Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Metabolic engineering for 4-aminophenylalanine production from lignocellulosic biomass by recombinant Escherichia coli†
Hideo
Kawaguchi
 ab,
Shunsuke
Masuo
ab,
Shunsuke
Masuo
 c,
Keiko
Wakai
a,
Naoki
Takaya
c,
Keiko
Wakai
a,
Naoki
Takaya
 c,
Tomohisa
Hasunma
c,
Tomohisa
Hasunma
 ab,
Tatsuo
Kaneko
ab,
Tatsuo
Kaneko
 d,
Satoshi
Okada
e,
Takashi
Sazuka
d,
Satoshi
Okada
e,
Takashi
Sazuka
 e,
Chiaki
Ogino
e,
Chiaki
Ogino
 *f and
Akihiko
Kondo
afg
*f and
Akihiko
Kondo
afg
aGraduate School of Science, Technology and Innovation, Kobe University, 1-1 Rokkodai, Nada, Kobe 657-8501, Japan
bEngineering Biology Research Center, Kobe University, 1-1 Rokkodai, Nada, Kobe 657-8501, Japan
cFaculty of Life and Environmental Sciences, Microbiology Research Center for Sustainability, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8572, Japan
dEnergy and Environmental Area, Graduate School of Advanced Science and Technology, Japan Advanced Institute of Science and Technology, 1-1 Asahidai, Nomi, Ishikawa 923-1292, Japan
eBioscience and Biotechnology Center, Nagoya University, Furo, Chikusa, Nagoya 464-8601, Japan
fGraduate School of Engineering, Kobe University, 1-1 Rokkodai, Nada, Kobe 657-8501, Japan. E-mail: ochiaki@port.kobe-u.ac.jp; Fax: +81-78-803-6192; Tel: +81-78-803-6193
gBiomass Engineering Research Division, RIKEN, 1-7-22 Suehiro, Turumi, Yokohama, Kanagawa 230-0045, Japan
First published on 27th May 2023
Abstract
To synthesize a high-performance biopolyimide bioplastic from lignocellulosic feedstock, Escherichia coli was metabolically engineered to produce 4-amino-L-phenylalanine (4APhe) as a diamine monomer. A high-biomass sorghum cultivar was used as the model lignocellulosic feedstock, and the enzymatic hydrolysate was used as a substrate for 4APhe production in fed-batch culture. When using the ldh mutant strain, HKE6027, 4APhe production from glucose was increased by over three-fold compared to the parent strain. This increase was due to the disruption of biosynthetic pathways that produce either acetate or lactate as by-products. Comparative metabolomic analysis revealed increased flux in both the pentose phosphate and shikimate pathways in HKE6027, resulting in the highest yields. However, 5.7 g L−1 of 4APhe was produced from enzymatic hydrolysis by HKE6027, which was 24% lower than that obtained from glucose. The concentrations of 14 potential fermentation inhibitors present in the enzymatic hydrolysate were determined, and their inhibitory effects on both cell growth and 4APhe production were examined. The addition of groups of potential inhibitors present in the enzymatic hydrolysate of sorghum bagasse showed that benzaldehyde- and cinnamic acid derivatives inhibited 4APhe fermentation at low concentrations (half-maximum inhibitor concentration IC50 = 16 mg L−1), whereas furfural and 5-hydroxymethylfurfural, which are well-known fermentation inhibitors, inhibited 4APhe fermentation at relatively high concentrations (IC50 = 640 mg L−1). These results provide insight into the design of metabolic pathways tailored for the utilization of lignocellulosic biomass to produce aromatic compounds through the shikimate pathway.
Sustainability spotlightThis study produced a starting material for the synthesis of polyimide from an inedible and renewable feedstock of lignocellulosic biomass using microbial fermentation. This process can produce an organic molecule from plant biomass for CO2 fixation and convert it into a high-performance bioplastic, resulting in the long-term mitigation of greenhouse gas emissions in the industrial sector. Therefore, this study contributes to the circular economy and alternative plastic raw material production for a sustainable society. Furthermore, this work aligns with goals 12 of “Responsible Consumption and Production” and 13 of “Climate Action” of the United Nation's Sustainable Development Goals. |
Introduction
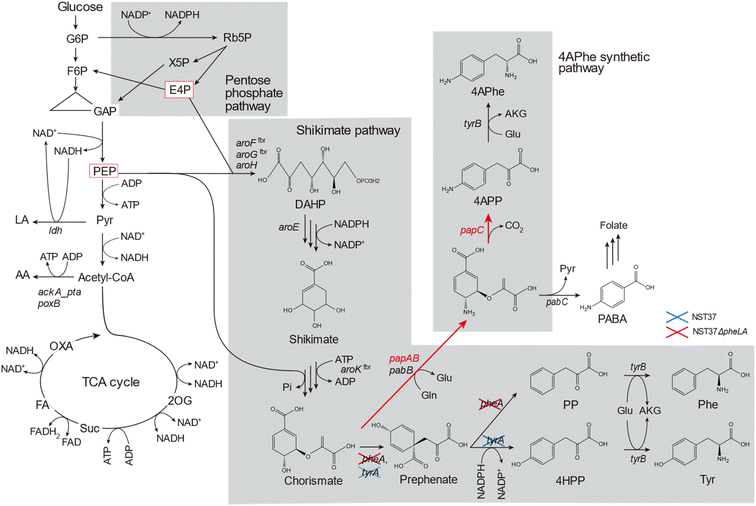
Bioplastics are becoming increasingly popular as alternatives to petroleum-derived plastics due to their bio-based or biodegradable nature.1,2 Bioplastics made from renewable biomasses, such as lignocellulosic feedstocks, have the potential to mitigate carbon dioxide emissions. This is especially important as global plastic production has risen from 1.5 million tons in the 1950s to 335 million tons in 2016.3 Currently, some bioplastics, such as poly(lactic acid) (PLA) and poly(butylene succinate) (PBS) are commercially available; however, the lower limits of thermostability (glass transition temperature, Tg ≤ 70 °C)4 and mechanical strength (tensile strength, σ ≤ 50 MPa)5 of these bio-based aliphatic polyesters compared to those of petroleum-based plastics hinder their extensive utilization. To replace petroleum-based plastics with bioplastics, new molecular structures must be designed to improve the physicochemical limitations of bioplastics.4-Amino-L-phenylalanine (4APhe) is a metabolic intermediate in the biosynthesis of chloramphenicol and pristinamycin I in Streptomyces venezuelae and S. pristinaespiralis, respectively.6,7 In the biosynthesis process involving S. pristinaespiralis, the chorismate metabolic intermediate in the shikimate pathway is converted into 4-aminophenylpyruvate (4APP) in a three-step enzymatic reaction encoded by the papABC genes. The resulting 4APP is then transformed into 4APhe by transaminase7 (Fig. 1). Recently, 4APhe has been used as a diamine monomer to synthesize unique high-performance bio-based polymers, such as gelable polypeptide8 and particulate polyimide.9 These biopolymers exhibit significantly higher thermostability (Tg > 300 °C) compared to PLA and PBS and are comparable to commercially available petroleum-based plastics.
Lignocellulose is mainly composed of cellulose, hemicellulose, and lignin,10 which are renewable and non-edible bioresources that can serve as alternative feedstocks for the synthesis of sustainable plastics in society.1 The hydrolysate of lignocellulosic biomass, which contains glucose released from cellulose, is a preferred carbon source for fermentation. However, this hydrolysate also contains organic contaminants derived from the decomposition of cellulose, hemicellulose, and lignin, which can act as fermentation inhibitors that hinder cell growth and metabolism during fermentation.1,11,12 Although a metabolically engineered Escherichia coli strain expressing papABC derived from Pseudomonas fluorescens was developed and found to successfully produce 4APhe from glucose,13–16 it was not able to produce 4APhe from lignocellulosic biomass (Table 1).
| Strain | Substrate | Process | Titer (g L−1) | Reference |
|---|---|---|---|---|
| Recombinant Escherichia coli | Glucose | Fed-batch | 2.2 | Tateyama et al., (2019)14 |
| Recombinant E. coli | Glucose | Fed-batch | 4.4 | Masuo et al., (2019)16 |
| Recombinant E. coli | Glucose | Fed-batch | 2.9 | Minakawa et al., (2019)15 |
| Recombinant E. coli | Glucose | Fed-batch | 7.4 | This study |
| Recombinant E. coli | Enzymatic hydrolysate of sorghum bagasse | Fed-batch | 5.7 | This study |
In this study, we aimed to produce 4APhe from lignocellulosic biomass. To achieve this, we developed metabolically engineered E. coli strains that can eliminate by-product formation pathways. We compared their metabolic profiles during 4APhe production from glucose to determine the limiting step for 4APhe production (Fig. 1). Using the recombinant strain with the highest 4APhe yield, we performed fed-batch culture for 4APhe production from the enzymatic hydrolysate of pretreated sorghum bagasse as a model lignocellulosic biomass. Additionally, we identified the types of molecules present in the hydrolysate of pretreated sorghum bagasse that inhibit 4APhe fermentation while not affecting cell growth.
Experimental
Microorganisms and media
The bacterial strains and plasmids used in this study are listed in Table 2. For genetic manipulation, E. coli strains were grown at 37 °C in Luria–Bertani (LB) medium.17 The E. coli strain NST37ΔpheLA (DE3), which has feedback-resistant alleles of aroGfbr encoding 3-deoxy-D-arabino-heptulosonate 7-phosphate synthetase and lacks a pheLA locus both in the leader peptide for pheA expression (pheL) and chorismate mutase (pheA),13 was used as a host strain to develop a metabolically engineered strain for 4APhe production. For 4APhe production, modified M9 medium containing the following was used: 6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 10 g (NH4)2SO4, 0.5 g MgSO4·7H2O, 15 mg CaCl2, 50 mg thiamine-HCl, 5.0 g peptone, 2.5 g yeast extract, 20 mg phenylalanine (Phe), 20 mg tyrosine (Tyr), 20 mg tryptophan (Trp), 4 mg ZnSO4·7H2O, 2.2 mg H3BO3, 1 mg MnCl2·4H2O, 1 mg FeSO4·7H2O, 0.32 mg CoCl2·6H2O, 0.32 mg CuSO4·5H2O, 0.22 mg (NH4)6Mo7O24·4H2O, and 10 mg EDTA per liter, supplemented with 40 g L−1 of either glucose or enzymatic hydrolysate of sorghum bagasse as the carbon source, unless otherwise described. When appropriate, media were supplemented with 50 μg mL−1 ampicillin, 50 μg mL−1 streptomycin, and 34 μg mL−1 chloramphenicol for E. coli.| Name | Relevant characteristics | Reference or source |
|---|---|---|
| a 4APhe, 4-amino-L-phenylalanine; DAHP, 3-deoxy-D-arabino-heptulosonate 7-phosphate. | ||
| Strains | ||
| Escherichia coli NST37 (ATCC 31882) | aroF fbr, aroGfbr, tyrA, tyrR, pheAfbr, pheAo, trpE | ATCC |
| E. coli NST(DE3)ΔpheLA | aroF fbr, aroGfbr, tyrA, tyrR, pheAfbr, pheAo, trpE, dcm(DE3)ΔpheLA | Tateyama, et al. (2016)14 |
| HKE1002 | E. coli NST(DE3)ΔpheLA bearing pACYC-aroG4, pET-pfpapA and pCDF-pfpapBC | Masuo, et al. (2016)13 |
| HKE6042 | Mutant of Δldh of strain E. coli NST(DE3)ΔpheLA | This work |
| HKE6021 | Mutant of ΔackA_ptaΔpoxB of strain E. coli NST(DE3)ΔpheLA | This work |
| HKE6054 | Mutant of ΔldhΔackA_ptaΔpoxB of strain E. coli NST(DE3)ΔpheLA | This work |
| HKE6027 | HKE6042 bearing pACYC-aroG4, pET-pfpapA and pCDF-pfpapBC | This work |
| HKE6046 | HKE6021 bearing pACYC-aroG4, pET-pfpapA and pCDF-pfpapBC | This work |
| HKE6057 | HKE6054 bearing pACYC-aroG4, pET-pfpapA and pCDF-pfpapBC | This work |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Plasmids | ||
| pET-pfpapA | Ampr; E. coli vector harboring papA gene derived from Pseudomonas fluorescens for 4APhe biosynthesis | Masuo, et al. (2016)13,14 |
| pCDF-pfpapBC | Smr, E. coli vector harboring papBC genes derived from P. fluorescens for 4APhe biosynthesis | Masuo, et al. (2016)13,14 |
| pACYC-aroG4 | Cmr; E. coli vector harboring the aroG4 gene encoding the feedback-resistant isozyme DAHP synthase | Masuo, et al. (2016)13,14 |
Preparation of the enzymatic hydrolysate of sorghum bagasse
The hybrid sorghum cultivar Tentaka18 was grown in 2020 in an experimental field at Nagoya University, Japan. Whole plants were harvested at the heading stage and fully dried in a greenhouse. After removal of the panicles, the culms were ground into a fine powder using a blender (WB-1; TGK, Hachioji, Japan) fitted with a 2 mm screen. A solid extract of sorghum bagasse was prepared by treating it with dilute acid as described previously.12 For preparing the enzymatic hydrolysate of the pretreated sorghum bagasse, the commercially available cellulase, enzyme blend (Sigma-Aldrich, Burlington, MA, USA) was used following a previous method.12 Briefly, sorghum bagasse was enzymatically hydrolyzed using the cellulase enzyme blend at a load of 50 filter paper units per gram of dry biomass. After 72 h of incubation at 50 °C, the hydrolysate was collected and subjected to centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C, 15 min); the resulting supernatant was aseptically filtered for compositional analysis with 4APhe fermentation as the carbon source.
000g, 4 °C, 15 min); the resulting supernatant was aseptically filtered for compositional analysis with 4APhe fermentation as the carbon source.
DNA manipulation
PrimeSTAR Max DNA Polymerase (Takara Bio, Kusatsu, Japan) was used for polymerase chain reaction (PCR) to amplify DNA fragments, according to the manufacturer's instructions. The PCR fragments were purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). Electroporation was used to transform E. coli, as previously described.19 Plasmid DNA was isolated from E. coli, as per a previously described procedure.20Construction of metabolically engineered E. coli strains for 4APhe production
E. coli strain NST37ΔpheLA(DE)13 (Table 2) was metabolically engineered via homologous recombination to suppress by-product formation by lambda Red recombination21 using a Quick & Easy E. coli Gene Deletion Kit (Gene Bridges GmbH, Heidelberg, Germany) according to the manufacturer's instructions. To inactivate the ldhA and poxB genes and ackA-pta locus, pairs of primers 1 and 2, 5 and 6, and 3 and 4, respectively, were used to generate linear DNA fragments for homologous recombination (Table 3). The resulting deletion mutants of HKE6042, HKE6021, and HKE6054 (Table 2) were transformed by electroporation with the plasmids pACYC-aroG4, pCDF-pfpapBC, and pET-pfpapA for 4APhe biosynthesis (Fig. 1). Transformants were selected based on their resistance to ampicillin, chloramphenicol, and streptomycin and subsequently screened for their ability to produce 4APhe from glucose as the sole carbon source, thereby yielding recombinant strains of HKE6027, HKE6046, and HKE6057 and eliminating the biosynthetic pathway of lactate, acetate, or both of them (Table 2).| Name | Target gene | Sequence (5′–3′)a |
|---|---|---|
| a The homology arm for homologous recombination with the red/ET system is indicated with underlines. | ||
| Primer 1 | ldh | AACGTCGACCTTGACGCGGCAAAAGAACTGGGGCTGAAAGTAGTCCGTG![[T with combining macron]](https://www.rsc.org/images/entities/char_0054_0304.gif) AATTAACCCTCACTAAAGGGCGG AATTAACCCTCACTAAAGGGCGG |
| Primer 2 | ldh | GCAGACAGGCGACGGAATACGTCATCCTGGATCACGTCGTTGGATTTAT![[C with combining macron]](https://www.rsc.org/images/entities/char_0043_0304.gif) TAATACGACTCACTATAGGGCTCG TAATACGACTCACTATAGGGCTCG |
| Primer 3 | ackA_pta | TGGCTCCCTGACGTTTTTTTAGCCACGTATCAATTATAGGTACTTCCATḠAATTAACCCTCACTAAAGGGCGG |
| Primer 4 | ackA_pta | ACGCTCTTTCAGCTGTTCCGGGGTGTCAGTGCCCTGAGACATAACGAAGĀTAATACGACTCACTATAGGGCTCG |
| Primer 5 | poxB | CTCTCTGAACGGTCTTAGTGACAGTCTTAATCGCATGGGCACCATCGAG![[T with combining macron]](https://www.rsc.org/images/entities/char_0054_0304.gif) AATTAACCCTCACTAAAGGGCGG AATTAACCCTCACTAAAGGGCGG |
| Primer 6 | poxB | TATACAGGCTGAAACCTTTGGCCTGTTCGAGTTTGATCTGCGGTGGAATḠTAATACGACTCACTATAGGGCTCG |
4APhe fermentation by recombinant E. coli strains
Comparative 4APhe production using four recombinant E. coli strains was conducted in batch culture with glucose (40 g L−1) as the sole carbon source. Recombinant E. coli strains were grown aerobically until the late log phase in a 10 mL test tube containing LB broth with glucose (10 g L−1) under constant agitation (180 rpm) for 18 h at 30 °C. The pre-culture was inoculated into 90 mL of modified M9 medium containing glucose (40 g L−1) in a 200 mL jar fermentor Bio Jr.8 BJR-25NA1S-8M (ABLE Co. & Biott Co., Tokyo, Japan) to obtain a cell concentration corresponding to an optical density at 600 nm (OD600) of 0.2. The cultivation conditions were as follows: temperature, 30 °C; pH, 7.2, maintained by the addition of ammonia; stirrer speed, 600 rpm; and aeration with compressed air, 1.0 vvm (all unless indicated otherwise). After 5 h of cultivation, 80 μM isopropyl-β-D-1-thiogalactopyranoside (0.8 mM, final concentration) was added to each of the four culture vessels. For the fed-batch culture, either glucose or enzymatic hydrolysate of sorghum bagasse was used as the sole carbon source and added to each culture batch to obtain an initial glucose concentration of 30 g L−1. These carbon sources were added to the culture batches four times in increments of 20 g L−1 glucose during the cultivation period. To identify fermentation inhibitors for 4APhe production, potential fermentation inhibitors detected in the enzymatic hydrolysate of sorghum bagasse were added to the fermentation medium at various concentrations. Modified M9 medium (2 mL) containing glucose as the sole carbon source was added to a Microplate 24 Deep-well (Porvair Science, Norfolk, UK), and the potential inhibitors were added at various concentrations (0 to 1000 mg L−1) before inoculation. Pre-culture (20 μL) was transferred to the medium and incubated under constant agitation (180 rpm) for 28 h at 30 °C in a BioShaker M BR-022UP (TAITEC, Saitama, Japan) after being sealed with the gas-permeable film Breathe-EASIER (Diversifie Biotech, Dedham, MA, USA).Metabolomic analysis
Metabolomic analysis of E. coli cells was conducted as previously described.12 Major metabolites of the central metabolic pathways (e.g., glycolysis, pentose phosphate pathway [PPP], tricarboxylic acid [TCA] cycle, and shikimate pathway) were analyzed using an ion-pair liquid chromatography-tandem mass spectrometry (LC-MS/MS) method.22 Dried cell extracts were dissolved in 50 μL MilliQ water for LC-MS/MS profiling and quantification of 30 intracellular E. coli metabolites. The following metabolites were analyzed: sugar phosphates (glucose-6-phosphate [G6P], fructose-6-phosphate [F6P], fructose-1,6-bisphosphate [FBP], dihydroxyacetone phosphate [DHAP], glyceraldehyde-3-phosphate [GAP], 2- and 3-phosphoglycerate [2PG + 3PG], phosphoenolpyruvate [PEP], 6-phospho-D-gluconate [6PG], ribulose-5-phosphate [Rib5P], ribose-5-phosphate [R5P], xylose-5-phosphate [X5P], erythrose-4-phosphate [E4P], and sedoheptulose-7-phosphate [S7P]); organic acids (aconitate [Aco], citrate [Cit], fumarate [Fum], isocitrate [IsoCit], lactate [LA], malate [Mal], oxaloacetate [OXA], 2-oxoglutarate [AKG], pyruvate, and succinate [Suc]); shikimate derivatives (4-aminobenzoic acid [PABA], 4APhe, 4APP, and shikimate-3-phosphate [Sh3P]); nucleotides (adenosine diphosphate [ADP] and adenosine triphosphate [ATP]); coenzymes (acetyl-CoA [AcCoA], oxidized and reduced nicotinamide adenine dinucleotide [NAD+ and NADH, respectively], oxidized and reduced NAD-phosphate [NADP+ and NADPH, respectively]); and amino acids (glutamate [Glu], glutamine [Gln], Phe, Trp, and Tyr). Metabolites were quantified as previously described23 using an Agilent 1200 series MS and Agilent 6460 with a Jet Stream Technology liquid chromatography with tandem mass spectrometry (LC-MS/MS) system (Agilent Technologies, Waldbronn, Germany) equipped with a Maestro C18 column (2.1 × 150 mm, 3 μm particle size; Shimadzu, Kyoto, Japan). For quantitative metabolomics, E. coli cells were subjected to cold methanol quenching followed by metabolite extraction from the cells, as previously described.12Analytical procedures
Culture samples were centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C, 10 min), and the concentrations of glucose, 4APhe and its derivative PABA, and organic acids (acetate, lactate, and Suc) in the resulting supernatants were measured. Glucose concentration was assayed enzymatically using the Glucose CII test kit (Wako, Osaka, Japan). The concentrations of 4APhe and PABA were determined via high-performance liquid chromatography (HPLC) using an LC-20A apparatus (Shimadzu, Kyoto, Japan) equipped with a COSMOSIL(R) HILIC Packed Column (4.6 mm I.D. × 250 mm; Nacalai Tesque, Tokyo, Japan) and an SPD-M20A diode array detector (Shimadzu). The column oven temperature was set at 30 °C, and the mobile phase was acetonitrile/10 mM ammonium acetate = 70
000g, 4 °C, 10 min), and the concentrations of glucose, 4APhe and its derivative PABA, and organic acids (acetate, lactate, and Suc) in the resulting supernatants were measured. Glucose concentration was assayed enzymatically using the Glucose CII test kit (Wako, Osaka, Japan). The concentrations of 4APhe and PABA were determined via high-performance liquid chromatography (HPLC) using an LC-20A apparatus (Shimadzu, Kyoto, Japan) equipped with a COSMOSIL(R) HILIC Packed Column (4.6 mm I.D. × 250 mm; Nacalai Tesque, Tokyo, Japan) and an SPD-M20A diode array detector (Shimadzu). The column oven temperature was set at 30 °C, and the mobile phase was acetonitrile/10 mM ammonium acetate = 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 (v/v) at a flow rate of 1.25 mL min−1. 4APhe and PABA were detected at wavelengths of 241 and 270 nm, respectively, and the concentrations of 4APhe and PABA were calculated based on standard curves of standard substances. Cell mass was estimated by measuring optical density at 600 nm (OD600) using a spectrophotometer (U-3010; Hitachi, Tokyo, Japan). Suc concentration was determined via HPLC, as described previously.24 To improve the selectivity and accuracy of analysis for trace organic acids in contaminants derived from lignocellulosic biomass, the concentrations of acetate and lactate were enzymatically determined using F-kit acetic acid and D-lactic acid/L-lactic acid (Roche, Penzberg, Bavaria, Germany), respectively, according to the manufacturer's instructions. The concentrations of potential fermentation inhibitors dissolved in the enzymatic hydrolysate of sorghum bagasse were determined via gas chromatography/mass spectrometry, as described previously.24 Based on GC/MS analysis, 18 potential inhibitors were selected based on their abundance, inhibition strength, and/or structure from previous reports,10–12,25,26 and the concentration of these compounds in the enzymatic hydrolysate of sorghum bagasse was determined.
30 (v/v) at a flow rate of 1.25 mL min−1. 4APhe and PABA were detected at wavelengths of 241 and 270 nm, respectively, and the concentrations of 4APhe and PABA were calculated based on standard curves of standard substances. Cell mass was estimated by measuring optical density at 600 nm (OD600) using a spectrophotometer (U-3010; Hitachi, Tokyo, Japan). Suc concentration was determined via HPLC, as described previously.24 To improve the selectivity and accuracy of analysis for trace organic acids in contaminants derived from lignocellulosic biomass, the concentrations of acetate and lactate were enzymatically determined using F-kit acetic acid and D-lactic acid/L-lactic acid (Roche, Penzberg, Bavaria, Germany), respectively, according to the manufacturer's instructions. The concentrations of potential fermentation inhibitors dissolved in the enzymatic hydrolysate of sorghum bagasse were determined via gas chromatography/mass spectrometry, as described previously.24 Based on GC/MS analysis, 18 potential inhibitors were selected based on their abundance, inhibition strength, and/or structure from previous reports,10–12,25,26 and the concentration of these compounds in the enzymatic hydrolysate of sorghum bagasse was determined.
Statistical analysis
Differences in metabolite concentrations and cell densities between fermentation media were compared using paired Student's t-tests. Results with p values <0.05 were considered statistically significant.Results and discussion
Metabolic engineering for improved 4APhe production by recombinant E. coli
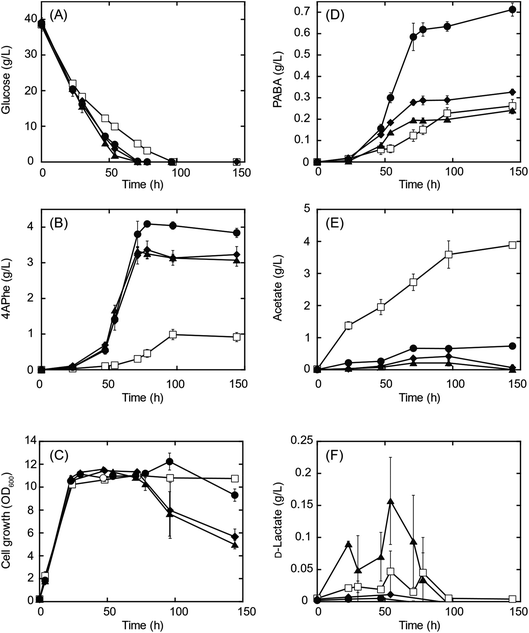
A recombinant E. coli strain HKE1002, expressing papABC derived from P. fluorescens SBW25 in E. coli NST37(DE3)ΔpheLA,13 was used as the parent strain for 4APhe production from glucose (Table 2). Using this strain, three metabolically engineered strains were developed to improve 4APhe production by eliminating the formation of acetate and lactate by-products (Table 2). Lactate formation was completely lost because of the elimination of the ldh gene encoding lactate dehydrogenase from the chromosome in the recombinant strains (HKE6027 and HKE6057); transient lactate accumulation was significantly increased by the disruption of ΔackA_ptaΔpoxB (HKE6046), encoding acetate kinase, phosphate acetyltransferase, and pyruvate oxidase,27 compared to that seen with the parent strain (HKE1002) (Fig. 2). Acetate accumulation after 96 h of cultivation was significantly lower in a culture with either of the three recombinant strains (<1.5 g L−1) than in a culture with the parent strain (HKE1002) (3.6 g L−1). All three recombinant strains consumed the entire available glucose in the culture media after 78 h of cultivation, whereas the parent strain consumed the entire available glucose after 96 h of cultivation. Furthermore, 4APhe concentration after 78 h of cultivation was more than three-fold higher in the culture with a recombinant strain than in that with the parent strain (Fig. 2B).Of the recombinant strains, the Δldh strain (HKE6027) showed the highest 4APhe yield (4.1 g L−1), followed by that seen with the ΔldhΔackA_ptaΔpoxB (HKE6057) and ΔackA_ptaΔpoxB (HKE6046) strains. In addition to 4APhe, another chorismate derivative of PABA28 also accumulated in the culture media as a by-product (Fig. 1). PABA accumulation after 78 h of cultivation was two-fold higher in the culture with the Δldh strain (HKE6027) (620 mg L−1) than in that with the other strains, including the parent strain (<300 mg L−1) (Fig. 2D). Consequently, the total amounts of 4APhe and PABA after 78 h of cultivation were 0.6 (with HKE1002), 4.7 (with HKE6027), 3.4 (with HKE6046), and 3.9 g L−1 (with HKE6057), respectively. In a previous study, 4.4 g L−1 of 4APhe was produced in glucose-fed batch culture using a recombinant E. coli strain without eliminating the acetate and lactate biosynthesis pathway.13 In the present study, a comparable concentration of 4APhe (4.1 g L−1) was produced from glucose in the bath culture using HKE6027 (Δldh) upon eliminating the lactate biosynthesis pathway (Fig. 2B). These results suggest that carbon flux via chorismite in the shikimate pathway was increased by the disruption of the Δldh or ΔackA_ptaΔpoxB strains and that further enhanced flux was not observed with the combined strain ΔldhΔackA_ptaΔpoxB. A small amount of D-lactate (0.15 g L−1) was produced by the parent strain (HKE1002) whereas its accumulation reduced significantly in the culture of the Δldh strain (HKE6027) (Fig. 2F). The concentration of L-lactate was below the detection limit under all cultivation conditions (data not shown).
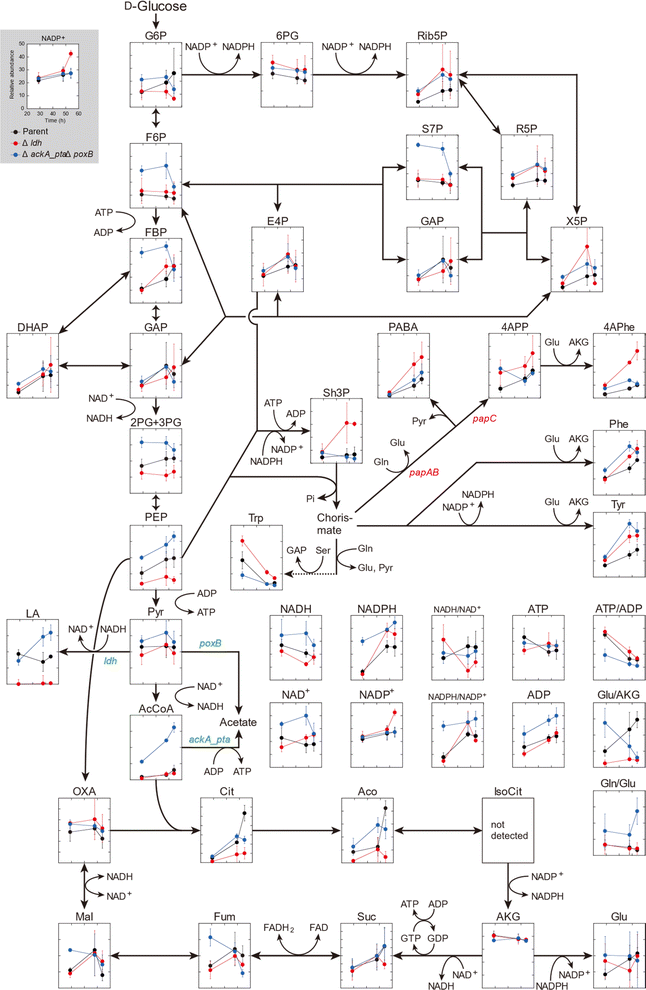
Comparative metabolome analysis using three 4APhe-producing strains
The metabolic profiles of the three metabolically engineered strains during 4APhe production were compared, thereby revealing metabolic shifts associated with enhanced 4APhe production (parent, Δldh, and ΔackA_ptaΔpoxB strains). Time-course metabolomics of the central metabolic pathway clearly showed strain-specific profiles associated with improved 4APhe productivity (Fig. 3). In the PPP, which plays a major role in NADPH regeneration in E. coli cells, intracellular levels of 6PG, Rib5P, R5P, and NADPH were relatively higher in both HKE6027 (Δldh) and HKE6046 (ΔackA_ptaΔpoxB) strains with higher 4APhe productivity than in the parent strain (HKE1002). In the glycolytic pathway, the levels of F6P and 2PG + 3PG after 29 and 48 h of cultivation were significantly higher in HKE6046 (ΔackA_ptaΔpoxB) than those in the other two strains. Compared to the parent strain with low 4APhe productivity, in addition, PEP levels were significantly higher in HKE6046 (ΔackA_ptaΔpoxB) and significantly lower in HKE6027 (Δldh). Knockout of the acetate synthesis pathway exerted a significant impact on pyruvate metabolism. Lactate levels after 48 h of cultivation and AcCoA levels during 29 to 59 h of cultivation were significantly higher in HKE6046 (ΔackA_ptaΔpoxB) than in the other two strains. In addition, compared to the other two strains, HKE6046 (ΔackA_ptaΔpoxB) showed significantly higher ADP levels and comparable ATP levels. Consequently, the ATP/ADP ratio was significantly lower in HKE6046 than in the other two strains during 29 to 59 h of cultivation.For 4APhe production from glucose, HKE6027 (Δldh) showed the highest productivity, followed by HKE6057 (ΔldhΔackA_ptaΔpoxB) and HKE6046 (ΔackA_ptaΔpoxB) (Fig. 2). HKE6027 (Δldh) exhibited distinct metabolic profiles. After 48 h of cultivation, the intracellular levels of Sh3P and 4APhe were significantly higher and PABA and 4APP levels were markedly higher than those of other strains, whereas the PEP level was significantly lower, suggesting an increased flux toward 4APhe biosynthesis via the shikimate pathway. In contrast, HKE6046 (ΔackA_ptaΔpoxB), which eliminates the acetate biosynthesis pathway, accumulated glycolytic intermediates including F6P, FBP, 2PG or 3PG, PEP, pyruvate, and AcCoA, in addition to having a markedly low ATP/ADP ratio (Fig. 3). Although the intracellular levels of PEP and the NADPH/NAPD+ ratio, which serve as precursors and cofactors to synthesize shikimate for 4APhe production, respectively,29 were the highest of all intracellular metabolite levels in all three strains, 4APhe production by HKE6046 (ΔackA_ptaΔpoxB) was lower than that by HKE6027 (Δldh) (Fig. 2). Unlike HKE6027 (Δldh), HKE6046 showed comparable levels of intracellular metabolites, including Sh3P, PABA, 4APP, and 4APhe, in the shikimate pathway and subsequent 4APhe biosynthesis with the parent strain. In E. coli, acetate kinase encoded by the ackA gene is responsible for ATP generation,30 and ATP is a cofactor for Sh3P synthesis by shikimate kinase in the shikimate pathway.31 These results suggest that the knockout of ackA_pta genes results in the overflow of AcCoA and its upstream metabolites and reduces ATP generation required for the shikimate pathway, leading to attenuated 4APhe production by HKE6046. Compared to the parent strain, the ldh mutant showed four-hold higher 4APhe production (Fig. 2B) and significantly lower intracellular PEP levels with comparable pyruvate levels (Fig. 3). These results suggest that more PEP was utilized for 4APhe production rather than acetate production after the deletion of ldh.
Fed-batch culture for 4APhe production from the enzymatic hydrolysate of lignocellulosic biomass
The concentrations of glucose and potential fermentation inhibitors in the enzymatic hydrolysate of sorghum bagasse were determined to utilize the enzymatic hydrolysate as the sole carbon source for 4APhe production. The enzymatic hydrolysate contained glucose and xylose at concentrations of 160 and 0.7 g L−1, respectively (Table 4). Of the potential fermentation inhibitors, sinapinic acid was the most abundant (3136 mg L−1), followed by 5-hydroxymethylfurfural (5HMF), trans-ferulic acid, and syringic acid at concentrations >1000 mg L−1 (Table 4). In addition, major fermentation inhibitors of furfural and p-coumaric acid were also present at concentrations of 227 and 525 mg L−1, respectively, whereas the concentrations of other potential inhibitors, such as benzaldehydes of 4-hydroxybenzaldehyde, vanillin, and syringaldehyde, were relatively low, ranging from 21 to 83 mg L−1. The enzymatic hydrolysate of sorghum bagasse was used as the sole carbon source for subsequent 4APhe fermentation.| Component | Unit | Concentrationa |
|---|---|---|
| a Concentrations of monosaccharides and potential inhibitors are presented as the average ± standard deviation calculated from the results of triplicate individual experiments. | ||
| Monosaccharides | ||
| Glucose | g L−1 | 160.1 ± 13.7 |
| Xylose | g L−1 | 0.7 ± 0.1 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Potential inhibitors | ||
| Furfural | mg L−1 | 227.3 ± 73.8 |
| 5-Hydroxymethylfurfural | mg L−1 | 2728.3 ± 339.9 |
| 5-Methyl-2-furaldehyde | mg L−1 | 66.6 ± 37.8 |
| 4-Hydroxybenzaldehyde | mg L−1 | 30.9 ± 14.3 |
| Vanillin | mg L−1 | 21.1 ± 2.7 |
| Syringaldehyde | mg L−1 | 82.8 ± 1.3 |
| Phenylacetaldehyde | mg L−1 | 6.3 ± 2.7 |
| Benzoic acid | mg L−1 | 337.1 ± 101.7 |
| p-Hydroxybenzoic acid | mg L−1 | 363.2 ± 106.6 |
| Syringic acid | mg L−1 | 1224.0 ± 313.8 |
| Sinapinic acid | mg L−1 | 3135.5 ± 553.5 |
| p-Coumaric acid | mg L−1 | 525.4 ± 65.0 |
| trans-Ferulic acid | mg L−1 | 1370.1 ± 106.9 |
| Levulinic acid | mg L−1 | 311.5 ± 177.8 |
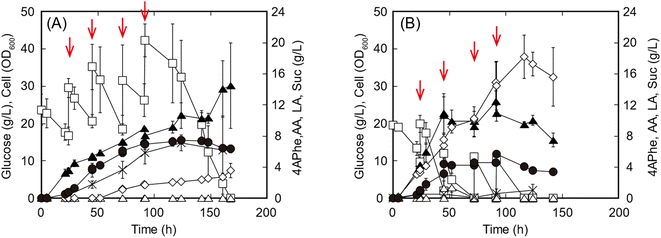
In the fed-batch culture for 4APhe production, either glucose or the enzymatic hydrolysate of sorghum bagasse was fed as the sole carbon source to increase the glucose concentration from 10 to 15 g L−1 in the culture at each feeding point. Within 48 h of cultivation, both glucose consumption and cell growth were higher in cultures with the enzymatic hydrolysate than in those with pure glucose (Fig. 4). Within 45 h of cultivation before the second feeding, volumetric productivity of 4APhe was 0.09 and 0.08 g h−1 L−1 from glucose and enzymatic hydrolysate of sorghum bagasse, respectively, while its yields based on the amount of glucose consumed were 0.098 and 0.103 g 4APhe/g glucose from glucose and the enzymatic hydrolysate, respectively. After 142 h of cultivation, the glucose-fed culture yielded 7.4 g L−1 of 4APhe and accumulated 2.3 and 7.1 g L−1 of Suc and acetate, respectively, as by-products (Fig. 4A). During this period, the cell concentration in each culture continuously increased and reached an OD600 of 21.3. However, in the culture with sorghum hydrolysate, cell growth and 4APhe production almost ceased after 48 h of cultivation, although the sorghum hydrolysate was fed even after that (Fig. 4B). Consequently, the 4APhe concentration transiently reached 5.7 g L−1 after 91.5 h of cultivation and then gradually decreased. In the culture with sorghum hydrolysate, Suc production was significantly increased compared with that seen in the culture with glucose, and the concentrations of Suc and acetate were 18.2 and 1.0 g L−1, respectively, after 116.5 h of cultivation. Global efforts are increasingly being made to use second-generation feedstock.22 Second-generation feedstocks of lignocellulosic biomass predominantly contain glucose followed by xylose with trace amounts of other monosaccharides such as arabinose and mannose;10 diluted acid-pretreated sorghum bagasse contains 60% and 7% glucose and xylose as dry biomass, respectively,12 and adaptive mutant of E. coli can simultaneously utilize both xylose and glucose along with arabinose.32 These results suggest that the metabolic engineering of E. coli for the co-utilization of hexose and pentose could improve 4APhe in further studies.
In previous studies, 4.4 g L−1 of 4APhe was produced from glucose using recombinant E. coli (Table 1).13,14 In the present study, a high concentration (5.7 g L−1) of 4APhe was obtained from lignocellulosic hydrolysate by a metabolically engineered E. coli strain that eliminated by-product synthesis. However, 4APhe yield from the lignocellulosic hydrolysate was 24% less than that from pure glucose. The enzymatic hydrolysate of sorghum bagasse contained various contaminants, such as furfural, 5HMF, p-coumaric acid, and trans-ferulic acid, which served as fermentation inhibitors for E. coli in previous studies33–35 (Table 4). These results suggest that 4APhe production from the enzymatic hydrolysate of sorghum bagasse by metabolically engineered E. coli was subjected to fermentation inhibition.11,36
In previous studies, methods for the purification of 4APhe14 and the direct use for the synthesis of high-performance polymer37 have been developed. For the downstream process, a higher concentration of 4APhe after fermentation can achieve higher recovery efficiency for the following polymer synthesis. In the present study, over 2.2 g L−1 of 4APhe that was required for purification14 was produced from lignocellulosic biomass (Table 1). Although annual production of plastics globally is over millions of tonnes,2 most of them are commodity plastics, and the share of engineering and high-performance plastics that are synthesized from aromatic monomers, e.g. 4APhe, is only 7%.38 Based on the share of the engineering plastics, their production capacity is estimated at 15 kg per h per plant.39 Due to the low demand and high price of the engineering and high-performance plastics which require 4APhe as an aromatic monomer, the product concentration of 4APhe demonstrated in the present study is expected to be economically viable.
Identification of fermentation inhibitors for 4APhe production
To identify the fermentation inhibitor(s) present in the enzymatic hydrolysate of sorghum bagasse, groups of potential inhibitors were individually or collectively added to the medium in a dose-dependent manner before starting 4APhe production from glucose. The potential fermentation inhibitors were selected from the results shown in Table 4 and classified into five groups based on their structures: group A, furan aldehydes (furfural, 5HMF, and 5-methyl-2-furaldehyde); group B, hydroxybenzaldehydes (4-hydroxybenzaldehyde, vanillin, and syringaldehyde); group C, (hydroxy)benzoic acids (benzoic acid, syringic acid, and p-hydroxybenzoic acid); group D, cinnamic acid derivatives (trans-ferulic acid and p-coumaric acid); and group E, aliphatic acid (levulinic acid). Group E consisted of only levulinic acid, and potential inhibitors of acetate and lactate25 were not detected in the enzymatic hydrolysate of sorghum bagasse. Dose–response curves were measured for the aldehydes and acids or their mixture, and half and full maximum inhibitory concentrations (IC50 and IC100, respectively) were determined for cell growth and 4APhe production (Table 5 and Fig. S1†).| Group of inhibitorsa (mg L−1) | Cell growthb | 4APhe productionb | Specific productivity of 4APheb | Concentrations of inhibitorc | |||
|---|---|---|---|---|---|---|---|
| IC50 | IC100 | IC50 | IC100 | IC50 | IC100 | ||
| a Group: A, furan aldehydes (furfural, 5HMF, and 5-methyl-2-furaldehyde); B, benzaldehydes (4-hydroxybenzaldehyde, vanillin, and syringaldehyde); C, benzoic acids (benzoic acid, syringic acid, and p-hydroxybenzoic acid); D, cinnamic acid derivatives (trans-ferulic acid and p-coumaric acid); E, levulinic acid; all, a mixture of all of the aforementioned compounds. Each group of inhibitors was individually added to the culture medium before inoculation in a dose-dependent manner, and cell growth and 4APhe production were measured after 72 h of cultivation. b IC100 is the concentration of inhibitors where cell growth or 4APhe production is completely suppressed. IC50 is the concentration of inhibitors where cell growth or 4APhe production is reduced by half. These values were determined from the results of dose–response curves of cell density and 4APhe concentration after 72 h of cultivation (shown in Fig. S1, ESI). c Initial concentration of inhibitors present in fed-batch culture with the enzymatic hydrolysate of sorghum bagasse for 4APhe production, as shown in Fig. 4B. | |||||||
| A | >1000 | >1000 | 640 | >1000 | 840 | >1000 | 377.5 |
| B | 240 | 1000 | 20 | 320 | 20 | >1000 | 16.8 |
| C | 240 | 320 | 80 | 320 | 160 | >1000 | 240.4 |
| D | 240 | 1000 | 20 | 640 | 20 | 640 | 236.8 |
| E | 600 | >1000 | 170 | 1000 | 240 | >1000 | 38.9 |
| Mixture | 90 | 320 | 20 | 160 | 45 | 160 | 910.4 |
Both hydroxybenzaldehydes and cinnamic acid derivatives exerted the highest inhibitory effects on 4APhe production (IC50 = 20 mg L−1), whereas the IC50 of these inhibitors regarding cell growth was relatively high (240 mg L−1). In 4APhe production, benzoic acids also demonstrated a notable inhibitory effect (IC50 = 80 mg L−1), whereas furan aldehydes, which are some of the major fermentation inhibitors, exhibited a relatively low inhibitory effect (IC50 = 640 mg L−1). A synergetic effect of these fermentation inhibitors was observed for cell growth only but not for 4APhe production. The concentrations of benzoic acids and cinnamic acid derivatives present in sorghum bagasse hydrolysate were more than three-fold higher than the IC50 values for 4APhe production, whereas the concentration of benzaldehydes in the hydrolysate was comparable to the IC50 values for 4APhe production. Different levels of inhibitory effects of potential fermentation inhibitors derived from lignocellulosic biomass on microbial production by E. coli cells have been reported previously.12,24,26 These results suggest that benzoic acid and cinnamic acid derivatives present in sorghum bagasse hydrolysate play a major role in the inhibition of 4APhe production from the hydrolysate.
Compared to pure glucose as the sole carbon source, the enzymatic hydrolysate of sorghum bagasse reduced 4APhe production by 23% and increased Suc formation (Fig. 4). Increased Suc production by an engineered E. coli strain was also observed in the presence of furfural present in lignocellulosic hydrolysate.40 Fermentation inhibition assay revealed that hydroxybenzaldehydes, cinnamic acid derivatives, and (hydroxy)benzoic acids served as strong inhibitors for 4APhe production, whereas the inhibitory effect of well-known fermentation inhibitors furan aldehydes, such as furfural and 5HMF,33 on 4APhe production was limited (Table 5). In a previous study, the cinnamic acid derivatives p-coumaric acid and trans-ferulic acid inhibited fermentation by E. coli.34 In the enzymatic hydrolysate of sorghum bagasse, the total concentrations of cinnamic acid derivatives and (hydroxy)benzoic acids were markedly higher than the IC50 values for 4APhe production, whereas the total concentrations of furan aldehydes and benzaldehydes were lower and comparable to the IC50 values (Tables 4 and 5). In addition, p-coumaric acid present in lignocellulosic hydrolysate inhibits the enzymatic activities of shikimate kinase from Sorghum bicolor,41 and chorismate mutase or prephenate dehydrogenase from Alcaligenes eutrophus42 is responsible for 4APhe production in the shikimate pathway, although the inhibitory effects of these enzymes have not been reported in E. coli. In previous studies, the effects of components of lignocellulosic hydrolysate on microbial activity were evaluated based on the levels of metabolic intermediates and gene expression as well as cell growth.11,12 These results suggest that benzoic acids and cinnamic acid derivatives are critical fermentation inhibitors for 4APhe production from the enzymatic hydrolysate of sorghum bagasse and that the inhibitory effect of furfural and 5HMF on 4APhe production by E. coli is limited, even after repeated feeding of the hydrolysate.
Conclusions
Microbial production of starting materials for the synthesis of bio-based polymers exclusively depends on the use of edible glucose but not non-edible lignocellulosic feedstocks. In this study, 4APhe, which serves as a diamine monomer to synthesize unique biopolyimides,9,43 was produced from the inedible feedstock of sorghum bagasse instead of glucose, as reported previously. Comparable metabolome analysis revealed specific metabolic profiles in E. coli cells with high 4APhe productivity, which was improved by metabolic engineering. Although 5.7 g L−1 of 4APhe produced from lignocellulosic biomass was the highest concentration recorded, it was 24% lower than that produced from glucose. Among the potential inhibitors present in the enzymatic hydrolysate of sorghum bagasse, benzaldehydes, and cinnamic acid derivatives inhibited 4APhe fermentation at low concentrations (IC50 = 16 mg L−1), whereas the predominant and well-known fermentation inhibitors furfural and 5HMF inhibited 4APhe fermentation at relatively high concentrations (IC50 = 640 mg L−1). These results provide insight into the design of metabolic pathways tailored for the utilization of lignocellulosic biomass to produce aromatic compounds via the shikimate pathway by attenuating fermentation inhibition.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the JST ALCA Program, Japan (Grant Number JPMJAL1010), the JST Mirai Program (Grant Number PMJMI17EG), and the New Energy and Industrial Technology Development Organization (NEDO), Japan (Grant Number JPNP18016). We gratefully acknowledge the contribution of Junko Imada, Kumiko Yoshihara, Sharif Moinul Hasan, Taeko Fukumoto, Takanobu Yoshida, and Yoshimi Hori toward technical support.Notes and references
- H. Kawaguchi, K. Takada, T. Elkasaby, R. Pangestu, M. Toyoshima, P. Kahar, C. Ogino, T. Kaneko and A. Kondo, Bioresour. Technol., 2022, 344, 126165 CrossRef CAS PubMed.
- S. A. Park, H. Jeon, H. Kim, S. H. Shin, S. Choy, D. S. Hwang, J. M. Koo, J. Jegal, S. Y. Hwang, J. Park and D. X. Oh, Nat. Commun., 2019, 10, 2601 CrossRef PubMed.
- R. Y. Getor, N. Mishra and A. Ramudhin, Resour., Conserv. Recycl., 2020, 163, 105094 CrossRef.
- B. Gupta, N. Revagade and J. Hilborn, Prog. Polym. Sci., 2007, 32, 455–482 CrossRef CAS.
- H. Kawaguchi, T. Hasunuma, C. Ogino and A. Kondo, Curr. Opin. Biotechnol., 2016, 42, 30–39 CrossRef CAS PubMed.
- J. He, N. Magarvey, M. Piraee and L. C. Vining, Microbiology, 2001, 147, 2817–2829 CrossRef CAS PubMed.
- V. Blanc, P. Gil, N. BamasJacques, S. Lorenzon, M. Zagorec, J. Schleuniger, D. Bisch, F. Blanche, L. Debussche, J. Crouzet and D. Thibaut, Mol. Microbiol., 1997, 23, 191–202 CrossRef CAS PubMed.
- T. Kaneko, M. A. Ali, I. Captain, P. Perlin and T. J. Deming, Polym. Chem., 2018, 9, 3466–3472 RSC.
- T. Hirayama, A. Kumar, K. Takada and T. Kaneko, ACS Omega, 2020, 5, 2187–2195 CrossRef CAS PubMed.
- S. K. Bhatia, S. S. Jagtap, A. A. Bedekar, R. K. Bhatia, A. K. Patel, D. Pant, J. Rajesh Banu, C. V. Rao, Y. G. Kim and Y. H. Yang, Bioresour. Technol., 2020, 300, 122724 CrossRef CAS PubMed.
- Y. Suo, W. Li, L. Wan, L. Luo, S. Liu, S. Qin and J. Wang, Appl. Microbiol. Biotechnol., 2023, 107, 327–339 CrossRef CAS PubMed.
- H. Kawaguchi, H. Teramura, K. Uematsu, K. Y. Hara, T. Hasunuma, K. Hirano, T. Sazuka, H. Kitano, Y. Tsuge, P. Kahar, S. Niimi-Nakamura, K. I. Oinuma, N. Takaya, S. Kasuga, C. Ogino and A. Kondo, Bioresour. Technol., 2015, 182, 169–178 CrossRef CAS PubMed.
- S. Masuo, S. M. Zhou, T. Kaneko and N. Takaya, Sci. Rep., 2016, 6, 25764 CrossRef CAS PubMed.
- S. Tateyama, S. Masuo, P. Suvannasara, Y. Oka, A. Miyazato, K. Yasaki, T. Teerawatananond, N. Muangsin, S. M. Zhou, Y. Kawasaki, L. B. Zhu, Z. M. Zhou, N. Takaya and T. Kaneko, Macromolecules, 2016, 49, 3336–3342 CrossRef CAS.
- H. Minakawa, S. Masuo, T. Kaneko and N. Takaya, Process Biochem., 2019, 77, 100–105 CrossRef CAS.
- S. Masuo, Y. Tsuda, T. Namai, H. Minakawa, R. Shigemoto and N. Takaya, Chembiochem, 2019, 21, 353–359 CrossRef PubMed.
- J. Sambrook and D. W. Russell, Molecular Cloning: a Laboratory Manual, Cold Spring Harbor Laboratory Press, New York, 3rd edn, 2001 Search PubMed.
- S. Hashimoto, T. Wake, H. Nakamura, M. Minamiyama, S. Araki-Nakamura, K. Ohmae-Shinohara, E. Koketsu, S. Okamura, K. Miura, H. Kawaguchi, S. Kasuga and T. Sazuka, Sci. Rep., 2021, 11, 4532 CrossRef CAS PubMed.
- K. A. Datsenko and B. L. Wanner, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6640–6645 CrossRef CAS PubMed.
- H. Kawaguchi, M. Sasaki, A. A. Vertes, M. Inui and H. Yukawa, Appl. Microbiol. Biotechnol., 2008, 77, 1053–1062 CrossRef CAS PubMed.
- T. Baba, T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner and H. Mori, Mol. Syst. Biol., 2006, 2, 0008 CrossRef PubMed.
- J. M. Buescher, S. Moco, U. Sauer and N. Zamboni, Anal. Chem., 2010, 82, 4403–4412 CrossRef CAS PubMed.
- H. Kato, Y. Izumi, T. Hasunuma, F. Matsuda and A. Kondo, J. Biosci. Bioeng., 2012, 113, 665–673 CrossRef CAS PubMed.
- H. Kawaguchi, Y. Katsuyama, D. Danyao, P. Kahar, S. Nakamura-Tsuruta, H. Teramura, K. Wakai, K. Yoshihara, H. Minami, C. Ogino, Y. Ohnishi and A. Kondo, Appl. Microbiol. Biotechnol., 2017, 101, 5279–5290 CrossRef CAS PubMed.
- J. R. M. Almeida, T. Modig, A. Petersson, B. Hahn-Hagerdal, G. Liden and M. F. Gorwa-Grauslund, J. Chem. Technol. Biotechnol., 2007, 82, 340–349 CrossRef CAS.
- G. Padmapriya, V. Dhivya, M. Vishal, Y. A. J. Roshni, Y. Akila and S. Ramalingam, J. Environ. Biol., 2021, 42, 1239–1248 CrossRef CAS.
- T. B. Causey, K. T. Shanmugam, L. P. Yomano and L. O. Ingram, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 2235–2240 CrossRef CAS PubMed.
- T. Kubota, A. Watanabe, M. Suda, T. Kogure, K. Hiraga and M. Inui, Metab. Eng., 2016, 38, 322–330 CrossRef CAS PubMed.
- S. Noda, T. Shirai, S. Oyama and A. Kondo, Metab. Eng., 2016, 33, 119–129 CrossRef CAS PubMed.
- A. Matsuyama, H. Yamamoto and E. Nakano, J. Bacteriol., 1989, 171, 577–580 CrossRef CAS PubMed.
- X. Liu, J. Lin, H. Hu, B. Zhou and B. Zhu, World J. Microbiol. Biotechnol., 2014, 30, 2543–2550 CrossRef CAS PubMed.
- C. Dev, S. B. Jilani and S. S. Yazdani, Microb. Cell Fact., 2022, 21, 154 CrossRef CAS PubMed.
- A. L. Luhe, C. Y. Lim, H. Gerken, J. Wu and H. Zhao, Biotechnol. Appl. Biochem., 2015, 62, 32–36 CrossRef CAS PubMed.
- T. Mitani, K. Ota, N. Inaba, K. Kishida and H. A. Koyama, Biol. Pharm. Bull., 2018, 41, 208–212 CrossRef CAS PubMed.
- H. Kawaguchi, K. Uematsu, C. Ogino, H. Teramura, S. Niimi-Nakamura, Y. Tsuge, T. Hasunuma, K. Oinuma, N. Takaya and A. Kondo, Biochem. Eng. J., 2014, 88, 188–194 CrossRef CAS.
- A. Fehér, C. Feher, M. Rozbach and Z. Barta, Chem. Biochem. Eng. Q., 2017, 31, 77–87 CrossRef.
- K. Takada, H. Shinagawa, Y. Morita, M. S. Grewal, K. Taya, A. Kumar and T. Kaneko, Chin. J. Polym. Sci., 2020, 38, 1117–1123 CrossRef CAS.
- D. K. Platt, Engineering and High Performance Plastics Market Report: A Rapra Market Report, Rapra Technolgy Limit, Shropshire, UK, 2003 Search PubMed.
- R. P. Babu, K. O'Connor and R. Seeram, Prog. Biomater., 2013, 2, 8 CrossRef PubMed.
- X. Wang, L. P. Yomano, J. Y. Lee, S. W. York, H. B. Zheng, M. T. Mullinnix, K. T. Shanmugam and L. O. Ingram, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4021–4026 CrossRef CAS PubMed.
- J. R. Bowen and T. Kosuge, Plant Physiol., 1979, 64, 382–386 CrossRef CAS PubMed.
- C. G. Friedrich, B. Friedrich and H. G. Schlegel, J. Bacteriol., 1976, 126, 723–732 CrossRef CAS PubMed.
- K. Takada, T. Noda, T. Kobayashi, T. Harimoto, M. Singh and T. Kaneko, Polym. J., 2021, 53, 1223–1230 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3su00053b |
| This journal is © The Royal Society of Chemistry 2023 |