Open Access Article
Open Access ArticleCluster effect through the oligomerisation of bioactive disaccharide AMOR on pollen tube capacitation in Torenia fournieri†
Akane G.
Mizukami‡
 *a,
Shuhei
Kusano
*a,
Shuhei
Kusano
 b,
Kumi
Matsuura-Tokita
a,
Shinya
Hagihara
b,
Kumi
Matsuura-Tokita
a,
Shinya
Hagihara
 b and
Tetsuya
Higashiyama
b and
Tetsuya
Higashiyama
 *ac
*ac
aDepartment of Biological Sciences, Graduate School of Science, The University of Tokyo, Tokyo 113-0033, Japan. E-mail: amizukami@g.ecc.u-tokyo.ac.jp; higashi@bs.s.u-tokyo.ac.jp
bRIKEN Center for Sustainable Resource Science, Saitama 351-0198, Japan
cInstitute of Transformative Bio-Molecules (WPI-ITbM), Nagoya University, Nagoya 464-8601, Japan
First published on 19th June 2024
Abstract
Arabinogalactan proteins (AGPs) are plant-specific glycoproteins involved in cellular mechanics and signal transduction. There has been major progress in understanding the structure, synthesis, and molecular functions of their carbohydrate chains; however, the mechanisms by which they function as signalling molecules remain unclear. Here, methyl-glucuronosyl arabinogalactan (AMOR; Me-GlcA-β(1,6)-Gal), a disaccharide structure at the end of AGP carbohydrate chains, was oligomerised via chemical synthesis. The biological activity of AMOR oligomers was enhanced via clustering of the carbohydrate chains. Furthermore, AMOR oligomers yielded a pollen tube morphology (i.e., callose plug formation) similar to that when cultured with native AMOR, suggesting it may be functionally similar to native AMOR.
Introduction
The plant cell wall is not simply a structural component that provides mechanical support and protection to plant cells, it also contains cellular signalling molecules. Arabinogalactan proteins (AGPs), a subfamily of hydroxyproline-rich proteins highly glycosylated with type II arabinogalactans (AGs), are involved in various signalling pathways related to environmental responses, development, and reproduction in plants.1–5 Type II AGs consist of a 1,3-β-galactan main chain with branches of 1,6-β-galactan side chains, which are further decorated with sugars such as arabinose, fucose, and glucuronic acid.5–9 Type II AGs are synthesised as a sugar moiety of AGPs in the secretory pathway of plant cells by enzymes specific to each step of glycosylation.6,8Many AGPs have been identified as signalling molecules.1,4,5 For example, the signalling molecules involved in plant reproduction include tobacco transmitting-tissue specific proteins, 120-kDa glycoprotein, and PELPIII for pollen tube growth and pollen–pistil interactions;10–14Arabidopsis thaliana (At)AGP18 for female gametogenesis;15,16 AtAGP6, AtAGP11, AtFLA3, rice MTR1, and AtAGP11 homolog Brassica campestris MF8 for pollen development;17–22 AtENODL11/12/13/14/15 for pollen tube reception by the ovule;23 and AtAGP4 (JAGGER) for polytubey block.24 However, the functions of AGs in AGPs remain elusive. Knockout of enzymes involved in the initial steps of AG glycosylation show severe defects in plant development and reproduction, suggesting that type II AGs are critical for the bioactivity of AGPs.25–28 Consistently, the distribution of AG structures visualised via immunostaining with monoclonal antibodies suggests tissue-specific accumulation of specific AG structures.3,28–30 However, the functions of specific AG structures are largely unknown.
The heterogeneity of native polysaccharides, including their branch structures, impedes structural analyses. It is difficult to elucidate the structure and structure–activity relationships of individual native glycans. Research based on the chemical synthesis of sugar chains is important for closing this knowledge gap. Methyl-glucuronosyl arabinogalactan (AMOR; Me-GlcA-β(1,6)-Gal) is an AG derived from unfertilised female ovule tissue. In the flowering plant Torenia fournieri, AMOR enables the response of pollen tubes to cysteine-rich LURE attractant peptides.31,32 A terminal disaccharide structure of AGs, 4-Me-GlcA-β(1,6)-Gal is necessary and sufficient for the bioactivity of AMOR. Native AMOR is likely a typical, high-molecular-weight AG of approximately 25 kDa based on a gel filtration study,31 which potentially contains branched structures with multiple 4-Me-GlcA-β(1,6)-Gal termini. The activity of the AMOR disaccharide has not been compared with other synthetic polysaccharides with multiple 4-Me-GlcA-β(1,6)-Gal termini on a single molecule.
As indicated by the structure–activity relationship of synthetic AMOR derivatives, it is possible to introduce modifications to the second sugar, galactose, without impacting its activity.32 However, the first sugar, methyl-glucuronic acid, cannot be modified without altering activity; for example, converting the terminal sugar to non-methylated glucuronic acid decreases the activity by approximately1000-fold, suggesting precise recognition by an unidentified receptor in pollen tubes.31,32 The synthesis of AMOR derivatives with modifications at the second sugar would facilitate important aspects of AMOR research, such as the production of branched AMOR oligomers, visualisation of AMOR, and crosslinking with receptor candidates.
In this study, we successfully prepared AMOR derivatives with introduced azide groups and produced various AMOR oligomers, including dimers, trimers, and tetramers. When comparing the activity of branched oligomeric AMOR to that of unbranched AMOR disaccharide (monomer), the AMOR oligomers exhibited higher activities, although the total amount of 4-Me-GlcA-β(1,6)-Gal was same in each fraction. The AMOR oligomers exhibited novel activity promoting callose plug formation in pollen tubes. The callose plug formation activity was also shown in native AMOR in ovule pre-cluture medium. Our results together suggest that the synthetic AMOR oligomers are structurally and functionally similar to native AMOR.
Results and discussion
Chemical synthesis of AMOR oligomers
Synthesis of AMOR oligomers
To synthesise AMOR oligomers, we first synthesised azide-tethered AMOR with protecting groups (Scheme 1). We prepared the glycosyl donor 3 from tetra-acetyl-glucopyranosyl-bromide 1. Koenigs–Knorr glycosylation of 1 and tetraethylene glycol monoazide with silver trifluoromethanesulfonate (AgOTf) afforded 2. We transformed the protecting groups of 2 in four steps to synthesise the benzoyl-protected glycosyl acceptor 3: the removal of acetyl groups; trityl protection of 6-OH; benzoyl protection; and the removal of the trityl group. Next, we synthesised the glycosyl donor 8 from benzylidene-protected glucopyranoside 4. Three protection and deprotection steps yielded 5: benzoyl protection; the removal of the benzylidene group; and trityl protection of 6-OH. The silver oxide-mediated methylation of 5 at 4-OH and the subsequent removal of the trityl group afforded 6. Oxidation of the C-6 position and esterification of the resulting carboxylic group generated the glucuronic acid derivative 7. The removal of the allyl group and trichloroacetimidation completed glycosyl donor synthesis. Glycosylation of 3 and 8 were performed using trimethylsilyl trifluoromethanesulfonate (TMSOTf) at −20 °C in CH2Cl2, resulting in β-anomer selective formation of azide-tethered AMOR 9.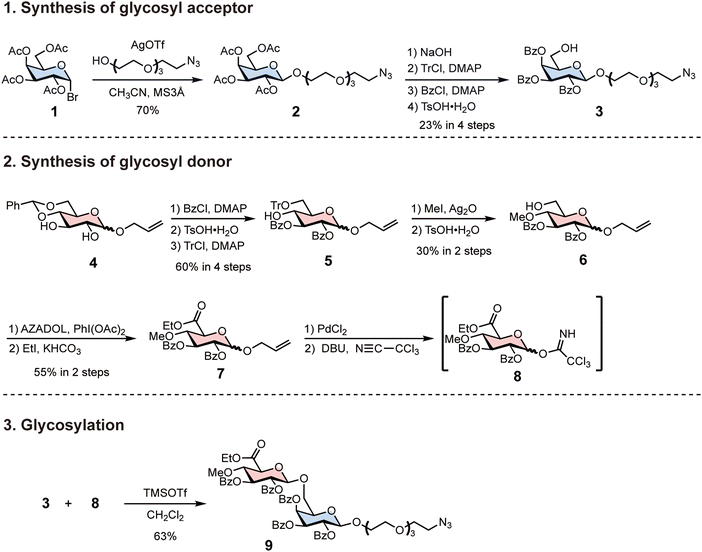 | ||
| Scheme 1 Synthesis of azide-tethered AMOR using protecting groups. Protecting the glycosyl donor with a benzoyl group increased the yield of β-anomer in the subsequent glycosylation reaction. | ||
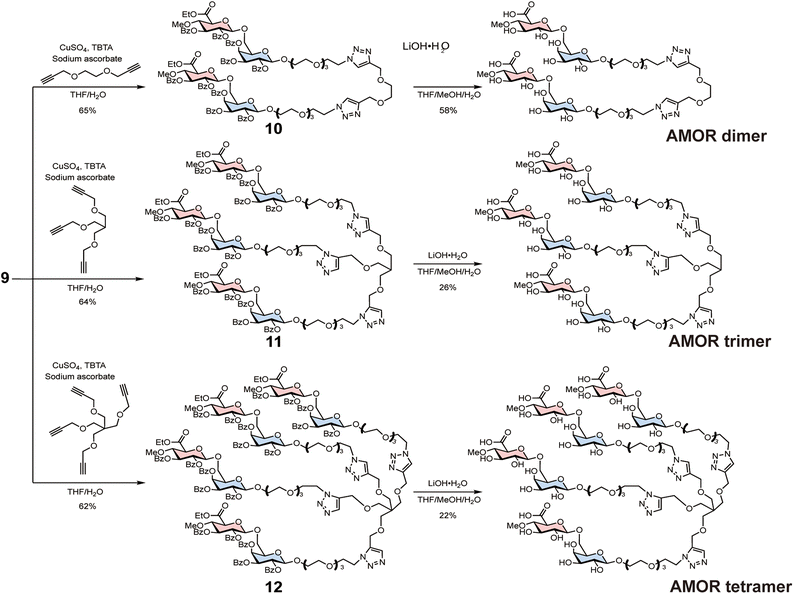
Subsequently, 9 was reacted with either bis-alkyne, tris-alkyne, or tetra-alkyne linkers to synthesise AMOR dimer (di-AMOR) 10, AMOR trimer (tri-AMOR) 11, and AMOR tetramer (tetra-AMOR) 12 with protecting groups, respectively (Scheme 2). The global deprotection of 10–12 was undertaken using LiOH·H2O to yield di-AMOR, tri-AMOR, and tetra-AMOR, respectively. The synthesised multivalent AMORs were characterised using 1H NMR and mass spectroscopy.
Comparison of AMOR monomer and oligomer activity
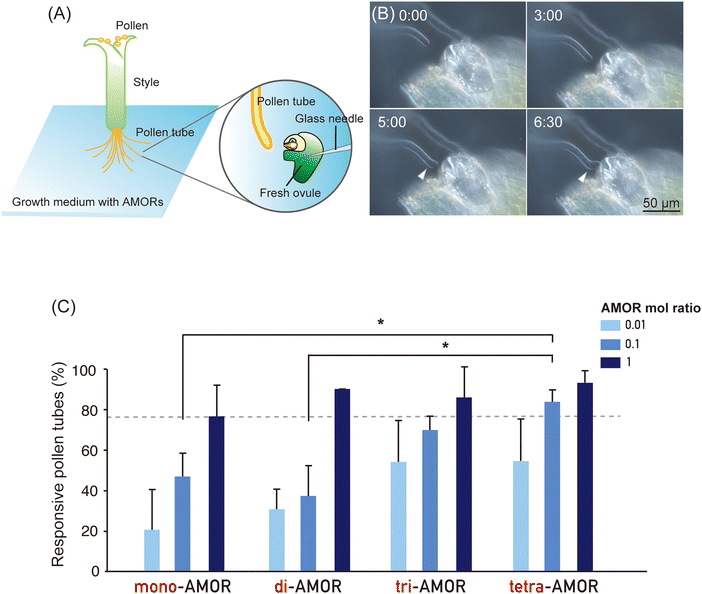
To investigate the impact of the cluster effect due to AMOR oligomerisation, we analysed the activity of AMOR monomer (mono-AMOR) and AMOR oligomers. To examine the response of pollen tubes, a freshly prepared ovule was placed using a glass needle near the tip of an elongating pollen tube growing through a cut style after overnight culture with chemically synthesised mono-AMOR or AMOR oligomers (Fig. 1A and B). Studies have shown that the AMOR activity of mono-AMOR saturates at 135 nM. Therefore, we measured AMOR activity at concentrations 10 and 100 times lower than that of mono-AMOR (Fig. 1C). Comparing equimolar concentrations of AMOR molecules, AMOR activity increased with the degree of oligomerisation. Thus, tetra-AMOR showed high AMOR activity at a low concentration of 3.38 nM, indicating that its structure readily clustered, leading to higher activity.Effect of AMORs on pollen tubes
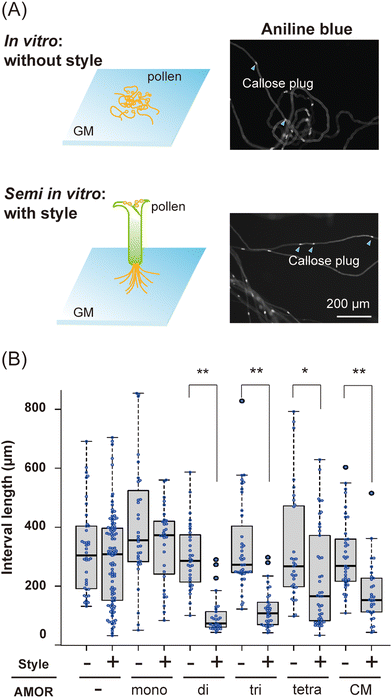
We next conducted a morphological analysis to investigate the effects of AMORs on pollen tube growth. Since brassinosteroids, which confer fertilization ability to pollen tubes, promote callose plug formation,33 the distance between callose plugs in in vitro and semi-in vitro pollen tubes elongating in the growth medium was measured using aniline blue staining (Fig. 2A). Pollen tubes cultured in medium without AMORs had significantly shorter distances between callose plugs when passing through the style (Fig. 2B). In contrast, the presence or absence of AMORs had no effect on the distance between callose plugs in pollen tubes that did not traverse the style (Fig. 2B). These findings reveal that callose plug synthesis in pollen tubes is actively regulated by AMORs only when passing through the style. Interestingly, when using growth medium pre-cultured with two-flowered ovules (=native AMOR), even when the pollen tubes did not pass through the style, compared to cases in which they passed through the style, there was a significant decrease in the distance between the callose plugs of the pollen tubes (Fig. 2B). These observations suggest that AMOR oligomers might functionally resemble native AMOR.In general, glycan–protein interactions are weak. However, high affinity is achieved through the cluster glycoside effect,34–36 though the physical basis for the enhancement is less clear. The increased bioactivity of AMOR oligomers might represent an enhanced affinity of AMOR with unidentified receptor proteins with multiple carbohydrate recognition domains (e.g., lectin domains) or the activation of such AMOR receptors by their clustering, as previously suggested for some receptors such as the epidermal growth factor receptor in mammals.37,38 The cell surface receptor chitin elicitor receptor kinase 1 of Arabidopsis thaliana (AtCERK1) directly binds chitin, which acts as a bivalent ligand to induce dimerization of AtCERK1.39 Synthetic AMOR oligomers could provide novel approaches to better understand native AMOR and identify the AMOR receptor via chemical crosslinking of interacting proteins.
Author contributions
A. G. M. and T. H. conceived of the idea; S. K. and S. H. synthesised and acquired the spectroscopic data of the AMOR and AMOR multimers; K. M. designed the callose plug formation experiment and contributed the development of the experimental protocols; A. G. M. performed the AMOR assay and pollen tube observation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Dr Erika Toda (The University of Tokyo, Japan), Dr Daichi Susaki (Yokohama City University, Japan), and Dr Satohiro Okuda (The University of Tokyo, Japan) for their helpful discussion on this work. We also thank Ryo Yoshida, Akiko Takahashi, and Shoko Namiki for their help in maintaining the plants and other invaluable assistance. This work was supported by the Japan Society for the Promotion of Science KAKENHI (grant no. 21K15119 to A. G. M., and 21K18235, 18H03997, 22H04980, and 22K21352 to T. H.) and the Japan Science and Technology Agency, Core Research for Evolutional Science and Technology (grant no. JPMJCR20E5 to T. H.).References
- M. Ellis, J. Egelund, C. J. Schultz and A. Bacic, Plant Physiol., 2010, 153, 403–419 CrossRef CAS PubMed
.
- D. T. A. Lamport, P. Varnai and C. E. Seal, Ann. Bot., 2014, 114, 1069–1085 CrossRef CAS PubMed
.
- A. M. Pereira, L. G. Pereira and S. Coimbra, Plant Reprod., 2015, 28, 1–15 CrossRef CAS PubMed
.
- A. M. Pereira, A. L. Lopes and S. Coimbra, Front. Plant Sci., 2016, 7, 1895 Search PubMed
.
- S. Su and T. Higashiyama, Plant Reprod., 2018, 31, 67–75 CrossRef CAS PubMed
.
- G. J. Seifert, Front. Plant Sci., 2020, 11, 563735 CrossRef PubMed
.
- J. Silva, R. Ferraz, P. Dupree, A. M. Showalter and S. Coimbra, Front. Plant Sci., 2020, 11, 610377 CrossRef PubMed
.
- K. Ghosh, D. Takahashi and T. Kotake, Carbohydr. Res., 2023, 529, 108828 CrossRef CAS PubMed
.
- A. Leszczuk, P. Kalaitzis, J. Kulik and A. Zdunek, BMC Plant Biol., 2023, 23, 45 CrossRef CAS PubMed
.
- A. Y. Cheung, H. Wang and H. M. Wu, Cell, 1995, 82, 383–393 CrossRef CAS PubMed
.
- H. M. Wu, H. Wang and A. Y. Cheung, Cell, 1995, 82, 395–403 CrossRef CAS PubMed
.
- C. J. Schultz, K. Hauser, J. L. Lind, A. H. Atkinson, Z.-Y. Pu, M. A. Anderson and A. E. Clarke, Plant Mol. Biol., 1997, 35, 833–845 CrossRef CAS PubMed
.
- C. N. Hancock, L. Kent and B. A. McClure, Plant J., 2005, 43, 716–723 CrossRef CAS PubMed
.
- C. A. Eberle, N. O. Anderson and B. M. Clasen, Plant, 2013, 74, 805–814 CAS
.
- G. Acosta-Garciéa and J. P. Vielle-Calzada, Plant Cell, 2004, 16, 2614–2628 CrossRef PubMed
.
- E. Demesa-Arévalo and J. P. Vielle-Calzada, Plant Cell, 2013, 25, 1274–1287 CrossRef PubMed
.
- L. G. Pereira, S. Coimbra, H. Oliveira and L. Monteiro, Planta, 2006, 223, 374–380 CrossRef CAS PubMed
.
- B. Levitin, D. Richter, I. Markovich and M. Zik, Plant J., 2008, 56, 351–363 CrossRef CAS PubMed
.
- S. Coimbra, M. Costa, B. Jones, M. A. Mendes and L. G. Pereira, J. Exp. Bot., 2009, 60, 3133–3142 CrossRef CAS PubMed
.
- J. Li, M. Yu, L.-L. Geng and J. Zhao, Plant J., 2010, 64, 482–497 CrossRef CAS PubMed
.
- H. Tan, W. Liang, J. Hu and D. Zhang, Dev. Cell, 2012, 22, 1127–1137 CrossRef CAS PubMed
.
- S. Lin, H. Dong, F. Zhang, L. Qiu, F. Wang, J. Cao and L. Huang, Ann. Bot., 2014, 113, 777–788 CrossRef CAS PubMed
.
- Y. Hou, X. Guo, P. Cyprys, Y. Zhang, A. Bleckmann, L. Cai, Q. Huang, Y. Luo, H. Gu, T. Dresselhaus, J. Dong and L.-J. Qu, Curr. Biol., 2016, 26, 2343–2350 CrossRef CAS PubMed
.
- A. M. Pereira, A. L. Lopes and S. Coimbra, Plant Signaling Behav., 2016, 11, e1209616 CrossRef PubMed
.
- M. Ogawa-Ohnishi and Y. Matsubayashi, Plant J., 2015, 81, 736–746 CrossRef CAS PubMed
.
- D. Kaur, M. A. Held, M. R. Smith and A. M. Showalter, BMC Plant Biol., 2021, 21, 590 CrossRef CAS PubMed
.
- D. Kaur, D. Moreira, S. Coimbra and A. M. Showalter, Front. Plant Sci., 2022, 13, 935413 CrossRef PubMed
.
- A. L. Lopes, D. Moreira, A. M. Pereira, R. Ferraz, S. Mendes, L. G. Pereira, L. Colombo and S. Coimbra, Ann. Bot., 2023, 131, 827–838 CrossRef CAS PubMed
.
- M. Juranić, M. R. Tucker, C. J. Schultz, N. J. Shirley, J. M. Taylor, A. Spriggs, S. D. Johnson, V. Bulone and A. M. Koltunow, Plant Physiol., 2018, 177, 1027–1049 CrossRef PubMed
.
- C. M. Lara-Mondragón and C. A. MacAlister, Methods Cell Biol., 2020, 160, 215–234 Search PubMed
.
- A. G. Mizukami, R. Inatsugi, J. Jiao, T. Kotake, K. Kuwata, K. Ootani, S. Okuda, S. Sankaranarayanan, Y. Sato, D. Maruyama, H. Iwai, E. Garénaux, C. Sato, K. Kitajima, Y. Tsumuraya, H. Mori, J. Yamaguchi, K. Itami, N. Sasaki and T. Higashiyama, Curr. Biol., 2016, 26, 1091–1097 CrossRef CAS PubMed
.
- J. Jiao, A. G. Mizukami, S. Sankaranarayanan, J. Yamguchi, K. Itami and T. Higashiyawma, Plant Physiol., 2017, 173, 354–363 CrossRef CAS PubMed
.
- K. Matsuura-Tokita, T. Suzuki, Y. Kimata, Y. Takebayashi, M. Ueda, T. Nakano, H. Sakakibara, A. Nakano and T. Higashiyama, Brassinosteroids function as the plant male and female reproductive hormone coordinating gene expression, bioRxiv, 2024 DOI:10.1101/2024.05.07.592278
.
- Y. C. Lee and R. T. Lee, Acc. Chem. Res., 1995, 28, 321–327 CrossRef CAS
.
- R. T. Lee and Y. C. Lee, Glycoconjugate J., 2000, 17, 543–551 CrossRef CAS PubMed
.
- J. J. Lundquist and E. J. Toone, Chem. Rev., 2002, 102, 555–578 CrossRef CAS PubMed
.
- Y. Wang, J. Gao, X. Guo, T. Tong, X. Shi, L. Li, M. Qi, Y. Wang, M. Cai, J. Jiang, C. Xu, H. Ji and H. Wang, Cell Res., 2014, 24, 959–976 CrossRef CAS PubMed
.
- J. Gao, Y. Wang, M. Cai, Y. Pan, H. Xu, J. Jiang, H. Ji and H. Wang, Nanoscale, 2015, 7, 2511–2519 RSC
.
- T. Liu, Z. Liu, C. Song, Y. Hu, Z. Han, J. She, F. Fan, J. Wang, C. Jin, J. Chang, J.-M. Zhou and J. Chai, Science, 2012, 336, 1160–1164 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00032c |
| ‡ Present address: Division of Liberal Arts and Sciences, Aichigakuin University, Aichi 470-0195, Japan. |
| This journal is © The Royal Society of Chemistry 2024 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: