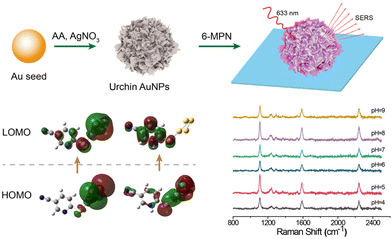
Design and performance of pH-responsive cyano-Raman label SERS probes based on single urchin Au nanoparticles†
Yang
Zhou
,
Zejie
Yu
,
Qirong
Zhou
,
Jiachang
Chen
,
Miaomiao
Cai
,
Yi
Wang
* and
Lei
Zhang
 *
*
State Key Laboratory of Organic Electronics and Information Displays & Institute of Advanced Materials (IAM), Nanjing University of Posts & Telecommunications, Nanjing 210023, China. E-mail: iamlzhang@njupt.edu.cn
First published on 7th November 2023
Abstract
A cyano-Raman label pH-responsive SERS probe was constructed by immobilizing 6-MPN molecules onto the surface of a single urchin Au nanoparticle (AuNP). The effects of different conditions on the synthetic materials were investigated and the optical properties of the single nanoparticles were evaluated. The peak-strength ratio of SERS probes at 1589 cm−1 and 2240 cm−1 exhibited a linear relationship in the pH range 4–7. The properties and stability of the probe were also verified by the acid–base cycle and ion interference tests.
Introduction
Surface-enhanced Raman scattering (SERS) is a technique for enhancing Raman signals based on the molecular enhanced scattering effect of chemical adsorption onto the surface of metal nanostructures.1 SERS technology can be used for high sensitivity analysis and detection, including the measurement of pH value.2 SERS probes can provide very high sensitivity, can detect very low concentration of analytes, and can achieve single-molecule-level detection.3 At the same time, SERS probes have high selectivity and can accurately identify and analyze target molecules in complex sample matrices. This can be achieved by selecting specific functionalized molecules that interact specifically with a particular analyte when it is present. SERS probes can be combined with a variety of analytical techniques, such as spectroscopy,4,5 electrochemistry and imaging techniques.6,7 Due to the high sensitivity and selectivity of SERS probes, they can also be used to image and visualize the distribution and transfer process of target molecules in real time. This is useful in areas such as biological research,8,9 tissue engineering and drug delivery.10Though the electromagnetic enhancement model of SERS has been widely used in developing novel analytical methods, chemical enhancement, as an important branch of SERS research, plays an irreplaceable role in most SERS research studies, especially in the field of pH sensing. The pH value is an indicator to describe the acid–base properties of a solution, due to the protonation process of Raman-labeled molecules that occurs during pH sensing, which leads to a change in their charge distribution, giving rise to interesting electron transfer phenomena. And this process inevitably causes the Raman characteristic peaks of Raman-labeled molecules to be enhanced or attenuated, or even to disappear, or generates new Raman characteristic peaks, which are often used for detection and analysis in the field of chemistry, serving as an important parameter in the chemical reactions of various materials. For example, by measuring the pH of water, the extent of ecosystem damage caused by acidification can be determined.11,12 At present, the main methods for detecting the pH value are colorimetry,13,14 ultraviolet–visible (UV-vis) region spectroscopy,15,16 potential measurements,17 fluorescence spectroscopy18 and SERS probes.19–21
The main principle of pH-responsive SERS probes is based on the relationship between the structure and activity of probe molecules and the pH value of the solution. Such probes generally consist of two main components: gold nanoparticles as a SERS signal enhancing base22 and pH-sensitive molecules that respond to pH changes. The pH-sensitive molecules are adsorbed on the surface of gold nanoparticles. When the pH value changes, the electronic structure or the molecular conformation of the molecules changes due to chemical enhancement, resulting in different characteristic peaks of SERS signals.
Regardless of the kind of SERS probe, the key is to select appropriate pH-sensitive molecules and combine them with gold nanoparticles to establish a clear correlation between the enhanced effect of SERS signals and the change in the characteristic spectral peak.23 Representative studies of pH-sensitive molecules include those on 4-aminobenzene mercaptan (4-ATP),24 2-aminobenzene mercaptan (2-ABT), 4-mercaptobenzoic acid (4-MBA),25 and 4-mercaptopyridine (4-MPy).26 By analyzing the SERS signal measured, the pH value of the solution can be inferred indirectly. The study of pH SERS probes revealed that the pH sensitive molecules selected to achieve a pH response should have amino acid groups, carboxyl groups and pyridine rings (Scheme 1).
In this paper, we successfully designed and prepared a pH response SERS probe on a single nanoparticle which was constructed by immobilizing a cyano-Raman tag molecule (6-mercaptonicotinonitrile: 6-MPN) on the surface of urchin AuNPs. In the experiments, urchin AuNPs were formed on small-sized Au species by the seed growth method in an AgNO3 solution under acidic conditions. The interaction force of the sulfhydryl group of the 6-MPN molecule with urchin AuNPs was utilized to form the urchin AuNPs-6-MPN pH SERS probe. And this nanoscale SERS probe may offer an alternative method for monitoring pH values in confined spaces, such as by entering cancer cells by endocytosis.
Experimental
Detailed synthesis methods are available in the ESI.† Urchin AuNPs were formed on small size Au species by the seed growth method under acidic and AgNO3 regulated conditions, and then a single nanoparticle urchin AuNPs-6-MPN pH-responsive SERS probe was constructed using the strong interaction between the sulfhydryl group of the 6-MPN molecule and gold.Synthesis of urchin AuNPs
AuNP seeds with a diameter of 20 nm were synthesized by the kinetically controlled seeded growth method,27 and then urchin AuNPs were obtained using AuNP seeds of 20 nm. 10 mL of ultra-pure water, 100 μL of 1% (wt%) HAuCl4 solution, 10 μL of 1 mol L−1 of HCl, 20 μL of Au seed solution and 100 μL of 2 wt% sodium dodecyl sulfate were added to a glass bottle in sequence. Then, under intense agitation, 100 μL of AgNO3 and 50 μL of 0.1 mol ascorbic acid (AA) solution were added at the same time, and the reaction was completed after continuous agitation for 10 min.Preparation of SERS substrates
1 mL of urchin AuNPs was centrifuged and redissolved 3 times, and then divided into two groups. Urchin AuNPs were dripped onto ITO glass, and the SERS substrate was prepared after evaporation and drying. Another 1 mL of urchin AuNPs was taken next; the ITO glass sheet was placed in a centrifugal tube, the urchin AuNPs were added, and the ITO glass sheet was immersed in ultra-pure water and sonicated for 15 min. The ITO glass was blow-dried with N2 and a single nanoparticle SERS substrate was prepared.Synthesis of the urchin AuNP-6-MPN probe
0.001 g of 6-MPN was dissolved in 1 ml of ethanol and diluted with water to prepare 6-MPN molecular solutions of different concentrations. Drops of 100 μL molecular solution were added to the SERS substrate, followed by incubation at room temperature for 2 h; then, ITO glass was rinsed with water and dried with N2. Another 1 mL of urchin AuNPs was added to a centrifuge tube; then, 20 μL of the 6-MPN solution was added and the mixture was incubated in a shaker at 25 °C and 300 rpm for 4 h. After centrifugation and re-dissolution, the modified sea urchin AuNP solution was added into a centrifuge tube with ITO glass slides, sonicated for 10 minutes, then dried with N2 and tested.Results and discussion
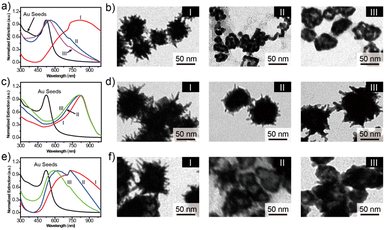
The optical properties of the synthesized Au seeds were characterized by UV-vis technology. As shown in Fig. 1a, the peak of the extinction spectrum of the Au seeds was located at 523 nm. The morphology and size of Au seeds were characterized by TEM, as shown in Fig. 1b. The morphology was relatively uniform and the average diameter of the Au seeds was measured to be 20 ± 5 nm by ImageJ software. The characterization analysis of urchin AuNPs is shown in Fig. 1c. After the addition of AgNO3 and AA, the peak of the extinction spectrum was red-shifted to 780 nm and the full width at half maximum (FWHM) expanded to about 320 nm with the growth of urchin AuNPs. The morphology and size of the prepared urchin AuNPs were characterized using TEM. As shown in Fig. 1d, the nanoparticles were urchin shaped, with lots of burrs around them, and their diameter was up to 85 nm.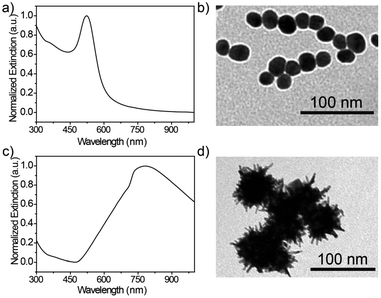 | ||
| Fig. 1 (a) Characterization of the AuNPs and urchin AuNPs, (b) UV-vis and TEM images of Au seeds, and (c) and (d) UV-vis and TEM images of urchin AuNPs. | ||
Effects of the different preparation conditions of urchin AuNPs on their morphology
There are three main factors that affect the morphology of urchin AuNPs: pH, Au seed concentration and silver nitrate concentration. Zhang28et al. found that the morphology of gold nanoparticles prepared by the seed growth method was closely related to the pH value in the reaction solution and claimed that the influence of pH was mainly due to the regulation of the reduction potential of the reducing agent, thus determining the deposition mode of gold atoms on the seed surface. Therefore, the influence of different pH values on the synthesized urchin AuNPs was explored in the experiment. As shown in Fig. 2a, the extinction peaks of urchin AuNPs synthesized at pH 5, 7 and 10 are located at 812, 563 and 542 nm, respectively. In addition, the TEM results indicate that acidic conditions are more favorable for the formation of burr-shaped urchin AuNPs(Fig. 2b), while the burr disappeared in those formed under neutral or alkaline conditions. Urchin AuNPs synthesized at pH 5 have more spikes and provide more “hot spots”.Ag+ is necessary for the synthesis of urchin AuNPs. Without Ag+, polydisperse rods and globules will be formed. Yuan29et al. believe that the main role of Ag+ is not to form Ag branches, but to assist in the anisotropic growth of Au branches. The absorption peaks were located at about 826, 813 and 807 nm in the presence of AgNO3 with 20, 30 and 40 mM concentrations, respectively (Fig. 2c). As shown in Fig. 2d, TEM was used to characterize the morphology of the synthesized urchin AuNPs at different AgNO3 concentrations. The concentration of AgNO3 affects the number of burrs on the surface of urchin AuNPs; the extinction peaks are blue-shifted when the Ag+ concentration is higher, but the number of burrs on the surface of the nanoparticles decreases, tending to form a gold shell. Thus, 20 mM concentration of AgNO3 was chosen for the synthesis of urchin AuNPs.
The concentration of Au seeds is the most important factor in the synthesis process. The absorption peaks of urchin AuNPs synthesized by adding 20, 100 and 200 μL of approximately 5 nM Au seeds were located at 743, 621 and 595 nm, respectively. Obviously, the extinction spectra blue-shift to shorter wavelengths with the increasing Au seed concentration (Fig. 2e). Excessive Au seed dosage will lead to insufficient coupling substitution between gold and silver during the synthesis process, which will result in insufficient branching of Au nanoparticles, producing insufficient burrs and then forming a gold-shell structure (Fig. 2f). It could be inferred that more Au seeds led to a notable decrease of Ag atoms on the surface of AuNPs, making it difficult to form the burr structure. And the core–shell sphere structure exhibited a certain interaction capacity with higher-energy light, but this was disadvantageous for SERS detection. Therefore, the condition of a 20 μL Au seed dosage was chosen to synthesize urchin AuNPs in the experiment.
Characterization of the SERS probe
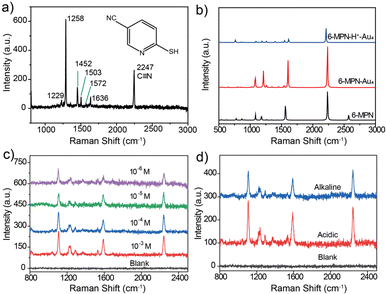
As shown in Fig. 3a, the molecular structure of 6-MPN contains three groups, that is, a pyridine ring, a sulfhydryl group and a cyano group, which meets the requirements of a pH SERS probe. The Raman spectrum of the 6-MPN molecule was recorded (Fig. 3a) by spectral microscopy. The Raman peak located at 2247 cm−1 corresponds to the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N stretching vibration, the 1572 and 1636 cm−1 vibrational peaks correspond to the antisymmetric stretching and the deprotonated C
N stretching vibration, the 1572 and 1636 cm−1 vibrational peaks correspond to the antisymmetric stretching and the deprotonated C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N symmetric stretching vibration of protonated C
C–N symmetric stretching vibration of protonated C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N, and the 1452 and 1503 cm−1 vibrational peaks correspond to the symmetric stretching vibration of protonated C
C–N, and the 1452 and 1503 cm−1 vibrational peaks correspond to the symmetric stretching vibration of protonated C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N. The 1503 cm−1 vibrational peak correspond to the C
C–N. The 1503 cm−1 vibrational peak correspond to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations, the 1288 cm−1 vibrational peaks correspond to the C–N stretching vibrations, and the 1229 cm−1 vibrational peak correspond to the C–H in-plane bending vibrations. The Raman spectra of the 6-MPN molecule, the 6-MPN molecule linked with 4 Au atoms by Au–S covalent bonds (6-MPN-Au4) and 6-MPN-Au4 with protonation of the N atoms (6-MPNH-Au4) were simulated by the DFT method (Fig. 3b). The vibrational peaks of the cyano group were found to be consistent before and after the calculation, while the calculated positions of the C
C stretching vibrations, the 1288 cm−1 vibrational peaks correspond to the C–N stretching vibrations, and the 1229 cm−1 vibrational peak correspond to the C–H in-plane bending vibrations. The Raman spectra of the 6-MPN molecule, the 6-MPN molecule linked with 4 Au atoms by Au–S covalent bonds (6-MPN-Au4) and 6-MPN-Au4 with protonation of the N atoms (6-MPNH-Au4) were simulated by the DFT method (Fig. 3b). The vibrational peaks of the cyano group were found to be consistent before and after the calculation, while the calculated positions of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N ring deformation symmetry stretching vibration and C–N stretching vibration peaks were different from the actual measurements, which may be attributed to the lack of purity of the drug, but the subsequent SERS spectral signals were consistent with the calculation.
C–N ring deformation symmetry stretching vibration and C–N stretching vibration peaks were different from the actual measurements, which may be attributed to the lack of purity of the drug, but the subsequent SERS spectral signals were consistent with the calculation.
By modifying the blank SERS substrate with 6-MPN molecules at different concentrations (10−3, 10−4, 10−5 and 10−6 M), it was found that the intensity of the characteristic peaks of 6-MPN gradually increased with the increase in modification concentration. The SERS substrate modified with 10−4 M 6-MPN molecules was selected for the acquisition of spectra under acid–base conditions. As shown in Fig. 3c, the spectra of the blank SERS substrate and the 6-MPN-modified SERS substrate were acquired under acid–base conditions. It was found that the ratio of the peak intensity of the characteristic peak at 1589 cm−1 to that of the cyanide peak at 2240 cm−1 is slightly higher under acidic conditions than under alkaline conditions (Fig. 3d). Based on this, it is possible to design a probe for pH detection and analysis.
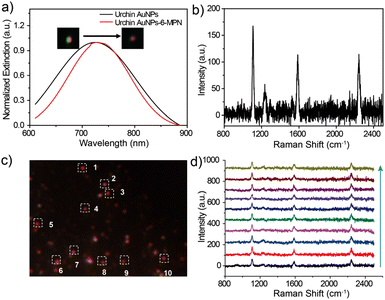
After careful evaluation of the relationship between the SERS scattering signal and the LSPR scattering spectra, it was found that the maximum intensity of Raman spectra could be obtained and kept at a certain level on the urchin AuNPs-6-MPN probes, which had the same LSPR scattering peak position located at about 727 nm. The DFM images of urchin AuNPs before and after the modification with 6-MPN molecules are shown in the inset of Fig. 4a, and the scattering spot of urchin AuNPs modified by 6-MPN exhibited a red color. In addition, the peak of the LSPR scattering spectra of urchin AuNPs-6-MPN was located in the range from 724 to 727 nm. In addition, the SERS spectral signals of the urchin AuNPs-6-MPN probes were consistent with those detected by probes constructed on SERS substrates (Fig. 3d), as shown in Fig. 4b. As 6-MPN and urchin AuNPs are incubated to construct the probe, 6-MPN molecules are randomly modified on the hot spot regions of urchin AuNPs. To validate the effectiveness of nanoparticles as SERS pH nanoprobes, ten urchin AuNPs-6-MPN SERS probes were selected for detection under the same dark-field imaging conditions (Fig. 4c). The SERS spectra of these urchin AuNPs-6-MPN SERS probes were acquired as in (Fig. 4d). The SERS signals of the urchin AuNPs-6-MPN SERS probes were all acquired, and the characteristic peaks of the molecules were stronger at 1229 cm−1, 1589 cm−1 and 2240 cm−1.
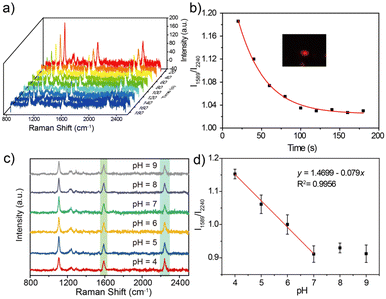
Detection application of the SERS probe
Urchin AuNPs-6-MPN SERS probes were modified on ITO glass slides to examine the pH response of an individual nanoparticle. Individual nanoparticles with red scattering were picked out under a dark-field microscope (as illustrated in Fig. 5b). The response time of the probe under a pH buffer was first tested. SERS spectra of urchin AuNPs-6-MPN SERS probes at pH 4 were recorded. Then a pH 9 buffer solution was added dropwise. SERS spectra were recorded at 20-second intervals after dropping the buffer solution (Fig. 5a). After fitting the characteristic peak intensities in the spectra, it was found that the ratios of Raman peak intensities (RPI) at 1589 cm−1 and 2240 cm−1 varied greatly within 60 s, and stabilized after 140 s (Fig. 5b). Therefore, the response time was set at 140 s for the acquisition of spectral signals.The responsiveness of the urchin AuNPs-6-MPN SERS probe was tested at different buffers with pH = 4, 5, 6, 7, 8 and 9. SERS spectra of the probes were recorded (Fig. 5c). The peak intensities of the collected spectra at 1589 cm−1 and 2240 cm−1 were fitted to obtain the fitted curves of RPI along with equations and correlation coefficients (Fig. 5d). It was found that the RPI of the urchin AuNPs-6-MPN SERS probe decreased with the increase in the pH value of the buffer. The decrease trend of the RPI slowed down after the pH value exceeded 8 and was further fitted to the buffer pH range of 4–7.
Stability detection of urchin AuNPs-6-MPN SERS probes
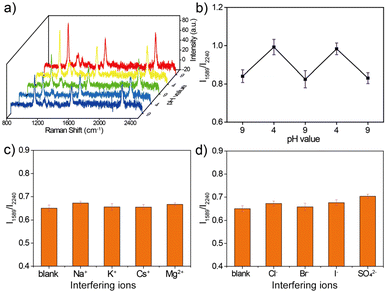
The stability of urchin AuNPs-6-MPN SERS probes during pH detection was tested at pH values 4 and 9. As shown in Fig. 6a, the single-particle SERS probe was sequentially cycled in pH = 9 and pH = 4 buffer solutions to collect SERS spectra. The characteristic peaks of the acquired cyclic SERS spectra were fitted to obtain the peak intensity information, and the RPI were plotted as line graphs (Fig. 6b). Urchin AuNPs-6-MPN SERS probes showed reciprocal cyclic changes of the RPI in the pH range of 4–9, which demonstrated that the SERS probe could realize the repeatable detection of the pH value of the environmental samples to be tested in a certain period of time. This indicates that the SERS probe is capable of reproducing the pH value of the environmental samples to be measured within a certain period of time.Cl−, Br−, I−, SO42−, Na+, K+, Cs+, and Mg2+ were selected to verify the effects of anions and cations on the urchin AuNPs-6-MPN SERS probes. In the presence of the different anions and cations used, the RPI were analyzed and plotted on histograms (Fig. 6c and d), as shown in the SERS spectrum in the ESI (Fig. S2†). The RPI of the urchin AuNPs-6-MPN SERS probes show relatively small variations and stable performance under the interference of anions and cations. It can be seen that the urchin AuNPs-6-MPN SERS probe has an excellent anti-interference ability for pH detection and can be further applied to obtain a stable and reliable pH signal of the analyte.
Urchin AuNPs have a unique LSPR scattering peak (Fig. 4a). This peak is at around 720 nm, and the LSPR peak is red-shifted after modification with the 6-MPN molecule. By measuring the LSPR peaks of the particles under a dark-field microscope, the particles with peak patterns matching those of urchin AuNPs were selected for the subsequent pH assay, and the stability, reproducibility and selectivity of the probes were good.
Conclusions
In this paper, a novel pH-responsive SERS probe on a single-particle level was synthesized by self-assembly of 6-MPN on the surface of urchin AuNPs. Firstly, we chose AgNO3 at a concentration of 20 mM to modulate the synthesis of urchin AuNPs with a diameter of 85 nm at pH 5. Then the urchin AuNPs-6-MPN SERS probes were constructed by modifying urchin AuNPs with 6-MPN molecules at a concentration of 10−4 M. This probe is stable and repeatable over a long period of time and has good immunity to interference. A linear relationship was found between the RPI at 1589 cm−1 and 2240 cm−1 in the SERS probe between pH 4 and 7, which just coincides with the pH range of the microenvironment of tumor cells and thus holds promise for further application in real-time dynamic detection of the tumor microenvironment.Author contributions
Yang Zhou: conceptualization, data curation, validation, formal analysis, investigation, and writing – original draft. Zejie Yu: writing – original draft. Qirong Zou: data curation. Jiachang Chen: software and investigation. Miaomiao Cai: investigation and validation. Yi Wang and Lei Zhang: funding acquisition, resources, methodology, writing – review & editing, project administration, supervision and conceptualization. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are grateful for financial support from the National Natural Science Foundation of China (62075103, 22374081, 22204081), the Natural Science Foundation of Jiangsu Province (BK20211271) and State Key Laboratory of Analytical Chemistry for Life Science (SKLACLS2210).References
- Y. Liu, H. Zhou, Z. Hu, G. Yu, D. Yang and J. Zhao, Biosens. Bioelectron., 2017, 94, 131–140 CrossRef PubMed.
- X. L. Huang, J. B. Song, B. C. Yung, X. H. Huang, Y. H. Xiong and X. Y. Chen, Chem. Soc. Rev., 2018, 47, 2873–2920 RSC.
- X. Qiao, B. Su, C. Liu, Q. Song, D. Luo, G. Mo and T. Wang, Adv. Mater., 2018, 30, 1702275 CrossRef PubMed.
- A. I. Perez-Jimenez, D. Lyu, Z. Lu, G. Liu and B. Ren, Chem. Sci., 2020, 11, 4563–4577 RSC.
- S. Schluecker, Angew. Chem., Int. Ed., 2014, 53, 4756–4795 CrossRef PubMed.
- J.-F. Li, Y.-J. Zhang, S.-Y. Ding, R. Panneerselvam and Z.-Q. Tian, Chem. Rev., 2017, 117, 5002–5069 CrossRef CAS PubMed.
- H. Zhou, J. Zhang, B. Li, J. Liu, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2021, 93, 6120–6127 CrossRef CAS PubMed.
- M. F. Cardinal, E. V. Ende, R. A. Hackler, M. O. Mcanally, P. C. Stair, G. C. Schatz and R. P. Van Duyne, Chem. Soc. Rev., 2017, 46, 3886–3903 RSC.
- S. Yang, X. Dai, B. B. Stogin and T.-S. Wong, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 268–273 CrossRef CAS PubMed.
- S. Laing, L. E. Jamieson, K. Faulds and D. Graham, Nat. Rev. Chem., 2017, 1, 0060 CrossRef CAS.
- Z. Liu, Z. Liu, L. Wu, Y. Li, J. Wang, H. Wei and J. Zhang, Environ. Pollut., 2022, 303, 119094 CrossRef CAS.
- H. Wei, W. Liu, J. Zhang and Z. Qin, Environ. Pollut., 2017, 220, 460–468 CrossRef CAS.
- K. D. Gong, R. R. Jones, K. J. Li, G. J. Xu, H. Y. Cheng, Y. Q. Feng, V. K. Valev and L. W. Zhang, Sens. Actuators, B, 2021, 346, 130521 CrossRef CAS.
- X. M. Nie, X. Wang, Y. P. Tian, Z. Y. Chen, S. Chen, X. Meng and M. B. Fan, Sustainable Mater. Technol., 2021, 30, e00332 CrossRef CAS.
- W. H. Skinner, M. Chung, S. Mitchell, A. Akidil, K. Fabre, R. Goodwin, A. A. Stokes, N. Radacsi and C. J. Campbell, Anal. Chem., 2021, 93, 13844–13851 CrossRef.
- S. Sloan-Dennison, S. Laing, D. Graham and K. Faulds, Chem. Commun., 2021, 57, 12436–12451 RSC.
- Y. Ohno, K. Maehashi, Y. Yamashiro and K. Matsumoto, Nano Lett., 2009, 9, 3318–3322 CrossRef PubMed.
- L. Zhang, Q. Q. Zhao, Z. T. Jiang, J. J. Shen, W. B. Wu, X. F. Liu, Q. L. Fan and W. Huang, Biosensors, 2021, 11, 282 CrossRef.
- M. K. Hossain, Spectrochim. Acta, Part A, 2020, 242, 118759 CrossRef.
- L. Montali, M. M. Calabretta, A. Lopreside, M. D'elia, M. Guardigli and E. Michelini, Biosens. Bioelectron., 2020, 162, 112232 CrossRef PubMed.
- X. Zhou, Z. W. Hu, D. T. Yang, S. X. Xie, Z. J. Jiang, R. Niessner, C. Haisch, H. B. Zhou and P. H. Sun, Adv. Sci., 2020, 7, 2001739 CrossRef PubMed.
- J. Reguera, J. Langer, D. Jimenez de Aberasturi and L. M. Liz-Marzan, Chem. Soc. Rev., 2017, 46, 3866–3885 RSC.
- J. Wang, D. W. Liang, Q. Q. Jin, J. Feng and X. J. Tang, Bioconjugate Chem., 2020, 31, 182–193 CrossRef.
- J. E. Park, N. Yonet-Tanyeri, E. Vander Ende, A. I. Henry, B. E. P. White, M. Mrksich and R. P. Van Duyne, Nano Lett., 2019, 19, 6862–6868 CrossRef PubMed.
- N. Hao, J. W. Lu, Z. Zhou, R. Hua and K. Wang, ACS Sens., 2018, 3, 2159–2165 CrossRef.
- C. Long, L. W. He, F. Y. Ma, L. Wei, Y. X. Wang, M. A. Silver, L. H. Chen, Z. Lin, D. X. Gui, D. W. Juan, Z. F. Chai and S. A. Wang, ACS Appl. Mater. Interfaces, 2018, 10, 15364–15368 CrossRef PubMed.
- N. G. Bastús, J. Comenge and V. Puntes, Langmuir, 2011, 27, 11098–11105 CrossRef.
- X. Zhang, R. Gallagher, D. He and G. Chen, Chem. Mater., 2020, 32, 5626–5633 CrossRef CAS.
- H. Yuan, C. G. Khoury, H. Hwang, C. M. Wilson, G. A. Grant and T. Vo-Dinh, Nanotechnology, 2012, 23, 075102 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the chemicals, instrumentation models, and probe synthesis methods. See DOI: https://doi.org/10.1039/d3an01678a |
| This journal is © The Royal Society of Chemistry 2024 |