Open Access Article
Open Access ArticleGraphitic carbon nitride with Cu2+ and triazole group co-doping for enhanced peroxidase-like activity and its application for glutathione detection†
Xiaotao
Liu
,
Xueyi
Zheng
,
Chunqiu
Xia
and
Liangqia
Guo
 *
*
Ministry of Education Key Laboratory for Analytical Science of Food Safety and Biology, Fujian Provincial Key Laboratory of Analysis and Detection Technology for Food Safety, College of Chemistry, Fuzhou University, Fuzhou 350116, China. E-mail: lqguo@fzu.edu.cn
First published on 23rd June 2023
Abstract
Nanozymes have attracted great interest in the field of biotechnology, (bio)sensing, and environmental monitoring. Herein, a co-doping strategy is proposed to enhance the peroxidase-like activity of graphitic carbon nitride (g-C3N4). Cu2+ and triazole group co-doped graphitic carbon nitride (g-C3N5-Cu2+) was derived from thermal polymerization of 3-amino-1,2,4-triazole and then coordination with Cu2+. g-C3N5-Cu2+ nanosheets (NSs) could catalyze the oxidation reaction of 3,3′,5,5′-tetramethylbenzidine (TMB) by H2O2 under neutral conditions, yielding 10.9-fold better catalytic activity than g-C3N4 NSs. Reactive oxygen species scavenging experiments indicated that ˙OH played a crucial role during the catalytic process. When glutathione (GSH) was introduced, the absorbance of the system decreased because of the reduction of the oxidation product of TMB by GSH. The colorimetric method demonstrated a sensitive response for GSH with a linear range of 0.8–33.3 μmol L−1 and a limit of detection of 0.3 μmol L−1. The sensitivity is higher than most other peroxidase mimetic-based colorimetric methods for GSH. Finally, the feasibility for the detection of GSH in HeLa cells was demonstrated.
1. Introduction
Natural enzymes have been extensively applied in the fields of medicine, chemistry, food, environment and agriculture due to their superior catalytic efficiency and substrate specificity.1 Nevertheless, their catalytic activities are susceptible to environmental conditions and their preparation and purification are expensive.2,3 To surmount the shortage of natural enzymes, nanozymes, with unique advantages including high activity and stability, low cost, and easy scaled-up and modification, have provoked wide interest in the fields of biosensing and biotherapy.4–6 Since 2007, when Fe3O4 nanoparticles were first reported to have peroxidase-like activity,7 more and more nanozymes have been discovered, including carbon-based nanomaterials,8 transition metal nanomaterials,9 precious metal nanomaterials, etc.10Graphitic carbon nitrides (g-C3N4), as a carbon-based material, are of particular importance due to their distinctive properties such as high physicochemical stability, tunability of band gaps, nontoxicity and biocompatibility.11–14 Since the first discovery of peroxidase-like activity,15 g-C3N4 as a peroxidase mimetic has attracted more and more attention. Since then, many methods such as modification of gold NPs,16 complexation of precious metal ruthenium ions,17 decoration with Au-Ni bimetallic NPs,18 functionalization of Cu2+ modified carbon dots,19,20 and hybridization of Cu (ref. 21) with g-C3N4 were proposed to improve its peroxidase-like activity. Despite great progress on g-C3N4-based peroxidase mimetics being achieved, the complex and cumbersome synthesis method,16–20 or the utilization of precious metals16–18 was involved.
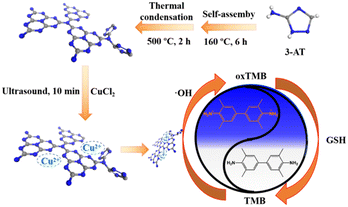
Herein, we proposed a co-doping strategy to enhance the peroxidase-like activity of g-C3N4. Cu2+ and triazole group co-doped g-C3N4 (denoted as g-C3N5-Cu2+) was derived from thermal polymerization of 3-amino-1,2,4-triazole (3-AT) and then coordination with Cu2+. The catalytic activity of the g-C3N5-Cu2+ nanozyme was 10.9-fold greater than that of g-C3N4. The catalytic kinetics and mechanism of the g-C3N5-Cu2+ nanozyme were investigated by steady-state kinetic experiments and reactive oxygen species (ROS) scavenging experiments, respectively. Finally, the g-C3N5-Cu2+ nanozyme was applied for the colorimetric detection of glutathione (GSH) in HeLa cells (Scheme 1).
2. Experimental
2.1. Chemical and materials
3-AT, cupric chloride (CuCl2), NaN3, GSH, methionine (Met), histidine (His), alanine (Ala), glycine (Gly), glutamate (Glu), cystine (Cys) and phenylalanine (Phe) were obtained from Sangon Biotech (Shanghai) Co., Ltd. Hydrogen peroxide (H2O2), disodium oxalate (Na2C2O4), terephthalic acid (TA), thiourea, and 3,3′,5,5′-tetramethylbenzidine (TMB) were bought from Aladdin Co., Ltd. Superoxide dismutase (SOD) was purchased from Worthington Biochemical Corporation. The commercial GSH kit was provided by Beyotime Biotechnology Co., Ltd.2.2. Preparation of g-C3N5-Cu2+ nanosheets
Under N2 atmosphere, 2 g 3-AT was self-assembled at 160 °C for 6 h, and then polymerized at 500 °C for 2 h. After cooling, 0.2 g g-C3N5 powder was treated ultrasonically in 50 mL water for 24 h. The precipitation between 8000 rpm to 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm was collected. After re-dispersion of the precipitate in water, the concentration of g-C3N5 NSs was adjusted to 1.0 mg mL−1. Similarly, g-C3N4 NSs were obtained via thermal polymerization of melamine15 and then ultrasonic exfoliation.
000 rpm was collected. After re-dispersion of the precipitate in water, the concentration of g-C3N5 NSs was adjusted to 1.0 mg mL−1. Similarly, g-C3N4 NSs were obtained via thermal polymerization of melamine15 and then ultrasonic exfoliation.
To 3 mL g-C3N5 NSs (1.0 mg mL−1), 1 mL CuCl2 solution (16 mmol L−1) was added. After ultrasonic treatment for 10 min, free Cu2+ in the mixture was removed by washing with water. The dispersion of g-C3N5-Cu2+ NSs was adjusted to 1.0 mg mL−1. Similarly, g-C3N4-Cu2+ NSs were also prepared.
2.3. Colorimetric detection of GSH
To 200 μL of Tris-HCl buffer (50 mmol L−1, pH 6.5), 50 μL of TMB (10 mmol L−1), 50 μL of g-C3N5-Cu2+ NSs (1.0 mg L−1), 200 μL of H2O2 (400 μmol L−1) and 100 μL of GSH with different concentrations were added. After incubation at 50 °C for 50 min, g-C3N5-Cu2+ NSs was removed from the mixture by centrifugation. The absorption spectrum of the mixture was recorded.3. Results and discussion
3.1. Characterization of g-C3N5-Cu2+ nanosheets
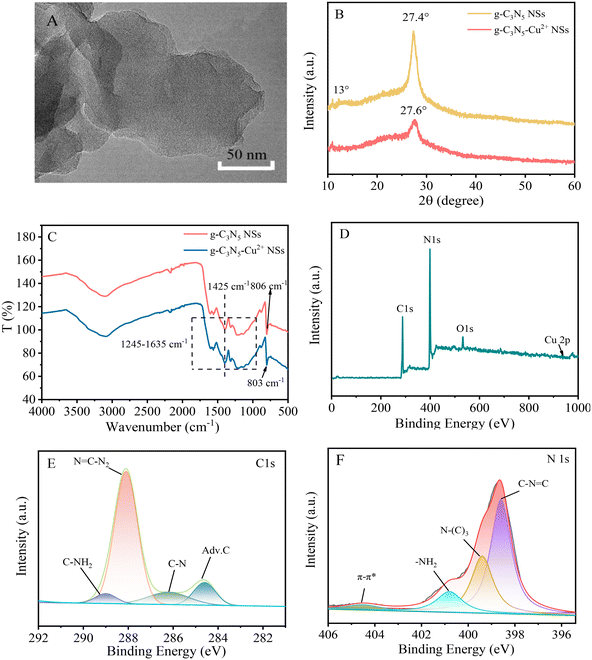
g-C3N5-Cu2+ NSs were derived from thermal polymerization of 3-AT at 500 °C under nitrogen atmosphere and then coordination with Cu2+. 3-AT was chosen as the precursor, which can incorporate triazole groups into g-C3N4 after thermal condensation.22 As shown in the TEM image (Fig. 1A), the lateral size of g-C3N5-Cu2+ NSs is less than 150 nm. The most probable size distribution of g-C3N5-Cu2+ NSs is about 165 nm (Fig. S1A†). In the XRD patterns (Fig. 1B), the peak assigned to the (002) plane of g-C3N5-Cu2+ exhibits a right-shift from 27.4° to 27.6° after doping with Cu2+, which may be caused by the crystal lattice distortion after Cu2+ coordination.23 The diffraction peak at 13.3° disappears, indicating the decreasing size of the conjugate plane.24 In the FT-IR spectra (Fig. 1C), the peaks at 803 cm−1 and 1245–1635 cm−1 feature the typical absorption of (tri-s-)triazine ring and aromatic carbon nitride heterocyclic ring, respectively.25,26 It's worth noting that there is a peak in 1425 cm−1 which belongs to the ring breathing modes of triazole groups.22 The above results indicate that there are triazole groups in the carbon nitride network. As observed from the survey XPS spectrum (Fig. 1D), g-C3N5-Cu2+ NSs are composed of C, N, O and Cu elements. The presence of the O element may be ascribed to the adsorption of H2O and O2 in the air on the surface of the material.27 The atom ratio of C/N in g-C3N5-Cu2+ NSs was calculated to be 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.7, which is higher than the ratio in g-C3N4 NSs.28 Due to the low content of Cu2+, the Cu element does not appear in the survey spectrum. In the C 1s core-level spectrum there are four peaks with binding energies (BEs) of 284.6, 286.1, 288.1, and 289 eV (Fig. 1E), which belong to the adventitious carbon (adv. C), C–N, N
4.7, which is higher than the ratio in g-C3N4 NSs.28 Due to the low content of Cu2+, the Cu element does not appear in the survey spectrum. In the C 1s core-level spectrum there are four peaks with binding energies (BEs) of 284.6, 286.1, 288.1, and 289 eV (Fig. 1E), which belong to the adventitious carbon (adv. C), C–N, N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N2, and C–NH2, respectively. There are three peaks with BEs of 398.6, 400, 404.8 eV in the N 1s core-level spectrum (Fig. 1F), which belong to C–N
C–N2, and C–NH2, respectively. There are three peaks with BEs of 398.6, 400, 404.8 eV in the N 1s core-level spectrum (Fig. 1F), which belong to C–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N–(C)3, and the heterocyclic compound charge effect or positive charge delocalization (π–π*), respectively. In the Cu 2p core-level XPS spectrum of g-C3N5-Cu2+ NSs (Fig. S1B†), there are three peaks at 932.3 eV, 942.0 eV and 952.3 eV, corresponding to Cu 2p3/2, satellite and Cu 2p1/2 characteristic peaks of Cu2+,29 indicating the adsorption of Cu2+ in g-C3N5 NSs. The energy-dispersive spectroscopy (EDS) mapping in Fig. S1C–F† further confirms that the C, N, O, Cu atoms are well distributed in the frameworks of g-C3N5-Cu2+ NSs; the mass of Cu in g-C3N5-Cu2+ NSs is about 1.4% by ICP-MS. For comparison, g-C3N4-Cu2+ NSs were also characterized in Fig. S2 and S3.† The morphology and chemical components of g-C3N4-Cu2+ NSs are similar with g-C3N5-Cu2+ NSs except for the absence of triazole groups.
C, N–(C)3, and the heterocyclic compound charge effect or positive charge delocalization (π–π*), respectively. In the Cu 2p core-level XPS spectrum of g-C3N5-Cu2+ NSs (Fig. S1B†), there are three peaks at 932.3 eV, 942.0 eV and 952.3 eV, corresponding to Cu 2p3/2, satellite and Cu 2p1/2 characteristic peaks of Cu2+,29 indicating the adsorption of Cu2+ in g-C3N5 NSs. The energy-dispersive spectroscopy (EDS) mapping in Fig. S1C–F† further confirms that the C, N, O, Cu atoms are well distributed in the frameworks of g-C3N5-Cu2+ NSs; the mass of Cu in g-C3N5-Cu2+ NSs is about 1.4% by ICP-MS. For comparison, g-C3N4-Cu2+ NSs were also characterized in Fig. S2 and S3.† The morphology and chemical components of g-C3N4-Cu2+ NSs are similar with g-C3N5-Cu2+ NSs except for the absence of triazole groups.
3.2. Peroxidase-like activity of g-C3N5-Cu2+ nanosheets
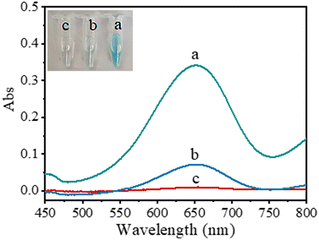
The peroxidase-like activity of g-C3N5-Cu2+ NSs was studied with TMB as the substrate. When TMB was mixed with g-C3N5-Cu2+ NSs and H2O2, the mixed solution turned to a blue color. A typical absorption peak at 652 nm originating from the oxidation product of TMB (oxTMB) is observed (line a Fig. 2a). However, without g-C3N5-Cu2+ NSs or H2O2, the color of the mixed solutions does not change (lines b and c in Fig. 2a). These results indicate that g-C3N5-Cu2+ NSs catalyze the oxidation of TMB by H2O2, revealing their peroxidase-like activity. The effect of the concentration of Cu2+ coordinated with g-C3N5 NSs on the catalytic activity was investigated (Fig. S4†). The catalytic activity of g-C3N5-Cu2+ NSs is enhanced with the increase of Cu2+ concentration and reaches a peak at 4 mmol L−1 Cu2+.To prove the vital role of Cu2+ and triazole groups on the catalytic activity of g-C3N5-Cu2+ NSs, the catalytic activities of g-C3N5-Cu2+, g-C3N4-Cu2+ NSs, g-C3N5 NSs, and g-C3N4 NSs were compared. As shown in Fig. 3, the peroxidase-like activity is in the order of g-C3N5-Cu2+ NSs > g-C3N4-Cu2+ NSs > g-C3N5 NSs > g-C3N4 NSs. The catalytic activity of g-C3N5-Cu2+ NSs is 3.4-fold greater than that of g-C3N4-Cu2+ NSs, 7.2-fold greater than that of g-C3N5 NSs and 10.9-fold greater than that of g-C3N4 NSs. These phenomena indicate that Cu2+ and triazole groups synergistically boost the peroxidase-like activity of g-C3N4 NSs.
3.3. Catalytic kinetics and mechanism
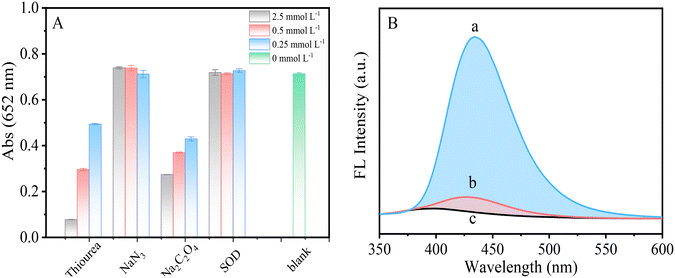
To investigate the catalytic performance of g-C3N5-Cu2+ NSs, the steady-state kinetics of g-C3N5-Cu2+ NSs was investigated (Fig. S5†). The catalytic reactions between g-C3N5-Cu2+ NSs and TMB, and between g-C3N5-Cu2+ NSs and H2O2 followed the typical Michaelis–Menten behavior. The maximal reaction velocity (Vmax) as well as the Michaelis–Menten constant (Km) was deduced from the Lineweaver–Burk plot. The Vmax value of g-C3N5-Cu2+ NSs for the substrate H2O2 was calculated to be 1.8 × 10−6 mol L−1 s and is higher than those of HRP and some other peroxidase mimetics (Table S1†).15,30–32 These results indicated that g-C3N5-Cu2+ NSs showed a higher reaction rate for the substrate H2O2 than those of HRP and some other peroxidase mimetics.The catalytic mechanism of g-C3N5-Cu2+ NSs was further investigated by reactive oxygen species scavengers. Three concentrations of thiourea, NaN3, SOD, and NaC2O4 were used to capture ˙OH, 1O2, ˙O2− and H2O2, respectively.33,34 As shown in Fig. 4A, NaN3 and SOD have almost no effect on the catalytic reaction, indicating that 1O2 and ˙O2− are not produced during the catalytic reaction. However, with the increasing concentration of thiourea and NaC2O4, the absorbance of the system gradually decreased, indicating that ˙OH and H2O2 play a crucial role in the catalytic reaction. To further confirm the production of ˙OH in the system, TA was commonly used to verify the presence of ˙OH (Fig. 4B). ˙OH can oxidize TA to generate strongly fluorescent 2-hydroxyterephthalic acid.35 In the presence of only H2O2, the fluorescence signal is very low. However, the fluorescence signal is significantly enhanced in the presence of both g-C3N5-Cu2+ NSs and H2O2. The results further verified that ˙OH are produced in the systems. From these results we can infer that g-C3N5-Cu2+ NSs catalyze H2O2 to form ˙OH, which oxidizes TMB to oxTMB.
3.4. Colorimetric detection of H2O2 and GSH
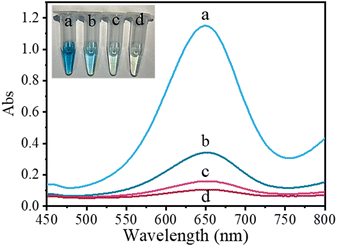
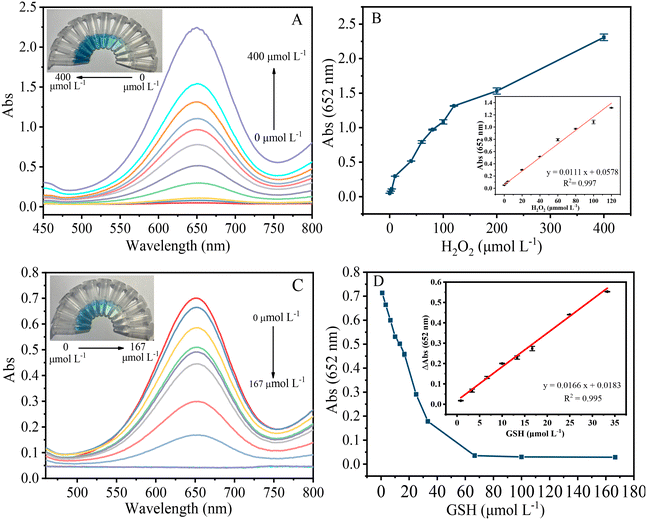
The effects of pH, g-C3N5-Cu2+ NS concentration, TMB concentration, incubation time, and reaction temperature on the absorbance of the system were first investigated. The experimental conditions were optimized at pH 6.5, 50 °C reaction temperature, 0.1 mg mL−1 g-C3N5-Cu2+ NSs and 1.0 mmol L−1 TMB (Fig. S6†). It is worth mentioning that g-C3N5-Cu2+ NSs can maintain relatively high catalytic activity over a wide pH range (2.0 to 6.5), which is beneficial for its application. This high peroxidase-like activity of g-C3N5-Cu2+ NSs over a wide pH may be explained by the triazole groups in g-C3N5-Cu2+ NSs favoring the adsorption of TMB molecules via π–π stacking.Thus, a facile colorimetric method for H2O2 was constructed via the g-C3N5-Cu2+ NSs-TMB system. The absorbance is gradually enhanced with the increase of H2O2 concentration (Fig. 5A). There is a linear range between the absorbance and H2O2 over 0–120 μmol L−1 (Fig. 5B). The limit of detection (LOD) of 0.32 μmol L−1 was achieved by 3σ/k, where σ and k are the standard deviation of 12 repeated detections of the blank solution and the slope of the linear equation, respectively. g-C3N5-Cu2+ NSs were further applied to construct a colorimetric method for GSH via the reduction of oxTMB. The absorbance gradually dropped with the increase of GSH concentration (Fig. 5C). The blue color of the mixed solution gradually fades. The linear detection range for GSH over 0.8–33.3 μmol L−1 is obtained (Fig. 5D). The LOD for GSH is calculated to be 0.25 μmol L−1 by 3σ/k. As shown in Table S2,† the LOD is lower than those colorimetric methods catalyzed by other peroxidase mimetics such as carbon nanodots,36 CuS–polydopamine–Au composites,37 Fe–Ne–C single atom nanozymes,38 carbon nanoparticles,39etc.
3.5. Selectivity toward GSH
To probe the selectivity of this colorimetric method for GSH, the absorbance changes on addition of possible interferents such as Met, His, Ala, Gly, Glu, Cys, and Phe were compared with that of GSH. The concentration of GSH and Cys was set to 16.7 μmol L−1 while other amino acids were set to 10-fold concentrations of GSH. Little absorbance change was observed except for GSH and Cys (Fig. S7†). Although Cys can cause relatively large absorbance changes with GSH, the concentration of Cys (about 60–200 μmol L−1) in cells is much lower than that of GSH (about 1–10 mmol L−1).40 When cellular GSH level was detected, the dilution of cell lysate can overcome the interference caused by Cys.3.6. Application
The colorimetric method was applied for the detection of GSH in HeLa cells. Cell culture and pretreatment details of HeLa cells are provided in the ESI.† The results are listed in Table S3.† The GSH concentration in the lysate of HeLa cells was measured as 4.6 mmol L−1 by the colorimetric method, close to the value (5.2 mmol L−1) assayed by the commercial GSH assay kit (Fig. S8†). This colorimetric method also demonstrated excellent recovery (100.4–103%) and relative standard derivation (0.6–2.1%) for the detection of GSH in cell lysate.4. Conclusions
g-C3N5-Cu2+ NSs were successfully prepared by thermal polymerization of 3-AT and coordination with Cu2+. The catalytic activity of g-C3N5-Cu2+ NSs is 3.4-fold greater than that of g-C3N4-Cu2+ NSs without triazole groups, and 10.9-fold greater than that of g-C3N4. The study of the catalytic mechanism reveals that ˙OH plays a crucial role during the catalytic process. Finally, g-C3N5-Cu2+ NSs were utilized to construct a sensitive and selective detection method for GSH in cell lysate. This study exploited a new strategy to rationally design effective bio-inspired nanozymes. It is envisioned that g-C3N5-Cu2+ NSs hold great potential to replace horseradish peroxidase for chemical sensing and biotechnology applications.Author contributions
Xiaotao Liu: conceptualization, methodology, investigation, and writing – original draft. Xueyi Zheng: validation and investigation. Chunqiu Xia: validation and investigation. Liangqia Guo: conceptualization, funding acquisition, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21874023, 22176034).Notes and references
- G. Wulff, Chem. Rev., 2001, 102, 1–28 CrossRef PubMed.
- A. Ashraf, Z. Bytesnikova, J. Barek, L. Richtera and V. Adam, Biosens. Bioelectron., 2021, 19, 113494 CrossRef PubMed.
- J. Xie, X. Zhang, H. Wang, H. Zheng and Y. Huang, TrAC, Trends Anal. Chem., 2012, 39, 114–129 CrossRef CAS.
- D. Jiang, D. Ni, Z. Rosenkrans, P. Huang, X. Yan and W. Cai, Chem. Soc. Rev., 2019, 48, 3683–3704 RSC.
- J. Wu, X. Wang, Q. Wang, Z. Lou, S. Li, Y. Zhu, L. Qin and H. Wei, Chem. Soc. Rev., 2019, 48(4), 1004–1076 RSC.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119(6), 4357–4412 CrossRef CAS.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577 CrossRef CAS PubMed.
- P. Zhang, D. Sun, A. Cho, S. Weon, S. Lee, J. Lee, J. Han, D. Kim and W. Choi, Nat. Commun., 2019, 10, 940 CrossRef PubMed.
- Y. Sang, Y. Huang, W. Li, J. Ren and X. Qu, Chem. – Eur. J., 2018, 24(28), 7259–7263 CrossRef CAS PubMed.
- Y. Xu, J. Fei, G. Li, T. Yuan, X. Xu and J. Li, Angew. Chem., Int. Ed., 2019, 58(17), 5572–5576 CrossRef CAS PubMed.
- S. Cao, J. Low, J. Yu and M. Jaroniec, Adv. Mater., 2015, 27(13), 2150–2176 CrossRef CAS PubMed.
- Q. Hao, G. Jia, W. Wei, A. Vinu, Y. Wang, H. Arandiyan and B. Ni, Nano Res., 2020, 13, 18–37 CrossRef CAS.
- Z. Zhou, Y. Zhang, Y. Shen, S. Liu and Y. Zhang, Chem. Soc. Rev., 2018, 47, 2298–2321 RSC.
- Y. Dong, Q. Wang, H. Wu, Y. Chen, C. Lu, Y. Chi and H. Yang, Small, 2016, 12(39), 5376–5393 CrossRef CAS PubMed.
- T. Lin, L. Zhong, J. Wang, L. Guo, H. Wu, Q. Guo, F. Fu and G. Chen, Biosens. Bioelectron., 2014, 59, 89–93 CrossRef CAS PubMed.
- N. Wu, Y. Wang, F. Guo, H. Wen, T. Yang and J. Wang, Anal. Chim. Acta, 2019, 1091, 69–75 CrossRef CAS PubMed.
- W. Den, Y. Peng, H. Yang, Y. Tan, M. Ma, Q. Xie and S. Chen, ACS Appl. Mater. Interfaces, 2019, 11(32), 29072–29077 CrossRef PubMed.
- G. Darabdhara, J. Bordoloi, P. Manna and M. Das, Sens. Actuators, B, 2019, 285, 277–290 CrossRef CAS.
- M. González, W. Liao, R. Cazelles, S. Wang, X. Yu, V. Gutkin and I. Willner, ACS Nano, 2017, 11, 3247–3253 CrossRef.
- Q. Li, D. Yang, Q. Yin, W. Li and Y. Yang, ACS Appl. Nano Mater., 2022, 5, 1925–1934 CrossRef CAS.
- T. Dang, N. Heo, H. Cho, S. Lee, M. Song, H. Kim and M. Kim, Microchim. Acta, 2021, 188, 293 CrossRef CAS PubMed.
- G. Man, S. Talapaneni, K. Lakhi, H. Ilbeygi, U. Ravon, K. Bahily, T. Mori, D. Park and A. Vinu, Angew. Chem., Int. Ed., 2017, 56, 848–8485 Search PubMed.
- S. Hu, X. Qu, J. Bai, P. Li, Q. Li, F. Wang and L. Song, ACS Sustainable Chem. Eng., 2017, 5, 6863–6872 CrossRef CAS.
- C. Huang, Y. Wen, J. Ma, D. Dong, Y. Shen, S. Liu, H. Ma and Y. Zhang, Nat. Commun., 2021, 12, 320 CrossRef CAS PubMed.
- Z. Song, T. Lin, L. Lin, S. Lin, F. Fu, X. Wang and L. Guo, Angew. Chem., Int. Ed., 2016, 55, 2773–2777 CrossRef CAS PubMed.
- X. Wang, S. Blechert and M. Antonietti, ACS Catal., 2012, 2, 1596–1606 CrossRef CAS.
- X. Liu, H. Zhang, Z. Cai and L. Guo, Spectrochim. Acta, Part A, 2022, 268, 120685 CrossRef CAS PubMed.
- W. Ong, L. Tian, Y. Ng, S. Yong and S. Chai, Chem. Rev., 2016, 116(12), 7159–7329 CrossRef CAS.
- C. Y. Hu, Z. W. Jiang, C. Z. Huang and Y. F. Li, Microchim. Acta, 2021, 188, 272 CrossRef CAS.
- N. Wu, Y. Wang, X. Wang, F. Guo, H. Wen, T. Yang and J. Wang, Anal. Chim. Acta, 2019, 1091, 69–75 CrossRef CAS PubMed.
- G. Darabdhara, J. Bordoloi, P. Manna and M. Das, Sens. Actuators, B, 2019, 285, 277–290 CrossRef CAS.
- T. Dang, N. Heo, H. Cho, S. Lee, M. Song, H. Kim and M. Kim, Microchim. Acta, 2021, 188, 293 CrossRef CAS PubMed.
- C. Liu, D. Kong, P. Hsu, H. Yuan, H. Lee, Y. Liu, H. Wang, S. Wang, K. Yan and D. Lin, Nat. Nanotechnol., 2016, 11, 1098–1104 CrossRef CAS PubMed.
- C. Lu, F. Zhou, S. Wu, L. Liu and D. Xing, Antioxid. Redox Signaling, 2016, 24, 249–262 CrossRef CAS PubMed.
- Y. Li, T. Li, W. Chen and Y. Song, ACS Appl. Mater. Interfaces, 2017, 9, 29881–29888 CrossRef CAS PubMed.
- M. Shamsipur, A. Safavi and Z. Mohammadpour, Sens. Actuators, B, 2014, 199, 463–469 CrossRef CAS.
- Y. Wang, Y. Liu, F. Ding, X. Zhu, L. Yang, P. Zou, H. Rao, Q. Zhao and X. Wang, Anal. Bioanal. Chem., 2018, 410, 4805–4813 CrossRef CAS PubMed.
- W. Lu, S. Chen, H. Zhang, J. Qiu and X. Liu, J. Materiomics, 2022, 23, 52–8478 Search PubMed.
- L. Chen, X. Li, Z. Li, K. Liu and J. Xie, RSC Adv., 2022, 12, 595–601 RSC.
- Y. Yue, F. Huo, P. Ning, Y. Zhang, J. Chao, X. Meng and C. Yin, J. Am. Chem. Soc., 2017, 139, 3181–3185 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00072a |
| This journal is © The Royal Society of Chemistry 2023 |