also observed that more hydrophobic proteins were identified using OCAM compared to the Oligo-EG-chip (Fig. S8b†), which was attributed to the improved efficiency and recovery of proteome sample preparation achieved by OCAM through chemical immobilization.
To estimate the abundance of quantified proteins in single cells, we calculated the copy numbers and the distributions of protein abundances of 2905 proteins in single cells were shown in Fig. 2e. The dynamic range of protein expression within single-cells was over 5 orders of magnitude, with a median copy number of 1.46 × 105 copies per cell, and 28 transcription factors were confidently identified by comparing the identified proteins with a reported transcription factor database.28 We also identified 41 proteins with the abundances below 1 × 104 copies per cell in single-cells, among which the abnormal spindle-associated protein ASPM had the lowest abundance of 1433 copies per cell. All these results demonstrated that high identification sensitivity could be achieved by OCAM.
OCAM for single-cell proteome profiling of mouse oocyte maturation
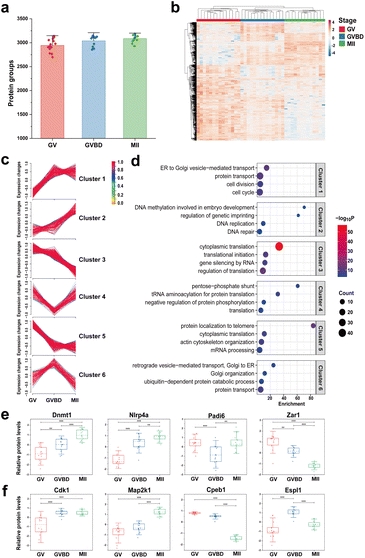
To provide the comprehensive understanding of cellular heterogeneity in oocyte maturation, in our study, 41 single mouse oocytes at three key stages during meiotic maturation, including 14 GV (germinal vesicle) oocytes, 14 GVBD (germinal vesicle breakdown) oocytes, and 13 MII (metaphase II) oocytes, were processed with OCAM, followed by DIA strategy for proteome quantification of single mouse oocytes. A total of 3457 protein groups were quantified (Table S2†), with an average of 2944, 3039, and 3085 proteins from single oocytes at GV, GVBD and MII stage, respectively (Fig. 3a), which greatly exceeded those achieved by the previously reported methods. For example, Li et al. for the first time identified 355 proteins from single mouse oocytes by employing the OAD chip.17 Recently, an efficient and simplified single-cell proteomics (ES-SCP) workflow was developed to realize the identification of more than 1500 proteins from single oocytes,29 which was a great advance in this field. More recently, Huang et al. conducted multi-omics analysis of mouse oocytes, including single-cell proteomics, translatomics and transcriptomics, and detected 1200 proteins per single oocyte.30To ensure the reliability of our quantitative analysis results, proteins identified in more than 90% samples of each group were retained for further analysis. Unsupervised hierarchical cluster analysis (HCA) revealed 1009 significant proteins (ANOVA test, p < 0.01) leading to the distinction of three stages (Table S3†), as shown in Fig. 3b. By using soft clustering analysis based on the fuzzy c-means algorithm,31 these differentially expressed proteins (DEPs) were further clustered into six groups with distinct patterns of expression kinetics (Fig. 3c). Cluster 1 and 2 consisted of 292 (29%) proteins with upregulated trends, whereas cluster 3 and 5 represent 508 proteins that have decreased expression, suggesting that more proteins (51%) were downregulated during oocyte meiosis (Fig. S9a†). Moreover, cluster 4 and 6 correspond to proteins (20%) that were transiently reduced or enriched only at the GVBD stage. GO analysis of DEPs revealed unique biological processes and functional features of each cluster (Fig. 3d). The cell cycle-related proteins (Cdk1, Plk1, Pik3c3, Kif23, Eif2ak4) displayed an upregulated tendency from the GV to MII stage. Our data showed that the expression of a large number of proteins involved in Golgi transportation was also increased, such as vesicle-fusing ATPase and syntaxin-binding proteins (Nsf, Scfd1, Scfd2, Stx5, Stx12), suggesting that vesicle-mediated transport is active during oocyte maturation. In addition, compared to immature oocytes, MII oocytes showed enhanced expression of proteins related to DNA replication and repair process. We also found that some epigenetic factors involved in DNA methylation (Dnmt1, Trim28, Zfp57) and histone modification (Rbbp7, Wdr5, Otub1) were upregulated in the MII oocytes, playing an important role in transcription regulation and early embryonic development.32
Notably, we identified many oocyte-specific proteins (ten Nlrp family proteins) and maternal effect proteins (such as Zar1, Ooep, Tle6, Npm2) that accumulate during oocyte maturation to support early embryo development.33 Among them, 38 differential maternal proteins were identified according to the published database.34 We noticed that the majority of maternal proteins (55%) were classified into cluster 1 and 2 (Fig. S9b†), such as Dnmt1 and Nlrp4a, which gradually increased in meiosis (Fig. 3e). We further mapped the DEPs to Kyoto Encyclopedia Genes and Genomes (KEGG) pathways, and proteins associated with the oocyte meiosis pathway showed distinct expression patterns (Fig. 3f). For example, the mitogen-activated protein kinase (Map2k1) and cyclin-dependent kinase (Cdk1) were upregulated during oocyte maturation, whereas the cytoplasmic polyadenylation element-binding protein (Cpeb1) was degraded for proper entry into the MII stage.35 We also observed upregulation of the separase Espl1 at the GVBD stage, which is responsible for cohesion degradation and chromosome segregation.36
It has been reported that fully grown GV oocytes become transcriptionally silent during the period before meiosis resumption until after fertilization.37 Among the differential proteins during oocyte maturation, we found 59 ribosomal proteins, the majority (98%) of which were downregulated (Fig. S9c†), consistent with the enrichment of mRNA processing and cytoplasmic translation of downregulated proteins in maturing oocytes. Interestingly, there was an alternative mechanism of translational regulation with a different temporal pattern in cluster 4. Although overall protein synthesis decreases during meiosis, active translation is also required for spindle formation and MII progression.38 We noticed that the expression of translation initiation factor Eif2a was significantly increased at the MII stage, which is essential for initiating protein synthesis by delivering initiator methionyl tRNA to the ribosome.39 In particular, we found that most aminoacyl-tRNA synthetases (Sars1, Wars1, Hars1, Kars1, Tars1) were upregulated in MII oocytes, suggesting that eIF2a-mediated tRNA aminoacylation contribute to the regulation of translation to support oocyte maturation. Together, these results indicated that oocyte maturation is a complex multifactorial regulatory process, and provided crucial insights into the proteome dynamics during oocyte maturation.
Single-cell multi-omics reveals distinct metabolic patterns during oocyte maturation
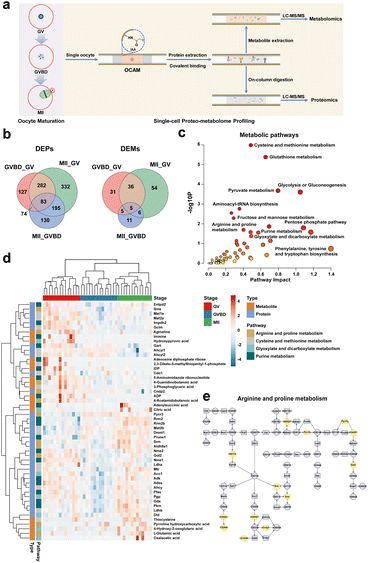
Due to the fact that OCAM only captures proteins via specific interaction with sulfhydryl groups, it could be used for simultaneous analysis of proteome and metabolome from the same single mouse oocytes. As shown in Fig. 4a, after cell lysis and protein covalent binding, 80% (v/v) acetonitrile was introduced into OCAM to elute metabolites, followed by on-column digestion of captured proteins. Finally, the metabolites and peptides collected from the same oocyte were subjected for LC-MS/MS analysis, respectively. Therefore, in our study, in addition to the proteome profiling of single mouse oocytes as mentioned above, we also analyzed the metabolites from the same oocytes. To improve the reliability of metabolite identification from single cells, we first performed background subtraction to eliminate matrix interferences by comparing with the features detected in the blank samples. To further eliminate the variations arising from sample preparation and LC-MS analysis, we also prepared reference samples containing a mixture of equal amounts of metabolite samples from GV, GVBD and MII oocytes, and analyzed them repeatedly under the same conditions used in the single-cell analyses. The data quality was further controlled by filtering out features with RSD > 20%, as reported in the large-scale metabolomics studies.40According to the criteria mentioned above, a total of 171 compounds were identified from single mouse oocytes (Table S4†), among which 111 differential metabolites during meiotic maturation were identified (Table S5†), based on the ANOVA test (p < 0.05) coupled with the variable importance in projection (VIP) analysis (VIP > 1.00). To better understand the proteomic and metabolomic differences, the differentially expressed proteins (DEPs) and differentially expressed metabolites (DEMs) between distinct stages were further analyzed (p < 0.05, |fold change| > 1.5), as shown in Fig. 4b. Both proteomic and metabolomic features showed the dramatic changes between GV and MII oocytes. However, GVBD and MII oocytes had the lowest number of DEMs, which reflected that these two stages are relatively closer to each other at the metabolome level, emphasizing the differences in proteome and metabolome profiles during oocyte maturation.
We further performed the integrated analysis of metabolomic and proteomic data to characterize the metabolic patterns of oocyte maturation. We mapped all differential proteins and metabolites between the three stages to KEGG pathways, revealing significant enrichment of distinct metabolic processes, including glutathione metabolism, glycolysis or gluconeogenesis, and various amino acid metabolism (Fig. 4c). Notably, some metabolic pathways (such as pentose phosphate pathway) were only represented by proteins, whereas other pathways were jointly represented by proteins and metabolites, such as arginine and proline metabolism, cysteine and methionine metabolism, purine metabolism and glyoxylate and dicarboxylate metabolism (Table S6†). This highlighted the benefits of complementary detection of proteins and metabolites. The HCA heatmap revealed notable quantitative proteo-metabolomic changes between the three stages (Fig. 4d), including 18 metabolites and 34 proteins involved in the representative metabolic pathways. Although 41 single oocytes were closely clustered into three groups, we noticed that there were two GVBD oocytes located between GV and MII oocytes, but closer to the MII stage. This may be related to the heterogeneity of individual oocytes within the same group, which cannot be discerned by bulk analysis.
To understand the characteristics of distinct metabolic patterns under multi-omic dimensions, we focused on the pathways jointly represented by proteins and metabolites. Arginine metabolism has been reported to play a critical role in embryonic development.41 Arginine and proline can be metabolized to produce creatine and polyamines, constituting the major pathway for polyamine synthesis.42 While previous work has shown that polyamines are essential for cytoplasmic maturation of the Xenopus oocyte,43 little is known about the effects of polyamines in mammalian oogenesis. As shown in Fig. 4e, ornithine decarboxylase (Odc1) catalyses the decarboxylation of L-ornithine to yield putrescine (C00134), which is further transformed into spermidine and spermine by spermidine synthase (Srm) and spermine synthase (Sms), respectively. Our results exhibited that the enzymes (Srm and Sms) involved in polyamine synthesis were upregulated in MII oocytes. We also found that the key enzyme (Pycr3) and metabolite (pyrroline hydroxycarboxylic acid, C04281) related to hydroxyproline synthesis showed significant accumulation during oocyte maturation. These results indicated the enhanced metabolism of arginine and proline during mouse oocyte meiosis.
Purine and pyrimidine nucleotides are important energy carriers and precursors for the synthesis of nucleotide cofactors.44 For the de novo synthesis of purine, the activated ribose-5-phosphate (R5P) is transformed into inosine monophosphate (IMP), then converted into guanosine or adenosine monophosphate, and eventually phosphorylated into nucleoside di- or tri-phosphates (such as ADP and ATP).45 Our data revealed that the abundances of some metabolites (ADP, IDP, inosine) involved in purine metabolism were significantly decreased during oocyte maturation, and several enzymes responsible for purine metabolism (Nme1, Entpd2, Gart) were downregulated in matured oocytes, which is consistent with the changes in metabolites (Fig. 4d). Moreover, adenylosuccinate synthase (Adss) was reported to participate in the maintenance of meiotic arrest in mouse oocytes,46 and we noticed the level of Adss was significantly reduced during meiotic resumption, and then increased in MII oocytes. Our results reflected the diversity of regulatory trends in nucleotide metabolism. Together, these results illustrated that complementary detection of proteomics and metabolomics in the same single cell is beneficial to comprehensive characterization of metabolic patterns in oocyte maturation.
Conclusions
In this study, we developed a novel on-capillary alkylation micro-reactor (OCAM) that greatly streamlines the processing procedures commonly used in single-cell proteomics and metabolomics. In contrast to existing methods of sample preparation, there are several advantages to using OCAM for single-cell analysis. First, proteins were covalently captured on the inner wall of the capillary micro-reactor. Therefore, it is easy to isolate proteins and metabolites from the same single cell. Second, the sample processing including cell lysis, protein reduction and covalent binding could be completed in a single step, and the entire processing procedures were performed in a small volume without any sample transfer, thus significantly improving the recovery of sample preparation. Third, the capillary micro-reactor could be integrated online with nanoLC-MS/MS system, beneficial to improve the detection sensitivity of trace sample. Furthermore, except for proteo-metabolome profiling, it can also be scalable to parallel measurement of proteome and transcriptome from the same single cells, beneficial for studying system biology at single cell level. To further improve the analysis throughput, an integrated platform for single-cell multi-omics analysis with combination of online single-cell processing, multi-plex isobaric barcoding, and ultra-fast LC separation will be established. With these advances, we anticipate that this strategy would become a valuable tool for better understanding cellular heterogeneity and deciphering molecular mechanism of oocyte maturation.Data availability
The mass spectrometry data reported in this study have been deposited to the ProteomeXchange Consortium via the JPOST repository47 with the dataset identifier PXD039345 and PXD045491.Author contributions
Huiming Yuan, Lihua Zhang and Yukui Zhang designed and supervised the research. Yingyun He, Xinxin Liu and Yu Liang conducted the experiments. Yingyun He, Huiming Yuan, Xiaozhe Zhang, Baofeng Zhao and Kaiguang Yang collected and analyzed the MS data. Yahui Ji, Jue Zhang and Shen Zhang collected the single HeLa cells and mouse oocytes. Yingyun He, Huiming Yuan and Lihua Zhang wrote the manuscript with contribution from all co-authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21974136, 21991083 and 32088101) and the Talent Innovation Support Program of Dalian (2019CT07).Notes and references
- D. Wang and S. Bodovitz, Trends Biotechnol., 2010, 28, 281–290 CrossRef CAS PubMed.
- X. Xu, J. Wang, L. Wu, J. Guo, Y. Song, T. Tian, W. Wang, Z. Zhu and C. Yang, Small, 2020, 16, e1903905 CrossRef.
- E. Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar, M. Goldman, I. Tirosh, A. R. Bialas, N. Kamitaki, E. M. Martersteck, J. J. Trombetta, D. A. Weitz, J. R. Sanes, A. K. Shalek, A. Regev and S. A. McCarroll, Cell, 2015, 161, 1202–1214 CrossRef CAS.
- C. Vogel and E. M. Marcotte, Nat. Rev. Genet., 2012, 13, 227–232 CrossRef CAS PubMed.
- E. K. Neumann, T. D. Do, T. J. Comi and J. V. Sweedler, Angew Chem. Int. Ed. Engl., 2019, 58, 9348–9364 CrossRef CAS PubMed.
- T. M. J. Evers, M. Hochane, S. J. Tans, R. M. A. Heeren, S. Semrau, P. Nemes and A. Mashaghi, Anal. Chem., 2019, 91, 13314–13323 CrossRef CAS PubMed.
- X. Chen, J. C. Love, N. E. Navin, L. Pachter, M. J. Stubbington, V. Svensson, J. V. Sweedler and S. A. Teichmann, Nat. Biotechnol., 2016, 34, 1111–1118 CrossRef CAS PubMed.
- T. Cajka and O. Fiehn, Anal. Chem., 2016, 88, 524–545 CrossRef CAS.
- S. S. Thakur, T. Geiger, B. Chatterjee, P. Bandilla, F. Frohlich, J. Cox and M. Mann, Mol. Cell. Proteomics, 2011, 10, M110 003699 CrossRef.
- N. Slavov, Curr. Opin. Chem. Biol., 2021, 60, 1–9 CrossRef CAS.
- C. S. Hughes, S. Foehr, D. A. Garfield, E. E. Furlong, L. M. Steinmetz and J. Krijgsveld, Mol. Syst. Biol., 2014, 10, 757 CrossRef PubMed.
- N. A. Kulak, G. Pichler, I. Paron, N. Nagaraj and M. Mann, Nat. Methods, 2014, 11, 319–324 CrossRef CAS PubMed.
- Y. Li, H. Yuan, M. Cheng, X. Zhu, K. Yang, W. Zhang, Z. Sui, C. Zhang, L. Zhang and Y. Zhang, Sci. Bull., 2022, 67, 1628–1631 CrossRef CAS PubMed.
- Y. Zhu, G. Clair, W. B. Chrisler, Y. Shen, R. Zhao, A. K. Shukla, R. J. Moore, R. S. Misra, G. S. Pryhuber, R. D. Smith, C. Ansong and R. T. Kelly, Angew Chem. Int. Ed. Engl., 2018, 57, 12370–12374 CrossRef CAS.
- Y. Cong, K. Motamedchaboki, S. A. Misal, Y. Liang, A. J. Guise, T. Truong, R. Huguet, E. D. Plowey, Y. Zhu, D. Lopez-Ferrer and R. T. Kelly, Chem. Sci., 2021, 12, 1001–1006 RSC.
- X. Shao, X. Wang, S. Guan, H. Lin, G. Yan, M. Gao, C. Deng and X. Zhang, Anal. Chem., 2018, 90, 14003–14010 CrossRef CAS PubMed.
- Z. Y. Li, M. Huang, X. K. Wang, Y. Zhu, J. S. Li, C. C. L. Wong and Q. Fang, Anal. Chem., 2018, 90, 5430–5438 CrossRef CAS PubMed.
- Y. Li, H. Li, Y. Xie, S. Chen, R. Qin, H. Dong, Y. Yu, J. Wang, X. Qian and W. Qin, Anal. Chem., 2021, 93, 14059–14067 CrossRef CAS PubMed.
- T. Fujii, S. Matsuda, M. L. Tejedor, T. Esaki, I. Sakane, H. Mizuno, N. Tsuyama and T. Masujima, Nat. Protoc., 2015, 10, 1445–1456 CrossRef CAS PubMed.
- H. Zhu, N. Wang, L. Yao, Q. Chen, R. Zhang, J. Qian, Y. Hou, W. Guo, S. Fan, S. Liu, Q. Zhao, F. Du, X. Zuo, Y. Guo, Y. Xu, J. Li, T. Xue, K. Zhong, X. Song, G. Huang and W. Xiong, Cell, 2018, 173, 1716–1727 CrossRef CAS PubMed.
- C. Lombard-Banek, J. Li, E. P. Portero, R. M. Onjiko, C. D. Singer, D. O. Plotnick, R. Q. Al Shabeeb and P. Nemes, Angew Chem. Int. Ed. Engl., 2021, 60, 12852–12858 CrossRef CAS PubMed.
- P. Zhao, Y. X. Feng, J. H. Wu, J. W. Zhu, J. L. Yang, X. X. Ma, Z. Ouyang, X. R. Zhang, W. P. Zhang and W. H. Wang, Anal. Chem., 2023, 95, 7212–7219 CrossRef CAS.
- C. Coman, F. A. Solari, A. Hentschel, A. Sickmann, R. P. Zahedi and R. Ahrends, Mol. Cell. Proteomics, 2016, 15, 1453–1466 CrossRef CAS PubMed.
- F. Guan, X. Xiang, Y. Xie, H. Li, W. Zhang, Y. Shu, J. Wang and W. Qin, Anal. Methods, 2021, 13, 1930–1938 RSC.
- P. Volpe and T. Eremenko-Volpe, Eur. J. Biochem., 1970, 12, 195–200 CrossRef CAS PubMed.
- S. Saveliev, M. Bratz, R. Zubarev, M. Szapacs, H. Budamgunta and M. Urh, Nat. Methods, 2013, 10, i–ii CrossRef.
- Z. Yang, Z. Zhang, D. Chen, T. Xu, Y. Wang and L. Sun, Anal. Chem., 2021, 93, 10568–10576 CrossRef CAS.
- S. A. Lambert, A. Jolma, L. F. Campitelli, P. K. Das, Y. Yin, M. Albu, X. Chen, J. Taipale, T. R. Hughes and M. T. Weirauch, Cell, 2018, 172, 650–665 CrossRef CAS.
- Q. Li, L. Mu, X. B. Yang, G. Wang, J. Liang, S. L. Wang, H. Zhang and Z. Li, J. Proteome Res., 2023, 22, 2067–2078 CrossRef CAS PubMed.
- J. Huang, P. Chen, L. Jia, T. Li, X. Yang, Q. Liang, Y. Zeng, J. Liu, T. Wu, W. Hu, K. Kee, H. Zeng, X. Liang and C. Zhou, Adv. Sci., 2023, 10, 2301538 CrossRef CAS.
- M. E. Futschik and B. Carlisle, J. Bioinf. Comput. Biol., 2005, 03, 965–988 CrossRef CAS.
- S. F. Wang, Z. H. Kou, Z. Y. Jing, Y. Zhang, X. Z. Guo, M. Q. Dong, I. Wilmut and S. R. Gao, Proc. Natl. Acad. Sci. U.S.A., 2010, 107, 17639–17644 CrossRef CAS.
- J. Zhang, Y. Liu, W. Yao, Q. Li, H. Liu and Z. Pan, Reproduction, 2018, 156, 23–33 CAS.
- P. Zhang, X. J. Ni, Y. Guo, X. J. Guo, Y. F. Wang, Z. M. Zhou, R. Huo and J. H. Sha, BMC Genomics, 2009, 10, 348 CrossRef PubMed.
- D. Setoyama, M. Yamashita and N. Sagata, Proc. Natl. Acad. Sci. U.S.A., 2007, 104, 18001–18006 CrossRef CAS PubMed.
- M. Kucej and H. Zou, Nucleus, 2010, 1, 4–7 CrossRef PubMed.
- Y. Q. Su, K. Sugiura, Y. Woo, K. Wigglesworth, S. Kamdar, J. Affourtit and J. J. Eppig, Dev. Biol., 2007, 302, 104–117 CrossRef CAS PubMed.
- A. Susor, D. Jansova, M. Anger and M. Kubelka, Cell Tissue Res., 2016, 363, 69–84 CrossRef CAS PubMed.
- B. A. Castilho, R. Shanmugam, R. C. Silva, R. Ramesh, B. M. Himme and E. Sattlegger, Biochim. Biophys. Acta, Mol. Cell Res., 2014, 1843, 1948–1968 CrossRef CAS.
- J. Godzien, V. Alonso-Herranz, C. Barbas and E. G. Armitage, Metabolomics, 2015, 11, 518–528 CrossRef CAS.
- G. Y. Wu, F. W. Bazer, M. C. Satterfield, X. L. Li, X. Q. Wang, G. A. Johnson, R. C. Burghardt, Z. L. Dai, J. J. Wang and Z. L. Wu, Amino Acids, 2013, 45, 241–256 CrossRef CAS PubMed.
- S. M. Morris, J. Nutr., 2004, 134, 2743s–2747s CrossRef CAS PubMed.
- Y. Zhou, C. Q. Ma, J. Karmouch, H. A. Katbi and X. J. Liu, Mol. Cell. Biol., 2009, 29, 1786–1795 CrossRef CAS.
- B. Roy, A. Depaix, C. Perigaud and S. Peyrottes, Chem. Rev., 2016, 116, 7854–7897 CrossRef CAS PubMed.
- J. P. Dewulf, S. Marie and M. C. Nassogne, Mol. Genet. Metab., 2022, 136, 190–198 CrossRef CAS.
- S. M. Downs, Biol. Reprod., 1994, 50, 1403–1412 CrossRef CAS PubMed.
- S. Okuda, Y. Watanabe, Y. Moriya, S. Kawano, T. Yamamoto, M. Matsumoto, T. Takami, D. Kobayashi, N. Araki, A. C. Yoshizawa, T. Tabata, N. Sugiyama, S. Goto and Y. Ishihama, Nucleic Acids Res., 2017, 45, D1107–D1111 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc05047e |
| This journal is © The Royal Society of Chemistry 2023 |