A computer vision enhanced smart phone platform for microfluidic urine glucometry†
Zhuolun
Meng
 ,
Muhammad
Tayyab
,
Muhammad
Tayyab
 ,
Zhongtian
Lin
,
Hassan
Raji
,
Zhongtian
Lin
,
Hassan
Raji
 and
Mehdi
Javanmard
*
and
Mehdi
Javanmard
*
Electrical and Computer Engineering, Rutgers University-New Brunswick, 94 Brett Road, Piscataway, NJ, USA. E-mail: mehdi.javanmard@rutgers.edu
First published on 8th December 2023
Abstract
Glucose is an important biomarker for diagnosing and prognosing various diseases, including diabetes and hypoglycemia, which can have severe side effects, symptoms, and even lead to death in patients. As a result, there is a need for quick and economical glucose level measurements to help identify those at potential risk. With the increase in smartphone users, portable smartphone glucose sensors are becoming popular. In this paper, we present a disposable microfluidic glucose sensor that accurately and rapidly quantifies glucose levels in human urine using a combination of colorimetric analysis and computer vision. This glucose sensor implements a disposable microfluidic device based on medical-grade tapes and glucose analysis strips on a glass slide integrated with a custom-made polydimethylsiloxane (PDMS) micropump that accelerates capillary flow, making it economical, convenient, rapid, and equipment-free. After absorbing the target solution, the disposable device is slid into the 3D-printed main chassis and illuminated exclusively with Light Emitting Diode (LED) illumination, which is pivotal to color-sensitive experiments. After collecting images, the images are imported into the algorithm to measure the glucose levels using computer vision and average RGB values measurements. This article illustrates the impressive accuracy and consistency of the glucose sensor in quantifying glucose in sucrose water. This is evidenced by the close agreement between the computer vision method used by the sensor and the traditional method of measuring in the biology field, as well as the small variation observed between different sensor performances. The exponential regression curve used in the study further confirms the strong relationship between glucose concentrations and average RGB values, with an R-square value of 0.997 indicating a high degree of correlation between these variables. The article also emphasizes the potential transferability of the solution described to other types of assays and smartphone-based sensors.
1. Introduction
Glucose, also known as blood sugar, is a crucial component of the human body and is the primary source of energy for cells. The blood carries glucose to all the human body's cells for energy. When glucose levels become abnormal, they can lead to various diseases, including diabetes. This condition occurs when blood sugar levels are too high and can cause serious long-term health problems.1,2 According to the Centers for Disease Control and Prevention, there are currently 37.3 million diabetes patients in the US, with an additional 96 million people aged 18 years or older having prediabetes which takes the total to 38% of the adult US population.3 Treating diabetes is costly, with an estimated total cost of 327 billion USD in the US in 2017, due to the many health problems associated with high blood glucose, such as heart disease, stroke, kidney disease, and eye problems.4 Even individuals without diabetes may experience issues with blood sugar that is too low or high. Maintaining a regular schedule of eating, physical activity, and taking medicines can help to manage blood sugar levels which in the normal range for glucose varies generally from 70 to 99 mg dL−1. The cost of glucose tests is critical for individuals at risk, and with the increasing popularity, convenience, and user-friendly nature of smartphones, they are becoming an essential tool for monitoring and diagnosing glucose levels in healthcare.In the last few decades, biomedical devices and sensors have been rapidly developing to meet the requirements and challenges of modern society. These devices and sensors can utilize small volumes of fluids for medical detection and diagnosis.5–12 Specifically, microfabrication technology has had a profound effect on the types of biosensors and has been introduced into biological assays.13–20 However, microfabrication techniques technically require a cleanroom environment to fabricate devices, which makes it difficult to operate equipment and reduce the price. As a result, PDMS was explored as a replacement for microfabrication, thanks to its advantages such as being low-cost, biocompatible, flexible, and having inert elastomers. PDMS is commonly used as the material to fabricate soft microfluidic channels, which can adjust their dimensions and shapes according to the design of molds. Cured PDMS is then treated by baking or oxide plasma to bond onto substrates and form microfluidic channels. However, bonding issues during the bonding of PDMS to other substrates are common.21–26 Additionally, PDMS needs time and temperature to cure and reach a solid state. Different molds are also required to fabricate microfluidic channels with various dimensions and shapes to obtain the expected PDMS channels.
In this paper, we present an alternative approach for fabricating biomedical devices using polymer materials and paper.27–29 These devices have desirable features such as disposability, ease of fabrication, and low risk of mutual contamination, making them ideal for use in the biomedical field.30–33 Specifically, we describe the design and fabrication of a glucose diagnostic sensor that utilizes colorimetric measurement based on a smartphone camera and computer vision algorithm; making it completely user-friendly and eliminating the need for large and expensive equipment for detection.10,34 Compared with the methods discussed in Yanyan Xia's review paper,33 we improved the accuracy of this sensor by utilizing external light sources and implementing background illumination control. These enhancements positively impacted the sensor's performance, in addition to incorporating a 3D-printed portable phone chassis.34 This approach satisfies the ASSURED criteria (Affordable, Sensitive, Specific, User-friendly, Rapid and Robust, Equipment-free, and Deliverable) established by the WHO.35 The design and fabrication process of the sensor is simple and easy. AutoCAD was used to design the sensor features, and a laser cutter was used to cut out the features on medical tapes, enabling easy shaping of the channels. The cost of each piece can be lowered to 0.24 USD per sensor, which is more economical than microfabrication and commercial glucose test kits. Compared to other similar approaches, this solution offers greater flexibility for application in various assays and contexts. The laser-cut channels can be conveniently reshaped to match the dimensions and shapes of the substrates.36,37 To enhance the flow speed and reaction rate in the channel and avoid solution waste, we used PDMS micropumps to control the volume and speed of the solution directly.37,38,46 To process a large number of pictures in a short time automatically,39,40 we utilized a computer vision method, and the automatic algorithm has been verified to replace the manual traditional measurement methods. This automatic customized detector has potential to be utilized in a wide range of assays effortlessly, providing advantages over the phone application and more convenient transferability over the cluster algorithm described in Uddin Jalal and Marzia Hoque Tania's papers.35,36 The regression model obtained from titration experiments can show the relationship between glucose concentrations and average RGB values.15,38,41,46
2. Methods and materials
2.1. Hardware components
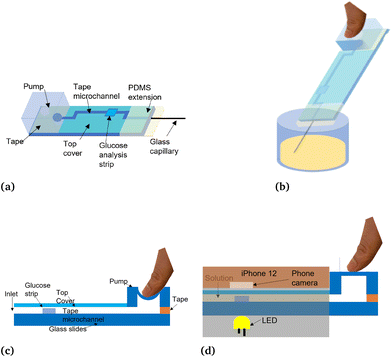
The hardware used for the smartphone-based glucose measurement platform includes a lens band (Easy-Macro Lens Band, Easy-Marco, MA, USA),42 which can reduce the depth of focus of the smartphone camera and further decrease the size of the main chassis, especially its height. Two LED diodes are used as illumination sources instead of the smartphone flash or other external sources for the purpose of eliminating the external light noise and consistently controlling the illumination magnitude. A customized 3D-printed chassis is used to house all components (weight: 178 g including LEDs, two coin batteries, one battery holder, chassis; size: 15.4 × 8.5 × 5.9 cm).38,41,43,44,46 An iPhone 12 (Apple, Inc., Cupertino, CA, USA) is utilized as the primary smartphone camera to conduct all experiments. The 3D-printed chassis was designed in AutoCAD and 3D printed using an Ultimaker S5 3D printer and Ultimaker Polylactic acid (PLA) as the filament material (Ultimaker, Utrecht, Netherlands). It is designed to fit the iPhone 12 (Apple, Inc., Cupertino, CA, USA) and acts as a whole black opaque phone holder, which can block the outside light noise that fluctuates with different environments.34,38,41,43,46In Fig. 1A, the main chassis is introduced, and a strips-window is designed to slide standard microscope glass slides (AmScope, United Scope LLC., CA, USA) where the glucose sensor is bonded. After absorbing the sucrose solution, glucose sensors are slid into the main chassis through the strip window which is shown in Fig. 1B.38,46
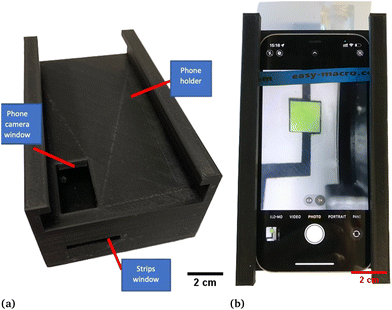 | ||
| Fig. 1 Smartphone-based glucose detection platform. (a) 3D-printed smartphone box. (b) Actual image that was taken after sliding the microfluidics device into the platform. | ||
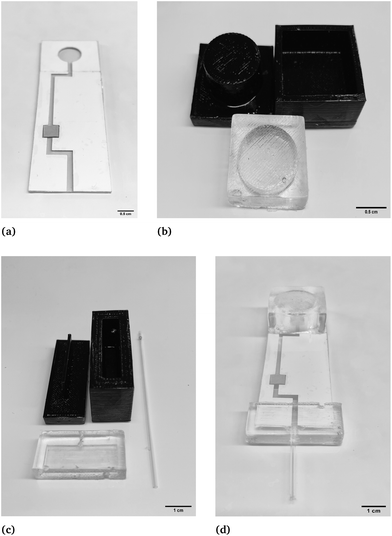
2.2. Microfluidic device fabrication
For this glucose sensor, there are in total three components consisting of a microfluidic channel, PDMS micropump, and PDMS extension part. The microfluidic channel was created using two medical-grade tapes. The double-sided adhesive tape (3M 9965 Diagnostic Microfluidics tape, 3M, MN, USA) was engraved using a laser cutter (Boss LS1420, MinPower (%)-1: 14, MaxPower (%)-1: 14, speed (mm s−1): 9, Boss Laser, FL, USA) to form the structure of the microfluidic channel. The 2D model was designed on AutoCAD. The glucose test strip (ACCUTEST Urine Reagent Strip, Accutest, CA, USA) was placed into the channel with the help of a hydrophilic cover (3M 9962 Diagnostic Microfluidics tape, 3M, MN, USA) that was designed on AutoCAD and engraved using a laser cutter (Boss LS1420, MinPower (%)-1: 14, MaxPower (%)-1: 14, speed (mm s−1): 9, Boss Laser, FL, USA) to assist the liquid flow. The actual microfluidic channel is shown in Fig. 2A. The inlet of the channels is located at the end of the device, and the height of the inlet is equal to the height of double-sided adhesive tapes, which is 86 μm.To accelerate the reaction of glucose strips through faster flow velocity, the PDMS micropump was used as an extremely powerful active solution. The disposable extension part is also clean and convenient for absorbing the solution into the sensor.37,38,46 To fabricate PDMS micropumps and the extension part, 3D-printed molds were used. However, the rough surface of the 3D-printed molds could potentially interfere with the PDMS curing process. To address this issue, acrylic lacquer (Acrylic lacquer, Rust-oleum) was sprayed onto the surface after 3D printing, forming a thin smooth acrylic layer over the coarse 3D-printed mold’s surface. This isolates the liquid PDMS from the coarse 3D-printed surfaces. The protocol for forming the acrylic layer involves three steps: first, spray acrylic to cover all surfaces and bake for 30 minutes; second, spray acrylic to cover all surfaces again and bake for 30 minutes; and third, let the mold bake for 60 minutes – baking is needed after spraying to ensure the solidity of the acrylic. To fabricate PDMS (Dow Chemical Company, Midland, MI, USA) structures that can feasibly support the application of a micropump and extension part, the ratio of elastomer and curing agent was adjusted from the normal 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 20
1 to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. This made the PDMS soft enough to press with enough strength to support the mechanical structure. Consequently, the molds, PDMS micropump, and extension part are shown in Fig. 2B and C.
1. This made the PDMS soft enough to press with enough strength to support the mechanical structure. Consequently, the molds, PDMS micropump, and extension part are shown in Fig. 2B and C.
In Fig. S2,† different configurations for diameters and heights of PDMS micropumps are shown, with the purpose of analyzing the relationship between theoretical and actual volumes micropumps can suck up and the effect of diameters and heights. For this purpose, theoretical volumes of the micropumps were calculated and compared to the actual volumes that the micropumps can introduce to the sensor. Regarding height, three dimensions of micropumps were fabricated with different heights of 1 cm, 0.7 cm, and 0.4 cm, all with a diameter of 2 cm, as shown in Fig. S2A.† From Fig. S2C,† it is evident that the 0.7 cm micropump was the ideal height for this application. Additionally, micropumps with different diameters of 2 cm, 1.5 cm, and 0.5 cm, all with a height of 0.7 cm were fabricated, as shown in Fig. S2B.† The density of dyed DI (deionized) water was measured by weighing the solution with a known volume to obtain the density, which is 1 g per cm3. The glucose sensor was weighed before and after micropumping to calculate the weight difference in terms of the micropumps’ capability.
Fig. S2† demonstrates that the effect of the diameter is larger than that of the height on the actual micropumps’ capability. To investigate the actual volume for micropumps, three PDMS micropumps were fabricated utilizing the same molds for each dimension. Three glucose sensors were assembled to introduce the dyed DI water into the devices. The weights were measured before and after sucking water into the device, and the actual volume of micropumps was calculated by dividing weight difference by the density. Nevertheless, the micropump is dysfunctional if the height of micropumps is too small since the structure cannot support the pressing force, which is shown in Fig. S2C,† and the actual volume for 0.4 cm height is 0. In Fig. S2C,† the ratio of theoretical and actual volume is about 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which is larger than that for micropumps with different diameters. To make the sensor more convenient to use, the extension part was fabricated as shown in Fig. 2C. The glass capillary is utilized as the guide for the solution, which increases the flexibility of this device. In Fig. 2D, the final biomedical device is shown after assembling all components.
1, which is larger than that for micropumps with different diameters. To make the sensor more convenient to use, the extension part was fabricated as shown in Fig. 2C. The glass capillary is utilized as the guide for the solution, which increases the flexibility of this device. In Fig. 2D, the final biomedical device is shown after assembling all components.
2.3. Experimental protocol
The first step to utilize this glucose sensor is to mount the PDMS micropump and PDMS extension onto the microfluidic channel. The assembled glucose sensor is shown conceptually in Fig. 3A. To operate and utilize this glucose sensor, the PDMS micropump is pressed to create a negative pressure environment inside the whole sensor. Next, the glass capillary is immersed into the container for sucrose solution or potential urine as shown in Fig. 3B. In Fig. 3C, the PDMS micropump is thumb-pressed to vent the air before dipping glass capillary into solution. Once the thumb is released, the solution is vacuumed into the channel to react with the glucose test strips. After absorbing enough solution, the device is slid into the phone chassis which as shown in Fig. 3D, and pictures are taken for the next step of colorimetric measurement.2.4. Automatic glucose measurement algorithm
To facilitate the convenience and allow accurate measurement of glucose sensor results in the R, G, and B channels, a combination of 3D printing technology and computer vision was implemented. Frames from the videos taken using an iPhone 12 positioned on the chassis were captured for glucose sensors following the recommended reading time of 30 seconds from the user manual. For automated glucose sensor measurement, an automated computer vision method, shown in Fig. 4, was used as a replacement for traditional manual methods. The computer vision workflow utilized YOLOv4, one of the most mature, accurate, and popular computer vision models available, to detect the target feature in one image and produce the metadata associated with the object which is glucose analysis strips in this application, such as locate target area with the coordinates of a rectangular box. Sample images of glucose analysis strips were picked randomly, manually annotated using Roboflow,45 and photos are prepared for training the detector, which can locate the glucose sensor from background and thereafter and provide the average RGB values for glucose test strips. To train the model fits for the photos, 40 sample photos were captured for training and validating. Then the target areas were manually selected, and the information was transferred into digital information, which is the relative YOLOV4 format coordinate in Roboflow. This provides the dataset for customized detector training in Google Colab. The .weights file was exported after training and was implemented into the TensorFlow platform on a desktop, which can automatically detect, crop, and send to the next step to provide measurement results.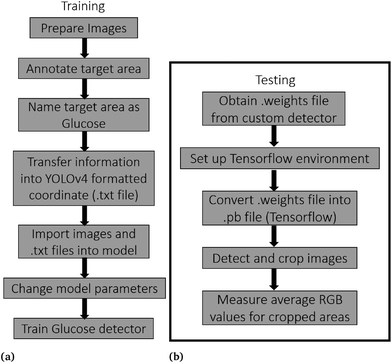 | ||
| Fig. 4 Flowcharts illustrate the steps of training and testing the glucose detection algorithm. (a) Training flowchart of glucose detector. (b) Testing flowchart of glucose detector. | ||
To quantitatively measure the colorimetric values, the average RGB value was selected as the parameter to reflect the concentration of glucose for the glucose test strips measurement. According to the user manual, the color of reacted analysis strips will shift from light to dark color as the concentration of glucose increases. Thus, the equation is avg RGB = (R + G + B)/3 which combines the red, green, and blue channels and interprets the inverse proportional relationship between average RGB values and glucose concentrations. This function was implemented using MATLAB.
3. Results
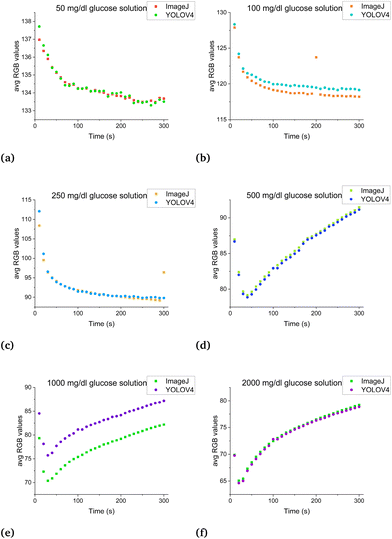
The comparison between manual (ImageJ) versus automatic (YOLOV4) measurements was shown in Fig. 5. The glucose measurement algorithm was able to automatically select the target area with 100% accuracy, as demonstrated as Glucosestrip 1.00 in the bottom picture. To quantify the relationship between ImageJ and YOLOV4 measurement results, 30 frames were captured from recorded videos with a 10-second interval over a 3-minute period. The plots in Fig. 6 show high coherence for glucose concentrations of 50, 100, 250, 500, 1000 and 2000 mg dL−1. Consequently, the computer vision method can be an efficient replacement of the conventional ImageJ method. The reason for deviation of Fig. 6E is because the position of the test strip is off from the horizontal line. Since the YOLOv4 can only detect targets in a square area, it will detect a small part of the boundary which affected the results a little. | ||
| Fig. 5 Comparison between ImageJ and glucose detector. (a) Manually selected target area through ImageJ software. (b) Automatically detected target area by glucose detector. | ||
The titration curve was obtained by plotting the glucose concentrations obtained from the glucometer (ANKOVO TD-4627, Shenzhen, China) against the average RGB values of the glucose sensors obtained from the automatic algorithm. Titration experiments were performed at six concentrations, and the average RGB values were measured at 30 seconds after reacting, which is the recommended reading time. The result shows an exponential relationship between glucose concentrations and the average RGB values, and the function is Y = 72.143 + 76.261 × exp(−0.00409X) and the R-square equals to 0.997, as shown in Fig. 7.
4. Statistical performance
In this section, the main results of the computer vision-related tests are presented statistically. Fig. 8a shows the correlation curve for YOLOv4 and ImageJ. The black curve represents the diagonal line with the formula Y = X. The regression line close association (R-square = 0.99166) with the artificial diagonal line validates the utility and accuracy of the computer vision method. In Fig. 8b, almost all data points are around the mean line, demonstrating the agreement between these two different methods. However, a few data points are around the −1.96 SD line due to the 1000 mg dL−1 glucose concentration shown in Fig. 6e. This error can be easily eliminated in the future. The statistical variation of the glucose detector over the mean average values is shown in Fig. 8c. Most percentage changes over average RGB values are lower than 5%. For the 2000 mg dL−1 concentration, the percentage changes are slightly higher, about 8%. The plot's small variation can prove that this glucose sensor and the automatic detector are steady and consistent. The disposable glucose sensor, when combined with the automatic measurement method, has shown to be a reliable and accurate tool for measuring glucose levels. Therefore, this approach is a promising candidate for glucose monitoring in various settings. | ||
| Fig. 8 Statistical performance. (a) Correlation curve of YOLOv4 and ImageJ. (b) The Bland–Altman plot. (c) Variation for replicates of different concentrations. | ||
5. Conclusions and discussion
The outcome presented in this paper illustrates the ability to use disposable tapes, PDMS, and glass slides to explore and fabricate innovative, accurate, and economical types of glucose sensors. Combining the PDMS extension part and micropump can bring advantages to assist the usage of glucose sensors. This glucose sensor is uncomplicated and flexible, even people without experience can operate it perfectly. The price of one glucose sensor can be lowered to 0.24 dollars per piece, which is even lower than the commercial glucose test kits. The retail price for one sensor is $0.24 USD. Based on bulk production, the estimated price can be $0.19 USD per sensor for large orders. However, the final price is subject to negotiations with the vendor and may vary based on factors such as shipping costs, taxes, and any other applicable fees. The improvement of control of the light environment from 3D printed chassis and LED diodes can provide a consistent and unnoisy light source for measurement. The results presented in this paper demonstrate the ability of the YOLOv4 automatic measurement algorithm to replace the traditional manual measurement method (which in this paper is ImageJ). Computer vision can accelerate the measurement process, and in the future, this algorithm will be transferred onto portable electronics through TensorFlow Lite. The results presented in this paper demonstrate the fundamental research and feasibility. One phone model was tested since it describes the methodology and algorithm using the average RGB value, which is independent of the phone models. For future work, ultimately, if and when this device gets used by users of various smartphone models and cameras, each different phone model will require its own custom phone chassis design (like any smartphone product and its respective accessories) and also will need to be recalibrated for its image response, as every camera model differs from another. The scope of this paper was fundamental research and demonstrating feasibility rather than showing universality across all smartphone models. Finally, the regression curve was given by different concentrations of glucose with a 0.997 R-square which demonstrates the confidence of this model.Scalable manufacturability and flexibility of the substrate material are key strengths of our device. By utilizing a laser cutter to create the channels, the device can be easily scaled to fit the dimensions of various substrates, which allows for efficient and cost-effective manufacturing. In addition, the device is compatible with a variety of sterile substrate materials, which provides flexibility for producers to use whatever substrate they have available. Strong transferability is a key strength of our device, thanks to our flexible manufacturing methods. The device can be easily redesigned to fit other assay candidates and smartphone candidates, which provides versatility and adaptability for users and manufacturers. In addition, the use of low-cost assay test strips makes our sensor a cost-effective diagnosis method that can be adapted to various assays. However, it's important to note that the sensor is color sensitive and requires the assay reaction to bring about obvious color changes for detection. Additionally, the recommended reading time for specific assays may need to be recalibrated. These potential challenges should be taken into account when considering the use of our device for different assay candidates. Overall, the combination of scalable manufacturability, strong transferability, and cost-effectiveness of our device makes it a highly adaptable and valuable solution for a range of applications.
6. Glucose sensor measurement algorithm
To train the YOLOv4 model used in the test strip analysis workflow (shown in Fig. 4), we analyzed 40 test strips (in the GitHub described in the data availability section) that were manually labeled (using Roboflow [https://roboflow.com]; Annotate—Roboflow) with the positions of each glucose test strip (i.e., each test was represented/encapsulated by an ‘information area’); each area was labeled according to what type of test it captured (i.e., the “group information” of each information area). These annotations were transformed into coordinates and used to train the YOLOv4 detector. The custom detectors were trained in Google colab with a clone of the YOLOv4 darknet from Github (https://www.github.com/AlexeyAB/darknet), implemented with the NVDIA CUDA GPU acceleration toolkit. After training, we obtained a weights file which includes all information about the custom detector and can predict the target areas with 99–100% accuracy. The areas covering the glucose strips that are extracted from each image are further processed by Tensorflow (https://www.Tensorflow.org), which is an open-source software library for machine learning and artificial intelligence, to detect the glucose analysis strips. To utilize the weights files/custom detector in a convenient manner on desktop computers, we created the Tensorflow GPU environment using the CUDA toolkit and the Anaconda environment (https://www.anaconda.com) for processing graphical input. To implement YOLOv4 using TensorFlow, we first converted the weights into the corresponding TensorFlow model files and then ran the custom models on desktop computers. We used computer terminal commands to crop the detected areas and saved these as new images for subsequent steps in image processing. The target areas extracted from each image by the trained YOLOv4 detector (i.e., the glucose strip from background) are imported into the colorimetric measurement software in MATLAB (https://www.mathworks.com/) to generate the average RGB values produced by the workflow.Data availability
The glucose strips dataset and workflow are available from https://github.com/zhuolunmeng/A-Smartphone-Based-Disposable-Microfluidic-Glucose-Sensor-Utilizing-Computer-Vision.Author contributions
Zhuolun Meng: conceptualization, methodology, formal analysis, investigation, data curation, writing – original draft, writing – review & editing, software, validation, visualization. Muhammad Tayyab: validation, writing – review & editing. Zhongtian Lin: validation, writing – review & editing. Hassan Raji: validation, writing – review & editing. Mehdi Javanmard: conceptualization, methodology, funding acquisition, project administration, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by the National Science Foundation under Grant 1711165, 1556253 and 1846740.References
- Blood sugar | blood glucose | diabetes, https://medlineplus.gov/bloodsugar.html.
- Blood glucose (sugar) test: Levels and amp; What They mean, https://my.clevelandclinic.org/health/diagnostics/12363-blood-glucose-test.
- National Diabetes Statistics Report, 2022, https://www.cdc.gov/diabetes/data/statistics-report/index.html.
- B. Zhou, Y. Lu, K. Hajifathalian, J. Bentham, M. Di Cesare, G. Danaei, H. Bixby, M. J. Cowan, M. K. Ali and C. Taddei, et al. , Lancet, 2016, 387, 1513–1530 CrossRef PubMed.
- T. G. Drummond, M. G. Hill and J. K. Barton, Nat. Biotechnol., 2003, 21, 1192–1199 CrossRef CAS PubMed.
- A. E. Kamholz, B. H. Weigl, B. A. Finlayson and P. Yager, Anal. Chem., 1999, 71, 5340–5347 CrossRef CAS PubMed.
- K. S. Kim and J.-K. Park, Lab Chip, 2005, 5, 657–664 RSC.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS PubMed.
- P. S. Dittrich, K. Tachikawa and A. Manz, Anal. Chem., 2006, 78, 3887–3908 CrossRef CAS PubMed.
- D. R. Reyes, D. Iossifidis, P.-A. Auroux and A. Manz, Anal. Chem., 2002, 74, 2623–2636 CrossRef CAS PubMed.
- S. R. Mahmoodi, P. Xie, D. P. Zachs, E. J. Peterson, R. S. Graham, C. R. Kaiser, H. H. Lim, M. G. Allen and M. Javanmard, Sci. Adv., 2021, 7, eabf4401 CrossRef PubMed.
- J. Mok, M. N. Mindrinos, R. W. Davis and M. Javanmard, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 2110–2115 CrossRef CAS PubMed.
- Z. Huang, X. Li, M. Martins-Green and Y. Liu, Biomed. Microdevices, 2012, 14, 873–883 CrossRef CAS PubMed.
- M. E. Piyasena and S. W. Graves, Lab Chip, 2014, 14, 1044–1059 RSC.
- D. B. Weibel, W. R. DiLuzio and G. M. Whitesides, Nat. Rev. Microbiol., 2007, 5, 209–218 CrossRef CAS PubMed.
- Z. Lin, S.-Y. Lin, P. Xie, C.-Y. Lin, G. M. Rather, J. R. Bertino and M. Javanmard, Sci. Rep., 2020, 10, 3015 CrossRef CAS PubMed.
- J. Sui, F. Foflonker, D. Bhattacharya and M. Javanmard, Sci. Rep., 2020, 10, 1251 CrossRef CAS PubMed.
- J. Sui, P. Xie, Z. Lin and M. Javanmard, Talanta, 2020, 215, 120791 CrossRef CAS PubMed.
- M. Tayyab, P. Xie, M. A. Sami, H. Raji, Z. Lin, Z. Meng, S. R. Mahmoodi and M. Javanmard, Sci. Rep., 2022, 12, 20119 CrossRef CAS PubMed.
- P. Xie, N. Song, W. Shen, M. Allen and M. Javanmard, Biomed. Microdevices, 2020, 22, 1–9 CrossRef PubMed.
- W. W. Y. Chow, K. F. Lei, G. Shi, W. J. Li and Q. Huang, Smart Mater. Struct., 2006, 15, S112 CrossRef CAS.
- J. El-Ali, P. K. Sorger and K. F. Jensen, Nature, 2006, 442, 403–411 CrossRef CAS PubMed.
- S. R. Quake and A. Scherer, Science, 2000, 290, 1536–1540 CrossRef CAS PubMed.
- A. R. Abate, D. Lee, T. Do, C. Holtze and D. A. Weitz, Lab Chip, 2008, 8, 516–518 RSC.
- D. C. Duffy, J. C. McDonald, O. J. Schueller and G. M. Whitesides, Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS PubMed.
- C. Iliescu, H. Taylor, M. Avram, J. Miao and S. Franssila, Biomicrofluidics, 2012, 6, 016505 CrossRef PubMed.
- P. Yager, T. Edwards, E. Fu, K. Helton, K. Nelson, M. R. Tam and B. H. Weigl, Nature, 2006, 442, 412–418 CrossRef CAS PubMed.
- H. Yu, Y. Lu, Y.-g. Zhou, F.-b. Wang, F.-y. He and X.-h. Xia, Lab Chip, 2008, 8, 1496–1501 RSC.
- J. Y. Zhang, J. Do, W. R. Premasiri, L. D. Ziegler and C. M. Klapperich, Lab Chip, 2010, 10, 3265–3270 RSC.
- H. Cho, J. Kim, C.-W. Jeon and K.-H. Han, Lab Chip, 2017, 17, 4113–4123 RSC.
- U. Eletxigerra, J. Martinez-Perdiguero and S. Merino, Sens. Actuators, B, 2015, 221, 1406–1411 CrossRef CAS.
- G. S. Fiorini and D. T. Chiu, BioTechniques, 2005, 38, 429–446 CrossRef CAS PubMed.
- Y. Xia, J. Si and Z. Li, Biosens. Bioelectron., 2016, 77, 774–789 CrossRef CAS PubMed.
- G. T. Smith, N. Dwork, S. A. Khan, M. Millet, K. Magar, M. Javanmard and A. K. E. Bowden, Lab Chip, 2016, 16, 2069–2078 RSC.
- M. H. Tania, K. T. Lwin, A. M. Shabut, M. Najlah, J. Chin and M. A. Hossain, Expert Syst. Appl., 2020, 139, 112843 CrossRef.
- U. M. Jalal, G. J. Jin and J. S. Shim, Anal. Chem., 2017, 89, 13160–13166 CrossRef CAS PubMed.
- M. K. Kanakasabapathy, M. Sadasivam, A. Singh, C. Preston, P. Thirumalaraju, M. Venkataraman, C. L. Bormann, M. S. Draz, J. C. Petrozza and H. Shafiee, Sci. Transl. Med., 2017, 9, eaai7863 CrossRef PubMed.
- Z. Meng, M. Tayyab, Z. Lin, H. Raji and M. Javanmard, Sensors, 2022, 23, 394 CrossRef PubMed.
- S. Jiang, M. Jafari, M. Kharbeche, M. Jalayer and K. N. Al-Khalifa, IEEE Trans. Intell. Transp. Syst., 2022, 23, 3169–3179 Search PubMed.
- S. Jiang, Y. Zhang, R. Liu, M. Jafari and M. Kharbeche, IEEE Trans. Intell. Transp. Syst., 2022, 23, 18237–18252 Search PubMed.
- Z. Meng, A. Williams, P. Liau, T. G. Stephens, C. Drury, E. N. Chiles, X. Su, M. Javanmard and D. Bhattacharya, Sci. Rep., 2022, 12, 14398 CrossRef CAS PubMed.
- Easy-Macro, Macro: Macro lens for iPhone, Android, or any mobile device, https://www.easy-macro.com/.
- S. Talebian and M. Javanmard, Talanta, 2021, 228, 122244 CrossRef CAS PubMed.
- Q. Mei, H. Jing, Y. Li, W. Yisibashaer, J. Chen, B. N. Li and Y. Zhang, Biosens. Bioelectron., 2016, 75, 427–432 CrossRef CAS PubMed.
- Give your software the power to see objects in images and video, https://roboflow.com/.
- Z. Meng, Biomed. Microdevices, 2023 DOI:10.1007/s10544-023-00682-y.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an01356a |
| This journal is © The Royal Society of Chemistry 2024 |