Open Access Article
Open Access ArticleMiltefosine impacts small molecule transport in Gram-positive bacteria†
Marea J.
Blake
 ,
Eleanor F.
Page
,
Madeline E.
Smith
and
Tessa R.
Calhoun
,
Eleanor F.
Page
,
Madeline E.
Smith
and
Tessa R.
Calhoun
 *
*
University of Tennessee, Knoxville, TN, USA. E-mail: trcalhoun@utk.edu
First published on 8th July 2024
Abstract
Miltefosine (MLT) is an alkylphosphocholine with clinical success as an anticancer and antiparasitic drug. Although the mechanism of action of MLT is highly debated, the interaction of MLT with the membrane, specifically lipid rafts of eukaryotes, is well-documented. Recent reports suggest MLT impacts the functional membrane microdomains in bacteria – regions of the membrane structurally and functionally similar to lipid rafts. There have been conflicting reports, however, as to whether MLT impacts the overall fluidity of cellular plasma membranes. Here, we apply steady-state fluorescence techniques, generalized polarization of laurdan and anisotropy of diphenylhexatriene, to discern how MLT impacts the global ordering and lipid packing of Staphylococcus aureus membranes. Additionally, we investigate how the transport of a range of small molecules is impacted by MLT for S. aureus and Bacillus subtilis by employing time-resolved second harmonic scattering. Overall, we observe MLT does not have an influence on the overall ordering and packing of S. aureus membranes. Additionally, we show that the transport of small molecules across the membrane can be significantly altered by MLT – although this is not the case for all molecules studied. The results presented here illustrate the potential use of MLT as an adjuvant to assist in the delivery of drug molecules in bacteria.
1 Introduction
Hexadecylphosphocholine, or miltefosine (MLT), is a synthetic alkylphospholipid (APL) that was originally developed as an anticancer agent1 and is currently in clinical use as an antiparastic drug.2 More recently, the antibacterial and antifungal activity of MLT has received attention.3–7 While only modest antibacterial activity has been observed, it has been suggested that MLT, and APLs more generally, could serve as a new framework for developing novel antibiotics or adjuvants.3 For the latter, it is imperative to investigate how the presence of MLT impacts small molecule transport through the membrane.Despite decades of research, the mechanism of action of MLT remains a point of debate and discussion. While there is agreement that the plasma membrane is compromised with MLT treatment, it is not clear if this is achieved via lipid or protein targets. It has been shown that MLT specifically targets membranes composed of lipid raft constituents of eukaryotic and fungal species, such as cholesterol and ergosterol, respectively.4,8–12 As such, some have reported that this lipid raft association results in MLT impacting the cell membrane fluidity,13–16 while others report this is not the case.10,11 For instances when membrane fluidity was altered, there is evidence that supports the presence of proteins is required. Specifically, Alonso et al. demonstrated that stratum corneum membranes had a significant increase in membrane fluidity due to MLT, but vesicles formed from the lipid extract of stratum corneum membranes had no change in fluidity.13 Additional mechanisms that have been proposed include miltefosine affecting the enzyme, cytochrome c oxidase,17,18 in eukaryotes and halting the cell's ability to synthesize phosphatidylcholine (PC),8,19,20 both cases leading to cell death. Most MLT research, however, has focused on eukaryotic systems or model membranes containing lipid species that are less common or nonexistent in bacteria. For example, MLT has been shown to significantly impact the presence of PC lipids,20 but most bacteria do not have a synthetic pathway for PC.21
In bacteria, MLT has been shown to interrupt aggregation of flotillin in Staphylococcus aureus cell membranes.6 Flotillin are scaffold proteins that have been proposed as crucial components for the formation of functional membrane microdomains6,22 although inconsistent results remain.23 It is not clear, however, if such a change in protein behavior would arise from direct MLT-protein interactions or, if instead, MLT alters the lipid environment preferred by the protein. In a recent paper, molecular dynamics simulations suggest that the lipids themselves are a major determinant in MLT passive transport into the membrane and may play a large role in its mechanism of action.10 When extending such questions to the potential use of MLT as an adjuvant species, it is necessary to consider how MLT's impact on the membrane alters small molecule transport through this important lipid boundary. In order to monitor transport dynamics, here, we implement time-resolved second harmonic scattering (trSHS).
SHS is a surface-specific spectroscopic technique capable of elucidating the organization of molecules adsorbed to surfaces in colloidal environments, such as that of living cell membranes.24 Previous studies have demonstrated SH-based approaches can be used to quantify molecule-membrane interactions within model systems,25–34 eukaryotic species,16,35–38 and bacterial cells.39–52 Specifically, we have implemented this technique to elucidate how structural moieties of small molecules facilitate transport through the Gram-positive membranes of living S. aureus and Bacillus subtilis.41
After employing the steady-state fluorescent assays, generalized polarization (GP) of laurdan and anisotropy of diphenylhexatriene (DPH), to determine the impact of MLT on global ordering and lipid packing of S. aureus membranes, we monitor the trSHS of small molecules (malachite green,43,44,49,53–55 FM 2-10,40,41 FM 1-43,41 and 4-Di-2-ASP41) with previously documented transport behavior to assess the impact of MLT. We also consider the role of membrane composition by extending trSHS studies to another Gram-positive bacterial species, B. subtilis. Although we do not observe a change in the overall ordering or packing of S. aureus membranes, we do observe significant alterations in the interactions of multiple small molecule species with the lipid bilayer. These results are not universal, however, leading to a discussion of the localized impact of MLT on the membrane and the potential relationship to the small molecule structures that are affected.
2 Materials and methods
2.1 Chemicals and cell treatment
Stock solutions of dye molecules FM 2-10 (Biotium), 4-Di-2-ASP (Sigma-Aldrich), and FM 1-43 (Biotium) were prepared in filter sterilized 80:20 Milli-Q water to DMSO to maintain a final DMSO concentration of 0.02% when introduced to cell solutions. Malachite green oxalate (Sigma-Aldrich) was prepared in sterile Milli-Q water. Stock solutions of laurdan (ThermoFisher Scientific), 1,6-diphenyl-1,3,5-hexatriene (diphenylhexatriene, Sigma-Aldrich), and miltefosine (Sigma-Aldrich) were prepared in DMSO.Single colonies of Staphylococcus aureus (ATCC27217) and Bacillus subtilis 168 grown on brain heart infusion (BHI) and Luria Bertani (LB) agar, respectively, were inoculated into liquid BHI and LB media, respectively. Cultures were grown anaerobically for S. aureus and aerobically at 250 RPM for B. subtilis overnight at 37 °C. Aliquots were then resuspended in fresh liquid media to an optical density at 600 nm (OD600) of 0.01 and allowed to grow to an OD600 of 0.2 under the same conditions. Cell samples were then filtered by centrifugation or vacuum filtration and washed twice with phosphate buffered saline (PBS). The membrane composition of both bacterial strains has been previously determined.56,57 For trSHS experiments, cells were resuspended in their respective media supplemented with 0.2% EC-OxyRase. For steady-state fluorescence experiments, cells were resuspended in PBS. Final cell suspensions were measured to be at an OD600 of 0.2. MLT was incubated with cells at room temperature for 10 minutes with FM 2-10, D289, and MG and for 30 minutes with MG and FM 1-43 immediately before experiments were conducted. While previous studies with MLT and S. aureus utilized longer exposure times,6 shorter incubation periods were chosen here to minimize potential impacts on the membrane composition.20
2.2 Time-resolved second harmonic scattering
TrSHS experiments were performed using home-built instruments previously described in detail elsewhere.40 Briefly, the output of a Ti:sapphire oscillator (80 MHz) was compressed to <100 fs and focused into a 2 mm quartz flow cell (Starna) that housed the cell samples. The resulting signals were then collected with a series of lenses and directed towards a photomultiplier tube (PMT) for detection. Filters were implemented to block any second harmonic (400 nm) produced by optics before the sample housing and fundamental light (800 nm) after the sample. In addition to SHS, two-photon fluorescence (TPF) was detected simultaneously by using a 450 longpass dichroic mirror to direct the TPF signals to second PMT. The second harmonic responses were selected using a 400/10 bandpass filter before the PMT. TPF responses were collected using a 625/50 bandpass filter. The power sent to the sample was controlled using a half-waveplate and polarizer. For FM 2-10 and 4-Di-2-ASP, the power was set to 35 mW and for experiments with MG and FM 1-43, the power was set to 100 mW. During the experiments, the samples were flowed through the laser excitation at a rate of >5 mL per minute.Immediately prior to trSHS experiments, the quartz flow cell was passivated to ensure there was no cell adsorption to the surface during the course of the trials. First, the sample chamber was sterilized with 1 M hydrochloric acid and rinsed with a copious amount of sterilized Milli-Q water. Then, we used an established bovine serum albumin (BSA)/glutaraldehyde cross-linking procedure, previously described.58 In short, 50 mM BSA (Sigma-Aldrich) was incubated for 10 minutes, rinsed with 10 mM TRIS buffer (corrected to pH 7.4) (Sigma-Aldrich), and then replaced with another 10 minute incubation of a 1% glutaraldehyde (Thermo-Fisher Scientific) solution. Finally, the flow cell was rinsed with TRIS buffer. All solutions were sterilized before performing the cross-linking procedure.
2.3 Generalized polarization measurements
Membrane order was assessed using generalized polarization (GP) measurements of laurdan.59 Laurdan is a fluorescent molecule that shifts its emission spectra due to the polarity of the surrounding environments. For instance, in environments with low polarity, such as membrane regions with more tightly packed lipids and limited water penetration, the emission maximum of laurdan is 440 nm. When laurdan is an environment of high polarity (e.g. a more disordered region), laurdan has an emission maximum shifted to 490 nm. The ratio of the emission intensities at 440 nm and 490 nm, or GP, can be calculated to quantify these environmental influences according to eqn (1),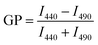 | (1) |
2.4 Steady-state fluorescence anisotropy
Another measure of membrane order used here is the steady-state fluorescence anisotropy of diphenylhexatriene (DPH). DPH preferentially localizes in the hydrophobic region of the membrane. As such, the orientation and rotational freedom of the DPH molecules in this region reports on the order of the lipid fatty acyl chains. This can be quantified by calculating the anisotropy, 〈r〉, of DPH: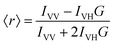 | (2) |
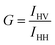 | (3) |
Anisotropy measurements were recorded with an Agilent Cary Eclipse fluorometer. After cells were treated with MLT, they were resuspended in PBS and incubated with 10 μM DPH for ∼45 minutes at 37 °C. Samples were excited at 350 nm and emission was collected at 430 nm. Measurements were taken at room temperature.
3 Results and discussion
3.1 MLT impact on global membrane order
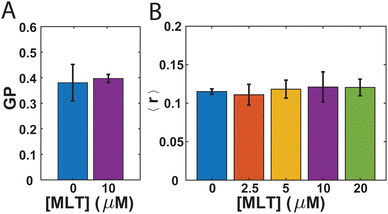
While there has been debate in the literature regarding the impact MLT has on the fluidity of the membranes it interacts with,10,11,13–16 no such studies have been performed with bacteria. Therefore, we first sought to investigate whether the presence of MLT alters the membrane order and lipid packing in S. aureus. In order to do so, we employ two steady-state fluorescence techniques: GP of laurdan and steady-state anisotropy of DPH. Both laurdan and DPH are fluorescent molecules commonly used as probes to assess membrane order.59,60 As the laurdan chromophore is polar, it reports on the presence of water near the junction between the lipid headgroup and tail with a higher GP value associated with a more disordered bilayer and increased water penetration.59,61 The nonpolar DPH molecule is expected to embed more deeply within the hydrophobic portion of the membrane and a lower DPH anisotropy value indicates higher membrane order.61,62 While many studies find similar trends between these different probe assays,60 occasional differences are noted.62Fig. 1A shows the GP values for laurdan in S. aureus membranes either with or without MLT incubation prior to the measurement. For the cells not exposed to MLT, the GP value obtained is 0.381 ± 0.07. This is consistent with values others have reported for exponential phase S. aureus cells,63 and differences in comparison to other studies64 likely arise from the drug resistance of our strain to tetracycline and the room temperature conditions of our measurements.61 In comparison to the cells exposed to MLT, there is no significant difference. This suggests that there is no overall decrease in the order of the S. aureus membrane due to the presence of MLT. The anisotropy of DPH adds further support to this conclusion. As can be seen in Fig. 1B, the DPH anisotropy values are found to be 0.115 ± 0.003 which is also consistent with previous literature values65 and remains unchanged over multiple MLT concentrations.
3.2 Small molecule organization and transport
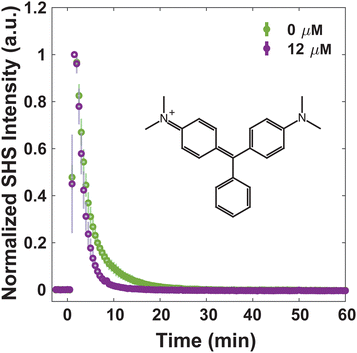
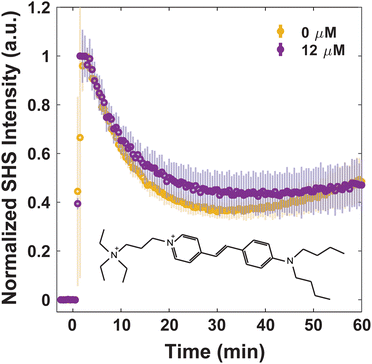
For consideration of MLT as an adjuvant to treat bacterial infections, understanding how MLT affects the interaction of small molecules with the membrane, and moreover their transport through it, is crucial. Four dye molecules with different established membrane interactions are studied here: MG, FM 2-10, 4-Di-2-ASP, and FM 1-43. MG is a small, positively charged molecule that has been extensively characterized with model liposomes27,31,32,66 as well as Escherichia coli43,49,53–55 and eukaryotic cells.37 In all previous work, MG has exhibited an SHS response consistent with a relatively rapid passive diffusion through lipid bilayers. Further, we have previously reported on the bacterial membrane interactions of the three styryl molecules listed where slight variances in their structures result in differences in their affinity for and translocation within the membranes of S. aureus.40,41 For instance, we have proposed that for molecules with short hydrophobic tails and small headgroups (FM 2-10 and 4-Di-2-ASP, respectively), the molecules have a greater propensity to remain in the outer leaflet of the membrane. After ∼1 hour, however, the molecules produce a rise in SHS signal indicative of further organization within the membrane.40,41 Further, we demonstrated that the introduction of molecules with larger branched tails (FM 1-43) results in a rapid decrease in SHS. This response is due to the molecules moving from the outer leaflet to the inner leaflet of the membrane.41Alterations to small molecule transport induced by MLT can be seen by examining the behavior of MG. Fig. 2 shows that at 0 μM MLT, the molecules rapidly diffuse through the hydrophobic portion of the membrane in agreement with MG SHS studies with both liposomal membranes27,31,32,66 as well as E. coli cells.43,49,53–55 After S. aureus cells are treated with 12 μM MLT (Fig. 2), the trSHS decreases more rapidly compared to untreated cells. While the alterations to FM 2-10 and 4-Di-2-ASP behavior, shown below, are apparent within 10 minutes of MLT incubation, significant changes for MG are observed after 30 minutes of MLT exposure (Fig. S1, ESI†). This additional time may arise from the delay in MLT accessing the inner membrane leaflet. These results indicate that MG may be experiencing a more disordered membrane environment to mediate quicker passive diffusion across the lipid bilayer.66
More complex dynamics are observed when moving to the styryl dye molecules which have a stronger association with the membrane environment. Past studies with FM 2-10 and 4-Di-2-ASP hypothesized that the molecule undergoes organization in the membrane of S. aureus. This hypothesis arises from the fact that the SHS signal in these samples rises over time and can be understood by examining the factors that contribute to the intensity of the SHS signal (ISHS). Specifically, ISHS = |χ(2)effEωEω|2 where Eω is the incoming electric field and χ(2)eff is the second-order effective susceptibility which can be further decomposed to χ(2)eff ∝ Ns〈β〉. Here, Ns is the population of probe molecules in the membrane while 〈β〉 is the orientational average of the hyperpolarizability. As we have previously determined, the population of cells throughout the duration of our measurements is static and subsequently, there is no increase in membrane probe population,40 therefore the SHS rise is attributed to 〈β〉. This term can increase through a change in the probe molecule's environment either due to differences in solvation or an overall increase in molecular alignment due to environmental rigidity.
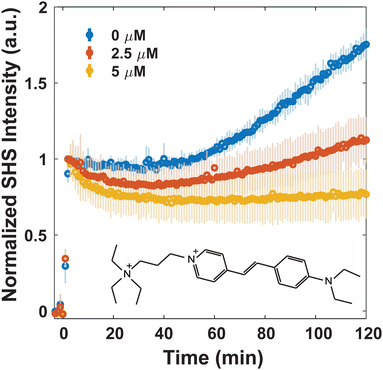
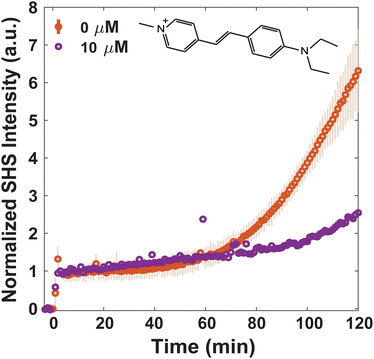
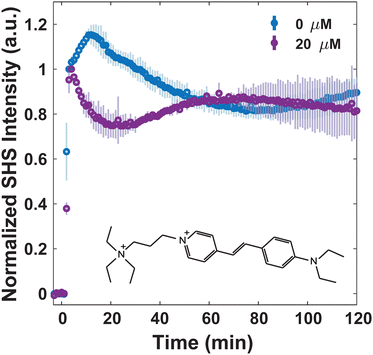
Since MLT has been shown to disrupt membrane regions of increased rigidity,8 the membrane interactions of FM 2-10 and 4-Di-2-ASP were studied here. As can be seen in Fig. 3, when no MLT is present, the SHS signal of FM 2-10 is relatively static over the first 40 minutes but then nearly doubles after two hours. This behavior, and especially the rise, are clearly altered by MLT. As the concentration of MLT increases from 0 to 2.5 to 5 μM, the SHS rise is diminished and eventually lost. There are no significant alterations to the relative change in long-time signal for concentrations higher than 5 μM (Fig. S2, ESI†). A reduced SHS rise is also observed for 4-Di-2-ASP after cells are treated with 10 μM MLT (Fig. 4).
Another change induced by MLT is also observed within the first 20 minutes. Whereas the SHS signal of FM 2-10 with no MLT present displays a static signal, cells treated with MLT exhibit an SHS decay after the initial adsorption of FM 2-10. Such a decrease is most often interpreted as a movement of some population of the probe molecule to the inner leaflet.42,43,49 Alternatively, in the same way a more rigid environment would induce a rising SHS signal, a more disordered environment can cause a reduction as the alignment of the molecules is decreased. More support for this argument can be seen by examining the trends in SHS intensity values. While Fig. 3 shows the time-resolved SHS signals normalized to the initial absorption peak to better differentiate the dynamic changes, Fig. 5 compares the initial SHS intensity after adsorption to that of the two-photon fluorescence signal taken at the same time. As FM 2-10 only exhibits significant fluorescence when embedded in the membrane environment, monitoring the fluorescence signal allows us to separately monitor the population contribution to the SHS signal. As can be seen in Fig. 5, the fluorescence intensity for different MLT concentrations remains unchanged, suggesting that the same number of probe molecules are adsorbed onto the membrane. In contrast, the SHS intensity shows an overall decreasing trend with increasing MLT concentration suggesting that 〈β〉 is decreasing over these conditions. Taken together, these data suggest that FM 2-10 in an MLT-altered membrane does not experience a more rigid environment over time and that, in fact, its initial lipid environment may be more disordered, potentially promoting translocation to the inner leaflet.
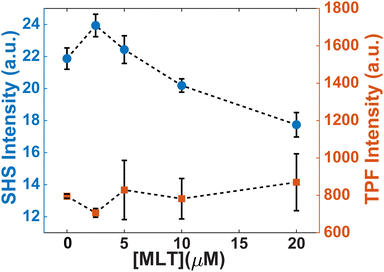 | ||
| Fig. 5 Initial intensities of trSHS (circles, left axis) and TPF (squares, right axis) signals of S. aureus with 16 μM FM 2-10. | ||
We have previously shown that the membrane composition itself plays a large role in how small molecules trespass the bacterial surface.41 Specifically, we observed that for FM 2-10 and 4-Di-2-ASP, where a rise over the two-hour time period is seen with S. aureus, a slight rise is followed by a decrease in SHS intensity over time for B. subtilis. This discrepancy was proposed to be attributed to the generally more fluid membrane of B. subtilis, which in turn facilitated translocation events, whereas S. aureus mediated molecule organization in the outer leaflet over the same time period.41 The fact that these membranes facilitate starkly different transport properties of FM 2-10 allows an opportunity to determine how translatable MLT's impact is for different species. Fig. 6 shows that when MLT is introduced to B. subtilis, FM 2-10 produces an immediate decrease in SHS intensity after initial adsorption, similar, albeit more rapid, to what was witnessed with S. aureus (Fig. 3). Following this first translocation event, the SHS intensity displays a slight increase over time and then remains relatively stagnant for the duration of the experiment.
Although the two cell species have different FM 2-10 transport in the absence of MLT, the final result after MLT incubation is similar. This could be due to MLT having the same mechanism of action for both cell types. While S. aureus and B. subtilis have differences in their overall phospholipid membrane composition,67 they do have similarities that could explain the analogous MLT results. For instance, both membranes have been shown to be equipped with flotillin scaffold proteins6,22,68 – proteins that are largely conserved across different domains of life and hypothesized to play a role in MLT's mechanism of action for bacteria.6,69 Additionally, both membrane species are known to moderate their membrane fluidity via isoprenoid-derived minor lipid species.70–72 For instance, S. aureus produces carotenoids, such as staphyloxanthin, and B. subtilis synthesizes hopanoids.57,73 Hopanoids in particular are structurally and functionally similar to that of ergosterol and cholesterol in eukaryotes.74–76 Subsequently, both of these sterols in eukaryotes are important constituents for MLT activity.4,8–12 Therefore, flotillin proteins and/or isoprenoid-derived molecules could be potential targets for MLT in the bacterial species studied here.
When attempting to understand how MLT does not affect laurdan and DPH behavior but does affect the behavior of MG, FM 2-10, and 4-Di-2-ASP, one may be quick to note the neutral forms of the former group of molecules vs. the ionic forms of the latter. This justification, however, is refuted by examining FM 1-43. Unlike the other three dyes measured with trSHS, FM 1-43 transport is unaffected by the presence of MLT. As can be seen in Fig. 7, after initial adsorption, the SHS signal from FM 1-43 decreases over 30 minutes before stabilizing. The signal from the MLT-affected cells exhibits the same behavior within the error of the measurement.
We propose two possible explanations for the differences in MLT's impact for these five different probe molecules. First, MLT may induce a more local effect to specific regions, i.e. domains, within the membrane that is limiting in its impact to only those molecules that are strongly associated with the same domains. This local impact is what has been proposed before for its behavior.6,8 The novelty of our measurements is then the suggestion that MG, FM 2-10, and 4-Di-2-ASP have a preferential association with such domains. Previous work has shown that both laurdan61,77 and DPH62 can partition into multiple different lipid phases promoting a more homogeneous distribution within the membrane. In contrast, styryl dyes have been seen to heterogeneously localize within bacterial membranes78 although this is not always the case.79 The heterogeneous distribution has been attributed to the cationic charge on the styryl dyes interacting with the negatively charged phosphatidylglycerol (PG) lipid headgroup.78 It is important to note, however, that such heterogeneous behavior was seen explicitly for FM 1-43 which we find to be unaffected by MLT.
As a second, not necessarily unrelated, explanation for the differing behavior, we consider the hydrophobic character of the probe molecules. As noted previously, both laurdan and DPH are neutral molecules predominantly interacting with lipids below the headgroup. Beyond those two molecules, the only other one not impacted by MLT was FM 1-43. In comparison to MG, FM 2-10, and 4-Di-2-ASP, FM 1-43 contains the largest hydrophilic moiety. As such it may be that those molecules with stronger interactions with the lipid tails and hydrophobic core of the membrane are less susceptible to MLT alterations.
4 Conclusions
The results of our trSHS experiments show that MLT does hold promise as an adjuvant species for small molecule antibiotics targeting Gram-positive bacteria. Our steady-state fluorescence measurements with laurdan and DPH show no evidence of a large-scale decrease in the membrane order of S. aureus cells induced by MLT. This result supports previous work by others10,11 but extends this finding to bacteria. In contrast, multiple small molecules exhibit altered interactions with the membranes after exposure to MLT. These alterations led to an overall higher propensity for the molecules to more efficiently traverse the lipid bilayer. While the lack of impact on FM 1-43 dynamics may seem limiting for MLT's use, it could alternatively be viewed as promising for future selectivity. Work remains, however, to better understand MLT's relationship with different small molecules and the mechanism by which MLT impacts bacterial membranes overall.Data availability
Data supporting this article is included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the University of Tennessee Bioanalytical Research Facility and Prof. Rajan Lamichhane for instrument use and the National Institute of General Medical Sciences (R35GM142928) for funding.References
- H. Eibl and C. Unger, Cancer Treat. Rev., 1990, 17, 233–242 CrossRef CAS PubMed.
- T. P. C. Dorlo, M. Balasegaram, J. H. Beijnen and P. J. de Vries, J. Antimicrob. Chemother., 2012, 67, 2576–2597 CrossRef CAS.
- D. Obando, F. Widmer, L. C. Wright, T. C. Sorrell and K. A. Jolliffe, Bioorg. Med. Chem., 2007, 15, 5158–5165 CrossRef CAS PubMed.
- Y. Wu, M. Wu, J. Gao and C. Ying, Front. Microbiol., 2020, 11, 854 CrossRef PubMed.
- M. Margaritova Zaharieva, A. Dimitrov Kroumov, L. Dimitrova, I. Tsvetkova, A. Trochopoulos, S. Mihaylov Konstantinov, M. Reinhold Berger, M. Momchilova, K. Yoncheva and H. Miladinov Najdenski, Biotechnol. Biotechnol. Equip., 2019, 33, 38–53 CrossRef.
- G. Koch, C. Wermser, I. C. Acosta, L. Kricks, S. T. Stengel, A. Yepes and D. Lopez, Cell Chem. Biol., 2017, 24, 845–857.e6 CrossRef CAS PubMed.
- D. Llull, L. Rivas and E. García, Antimicrob. Agents Chemother., 2007, 51, 1844–1848 CrossRef CAS PubMed.
- A. H. van der Luit, M. Budde, P. Ruurs, M. Verheij and W. J. van Blitterswijk, J. Biol. Chem., 2002, 277, 39541–39547 CrossRef CAS PubMed.
- Y. D. L. M. Zulueta Díaz and M. L. Fanani, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 1891–1899 CrossRef PubMed.
- M. Malta de Sá, V. Sresht, C. O. Rangel-Yagui and D. Blankschtein, Langmuir, 2015, 31, 4503–4512 CrossRef.
- B. M. Castro, A. Fedorov, V. Hornillos, J. Delgado, A. U. Acuña, F. Mollinedo and M. Prieto, J. Phys. Chem. B, 2013, 117, 7929–7940 CrossRef CAS PubMed.
- B. Heczková and J. P. Slotte, FEBS Lett., 2006, 580, 2471–2476 CrossRef PubMed.
- L. Alonso, S. A. Mendanha, C. A. Marquezin, M. Berardi, A. S. Ito, A. U. Acuña and A. Alonso, Int. J. Pharm., 2012, 434, 391–398 CrossRef CAS PubMed.
- K. S. Fernandes, P. E. N. de Souza, M. L. Dorta and A. Alonso, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 1–9 CrossRef CAS PubMed.
- K. Petit, M. Suwalsky, J. R. Colina, L. F. Aguilar, M. Jemiola-Rzeminska and K. Strzalka, Biochim. Biophys. Acta, Biomembr., 2019, 1861, 17–25 CrossRef CAS.
- R. A. Moreira, S. A. Mendanha, K. S. Fernandes, G. G. Matos, L. Alonso, M. L. Dorta and A. Alonso, Antimicrob. Agents Chemother., 2014, 58, 3021–3028 CrossRef.
- X. Zuo, J. T. Djordjevic, J. B. Oei, D. Desmarini, S. D. Schibeci, K. A. Jolliffe and T. C. Sorrell, Mol. Pharmacol., 2011, 80, 476–485 CrossRef CAS PubMed.
- J. R. Luque-Ortega and L. Rivas, Antimicrob. Agents Chemother., 2007, 51, 1327–1332 CrossRef CAS PubMed.
- W. J. van Blitterswijk and M. Verheij, Curr. Pharm. Des., 2008, 14, 2061–2074 CrossRef CAS PubMed.
- M. Rakotomanga, S. Blanc, K. Gaudin, P. Chaminade and P. M. Loiseau, Antimicrob. Agents Chemother., 2007, 51, 1425–1430 CrossRef CAS PubMed.
- M. Aktas, M. Wessel, S. Hacker, S. Klüsener, J. Gleichenhagen and F. Narberhaus, Eur. J. Cell Biol., 2010, 89, 888–894 CrossRef CAS PubMed.
- D. López and R. Kolter, Genes Dev., 2010, 24, 1893–1902 CrossRef PubMed.
- H. Strahl and J. Errington, Annu. Rev. Microbiol., 2017, 71, 519–538 CrossRef CAS PubMed.
- G. Gonella and H.-L. Dai, Langmuir, 2014, 30, 2588–2599 CrossRef CAS PubMed.
- L. Moreaux, O. Sandre and J. Mertz, JOSA B, 2000, 17, 1685–1694 CrossRef CAS.
- T. T. Nguyen and J. C. Conboy, Anal. Chem., 2011, 83, 5979–5988 CrossRef CAS PubMed.
- R. R. Kumal, H. Nguyenhuu, J. E. Winter, R. L. McCarley and L. H. Haber, J. Phys. Chem. C, 2017, 121, 15851–15860 CrossRef CAS.
- A. S. Dikkumbura, A. V. Aucoin, R. O. Ali, A. Dalier, D. W. Gilbert, G. J. Schneider and L. H. Haber, Langmuir, 2022, 38, 3852–3859 CrossRef CAS PubMed.
- P. Hamal, V. Subasinghege Don, H. Nguyenhuu, J. C. Ranasinghe, J. A. Nauman, R. L. McCarley, R. Kumar and L. H. Haber, J. Phys. Chem. B, 2021, 125, 10506–10513 CrossRef CAS PubMed.
- P. Hamal, H. Nguyenhuu, V. Subasinghege Don, R. R. Kumal, R. Kumar, R. L. McCarley and L. H. Haber, J. Phys. Chem. B, 2019, 123, 7722–7730 CrossRef CAS PubMed.
- E. C. Y. Yan and K. B. Eisenthal, Biophys. J., 2000, 79, 898–903 CrossRef CAS PubMed.
- A. Srivastava and K. B. Eisenthal, Chem. Phys. Lett., 1998, 292, 345–351 CrossRef CAS.
- Y. Ruan, P. Guha, S.-L. Chen, Q. Yuan and W. Gan, Chem. Phys., 2021, 548, 111250 CrossRef CAS.
- S. L. Chen, Y. Z. Liang, Y. Hou, H. Wang, X. Wu, W. Gan and Q. Yuan, Mater. Today Phys., 2019, 9, 100092 CrossRef.
- B. Li, J. Li, W. Gan, Y. Tan and Q. Yuan, Anal. Chem., 2021, 93, 14146–14152 CrossRef CAS PubMed.
- P. J. Campagnola, M.-D. Wei, A. Lewis and L. M. Loew, Biophys. J., 1999, 77, 3341–3349 CrossRef CAS PubMed.
- M. Sharifian Gh, M. J. Wilhelm, M. Moore and H.-L. Dai, Biochemistry, 2019, 58, 1841–1844 CrossRef CAS PubMed.
- J. Zeng, H. M. Eckenrode, H.-L. Dai and M. J. Wilhelm, Colloids Surf., B, 2015, 127, 122–129 CrossRef CAS PubMed.
- L. N. Miller, M. J. Blake, E. F. Page, H. B. Castillo and T. R. Calhoun, ACS Infect. Dis., 2021, 3088–3095 CrossRef CAS PubMed.
- L. N. Miller, W. T. Brewer, J. D. Williams, E. M. Fozo and T. R. Calhoun, Biophys. J., 2019, 117, 1419–1428 CrossRef CAS PubMed.
- M. J. Blake, H. B. Castillo, A. E. Curtis and T. R. Calhoun, Biophys. J., 2023, 122, 1735–1747 CrossRef CAS PubMed.
- E. F. Page, M. J. Blake, G. A. Foley and T. R. Calhoun, Chem. Phys. Rev., 2022, 3, 041307 CrossRef CAS PubMed.
- J. Zeng, H. M. Eckenrode, S. M. Dounce and H.-L. Dai, Biophys. J., 2013, 104, 139–145 CrossRef CAS PubMed.
- T. Wu, M. J. Wilhelm, Y. Li, J. Ma and H.-L. Dai, ACS Infect. Dis., 2022, 1124–1133 CrossRef CAS PubMed.
- M. J. Wilhelm, M. Sharifian Gh and H.-L. Dai, Biochemistry, 2015, 54, 4427–4430 CrossRef CAS PubMed.
- M. J. Wilhelm, M. Sharifian Gh, T. Wu, Y. Li, C.-M. Chang, J. Ma and H.-L. Dai, Biophys. J., 2021, 120, 2461–2470 CrossRef CAS PubMed.
- M. J. Wilhelm, J. B. Sheffield, M. Sharifian Gh, Y. Wu, C. Spahr, G. Gonella, B. Xu and H.-L. Dai, ACS Chem. Biol., 2015, 10, 1711–1717 CrossRef CAS PubMed.
- M. J. Wilhelm, M. Sharifian Gh and H.-L. Dai, J. Chem. Phys., 2019, 150, 104705 CrossRef PubMed.
- M. J. Wilhelm and H.-L. Dai, Chem. – Asian J., 2020, 15, 200–213 CrossRef CAS PubMed.
- M. J. Wilhelm, J. B. Sheffield, G. Gonella, Y. Wu, C. Spahr, J. Zeng, B. Xu and H.-L. Dai, Chem. Phys. Lett., 2014, 605–606, 158–163 CrossRef CAS.
- M. Sharifian Gh, M. J. Wilhelm and H.-L. Dai, ACS Med. Chem. Lett., 2018, 9, 569–574 CrossRef PubMed.
- M. Sharifian Gh, M. J. Wilhelm and H.-L. Dai, J. Phys. Chem. Lett., 2016, 7, 3406–3411 CrossRef CAS PubMed.
- M. Sharifian Gh, M. J. Wilhelm and H.-L. Dai, Biophys. Rep., 2024, 4, 100141 Search PubMed.
- D. Kumar, A. Gayen and M. Chandra, ACS Infect. Dis., 2023, 9, 2471–2481 CrossRef CAS PubMed.
- A. Gayen, D. Kumar, S. Matheshwaran and M. Chandra, Anal. Chem., 2019, 91, 7662–7671 CrossRef CAS PubMed.
- R. Boudjemaa, C. Cabriel, F. Dubois-Brissonnet, N. Bourg, G. Dupuis, A. Gruss, S. Lévêque-Fort, R. Briandet, M.-P. Fontaine-Aupart and K. Steenkeste, Antimicrob. Agents Chemother., 2018, 62(7) DOI:10.1128/aac.00023-18.
- A. Y. van Tilburg, P. Warmer, A. J. van Heel, U. Sauer and O. P. Kuipers, Microb. Biotechnol., 2022, 15, 1633–1651 CrossRef CAS PubMed.
- J. Hyeon Park, T. Naw Sut, J. A. Jackman, A. Rahim Ferhan, B. Kyeong Yoon and N.-J. Cho, Phys. Chem. Chem. Phys., 2017, 19, 8854–8865 RSC.
- A. G. Jay and J. A. Hamilton, J. Fluoresc., 2017, 27, 243–249 CrossRef PubMed.
- N. C. S. Mykytczuk, J. T. Trevors, L. G. Leduc and G. D. Ferroni, Prog. Biophys. Mol. Biol., 2007, 95, 60–82 CrossRef CAS PubMed.
- S. Vanounou, D. Pines, E. Pines, A. H. Parola and I. Fishov, Photochem. Photobiol., 2002, 76, 1–11 CrossRef CAS PubMed.
- C. S. López, H. A. Garda and E. A. Rivas, Arch. Biochem. Biophys., 2002, 408, 220–228 CrossRef PubMed.
- L. J. Bessa, M. Ferreira and P. Gameiro, Data Brief, 2018, 21, 128–132 CrossRef PubMed.
- K. Zhang, N. Yang, D. Teng, R. Mao, Y. Hao and J. Wang, Appl. Microbiol. Biotechnol., 2024, 108, 111 CrossRef CAS PubMed.
- L. J. Bessa, M. Ferreira and P. Gameiro, J. Photochem. Photobiol., B, 2018, 181, 150–156 CrossRef CAS PubMed.
- T. Wu, M. J. Wilhelm, J. Ma, Y. Li, Y. Wu and H.-L. Dai, Angew. Chem., Int. Ed., 2022, 61, e202205608 CrossRef CAS PubMed.
- N. Malanovic and K. Lohner, Pharmaceuticals, 2016, 9, 59 CrossRef PubMed.
- D. Lopez and G. Koch, Curr. Opin. Microbiol., 2017, 36, 76–84 CrossRef CAS PubMed.
- I. C. Morrow and R. G. Parton, Traffic, 2005, 6, 725–740 CrossRef CAS PubMed.
- J. R. Willdigg and J. D. Helmann, Front. Mol. Biosci., 2021, 8, 634438 CrossRef CAS PubMed.
- M. Bramkamp, EMBO J., 2022, 41, e110737 CrossRef CAS PubMed.
- L. M. Mitchison-Field and B. J. Belin, Curr. Opin. Microbiol., 2023, 74, 102315 CrossRef CAS PubMed.
- G.-D. López, E. Suesca, G. Álvarez Rivera, A. E. Rosato, E. Ibáñez, A. Cifuentes, C. Leidy and C. Carazzone, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2021, 1866, 158941 CrossRef PubMed.
- M. Rohmer, P. Bouvier and G. Ourisson, Proc. Natl. Acad. Sci. U. S. A., 1979, 76, 847–851 CrossRef CAS PubMed.
- J. P. Sáenz, E. Sezgin, P. Schwille and K. Simons, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14236–14240 CrossRef PubMed.
- J. P. Sáenz, D. Grosser, A. S. Bradley, T. J. Lagny, O. Lavrynenko, M. Broda and K. Simons, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11971–11976 CrossRef PubMed.
- M. Wenzel, N. O. E. Vischer, H. Strahl and L. W. Hamoen, Bio-Protoc., 2018, 8, e3063 CAS.
- I. Barák, K. Muchová, A. J. Wilkinson, P. J. O'Toole and N. Pavlendová, Mol. Microbiol., 2008, 68, 1315–1327 CrossRef PubMed.
- A. S. Johnson, S. van Horck and P. J. Lewis, Microbiology, 2004, 150, 2815–2824 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1 and S2. See DOI: https://doi.org/10.1039/d4cb00106k |
| This journal is © The Royal Society of Chemistry 2024 |