Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Towards a better understanding of the effect of protein conditioning layers on microbial adhesion: a focused investigation of fibronectin and bovine serum albumin layers on SiO2 surfaces
Maya
Rima
 a,
Christina
Villeneuve-Faure
a,
Christina
Villeneuve-Faure
 b,
Marvine
Soumbo
ab,
Fatima
El Garah
b,
Marvine
Soumbo
ab,
Fatima
El Garah
 a,
Ludovic
Pilloux
a,
Ludovic
Pilloux
 a,
Christine
Roques
a,
Christine
Roques
 *a and
Kremena
Makasheva
*a and
Kremena
Makasheva
 *b
*b
aLGC, University of Toulouse, CNRS, UTIII, INPT, Toulouse, France. E-mail: roques730@aol.com
bLAPLACE, University of Toulouse, CNRS, UTIII, INPT, Toulouse, France. E-mail: kremena.makasheva@laplace.univ-tlse.fr
First published on 22nd April 2024
Abstract
The interaction of foreign implants with their surrounding environment is significantly influenced by the adsorption of proteins on the biomaterial surfaces, playing a role in microbial adhesion. Therefore, understanding protein adsorption on solid surfaces and its effect on microbial adhesion is essential to assess the associated risk of infection. The aim of this study is to evaluate the effect of conditioning by fibronectin (Fn) or bovine serum albumin (BSA) protein layers of silica (SiO2) surfaces on the adhesion and detachment of two pathogenic microorganisms: Pseudomonas aeruginosa PAO1-Tn7-gfp and Candida albicans CIP 48.72. Experiments are conducted under both static and hydrodynamic conditions using a shear stress flow chamber. Through the use of very low wall shear stresses, the study brings the link between the static and dynamic conditions of microbial adhesion. The results reveal that the microbial adhesion critically depends on: (i) the presence of a protein layer conditioning the SiO2 surface, (ii) the type of protein and (iii) the protein conformation and organization in the conditioning layer. In addition, a very distinct adhesion behaviour of P. aeruginosa is observed towards the two tested proteins, Fn and BSA. This effect is reinforced by the amount of proteins adsorbed on the surface and their organization in the layer. The results are discussed in the light of atomic force microscopy analysis of the organization and conformation of proteins in the layers after adsorption on the SiO2 surface, as well as the specificity in bacterial behaviour when interacting with these protein layers. The study also demonstrates the very distinctive behaviours of the prokaryote P. aeruginosa PAO1-Tn7-gfp compared to the eukaryote C. albicans CIP 48.72. This underscores the importance of considering species-specific interactions between the protein conditioning layer and different pathogenic microorganisms, which appear crucial in designing tailored anti-adhesive surfaces.
I. Introduction
The tremendous effort dedicated to the understanding and advancement of innovative biomedical devices arises from their ability to improve and even to save patients’ lives. However, the implantation of these foreign materials into the human body poses unavoidable risks.1 According to the US Center for Prostheses Infection, biomedical devices are implicated in 50% of annual nosocomial infections, making them a major concern in healthcare systems worldwide, with serious clinical and economic burden.2 These infections are typically generated by microbial colonization mainly as a complex slimy “biofilm” leading to both clinical illness and device dysfunction.3 Indeed, upon their initial adhesion to the surface, the adhered microbes proliferate and excrete gelatinous polymeric substances, thus forming a resilient mature biofilm. The harm by this microbial community lies in its ability to escape the host's immune defense, as well as to tolerate high doses of antimicrobial agents.4The initial adhesion of microbes on foreign implants is strongly related to surface properties and surface conditioning by various components of corporal fluid, particularly by proteins.5 Indeed, protein adsorption is considered one of the first biological processes to occur upon implantation, resulting in protein layers with various conformations, orientations, and quantities and favoring cellular and microbial adhesion.6–8
Regarding microbial adhesion, adhesins, which are cell-surface components, have been recognized to be strongly involved in microbe–protein interactions, thus ensuring successful microbial colonization.9,10 Therefore, the type and the properties of protein conditioning layers, such as their roughness and hydrophobicity, play a key role in defining the infection risks associated with an implanted material.11
Among the various proteins found in the human body, fibronectin (Fn) is well known as the main mediator of microbial and cellular adhesion on foreign substrates. Fn is a dimeric, high-molecular weight (monomer weight: 230–270 kDa) multidomain glycoprotein present in two major forms: soluble plasma Fn and less-soluble cellular Fn, assembled into the extracellular matrix.12–14 While Fn adsorption on implants can be beneficial in terms of enhancing their bio-compatibility, particularly by promoting human cell migration, adhesion, and differentiation, this protein layer poses a serious risk of infection.15,16 Indeed, several studies have proved the high affinity between Fn and adhesins of many pathogenic microbes, leading to a wide variety of diseases.13
On the other hand, albumin (66.43 kDa) is known as the major plasma protein with a physiological concentration of 33 to 52 g L−1.17 In addition to its important metabolic role, albumin is also able to bind to the surface of implants upon their implantation. Despite its notable role in cell growth, especially of stem cells, as well as in tissue formation and regeneration, albumin has shown a significant anti-adhesion effect on microbes in several studies.18–21
In order to gain insight into the influence of the protein conditioning layer on microbial adhesion, and hence on the associated risk of infection, we experimentally investigated in this work the role of protein adsorption and conformation changes on SiO2-solid surfaces. The choice of SiO2 thin layers as a support is based on their well-established biocompatibility, classified by the US Food and Drug Administration (FDA) as “Generally Recognized as Safe” (GRAS). Despite the large number of studies devoted to exploring the biocompatibility of silicon oxides in vivo or in vitro, the reported results and the description of the physicochemical properties that drive or influence the biocompatibility of the SiO2 surface remain limited. Furthermore, many nanotechnology devices for biomedical applications involve SiO2 thin layers, in particular when biosensing and/or Complementary Metal Oxide Semiconductor (CMOS)-compatible bioelectronic devices are forecast.22 In addition, the SiO2 thin layers are well-known for their optically transparent properties in the visible range of the spectrum and can successfully be used as antireflective coatings, if properly dimensioned. The interest towards SiO2 thin layers is also based on their extensive application in plasmonics as a host matrix for biological studies, in microelectronics as diffusion or thin electrical insulating layers, among others.23,24 When talking about CMOS compatible devices, the development also involves the synthesis of thin SiO2 layers. Other than the thermal growth of SiO2 on Si-wafers, the plasma (electrical discharge) based deposition methods appear as very versatile ways of SiO2 layer deposition on Si-substrates,24 allowing a fine control over the thickness and the composition of the deposited SiO2 thin layers. Moreover, the plasma deposition of thin silica layers allows the development of strategies for surface coating of different medical implants, for example titanium-made dental implants.25 All these potential applications of SiO2 thin layers require a deeper understanding of the physicochemical properties and the basic principles of the complex material surface–microbiological interactions.26
Therefore, the main objective of this study was to explore the evolution of the adhesion and detachment profiles of opportunistic microorganisms (Pseudomonas aeruginosa and Candida albicans) on the surface of thin silica (SiO2) layers after conditioning with a thin layer of human plasma fibronectin (Fn) or of bovine serum albumin (BSA).
For P. aeruginosa, a major nosocomial pathogen widely associated with medical device-related infections,27 the studies were firstly performed under static conditions. Subsequently, the dynamic behavior of bacterial shear-induced detachment from the surfaces was evaluated in a broad spectrum of wall shear stresses. Atomic force microscopy (AFM) was employed to characterize the adsorption and organization of proteins to form conditioning layers on the SiO2 surfaces, as well as to localize adhered bacteria. We also investigated the effect of protein conditioning of the SiO2 surface on the adhesion of C. albicans, a yeast widely recognized for its important implication in nosocomial infections, leading to severe illnesses and even fatalities.28,29
II. Experimental
II.1 Proteins
The two proteins used in this study were human plasma fibronectin, usually existing as a dimer (Fn, MW: 450 kDa for the dimer) and bovine serum albumin (BSA, MW: 66.43 kDa), both purchased from Sigma-Aldrich, St Quentin Fallavier, France under a lyophilized powder form. In order to avoid possible interactions, water for injectable preparations (WIP, French Pharmaceutical Cooperation, Cooper, Melun, France) was used in the preparation of the protein solutions. The pH-value of the WIP was measured to 7.0 with a conductivity of 1.2 μS cm−1. The pH-value of the BSA stock solution was measured to 5.6. The pH-value of the Fn stock solution was measured to 7.5. For both proteins, all diluted solutions were prepared from a stock solution (1.0 g L−1) stored at −20 °C.II.2 Elaboration of thin thermal silica (SiO2) layers
The thin SiO2 layers (100 nm-thick) used in this study were thermally grown on pre-cleaned Si-substrates (Sil'tronix) at 1100 °C under a controlled slightly oxidizing atmosphere (N2 with 1% O2). The samples were pre-cut to always have an exposed surface of 1 cm2. The coupons were designed to fit into the shear-stress flow chamber.II.3 Preparation of thin protein conditioning layers on the SiO2 surfaces
The method used in the preparation of the thin protein conditioning layers on the SiO2 surface was dip coating.30 It allows a fine control over the homogeneity of the protein layer. Briefly, the SiO2/Si samples were immersed in 1.0 mL of protein solution at a defined protein concentration. After one hour of immersion at room temperature, the samples were rinsed with WIP to remove non-adsorbed proteins on the surface and air-dried.Molar and mass concentrations of the tested protein solutions are presented in Table 1. For the next sections and in all graphs, we refer to molar concentrations in solution, when describing the resulting protein conditioning layer. It should be noted that the choice of these concentrations was based on evaluating low and high ones while encompassing physiological levels. Given Fn's physiological concentrations range of 0.3–0.4 g L−1,12 both lower and slightly higher concentrations were examined in order to elucidate the correlation between Fn concentration, organization, and microbial adhesion. As for BSA, concentrations close to those of Fn were tested for comparison, alongside slightly higher concentrations reflecting BSA's elevated physiological levels (33–52 g L−1).17
| Proteins | Concentrations | |
|---|---|---|
| Molar (μM) | Mass (g L−1) | |
| Fibronectin (Fn) | 0.11 | 0.05 |
| 1.0 | 0.45 | |
| 2.0 | 0.90 | |
| Bovine serum albumin (BSA) | 0.75 | 0.05 |
| 2.0 | 0.13 | |
| 10.0 | 0.66 | |
| 100.0 | 6.60 | |
II.4 Microbial strains and culture conditions
The two microbial strains used in this study were the opportunistic bacterium P. aeruginosa (PAO1-Tn7-gfp, Tn7 chromosomal insertion of gfp, provided from the Department of Microbiology, University of Washington, Seattle, Washington, USA)31 and the yeast C. albicans CIP 48.72 (Collection of Pasteur Institute, Paris, France) preserved at −80 °C in a cryoprotective solution.Prior to each experiment, a first subculture of P. aeruginosa PAO1-Tn7-gfp was performed on tryptic soy agar (TSA, Sigma-Aldrich, St Quentin Fallavier, France) at 37 °C for 24 h. The inoculum used in each experiment came from an overnight second subculture in tryptic soy broth (TSB). To eliminate bacterial aggregates, the bacterial culture was filtered under vacuum on a sterile membrane filter (Durapore® Membrane Filter, 5.0 μm, Merck-Sigma, Saint-Quentin Fallavier, France) followed by centrifugation at 2400 rpm for 10 min at room temperature. The pellet was then rinsed twice with WIP to remove all the residual culture medium. The bacterial suspension was prepared in WIP and adjusted to an optical density of OD640 nm = 0.15 at 640 nm, corresponding to a concentration of 108 CFU mL−1. In all assays, the final concentration of the P. aeruginosa PAO1-Tn7-gfp suspension used was 107 CFU mL−1. The viability of the bacteria was monitored all over the experiment.
The same procedure was followed for C. albicans CIP 48.72 cultures, using Sabouraud agar (BioMérieux, Crapone, France) (30 °C, 48–72 h) and Sabouraud Dextrose liquid medium (OXOID, CM0145) (30 °C, 72 h) as culture media. Microbial culture was filtered through a sterile membrane filter (Isopore Membrane Filter, 10.0 μm, Merck Sigma, Saint-Quentin Fallavier, France). Yeasts were then recovered after centrifugation (1300 rpm, 3 min, room temperature), rinsed twice with WIP and adjusted to a final concentration of 107 CFU mL−1 (optical density at 600 nm, OD600 nm = 1.5). The viability of the yeast also was monitored all over the experiment.
II.5 Evaluation of the effect of protein conditioning layers on microbial adhesion/detachment
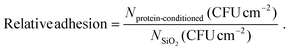
The results are presented as fraction of the adhered cells counted on proteins-conditioned coverslips (Nprotein-conditioned) relative to those adhered on non-conditioned ones (control, NSiO2), according to formula (1):
 | (1) |
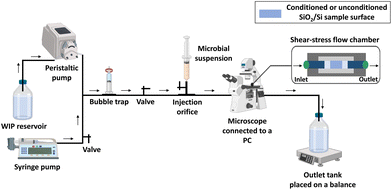
II.5.2.1 Shear-stress flow chamber experimental set-up. Evaluation of the shear-induced detachment of the two tested microorganisms on different non-conditioned and protein-conditioned surfaces of SiO2 thin layers was carried out under hydrodynamic conditions following the protocol described by Guillemot et al.,33 with some modifications. A commercially available shear-stress flow chamber (BST Model FC 71 Coupon Evaluation Flow Cell, BioSurface Technologies Corporation, USA) was used with a house-made customized coupon support, adapted to receive the tested SiO2/Si-samples and to ensure a uniform laminar flow. By including a syringe pump, our experimental arrangement was adapted to work with laminar flows at very low flow rates, in addition to the flow rates involved in the protocol of Guillemot et al. Such improvement gives the possibility to attain very low wall shear stresses. This is a very large step ahead to simulation of wall shear stresses comparable to those exerted on implants in the human body.34–36 A schematic representation of the experimental set-up is shown in Fig. 1.
First, the flow chamber in which the tested surface was inserted, was connected through tygon tubes (tubing size: 16, Masterflex™ Tygon™ E-Lab (E-3603) Pump Tubing, Thermo Fisher Scientific, Illkirch, France) to a syringe pump (CMA/100 Microinjection Pump) and a peristaltic one (Cole-Parmer 7550-50 Masterflex L/S), allowing the application of low and high flow rates, respectively. The wall shear stress (τp, Pa) induced by the different flow rates was calculated using formula (2):
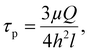 | (2) |
| Wall shear stress range | Flow rate | Wall shear stress τp (Pa) | |
|---|---|---|---|
| mL min−1 | m3 s−1 | ||
| Very low (low flow rates maintained with syringe pump) | 0.048 | 8.0 × 10−10 | 0.01 |
| 0.240 | 4.0 × 10−9 | 0.05 | |
| 0.480 | 8.0 × 10−9 | 0.1 | |
| 0.960 | 1.6 × 10−8 | 0.2 | |
| High (flow rates produced with peristaltic pump) | 24.0 | 4.0 × 10−7 | 5.0 |
| 37.8 | 6.3 × 10−7 | 7.9 | |
| 51.6 | 8.6 × 10−7 | 10.75 | |
| 66.0 | 1.1 × 10−6 | 13.75 | |
II.5.2.2 Detachment assay. The syringe and the tank connected to the peristaltic pump were filled with WIP, for the whole flow chamber experiment. Before starting the assay, the apparatus was pumped with WIP. Air bubbles were evacuated through the bubble trap and/or in the outlet tank. The output reservoir was placed on an electronic balance in order to weigh the outgoing fluid following flow application. The microbial suspension (107 CFU mL−1) was injected through the injection orifice and maintained under static condition for contact time of 1 h 30 min, allowing microbial settling and adhesion.
After the contact time, the number of initially sedimented cells (N0) was determined by microscopic observations. This initially observed cell count comprises both sedimented cells and adhered ones.
For P. aeruginosa PAO1-Tn7-gfp, the microscopic observations were made with a Zeiss-Axiotech epifluorescence microscope using a 20×/0.50 (Zeiss, EC Plan-Neofluar) objective, equipped with an HXP 120 C light source, and a digital camera (Zeiss AxioCam ICm 1), coupled to the ZEN software. For C. albicans CIP 48.72, a numerical optical microscope (Keyence VHX-1000) coupled to VHX 1.3.07 software was used. In order to evaluate the detachment profiles of the two microorganisms adhered on control SiO2 surfaces and on protein conditioned SiO2 surfaces, increasing wall shear stresses were applied each 3 min. This is the required time to get a constant number of cells remaining adhered on the surface. The number of cells remaining adhered on the surface (N) was determined in the observation area after the application of each shear stress. For comparison purposes, the cells remaining adhered are reported in percentage, according to formula (3):
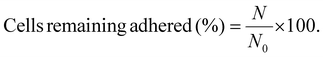 | (3) |
The detachment profiles, representing the percentage of cells remaining adhered on the surface, are plotted as a function of the wall shear stress.
II.5.2.3 Microbial viability control. Microbial viability upon injection (t0) as well as after 1 h 30 min of contact was controlled. For this purpose, the procedure described above (II.5.2.2) was followed while adding 1.0 μL of propidium iodide IP (1 mg mL−1, Invitrogen™, Thermo Fisher Scientific, Illkirch, France) and a mixture of Syto9 (5 mM, Invitrogen™, Thermo Fisher Scientific)/IP (1/1 vol.) to the injected suspension of P. aeruginosa PAO1-Tn7-gfp and C. albicans CIP 48.72, respectively. The percentage of live cells, with respect to t0, was then calculated.
II.6 Atomic force microscopy (AFM) analyses
In order to investigate the organization of adsorbed proteins forming the conditioning dehydrated Fn and BSA layers on the SiO2 surfaces, after contact with several protein concentrations in solution, as well as to localize the adhered bacteria on the stack (conditioning protein layer/SiO2 layer/Si-substrate), AFM microscopic analyses were performed. The dehydrated protein layers were directly studied. For the AFM observations of the adhered bacteria on the conditioned by protein layers SiO2 surfaces, the samples were prepared as described above (II.5.1). After rinsing, the samples with adhered microorganisms were air-dried.The AFM analyses in this work are based on topographic investigations. The topographic data were acquired with a Bruker Multimode mode 8 set-up in the Peak-Force Quantitative NanoMechanical (PF-QNM) mode. A SNL tip with a spring constant of 0.24 N m−1 and a curvature radius of around 5 nm was used to probe the soft materials (the protein layer of interest and the microorganisms). The peak force was set to 0.5 nN.30 For BSA-conditioned SiO2 surfaces, the average surface roughness (arithmetic Sa and quadratic Sq) was determined.
II.7 Statistical analysis
All values were expressed as mean ± standard deviation (SD) from three independent experiments. The corresponding statistical test (one- or two-way ANOVA followed by Dunnett's or Tukey's test for multiple comparisons) as well as all graphs were generated by using GraphPad Prism 10.2.1 for Windows (GraphPad Software, USA). Statistically significant values were defined as a p-value (* < 0.05, ** < 0.01, or *** < 0.001).III. Results
III.1 Evaluation of the effect of protein conditioning of SiO2 surfaces on P. aeruginosa PAO1-Tn7-gfp adhesion and detachment
III.1.2.1 Fn conditioning layer adsorbed on the surface of SiO2 thin layers. Prior to all experiments, the cell viability was assessed after 1 h 30 min of contact with non-conditioned and Fn-conditioned SiO2 surfaces. In both cases, the bacteria (100%) remain viable.
The number of initially sedimented bacteria (N0) for non-conditioned and Fn-conditioned SiO2 surfaces resulting from the contact with solutions with two different concentrations of Fn (0.11 and 1.0 μM) are summarized in (Table 3). For comparison reasons, Table 3 also reports the obtained results for the wall shear stresses required to detach 50% of the initially sedimented bacteria. The first observation to highlight is that no significant difference in the number of sedimented bacteria (N0) is observed under all tested conditions.
| SiO2 surfaces | Molar concentration of proteins in solution (μM) | N 0 (bacteria per cm2) | τ p50% (Pa) |
|---|---|---|---|
| Mean ± SD | |||
| Non-conditioned (control) | n/a | 1.3 ± 0.7 × 105 | 0.09 |
| Fn-conditioning layers | 0.11 | 1.8 ± 0.4 × 105 | 0.08 |
| 1.0 | 1.5 ± 1.1 × 105 | 8.00 |
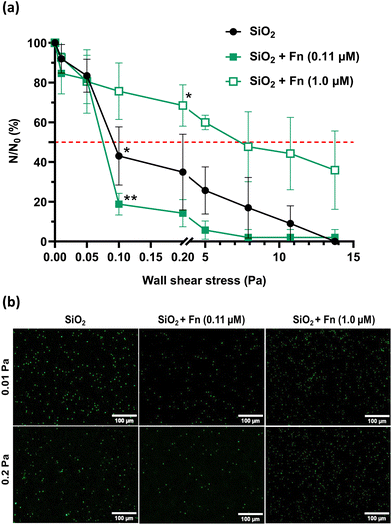
The detachment profiles of P. aeruginosa PAO1-Tn7-gfp under increasing wall shear stresses on bare SiO2/Si samples (control), and on the same surfaces conditioned with protein layers are presented in Fig. 3a. The results are reported according to the two well-distinguished stress ranges, with the first one of very low wall shear stresses (up to 0.2 Pa) and the second focusing on wall shear stresses in the range 5–13.75 Pa, called hereafter “high”.
Analysis of the recorded P. aeruginosa PAO1-Tn7-gfp detachment profiles in the very low wall shear stress range shows a very important observation (Fig. 3a and b). The first applied wall shear stress (0.01 Pa) is too low to exert any impact on the adhered cells. It only serves to evacuate those of the initially sedimented bacteria that have not adhered to the surface. Thus, the very low wall shear stress represents an experimental situation that can be compared to static conditions and the accounted number of cells can be considered representative for the initially adhered cells. This situation underlines the same trends as observed for static conditions (section III.1.1) i.e., there is a lower number of adhered cells for the SiO2 surface conditioned by Fn-protein layer resulting from contact with the lower protein concentration (0.11 μM), compared to the non-conditioned (bare) SiO2 surface. In the same way, when the SiO2 surface is conditioned by a Fn-protein layer resulting from larger protein concentration in solution (1.0 μM), the number of adhered bacteria is identical to the one representing the cell adhesion on the bare SiO2 surface. A slight increase in the wall shear stress, up to 0.05 Pa leads to detachment of the weakly adhered bacteria and thus to leveling out the number of cells remaining adhered on the surface (N) for the control and the two Fn-protein conditioned SiO2 surfaces.
Beyond wall shear stresses of 0.05 Pa, a significant detachment of adhered bacteria is observed on the non-conditioned SiO2 sample surface, resulting in a reduction of nearly 60% of the number of cells remaining adhered for 0.1 Pa. The wall shear stress required to detach 50% of the initially sedimented cells is at 0.09 Pa (τp50% = 0.09 Pa). When examining the SiO2 surface conditioned with the Fn protein layer adsorbed at low protein concentration in solution (0.11 μM), a significant, and even more pronounced detachment is observed at 0.1 Pa. Only 18.8 ± 5.5% of the bacteria remain adhered on the surface. τp50% is at 0.08 Pa in this case. Of particular interest is the obtained large number of cells remaining adhered on the SiO2 sample surface conditioned with the Fn-protein layer resulting from solution with a protein concentration of 1.0 μM for the same wall shear stress (0.1 Pa). The level of remaining adhered bacteria is as high as 75.7 ± 14.2% and is the largest one compared to the two other studied surfaces. For the upper limit of this very low shear stress range (0.2 Pa), the behavior remains unchanged for the three surfaces. Besides, the wall shear stress required to detach 50% of the initially sedimented cells in the case of conditioning of the SiO2 surface with protein layer resulting from 1.0 μM protein solution cannot be attained. The percentage of cells remaining adhered on this conditioned SiO2 surface is of 68.5 ± 10.4% (Fig. 3a and b).
As the wall shear stress is increased to the range of high wall shear stresses, a progressive decrease in the number of bacteria remaining adhered on the three surfaces is observed. A total detachment of the bacteria is achieved at 13.75 Pa for the non-conditioned SiO2 surface and for the SiO2 surface conditioned with Fn-layers resulting from contact with low protein concentration in solution (0.11 μM). The adhesion behavior of bacteria is again different for the SiO2 surface conditioned with Fn-layers resulting from contact with high protein concentration in solution (1.0 μM). A wall shear stress as high as 8.0 Pa is required to detach 50% of initially sedimented bacteria, which is 100 times higher than the wall shear stress needed for the non-conditioned SiO2 surfaces and for those conditioned with Fn layers at a low protein concentration in solution (0.11 μM). Total detachment of the bacteria is not achieved in this case.
III.1.2.2 BSA conditioning layer adsorbed on the surface of SiO2 thin layers. The effect of BSA conditioning layer adsorbed on SiO2 sample surfaces at two protein solution concentrations (0.75 and 10.0 μM) on the detachment profile of P. aeruginosa PAO1-Tn7-gfp was also investigated. In line with the studies performed for the Fn-protein conditioning of the SiO2 surfaces, prior to each experiment, the cell viability preservation was assessed after 1 h 30 min of contact with non-conditioned and BSA-conditioned SiO2 surfaces. In all cases, the majority of bacteria (95%) remain viable.
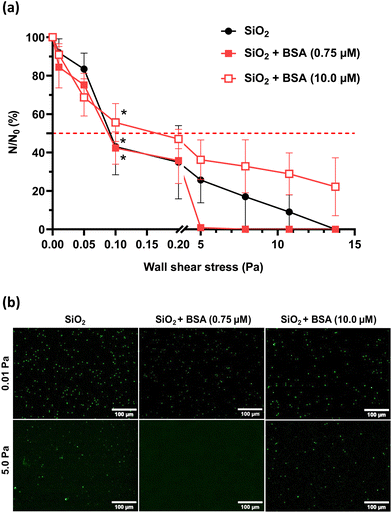
Results show no significant difference in the number of sedimented bacteria between the BSA-conditioned and the non-conditioned SiO2 surfaces (Table 4). For the very low shear stress range, similar detachment profiles are obtained for the non-conditioned SiO2 surface and the SiO2 surface conditioned with protein layer resulting from contact with low-concentration of BSA solution (0.75 μM): 42.3 ± 8.4% of bacteria remain adhered at 0.1 Pa and the 50% limit is attained at τp50% = 0.09 Pa (Fig. 4a).
| SiO2 surfaces | Molar concentration of proteins in solution (μM) | N 0 (bacteria per cm2) | τ p50% (Pa) |
|---|---|---|---|
| Mean ± SD | |||
| Non-conditioned (control) | n/a | 1.3 ± 0.7 × 105 | 0.09 |
| BSA-conditioning layers | 0.75 | 1.0 ± 0.3 × 105 | 0.09 |
| 10.0 | 1.1 ± 1.1 × 105 | 0.20 |
A high wall shear stress (τp = 5.0 Pa) induces a rapid and total detachment of the bacteria from the BSA conditioned SiO2 surface at low protein concentration (0.75 μM) conversely to a bare SiO2 surface (Fig. 4a). A total detachment from the bare SiO2 surface is only achieved at τp100% = 13.75 Pa. When the BSA solution concentration used for conditioning the SiO2 surface is increased to 10.0 μM, a doubled wall shear stress (τp100% = 0.2 Pa) is required to detach 50% of adhered bacteria (Table 4). The defined range of high wall shear stress is insufficient to detach all adhered bacteria under those conditions.
III.2 Evaluation of the effect of Fn and BSA protein conditioning of SiO2 surfaces on C. albicans CIP 48.72 adhesion and detachment under dynamic conditions
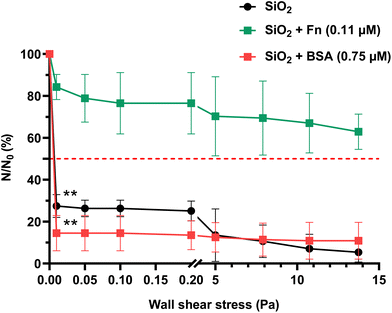
The same as for P. aeruginosa PAO1-Tn7-gfp, the cell viability of C. albicans CIP 48.72 was assessed after 1 h 30 min of contact with non-conditioned, Fn-conditioned (0.11 μM) and BSA-conditioned (0.75 μM) SiO2 surfaces. The following values were respectively recorded: 100%, 99.6% and 96.3%.Regarding the number of initially sedimented yeast cells (N0), no significant difference was noticed between the non-conditioned (control) and protein-conditioned SiO2 surfaces (Table 5). On the non-conditioned SiO2 surface, a significant reduction in the number of yeasts remaining adhered is observed when subjected to the application of the lowest wall shear stress (0.01 Pa). It results in the detachment of nearly 70% of the initial yeast population (27.5 ± 5.5% of remaining cells at 0.01 Pa, τp50% = 0.006 Pa). The adhesion of the remaining yeasts is maintained up to the highest value of the very low wall shear stress range (0.2 Pa) (Fig. 5). Change to the high wall shear stress range leads to a progressive detachment of the yeasts from the bare SiO2 surface, although some cells remain adhered even for the upper limit (τp = 13.75 Pa) of the applied wall shear stress.
| SiO2 surfaces | Molar concentration of proteins in solution (μM) | N 0 (yeasts per cm2) | τ p50% (Pa) |
|---|---|---|---|
| Mean ± SD | |||
| Non-conditioned (control) | n/a | 5.6 ± 0.2 × 104 | 0.006 |
| Fn-conditioning layer | 0.11 | 6.5 ± 0.3 × 104 | n/a |
| BSA-conditioning layer | 0.75 | 6.5 ± 0.4 × 104 | 0.007 |
The BSA-conditioned SiO2 surface with a protein layer resulting from contact with a solution with a protein concentration of 0.75 μM shows a rapid and dramatic detachment of the yeast cells with 14.5 ± 8.4% of cells remaining adhered at 0.01 Pa (τp50% = 0.007 Pa). It appears that the attachment of these residual yeasts on the BSA-conditioned SiO2 surface remains unchanged even though the wall shear stress is increased up to the highest applied value.
The detachment profile of C. albicans CIP 48.72 cells from Fn-conditioned SiO2 surfaces with a protein layer resulting from solution with concentration of 0.11 μM shows a very distinctive behavior in comparison with the recorded profiles of non-conditioned and BSA-conditioned SiO2 surfaces (Fig. 5). The percentage of cells remaining adhered at 0.01 Pa is as high as 84.3 ± 6.0% of the initial population. This is about 3 times higher than the recorded one on the non-conditioned SiO2 surfaces and more than 5 times higher than the one on the BSA-conditioned SiO2 surfaces. Even though the wall shear stress is increased up to the upper limit, the 50% detachment limit cannot be reached. At τp = 13.75 Pa, 62.89 ± 8.4% of the yeasts remain adhered.
III.3 Nanoscale analysis of P. aeruginosa PAO1-Tn7-gfp adhesion on SiO2 surfaces conditioned with Fn and BSA layers
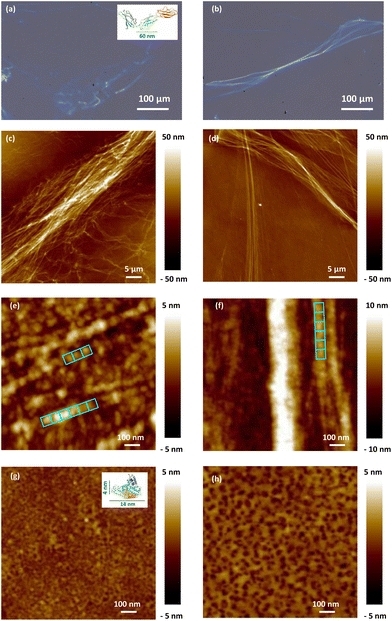 | ||
| Fig. 6 Optical images and PF-QNM surface topography images of Fn-dehydrated conditioning layers adsorbed on SiO2 surfaces using 0.11 μM (a, c and e) and 1.0 μM (b, d and f) protein solution concentrations. The aligned 60 nm × 60 nm blue squares represent the Fn protein domains. PF-QNM surface topography of the BSA-dehydrated conditioning layers adsorbed on SiO2 surfaces using 0.75 μM (g) and 10.0 μM (h) protein solution concentrations. The insets in figures (a) and (g) represent typical Fn and BSA structures and are for illustration purposes only, taken from UniProt.38 | ||
Regarding the Fn conditioning layer, resulting from contact with the low tested Fn solution concentration (0.11 μM), the protein fibril assemblies form branched and rather short fibers (Fig. 6c). When increasing the Fn solution concentration to 1.0 μM, the resulting protein layer on the SiO2 surface is composed of long and relatively straight protein fibers, well aligned in a bundle-like structure (Fig. 6d). These observations indicate modifications in the conformation of the adsorbed Fn in the layer based on the increased concentration of proteins in the solution employed during the SiO2 surface conditioning. As the concentration rises, there is a noticeable transition towards piling and expanding fibers, which may correspond to more significant Fn–Fn interactions.
Looking at the protein organization on the SiO2 surface at a higher resolution, the two Fn protein solution concentrations lead to fibers formed of small and aligned blocks, more visible at a low concentration, measuring around 60 nm × 60 nm, which corresponds to the dimensions of the Fn domain (type III) (Fig. 6e and f). Measurements of the fiber height indicate a value of 8.0 ± 1.0 nm and its multiples. The multiplication of this height can be attributed to either a pile-up of Fn molecules or to adoption of more complex conformations of the proteins in contact with the SiO2 surface.
For the dehydrated BSA conditioning layer, the results suggest a complete coverage of the SiO2 surface for the low (0.75 μM) and the high (10.0 μM) BSA solution concentrations (Fig. 6g and h). The surface topography of the adsorbed BSA conditioning layer is completely different from that of the Fn one, lacking any visible particular arrangement on the surface. The measured heights (<5 nm) suggest that the BSA proteins adsorb on the SiO2 surface in a “side-on” configuration, i.e., with their major axis parallel to the surface.30
Moreover, the surface roughness of the resulting protein layers increases with the protein concentration in solution. Indeed, considering a 1 μm × 1 μm surface, the BSA protein conditioning layer resulting from solution concentration of 0.75 μM displays a lower surface roughness (Sa = 0.4 nm, Sq = 0.5 nm) compared to the one obtained from 10.0 μM (Sa = 0.6 nm, Sq = 0.8 nm). This is a sign of an increased surface concentration of the BSA proteins in the layer.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 μm2 (7 zones × 2500 μm2 each). The number of adhered bacteria found in this area was 20 in total, with 17 bacteria located on Fn-protein covered regions and only 3 bacteria adhered on a SiO2 zone non-covered by Fn-proteins. All bacteria, adhered on the Fn-proteins were located on the Fn-fibers, as shown in Fig. 7a.
500 μm2 (7 zones × 2500 μm2 each). The number of adhered bacteria found in this area was 20 in total, with 17 bacteria located on Fn-protein covered regions and only 3 bacteria adhered on a SiO2 zone non-covered by Fn-proteins. All bacteria, adhered on the Fn-proteins were located on the Fn-fibers, as shown in Fig. 7a.
For the interaction of P. aeruginosa PAO1-Tn7-gfp with a BSA conditioning layer of the SiO2 surface from a solution concentration of 0.75 μM, the bacteria are systematically observed with their flagella (Fig. 7b). However, for a BSA conditioning layer resulting from high solution concentration (10.0 μM), a reorganization in the BSA protein layer surrounding the bacteria is detected. The latter is found extending to nearly 200 nm and with a height of 4 nm which corresponds to the BSA small dimension (Fig. 7c). In this case, the bacterial flagella are no visible.
IV. Discussion
Microbial adhesion followed by colonization of surfaces are widely recognized as being the initial stage of infections associated with biomedical devices. These steps depend on diverse physicochemical interactions, especially those involving a protein conditioning layer that spontaneously forms on indwelling medical devices immediately after their contact with a physiological fluid.39 Indeed, the type, composition, conformation, surface density, and orientation of the proteins within this layer can vary depending on the physicochemical properties of the surface.40 In this context, several studies have highlighted the significant correlation between the characteristics of the surface and those of the conditioning protein layer, and their ability to either promote or inhibit microbial adhesion.41,42 However, these studies consider complex body fluids, such as human saliva and human serum, which are known to contain mixtures of a large number of proteins. As identified by Kallas et al.,41 there are at least 10 most significant proteins, in addition to other constituents, as cations which are largely involved in the process. The richness of this media prevents from a fine description of the relative contribution of each constituent.In the present study, we have evaluated the effect of protein conditioning layers adhered on the surface of SiO2 thin layers, on the adhesion of P. aeruginosa PAO1-Tn7-gfp under static and dynamic conditions. The combination of these two complementary methods represents one of the unique aspects of this study, enabling the exploration of two key phenomena: initial adhesion in the static experiment and its strength in the dynamic one, depicted by the detachment profile.
The methodology applied in this work is for the evaluation of microbial detachment under dynamic conditions with increasing wall shear stresses. By combining all stresses into a single experiment, as performed in this work, uniform environmental factors across all rates were ensured, leading to more robust results compared to an experimental approach based on separate stress variations. In addition, the applied approach primarily focuses on investigating the impact of protein conditioning layers on microbial adhesion/detachment by comparing protein conditioned and non-conditioned SiO2 surfaces. Consequently, any potential microbial adaptation to increasing wall shear stresses would occur on both protein conditioned and non-conditioned samples.
Two proteins were considered, fibronectin and bovine serum albumin, aiming at to focus on the relative contribution of each specific type of protein and the protein organization in the layer. The selection of the two proteins was based on their specific properties: Fn was chosen for its well-known involvement in cell adhesion and biofilm formation,13,43 while BSA was selected as a model protein due to its physicochemical properties similar to those of human serum albumin, the major plasma protein.18 Dip coating method was applied for conditioning of the SiO2 surfaces since it naturally represents similarities with the immersion of implantable devices in corporal fluids. For covering a large range of experimental conditions, the SiO2 surfaces were brought to contact with protein solutions with varying concentrations. On the other hand, in our quest for a comprehensive understanding of the interaction between protein conditioning layer and microbial adhesion, we extended our investigation to C. albicans CIP 48.72, an opportunistic fungus known for its increased ability to form biofilms on implantable medical devices, leading to recalcitrant infections.44 To investigate microbial adhesion/detachment under dynamic conditions, a shear-stress flow chamber was used, employing a broad range of wall shear stresses. Variation of the wall shear stress is particularly interesting and enlightening, as the very low wall shear stresses simulate conditions representative of slow-flowing implants, such as ocular ones, while the high wall shear stresses are typical of high-flowing implants, similar to urinary catheters.34–36
Starting with the analysis of the results obtained with a non-conditioned SiO2 surface, a notable difference appeared between the two very different in nature microorganisms (Fig. 3a and 5). While P. aeruginosa PAO1-Tn7-gfp bacterium exhibited a gradual detachment when increasing the wall shear stress, C. albicans CIP 48.72 yeast displayed a swift detachment upon the application of the lowest wall shear stress (0.01 Pa). These findings highlight the diverse features displayed by the microbial adhesion processes according to the corresponding kingdom, genus, species, size and strain.
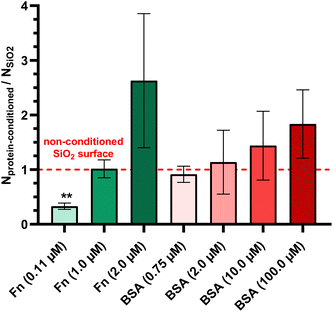
The adhesion/detachment process of P. aeruginosa PAO1-Tn7-gfp under static conditions was found dependent on the conditioning Fn-protein layer, with the latter being functional of the protein concentration in solution. In particular, the bacterial adhesion was observed identical for non-conditioned SiO2 surfaces and for SiO2 surfaces conditioned with protein layers resulting from high Fn concentration in solution (1.0 μM). When the SiO2 surfaces were conditioned with protein layers resulting from low Fn concentration in solution (0.11 μM), a significant reduction in bacterial adhesion was found (Fig. 2). This finding was confirmed under dynamic conditions for the very low wall shear stress (0.01 Pa) which could be considered comparable with the rinsing phase under the static condition experiment. It underlines the removal of non-adhered bacteria out of all sedimented ones on the surface (Fig. 3a). Moreover, it places the adhered bacteria on the three different surfaces (non-conditioned, Fn-conditioned after contact with low and high protein concentration solutions) in the same order as under static conditions. The observed differences in the adhesion of P. aeruginosa PAO1-Tn7-gfp under dynamic conditions can be attributed to either increased surface concentration of the Fn proteins in the layer resulting from contact with a higher protein concentration in solution or to a variety of conformations of the adsorbed Fn. Actually, the performed PF-QNM analysis revealed the link between the bacterial adhesion and the adopted conformation of Fn. It should be noted that the elasticity of the Fn proteins and its ability to adopt a wide variety of assemblies and conformations have been demonstrated in various studies.45,46 Our results are in line with these reports. They show a fibrillar organization of the Fn domains for the conditioning layers resulting from the two tested protein concentrations (Fig. 6c and d). However, at 0.11 μM Fn in solution, a fibrillar pattern with short branches is observed in the protein layer, suggesting an early arrangement of Fn in a “bead-on-a-string” structure with periodicity of 60 nm (Fig. 6e and f). Such chain-like assemblies of Fn have been observed for protein interaction with negatively charged surfaces.47 According to our results, the surface charge does not appear a necessary condition for their formation, although the electrostatic interactions are most likely at their origin. However, the results reported here confirm that such chain-like assemblies can be obtained only after protein adsorption on solid surfaces. The obtained protein layer is discontinuous due to its structuration in fibers. The protein surface concentration resulting after contact with the low Fn-concentration in solution is found: Γ = 1.32 μg cm−2.48 Besides, the refractive index of such a protein layer is measured of n = 1.62, which is a typical value for a polymer layer. Higher concentration of Fn in the solution leads to the formation of long and well aligned Fn fibers, suggesting also a higher protein surface concentration (multiple superposed fibers). Concerning the high Fn concentration in solution (1.0 μM), the extended length, as well as the structuring of the Fn fibers in bundles, can explain the enhanced bacterial adhesion obtained under this condition. In this context, Khan et al. demonstrated that Fn-proteins adsorbed in a fibrillar form promote the adhesion of the Gram-positive Staphylococcus epidermidis, in contrast to the folded protein form.49 The authors suggest that this dissimilar behavior arises from the position of the protein binding sites, which are exposed in the fibrillar Fn structure and buried in the folded one. Our results go beyond this demonstration. The adhesion of P. aeruginosa PAO1-Tn7-gfp also depends on the Fn-fiber organization on the SiO2 surface. It is reduced for Fn-protein layers with short and branched fibers, resulting from protein solution with a lower concentration and strengthened when occurring on fibrillar Fn-layers with long and aligned in bundle fibers, resulting from solution with a higher protein concentration.
Moreover, the PF-QNM surface topography studies of P. aeruginosa PAO1-Tn7-gfp adhered to Fn-conditioned SiO2 surfaces confirm the strong affinity of bacteria for Fn fibers, since a clear preference for adhesion to protein fibers was observed (Fig. 7a). The effect is more pronounced for Fn-layers with long and well aligned fibers, stemming from a high protein concentration in solution. Interestingly, the bacterial flagella are prominently visible on the Fn fibers, which suggests implication of this organelle in the bacteria–Fn interaction and thus in cell adhesion. In this context, various studies have highlighted the crucial role of flagella, a bacterial apparatus composed mainly of flagellin, in bacterial adhesion and invasion.50 In addition, the study conducted by Moraes et al. emphasized the substantial involvement of flagellin of an atypical enteropathogenic Escherichia coli in its interaction with Fn.51
Despite the scarcity of studies investigating P. aeruginosa PAO1 adhesion on Fn-conditioned SiO2 surfaces, the bacterial-Fn affinity has been demonstrated in some previous publications, both at a cellular scale and on inert surfaces. For instance, previous research has revealed a close association between P. aeruginosa PAO1 adhesion and the elevated deposition of fibronectin on cystic fibrosis airway epithelial cells.52 On the other hand, Arhin et al., have identified OprQ, an outer membrane protein, as Fn-binding protein (FnBP). The loss of the expression of this porin has been shown to hinder bacterial adhesion to Fn in vitro (wells coated after interaction with a protein solution at 10.0 μg mL−1).53,54
Concerning the effect of the Fn conditioning layer on the adhesion/detachment of C. albicans CIP 48.72, interestingly, unlike P. aeruginosa PAO1-Tn7-gfp, this yeast exhibits a strong adhesion to SiO2 surfaces conditioned with Fn protein layers, even at the lowest protein concentration in solution (0.11 μM) (Fig. 5). The difference in adhesion between these two microorganisms can be attributed to their distinct adhesion patterns involved in their respective behaviors. Regarding the C. albicans yeast, the fungal surface protein phosphoglycerate mutase 1 (Gpm1) has been shown to possess good affinity for Fn.55 Furthermore, the work of Rauceo et al. sheds light on another key player in C. albicans adhesion to Fn, namely Als5p (agglutinin-like protein 5), surface adhesin expressed by the fungus.56
On the other hand, the adhesion under static conditions of P. aeruginosa PAO1-Tn7-gfp on SiO2 surfaces shows a positive correlation with the conditioning protein layers, resulting from contact with increasing BSA concentrations in solution (Fig. 2). In this context, Yang et al. revealed that BSA adsorption on polyvinylidene fluoride microfiltration membrane enhances the adhesion of P. aeruginosa CMCC10104.57 This improvement in adhesion was attributed to the alteration of the membrane properties following BSA adsorption, notably an increase in hydrophobicity, thus favoring its proadhesive character. An increase in hydrophobicity of the SiO2 surfaces after BSA conditioning from increasing BSA-concentrations in solution was also demonstrated in our prevous work.30 Moreover, except for the very high protein concentration in solution (100.0 μM), for which the BSA proteins are “end-on” adsorbed on the SiO2 surface and the resulting protein layer is not a monolayer but adopts complex structuring, the two other protein layers, those resulting from BSA concentration in solution of 0.75 μM and 10.0 μM, are formed from “side-on” adsorbed BSA proteins on the SiO2 surface.30 They both are continuous monolayers as obtained by the AFM measurements, with an increase in their surface roughness (Fig. 6). The protein surface concentration is around Γ = 0.58 μg cm−2 for the protein layer resulting from BSA concentration in solution of 0.75 μM and almost doubled for the larger concentration in solution, keeping the same refractive index n = 1.61.48 However, a more focused investigation of the protein surface concentration, the fate of adsorbed on the surface proteins and the integrity of these thin protein conditioning layers is worthy, especially under dynamic conditions of microbial adhesion/detachment. Implementation of a quartz crystal balance would definitely benefit the analyses of these protein monolayers containing very small protein surface concentrations.
The interaction of P. aeruginosa PAO1-Tn7-gfp with these two protein conditioning layers was tested under dynamic conditions. The detachment profiles of P. aeruginosa PAO1-Tn7-gfp are similar as far as the wall shear stress is below 0.2 Pa. The difference in the adhesion of P. aeruginosa PAO1-Tn7-gfp becomes noticeable when the applied wall shear stress is 5.0 Pa and more, with a total bacteria detachment for the protein layer resulting from low BSA concentration in solution (Fig. 4a). This difference can be attributed to the surface roughness of the BSA protein layer, and more generally to the BSA layer structuring as demonstrated in Fig. 7b and c. The positive impact of the surface roughness on bacterial adhesion has been demonstrated in several studies although the reported outcomes remain contradictory and are shown to be dependent on the bacterial species as well as on the experimental conditions.58–61
Concerning the PF-QNM surface topography of P. aeruginosa PAO1-Tn7-gfp adhered to the SiO2 surface conditioned with a BSA layer, resulting from a solution with a protein concentration of 10.0 μM, the bacteria display a tendency to burrow into the BSA layer, thus modifying its local organization and rendering the bacterial flagella undiscernible in the contrary to the BSA layer obtained from solution with a lower protein concentration (0.75 μM) (Fig. 7b and c).
On the other hand, the investigation of the effect of BSA conditioning layer on C. albicans CIP 48.72 adhesion/detachment properties revealed a rapid and significant microbial detachment immediately after application of the lowest wall shear stress (0.01 Pa) (Fig. 5). This observation aligns with the fact that albumin is usually recognized as an eukaryotic cell adhesion-inhibiting protein on inert surfaces.18,62 Similarly, research conducted by Austermeier et al. reported that albumin has no impact on the initial adhesion of C. albicans SC5314 on intestinal epithelial cells.63 This can be attributed to the scarcity of adhesins involved in C. albicans adhesion to BSA, unlike those identified for adhesion to Fn. For instance, the agglutinin-like sequence Als1p, recognized for its role in C. albicans adhesion and biofilm formation, was found to have no part in adhesion to BSA.64 However, its implication in binding to Fn has been well established.65 These cumulative pieces of evidence consistently demonstrate the inhibitory effect of albumin on eukaryotic cell adhesion, including C. albicans, on various surfaces.
All these findings would naturally bring their value to eventual clinical trials, where the fundamental characteristic of this study will successfully draw the application perspectives. Moreover, the selected thin silica layers would find their implementation, given the biocompatibility properties of SiO2. The work here is performed with very thin SiO2 layers, of only 100 nm of thickness, that present a very flat surface and a hydrophilic character. They can be deposited, by a plasma (electrical discharge) based process24 on various surfaces, including titanium, which is largely applied for different kinds of medical implants.25 Thus, this study represents a part of a much broader project aiming at elaboration of antimicrobial surfaces involving SiO2 as a host matrix, to be applied as coating layers for implantable medical devices.
V. Conclusion
In the present work, we have investigated the effect of protein conditioning of the surface of SiO2 thin layers with dehydrated protein layers resulting from contact with protein solutions with different Fn and BSA concentrations on the adhesion and detachment of P. aeruginosa PAO1-Tn7-gfp and C. albicans CIP 48.72 under static and dynamic conditions.The novelty of this work resides in the improvement of the experimental arrangement that allowed the application of very low wall shear stresses, which in turn provided data necessary to perform analyses and to compare with static conditions, thus demonstrating a way to distinguish between the sedimented and adhered microorganisms in the initial phase of microbial adhesion, in relation to a specific protein conditioning layer present on the solid surface. The study successfully addresses the static and dynamic conditions of microbial adhesion and brings the link between the two.
Overall, this study provides new insights into the relationship between protein adsorption and microbial adhesion, which is crucial for the design of novel anti-adhesive surfaces. The obtained results show that the microbial adhesion critically depends on: (i) the presence of a protein layer conditioning the SiO2 surface, (ii) the type of protein and (iii) the protein conformation and organization in the conditioning layer. The presence of a protein conditioning layer on the SiO2 surface alters the microbial adhesion compared to a bare SiO2 surface. Moreover, very distinct behaviours are observed regarding the two tested proteins, Fn and BSA. This effect is reinforced by the amount of proteins adsorbed on the surface and their organization in the layer. While the former is closely related to the protein concentration in solution, the latter results from the dynamic processes of protein adsorption on the surface and the conformation change they undergo. For lower protein concentration in solution, the adsorption of Fn-proteins leads to protein fibril assemblies forming branched and rather short fibers, as demonstrated by PF-QNM analysis. Increasing the Fn protein concentration in solution entails the formation of long and relatively straight protein fibers, well aligned in a bundle-like structure. In both cases, the fibrillar patterns suggest an arrangement of the Fn domains in a “bead-on-a-string” structure with a periodicity of 60 nm and confirm the necessity of interaction with a solid surface for the appearance of such assemblies. A higher Fn-protein concentration in solution results in multiplication of the fiber height that can be attributed to a pile-up of the Fn molecules, thus providing conditions for stronger adhesion of P. aeruginosa PAO1-Tn7-gfp, mainly via binding of the bacterial flagella on the bundle of Fn fibers. The BSA-proteins condition the SiO2 surface differently since they organize in continuous layers, however with increased roughness when the protein concentration in solution is increased. This second condition allows the P. aeruginosa PAO1-Tn7-gfp flagella to be located underneath the BSA protein conditioning layer and thus to strengthen the bacterial adhesion.
The hydrodynamic method applied for the study of P. aeruginosa PAO1-Tn7-gfp adhesion reveals that higher wall shear stresses are required in order to detach 50% of the bacteria initially sedimented on SiO2 surfaces conditioned with protein layers resulting from contact with solution with a higher protein concentration, for either Fn or BSA protein. Complete detachment of the bacteria cannot be achieved even for the upper limit of the wall shear stress range. It is worth recalling that the high wall shear stresses applied in this study are similar to those in urinary catheters. It means that such medical devices require careful treatment.
Switching to the model yeast C. albicans CIP 48.72 in the study demonstrates the very different behaviour of the eukaryote species compared to prokaryote P. aeruginosa PAO1-Tn7-gfp ones and underlines the need for the investigation of different pathogenic microorganisms. Conditioning the SiO2 surface with a BSA protein layer leads to a much smaller number of C. albicans CIP 48.72 cells remaining adhered on the surface, while the presence of a Fn conditioning layer on the SiO2 surface provides conditions for a very strong adhesion of the yeasts. Although different degrees of the percentage of cells remain adhered, a complete detachment of C. albicans CIP 48.72 has not been achieved even for the highest applied wall shear stress in both cases.
In light of these results, further experiments are envisaged to reveal the microbial adhesion on solid surfaces via the interactions between proteins and microbes. Of particular interest are the examination of the protein adsorption and formation of protein conditioning layers on SiO2 surfaces from Fn/BSA protein mixtures and thus the evaluation of their effect on the microbial adhesion. Furthermore, it would also be interesting to explore the mechanisms underlying microbial adhesion to the two tested proteins through molecular approaches in order to enhance our understanding of species-specific microorganism–protein interactions.
Author contributions
MR (data curation, investigation, formal analyses, and writing – original draft), CV-F (data curation, investigation, formal analyses, and writing – review and editing), MS (data curation, investigation, formal analyses, and writing – review and editing), FEG (writing – review and editing), LP (writing – review and editing), CR (conceptualization, methodology, formal analyses, and writing – review and editing), and KM (conceptualization, methodology, investigation, formal analyses, writing – review and editing, and project administration).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by l'Agence Nationale de la Recherche in France, project ANR BENDIS (ANR-21-CE09-0008). The authors thank M. Parsek from the Department of Microbiology, University of Washington, Seattle, Washington, USA, for kindly providing the bacterium P. aeruginosa (PAO1-Tn7-gfp) used in this study. For the purpose of open access, the authors have applied a CC-BY public copyright license to any Author Accepted Manuscript (AAM) version arising from this submission.References
- M. M. Brigmon and R. L. Brigmon, Biomed. Mater. Diagn. Devices, 2022, 28, 1–8 Search PubMed.
- R. O. Darouiche, N. Engl. J. Med., 2004, 350, 1422–1429 CrossRef CAS PubMed.
- R. O. Darouiche, Clin. Infect. Dis, 2001, 33, 1567–1572 CrossRef CAS PubMed.
- Y. M. Wi and R. Patel, Infect. Dis. Clin. North Am., 2018, 32, 915–929 CrossRef PubMed.
- K. Vickery, H. Hu, A. S. Jacombs, D. A. Bradshaw and A. K. Deva, Healthc. Infect., 2013, 18, 61–66 CrossRef.
- Z. Othman, B. Cillero Pastor, S. van Rijt and P. Habibovic, Biomaterials, 2018, 167, 191–204 CrossRef CAS PubMed.
- M. J. Andersen, C. Fong, A. A. La Bella, J. J. Molina, A. Molesan, M. M. Champion, C. Howell and A. L. Flores-Mireles, eLife, 2022, 11, e75798 CrossRef CAS PubMed.
- L. Zhang, B. Casey, D. K. Galanakis, C. Marmorat, S. Skoog, K. Vorvolakos, M. Simon and M. H. Rafailovich, Acta Biomater., 2017, 54, 164–174 CrossRef CAS PubMed.
- C. Chagnot, M. A. Zorgani, T. Astruc and M. Desvaux, Front. Microbiol., 2013, 4, 303 Search PubMed.
- B. Singh, C. Fleury, F. Jalalvand and K. Riesbeck, FEMS Microbiol. Rev., 2012, 36, 1122–1180 CrossRef CAS PubMed.
- V. Nandakumar, S. Chittaranjan, V. M. Kurian and M. Doble, Polym. J., 2012, 45, 137–152 CrossRef.
- W. S. To and K. S. Midwood, Fibrog. Tissue Repair, 2011, 4, 21 CrossRef CAS PubMed.
- B. Henderson, S. Nair, J. Pallas and M. A. Williams, FEMS Microbiol. Rev., 2011, 35, 147–200 CrossRef CAS PubMed.
- C. J. Dalton and C. A. Lemmon, Cells, 2021, 10, 2443 CrossRef CAS PubMed.
- L. Parisi, A. Toffoli, B. Ghezzi, B. Mozzoni, S. Lumetti and G. M. Macaluso, Jpn. Dent. Sci. Rev., 2020, 56, 50–55 CrossRef PubMed.
- A. Miranda, D. Seyer, C. Palomino-Durand, H. Morakchi-Goudjil, M. Massonie, R. Agniel, H. Rammal, E. Pauthe and A. Gand, Front. Bioeng. Biotechnol., 2021, 9, 807697 CrossRef PubMed.
- S. Karger, Transfus. Med. Hemother., 2009, 36(6), 399–407 CrossRef.
- O. Kuten Pella, I. Hornyak, D. Horvathy, E. Fodor, S. Nehrer and Z. Lacza, Int. J. Mol. Sci., 2022, 23, 10557 CrossRef CAS PubMed.
- D. B. Horvathy, M. Simon, C. M. Schwarz, M. Masteling, G. Vacz, I. Hornyak and Z. Lacza, BioFactors, 2017, 43, 315–330 CrossRef CAS PubMed.
- T. J. Kinnari, L. I. Peltonen, P. Kuusela, J. Kivilahti, M. Kononen and J. Jero, Otol. Neurotol., 2005, 26, 380–384 CrossRef PubMed.
- T. V. Polyudova, D. V. Eroshenko and V. P. Korobov, AIMS Microbiol., 2018, 4, 165–172 CAS.
- B. Wang, C. Zhao, Z. Wang, K. A. Yang, X. Cheng, W. Liu, W. Yu, S. Lin, Y. Zhao, K. M. Cheung, H. Lin, H. Hojaiji, P. S. Weiss, M. N. Stojanovic, A. J. Tomiyama, A. M. Andrews and S. Emaminejad, Sci. Adv., 2022, 8, eabk0967 CrossRef CAS PubMed.
- J. R. Lakowicz, Plasmonics, 2006, 1, 5–33 CrossRef CAS PubMed.
- A. Pugliara, C. Bonafos, R. Carles, B. Despax and K. Makasheva, Mater. Res. Express, 2015, 2, 065005 CrossRef.
- B. Hussain, S. Khan, A. E. Agger, J. E. Ellingsen, S. P. Lyngstadaas, J. Bueno and H. J. Haugen, J. Funct. Biomater., 2023, 14, 394 CrossRef CAS PubMed.
- C. Liu, D. L. Steer, H. Song and L. He, J. Phys. Chem. Lett., 2022, 13, 1609–1616 CrossRef CAS PubMed.
- M. D. M. Cendra and E. Torrents, Biotechnol. Adv., 2021, 49, 107734 CrossRef CAS PubMed.
- M. Dadar, R. Tiwari, K. Karthik, S. Chakraborty, Y. Shahali and K. Dhama, Microb. Pathog., 2018, 117, 128–138 CrossRef CAS PubMed.
- J. P. Lopes and M. S. Lionakis, Virulence, 2022, 13, 89–121 CrossRef CAS PubMed.
- A. Scarangella, M. Soumbo, C. Villeneuve-Faure, A. Mlayah, C. Bonafos, M. C. Monje, C. Roques and K. Makasheva, Nanotechnology, 2018, 29, 115101 CrossRef CAS PubMed.
- M. J. Kirisits, L. Prost, M. Starkey and M. R. Parsek, Appl. Environ. Microbiol., 2005, 71, 4809–4821 CrossRef CAS PubMed.
- P. Khalilzadeh, B. Lajoie, S. El Hage, A. Furiga, G. Baziard, M. Berge and C. Roques, Can. J. Microbiol., 2010, 56, 317–325 CrossRef CAS PubMed.
- G. Guillemot, G. Vaca-Medina, H. Martin-Yken, A. Vernhet, P. Schmitz and M. Mercier-Bonin, Colloids Surf., B, 2006, 49, 126–135 CrossRef CAS PubMed.
- S. Baillif, E. Casoli, K. Marion, C. Roques, G. Pellon, D. J. Hartmann, J. Freney, C. Burillon and L. Kodjikian, Invest. Ophthalmol. Visual Sci., 2006, 47, 3410–3416 CrossRef PubMed.
- L. C. Gomes, R. Teixeira-Santos, M. J. Romeu and F. J. Mergulhão, in Urinary Stents, ed. F. Soria, D. Rako and P. de Graaf, Springer, Cham, 2022 Search PubMed.
- S. P. Gorman, C. P. Garvin, F. Quigley and D. S. Jones, J. Pharm. Pharmacol., 2003, 55, 461–468 CrossRef CAS PubMed.
- J. A. Espina, M. H. Cordeiro, M. Milivojevic, I. Pajic-Lijakovic and E. H. Barriga, J. Cell Sci., 2023, 136, jcs260985 CrossRef CAS PubMed.
- C. UniProt, Nucleic Acids Res., 2023, 51, D523–D531 CrossRef PubMed.
- A. H. Jesmer and R. G. Wylie, Front. Chem., 2020, 8, 604236 CrossRef CAS PubMed.
- K. Nakanishi, T. Sakiyama and K. Imamura, J. Biosci. Bioeng., 2001, 91, 233–244 CrossRef CAS PubMed.
- P. Kallas, H. Valen, M. Hulander, N. Gadegaard, J. Stormonth-Darling, P. O'Reilly, B. Thiede, M. Andersson and H. J. Haugen, Nanoscale, 2022, 14, 7736–7746 RSC.
- C. Wang, H. C. van der Mei, H. J. Busscher and Y. Ren, npj Biofilms Microbiomes, 2020, 6, 25 CrossRef CAS PubMed.
- S. A. Dieser, A. S. Fessia, A. R. Zanotti, C. G. Raspanti and L. M. Odierno, Microb. Pathog., 2019, 136, 103652 CrossRef CAS PubMed.
- G. Ramage, J. P. Martinez and J. L. Lopez-Ribot, FEMS Yeast Res., 2006, 6, 979–986 CrossRef CAS PubMed.
- V. Nelea, Y. Nakano and M. T. Kaartinen, Protein J., 2008, 27, 223–233 CrossRef CAS PubMed.
- S. Patel, A. F. Chaffotte, B. Amana, F. Goubard and E. Pauthe, Int. J. Biochem. Cell Biol., 2006, 38, 1547–1560 CrossRef CAS PubMed.
- V. Nelea and M. T. Kaartinen, J. Struct. Biol., 2010, 170, 50–59 CrossRef CAS PubMed.
- M. Soumbo, A. Scarangella, C. Villeneuve-Faure, C. Bonafos, C. Roques and K. Makasheva, IEEE 20th International Conference on Nanotechnology, 2020, 242–245. DOI:10.1109/NANO47656.2020.9183494.
- N. Khan, H. Aslan, H. Buttner, H. Rohde, T. W. Golbek, S. J. Roeters, S. Woutersen, T. Weidner and R. L. Meyer, eLife, 2022, 11, e76164 CrossRef CAS PubMed.
- J. Haiko and B. Westerlund-Wikstrom, Biology, 2013, 2, 1242–1267 CrossRef PubMed.
- C. T. Moraes, J. M. Polatto, S. S. Rossato, M. Izquierdo, D. D. Munhoz, F. H. Martins, D. C. Pimenta, M. J. Farfan, W. P. Elias, A. S. Barbosa and R. M. Piazza, BMC Microbiol., 2015, 15, 278 CrossRef PubMed.
- M. Badaoui, A. Zoso, T. Idris, M. Bacchetta, J. Simonin, S. Lemeille, B. Wehrle-Haller and M. Chanson, Cell Rep., 2020, 32, 107842 CrossRef CAS PubMed.
- A. Arhin and C. Boucher, Microbiology, 2010, 156, 1415–1423 CrossRef CAS PubMed.
- D. J. Vaca, A. Thibau, M. Schutz, P. Kraiczy, L. Happonen, J. Malmstrom and V. A. J. Kempf, Med. Microbiol. Immunol., 2020, 209, 277–299 CrossRef CAS PubMed.
- C. M. Lopez, R. Wallich, K. Riesbeck, C. Skerka and P. F. Zipfel, PLoS One, 2014, 9, e90796 CrossRef PubMed.
- J. M. Rauceo, R. De Armond, H. Otoo, P. C. Kahn, S. A. Klotz, N. K. Gaur and P. N. Lipke, Eukaryotic Cell, 2006, 5, 1664–1673 CrossRef CAS PubMed.
- S. Yang, Z. Song, P. Li, F. Sun, H. Zeng, W. Dong, X. Feng and N. Ren, Desalination, 2023, 545, 116151 CrossRef CAS.
- I. Yoda, H. Koseki, M. Tomita, T. Shida, H. Horiuchi, H. Sakoda and M. Osaki, BMC Microbiol., 2014, 14, 234 CrossRef PubMed.
- S. Zheng, M. Bawazir, A. Dhall, H. E. Kim, L. He, J. Heo and G. Hwang, Front. Bioeng. Biotechnol., 2021, 9, 643722 CrossRef PubMed.
- L. H. Kim and J. S. Vrouwenvelder, Membranes, 2019, 9, 162 CrossRef CAS PubMed.
- Y. W. Ji, Y. J. Cho, C. H. Lee, S. H. Hong, D. Y. Chung, E. K. Kim and H. K. Lee, Eye Contact Lens, 2015, 41, 25–33 CrossRef PubMed.
- H. Yamazoe and T. Tanabe, J. Biomed. Mater. Res., Part A, 2008, 86, 228–234 CrossRef PubMed.
- S. Austermeier, M. Pekmezovic, P. Porschitz, S. Lee, N. Kichik, D. L. Moyes, J. Ho, N. K. Kotowicz, J. R. Naglik, B. Hube and M. S. Gresnigt, mBio, 2021, 12, e00531 CrossRef CAS PubMed.
- V. Ho, P. Herman-Bausier, C. Shaw, K. A. Conrad, M. C. Garcia-Sherman, J. Draghi, Y. F. Dufrene, P. N. Lipke and J. M. Rauceo, mBio, 2019, 10, e01766–e01719 CrossRef CAS PubMed.
- D. S. Donohue, F. S. Ielasi, K. V. Goossens and R. G. Willaert, Mol. Microbiol., 2011, 80, 1667–1679 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |