Open Access Article
Open Access ArticleCaged aminoluciferin probe for bioluminescent immunoproteasome activity analysis†
Cody A.
Loy
 and
Darci J.
Trader
and
Darci J.
Trader
 *
*
Department of Pharmaceutical Sciences, University of California, Irvine, California 92617, USA. E-mail: dtrader@uci.edu
First published on 16th July 2024
Abstract
The immunoproteasome (iCP) can be expressed under inflammatory conditions, such as exposure to interferon-gamma (IFN-γ), that alerts the cell to begin generating iCP preferentially over the standard proteasome (sCP). With the iCP becoming a widely targeted isoform in a variety of diseases, there is a need to understand its activity and expression in cells and in vivo. Activity-based probes for the iCP have been developed but their application has been limited due to their difficult synthesis and cannot be used in tissues or whole animals. Our lab has previously demonstrated we can monitor iCP activity using a 4-mer peptide linked to a fluorophore and a peptoid. This was utilized in the development of the first cell-permeable iCP activity-based probe that did not include a covalent reactive moiety. Here, we demonstrate that this same peptide recognition sequence can be appended to aminoluciferin, caging it, until its interaction with the iCP. This probe should be applicable to monitor iCP activity in animal models where tumor or other tissue has been engineered to produce luciferase. We anticipate it could also be applied to observe iCP activity as tumors are formed in vivo.
Introduction
Elucidating the role of the proteasome and its isoforms is an endeavor that has experienced an increased interest in recent years, with the development of more selective inhibitors and the recent advancements of targeted protein degradation as a viable therapeutic option.1–8 The proteasome is a large, multi-catalytic complex that is responsible for the degradation of cytosolic and nuclear proteins.9 Under conditions of inflammation, newly synthesized proteasomes are formed, incorporating altered catalytic subunits that are homologous to the standard proteasome core particle (sCP). The immunoproteasome (iCP) has been discovered to have alerted cleavage preferences of protein/peptide substrates from the sCP due to these changes in catalytic subunits.9,10 This has led to the development of selective inhibitors to be generated that have shown to be advantageous over sCP inhibition in certain disease types.11Being able to effectively monitor these isoforms is of great interest, as the proteasome is responsible for several cellular functions from protein clearance, cell cycle regulation, MHC-I presentation, and cellular stress response. A variety of probes have been developed by us and others for different applications.12–17 These probes can be routinely used to study iCP activity utilizing purified iCP or in cell-based assays. However, there are still many questions about the activity of this proteasome isoform in animal models or tissue-based studies. Being able to study iCP activity in only a cellular environment has hindered our ability to understand the dynamics of this protein complex in relation to how far from the site of inflammation the iCP being expressed, how quickly is it being expressed, as well as better understanding its role after targeting with therapeutics. The current probes rely on fluorophores that require external excitation and have low tissue penetration. To overcome the current probe limitations, we describe here methodology to synthesize and apply an iCP-selective probe that has the potential for in vivo monitoring.
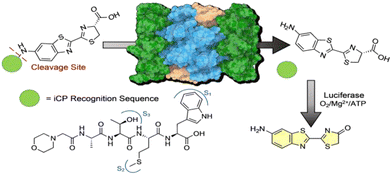
Bioluminescence imaging has proven to be a powerful tool for sensitive, reliable, and non-invasive imaging of a wide variety of targets and processes.18 One of the most common bioluminescence systems is the firefly luciferase–luciferin system, as it allows for simple imaging of complex processes while only requiring limited substrate.19–21 In addition, caged luciferin probes have been developed that serve as highly sensitive probes for detecting enzyme activity in an animal model.22–25 When the luciferin analogue is caged, it cannot interact with luciferase, and no signal is produced. In contrast, once the probe has been cleaved or uncaged by an enzyme, chemical reaction, or ultraviolet light, the free luciferin can interact with luciferase and signal can be detected through tissue.
To design this caged-amino luciferin iCP probe, we were inspired by the development of our previous cell permeable activity-based probe TBZ-1.26 This probe utilizes a 4-mer peptide sequence that is selective for the iCP over the sCP, appended to rhodamine 110 as the fluorophore. Upon interaction with the catalytic subunits of the iCP, the bond between our recognition peptide sequence and fluorophore was cleaved, generating a fluorescence signal that could be monitored over time and correlated to iCP activity.12,26 For this new probe, inspiration was taken from previously designed caged-bioluminescent probes for a variety of enzymes. The amino luciferin derivative of D-luciferin has a free amine that can form an amide bond to a peptide recognition sequence, caging the probe until released by the iCP, Fig. 1.
The probe described here was compared to commercially available non-selective proteasome probes. The results show that this probe was able to achieve similar sensitivity but gain the selectivity that is currently lacking. This new probe demonstrated an improvement in cleavage by the installment of a linker that limits steric hinderance at the active site of the iCP, allowing for more probe to be cleaved at a faster rate.
Results
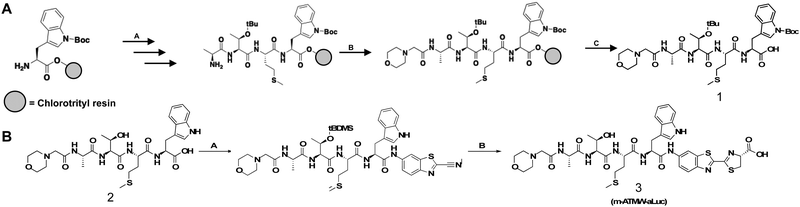
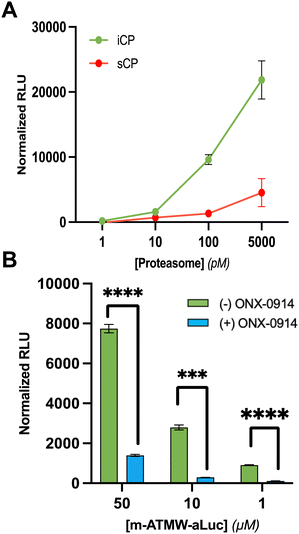
For the synthetic design of the probe a new route was needed to couple the amino luciferin moiety to the iCP recognition peptide sequence. The recognition peptide sequence can be generated in suitable quantities by solid phase peptide synthesis. Capping the N-terminus to prevent unwanted cleavage by proteases in the cell was accomplished using a two-step bromoacetic acid/DIC coupling followed by bromine displacement with morpholine (Scheme 1A). Once the peptide was synthesized and purified, the bioluminescent agent was coupled directly to the recognition sequence. As previously demonstrated the iCP is capable of cleaving bonds between the peptide and fluorophore.12 To accomplish this direct conjugation of the amino luciferin moiety to the peptide, the threonine's hydroxyl group needed to be re-protected as the t-butyl was removed after cleavage of the peptide from the resin. TBDMS-Cl with imidazole was used to protect the free hydroxyl of the threonine to ensure no reaction would occur at this site. The tryptophan contained no protecting group as we noted that if the Boc group remained it hindered the coupling abilities with the relatively weak nucleophile 6-amino-2-cyanobenzothiazole. After the Thr protecting group was installed, the coupling reagents TCFH and NMI were utilized, as these are used for unreactive anilines such as the 6-amino-2-cyanobenzothiazole.27 Lastly, D-cysteine was used to cyclize the cyanobenzothiazole to afford the final probe m-ATMW-aLuc, Scheme 1B.After completing the synthesis of this probe, we wanted to demonstrate the iCP could cleave the bond between the recognition peptide sequence and the amino luciferin. We utilized our established LC/MS assay where the probe is incubated with purified iCP, quenched at several time points using acetonitrile, and the amount of probe remaining was determined based on the control samples which did not contain iCP. Excitingly, it was determined the probe was being cleaved selectively by the iCP over the sCP over the course of 6 hours, Fig. S1 (ESI†). After confirmation that the probe was able to be accepted and cleaved by the iCP, we wished to assess its ability to generate a luminescent signal after interacting with the iCP in the presence of luciferase. Initially the probe was incubated with varying amounts of iCP and sCP to determine a limit of detection biochemically. After an hour of incubation with purified iCP or sCP, luciferin detection reagent (LDR) was added. This commercially available reagent contains the luciferase enzyme to convert free luciferin derivatives into their light emitting oxyluciferin products. This probe was selective for interacting with the iCP over the sCP at several concentrations of purified proteasome isoforms, Fig. 2A. The amount of luminescence was also dependent upon the amount of iCP included in the assay.
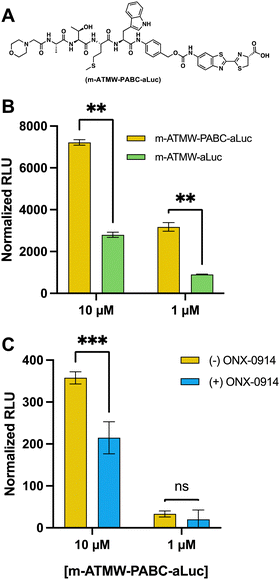
In order to ensure the luminescence values observed are due to iCP activity to release amino luciferin from the peptide, we utilized the iCP selective inhibitor ONX-0914. ONX-0914 was added to purified iCP as well as m-ATMW-aLuc for one hour. Upon addition of LDR and scanning on a plate reader, we were able to see significant reduction in the luminescent values observed in the ONX-0914 treated samples, Fig. 2B and Fig. S2 (ESI†). This result provides confidence that the amino luciferin must be released from the peptide by iCP activity to generate luminescent signal.
All of the previous experiments had been carried out using purified iCP and sCP, which does not necessarily correlate to the probe being successful when it enters the cellular environment. To assess the probe's ability to interact with iCP in cells, we needed to establish models that could demonstrate probe 3's ability to be cell permeable, interact with the iCP, and react with luciferase to generate a luminescent signal. Ramos (B-Cell Lymphoma) cells were selected as they were shown to have higher endogenous amounts of iCP without having to dose with a cytokine, Fig. S3 (ESI†). These cells do not express luciferase, so LDR was needed to observe any signal. To compliment the Ramos cell model, 4T1 cells were also selected. This cell line has been engineered to stable express luciferase and is widely used in a variety of mouse models, as well as are a model cell line for studying breast cancer tumor metastasis.28 The amounts of iCP expression in these cell lines was quantitated (with and without exposure to IFN-γ) by western blotting using an antibody to detect the iCP catalytic subunit Beta-5i without treatment. By western blotting the cell lysate, Ramos cells showed high endogenous expression of the iCP, while upon treatment of IFN-γ (20 ng mL−1) for 72 h, an increase in Beta-5i could be observed in 4T1 cells, Fig. S4 (ESI†).
Having established cellular models for testing the probe, we needed to ensure probe 3 was not causing any unwanted toxicity to the cell lines chosen at the relevant time points or concentrations. Both cell lines were dosed with probe at a range of concentrations and incubation times. At relevant concentrations and time points, there was no observed toxicity as anticipated, Fig. S5 and S6 (ESI†).
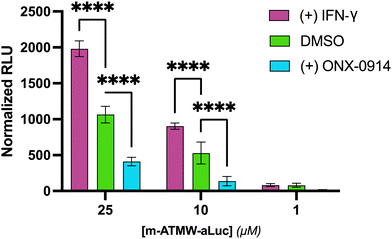
A dose dependent assay to determine probe concentration required to observe a signal in both cell lines was undertaken. In Ramos cells, which have a high amount of endogenous iCP, we were able to detect sufficient signal of probe 3 at 1 μM, Fig. S7 (ESI†). 4T1 cells express more iCP after exposure to IFN-γ. They were pre-dosed with 20 ng mL−1 of IFN-γ or DMSO for 72 h. The cells were then subjected to the same dosing protocol with the probe as the Ramos cells. A dose-dependent increase in luminescent signal in the 4T1 cells treated with IFN-γ was observed, indicating more active iCP correlates with an increase in iCP expression from the western blot analysis, Fig. 3. The 4T1 cells were also incubated with the iCP inhibitor ONX-0914. A reduction in luminescence occurred as the iCP was inhibited, limiting the amount of amino luciferin available as a luciferase substrate. These results highlights that the observed luminescence signal is dependent on iCP activity.
Next, we were interested to determine if the sensitivity of probe 3 compared to commercially available probes for the proteasome. Suc-LLVY-Amino luciferin (Suc-LLVY-aLuc) is commonly used to detect iCP or sCP activity in a variety of cell models or in vivo. This probe is not selective as the amino acid sequence LLVY can be recognized by both the sCP and iCP. We hoped that probe 3 would be able to generate similar signal levels as this commercially available option, but have improved selectivity that would be better suited for iCP analysis only. When incubating both probes with purified iCP for 1 h, we were excited to see that probe 3 retains almost the same sensitivity as the Suc-LLVY-aLuc probe, Fig. S8 (ESI†).
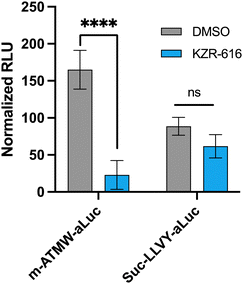
The 4T1 luciferin-expressing cell line after exposure to IFN-γ were dosed with 10 μM of either probe. Another iCP inhibitor, KZR-616, was utilized which has been noted to be a move selective inhibitor for the iCP over the sCP.29–32 Significant reduction in signal was observed using the m-ATMW-aLuc probe with co-treatment with KZR-616, but no significant change is observed with the Suc-LLVY-aLuc probe, Fig. 4. This result indicates the Suc-LLVY-aLuc probe produces signal after interacting with either the sCP and iCP, highlighting that it is not applicable for monitoring only iCP activity in cells. Having a nonselective probe could be hindering the ability to fully understand the role of iCP in a desired system.
To further improve the properties of this new iCP probe, with the long-term goal to be able to use concentrations applicable for in vivo activity studies, modifications of the scaffold were undertaken to improve the rate at which the amino luciferin is cleaved from the iCP recognition peptide. A hypothesis was formed that the cleavage rate of the uncaging of the amino luciferin moiety could be increased by separating the amino luciferin from the peptide recognition sequence using a self-immolative linker. Linkers are commonly employed in a variety of prodrugs and probes to limit any hinderance of the enzyme's active site and allow for improved cleavage.33–35 The para-aminobenzyl alcohol (PABC) linker is frequently used for conjugating molecules to peptides and could be incorporated into our synthesis easily.36,37 The iCP recognition peptide could be generated by solid phase peptide synthesis as before, and then incorporate the PABC linker before the introduction of the bioluminescent agent, Fig. 5A and Scheme S1 (ESI†).
To assess cleavage abilities of the iCP with and without the PABC linker, both probes were tested in a LCMS assay to quantitate how much aluc was being cleaved over time, Fig. S9 (ESI†). Once we had determined it could interact with the iCP and was able to release more of the aLuc substrate, we wanted to determine if this correlated to increased luminescent signal. Both probes were incubated with purified iCP or sCP for 1 h. After an hour, LDR was added, and the amount of luminescent signal was detected using a plate reader. The probe derivative that contained the linker had significantly more luminescent signal as compared the probe without the PABC linker, Fig. 5B. We were able to observe a reduction in luminescent signal when incubated with the iCP inhibitor, confirming our signal is directly related to the hydrolysis activity of the iCP, Fig. S10 (ESI†).
To demonstrate the cellular application of the m-ATMW-PABC-aLuc probe, 4T1 cells were incubated with iCP inhibitor ONX-0914 prior to treatment with probe. After the addition of LDR and scanning for luminescence, using 10 μM of the probe provided significant signal and that signal was reduced if the iCP was inhibited with ONX-0914, Fig. 5C. Using 1 μM of the probe in cells was too low of a concentration to detect a change when ONX-0914 was utilized.
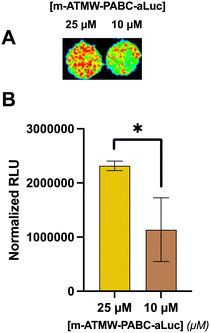
The long-term goal of this new probe is to use it in vivo to detect iCP activity in a variety of mouse models. However, the luminescent signal needs to be strong enough to be visualized through tissue, as well as remain intact without being cleaved by random proteases. Also, rather than a plate reader, luminescence would need to be detected using an EMCCD camera. Initially, probe 3 and probe 6 were incubated with 10% human serum at various time points to ensure no unwanted cleavage could be detected, Fig. S11 (ESI†). Then, 4T1 cells were dosed with varying amounts of the probe and with ONX-0914 to confirm signal detected by the camera was from iCP-mediated release of the aLuc from the probe. After imaging, similar results were obtained as those from the plate reader assay, Fig. S12 (ESI†). To mimic visualizing iCP activity through tissue, thin slices of turkey bacon were laid over wells of 4T1 cells after dosing with probe.38 This plate was imaged with the hope that signal could penetrate through the bacon in a similar fashion to how it could be used in vivo. With an exposure time of 5 min, significant signal over the background was detected using 25 μM and 10 μM of the probe, Fig. 6. Although using LDR can be used in these cellular and tissue studies, further refinement to the probe may be necessary before in vivo testing is possible.
Conclusion
Bioluminescent activity probes to monitor enzyme activity have an advantage over fluorescent ones, as they can provide significant signal over noise ratios and can be visualized through tissue. The iCP plays a variety of important roles including generating antigenic peptides and its expression can indicate an inflammatory response by the immune system. There is a need to develop improved tools of monitoring this proteasome isoform to increase our knowledge of the role the iCP plays in cancer and autoimmune diseases. Having a probe that has the potential to be utilized in an in vivo setting could allow for a better understanding of how quickly the iCP is being expressed after exposure to inflammatory cytokines, and how far from the site of inflammation it is being expressed, two pieces of knowledge which are currently lacking.The luminescent activity probe for the iCP described here includes a self-immolative linker conjugated to aLuc. After interacting with the iCP and decomposition of the linker, aLuc can be released to interact with luciferase. This “caged” aLuc probe is applicable to monitor iCP activity in biochemical and in cellulo assays. Signal generated from uncaging the aLuc by the iCP was also visualized using a EMCCD camera. Although this probe is shown to be effective in the assays described above, we believe further refinement and improvement is needed before actual in vivo applications can be conducted. The development of NanoLuc, which is significantly brighter than firefly luciferase, could potentially help achieve better signal, with less amount of probe. Swapping the amino luciferin moiety for a different, more sensitive bioluminescent substrate would greatly benefit this technology. There are also other potential applications that could be investigated with this probe by multiplexing with other enzymes to better elucidate how the iCP is working in correlation with other cellular processes.
Data availability
Data for this manuscript is available within the text or the ESI.†Conflicts of interest
Prof. Trader is a shareholder and consultant for Booster Therapeutics, GmbH. Other authors declares no conflict of interest.Acknowledgements
This work was supported by a start-up package from the UCI School of Pharmacy and the UCI Chao Family Comprehensive Cancer Center (P30CA062203). It was also supported by a NIH-NIAID grant (1R01AI50847).References
- E. M. Huber, M. Basler, R. Schwab, W. Heinemeyer, C. J. Kirk, M. Groettrup and M. Groll, Immuno- and Constitutive Proteasome Crystal Structures Reveal Differences in Substrate and Inhibitor Specificity, Cell, 2012, 148(4), 727–738, DOI:10.1016/j.cell.2011.12.030.
- I. Sosič, M. Gobec, B. Brus, D. Knez, M. Živec, J. Konc, S. Lešnik, M. Ogrizek, A. Obreza, D. Žigon, D. Janežič, I. Mlinarič-Raščan and S. Gobec, Nonpeptidic Selective Inhibitors of the Chymotrypsin-Like (B5 i) Subunit of the Immunoproteasome, Angew. Chem., Int. Ed., 2016, 55(19), 5745–5748, DOI:10.1002/anie.201600190.
- D. A. Ferrington and D. S. Gregerson, in Immunoproteasomes, Progress in Molecular Biology and Translational Science, Elsevier, 2012, vol. 109, pp. 75–112 Search PubMed.
- I. Churcher, Protac-Induced Protein Degradation in Drug Discovery: Breaking the Rules or Just Making New Ones, J. Med. Chem., 2018, 61(2), 444–452, DOI:10.1021/acs.jmedchem.7b01272.
- N. Vigneron and B. J. V. D. Eynde, Proteasome Subtypes and the Processing of Tumor Antigens: Increasing Antigenic Diversity, Curr. Opin. Immunol., 2012, 24(1), 84–91, DOI:10.1016/j.coi.2011.12.002.
- P. V. D. Bruggen and B. J. V. D. Eynde, Processing and Presentation of Tumor Antigens and Vaccination Strategies, Curr. Opin. Immunol., 2006, 18(1), 98–104, DOI:10.1016/j.coi.2005.11.013.
- J. Kast, Immunoproteasome Deficiency in Non-Small Cell Lung Cancer and Its Relevance to Immunotherapy, J. Thorac. Dis., 2016, 8(9), E1082–E1086, DOI:10.21037/jtd.2016.08.80.
- M. Basler and M. Groettrup, On the Role of the Immunoproteasome in Protein Homeostasis, Cells, 2021, 10(11), 3216, DOI:10.3390/cells10113216.
- O. Coux, K. Tanaka and A. L. Goldberg, Structure and Functions of the 20s and 26s Proteasomes, Annu. Rev. Biochem., 1996, 65(1), 801–847, DOI:10.1146/annurev.bi.65.070196.004101.
- X. Huang, B. Luan, J. Wu and Y. Shi, An Atomic Structure of the Human 26S Proteasome, Nat. Struct. Mol. Biol., 2016, 23(9), 778–785, DOI:10.1038/nsmb.3273.
- C. J. Kirk, T. Muchamuel, J. Wang and R. A. Fan, Discovery and Early Clinical Development of Selective Immunoproteasome Inhibitors, Cells, 2021, 11(1), 9, DOI:10.3390/cells11010009.
- B. L. Zerfas and D. J. Trader, Monitoring the Immunoproteasome in Live Cells Using an Activity-Based Peptide–Peptoid Hybrid Probe, J. Am. Chem. Soc., 2019, 141(13), 5252–5260, DOI:10.1021/jacs.8b12873.
- J. Gan, Y. Leestemaker, A. Sapmaz and H. Ovaa, Highlighting the Proteasome: Using Fluorescence to Visualize Proteasome Activity and Distribution, Front. Mol. Biosci., 2019, 6, 14, DOI:10.3389/fmolb.2019.00014.
- G. de Bruin, B. T. Xin, M. Kraus, M. van der Stelt, G. A. van der Marel, A. F. Kisselev, C. Driessen, B. I. Florea and H. S. Overkleeft, A Set of Activity-Based Probes to Visualize Human (Immuno)Proteasome Activities, Angew. Chem., Int. Ed., 2016, 55(13), 4199–4203, DOI:10.1002/anie.201509092.
- C. R. Berkers, F. W. B. van Leeuwen, T. A. Groothuis, V. Peperzak, E. W. van Tilburg, J. Borst, J. J. Neefjes and H. Ovaa, Profiling Proteasome Activity in Tissue with Fluorescent Probes, Mol. Pharm., 2007, 4(5), 739–748, DOI:10.1021/mp0700256.
- D. S. Hewings, J. A. Flygare, I. E. Wertz and M. Bogyo, Activity-Based Probes for the Multicatalytic Proteasome, FEBS J., 2017, 284(10), 1540–1554, DOI:10.1111/febs.14016.
- M. Verdoes, B. I. Florea, V. Menendez-Benito, C. J. Maynard, M. D. Witte, W. A. van der Linden, A. M. C. H. van den Nieuwendijk, T. Hofmann, C. R. Berkers, F. W. B. van Leeuwen, T. A. Groothuis, M. A. Leeuwenburgh, H. Ovaa, J. J. Neefjes, D. V. Filippov, G. A. van der Marel, N. P. Dantuma and H. S. Overkleeft, A Fluorescent Broad-Spectrum Proteasome Inhibitor for Labeling Proteasomes In Vitro and In Vivo, Chem. Biol., 2006, 13(11), 1217–1226, DOI:10.1016/j.chembiol.2006.09.013.
- D. M. Close, T. Xu, G. S. Sayler and S. Ripp, In Vivo Bioluminescent Imaging (BLI): Noninvasive Visualization and Interrogation of Biological Processes in Living Animals, Sensors, 2010, 11(1), 180–206, DOI:10.3390/s110100180.
- M. Deluca, Firefly Luciferase, in Advances in Enzymology - and Related Areas of Molecular Biology, ed. A. Meister, Wiley, 1976, vol. 44, pp. 37–68 DOI:10.1002/9780470122891.ch2.
- K. V. Wood, The chemical mechanism and evolutionary development of beetle bioluminescence, Photochem. Photobiol., 1995, 62(4), 662–673, DOI:10.1111/j.1751-1097.1995.tb08714.x.
- N. Lembert, Firefly Luciferase Can Use L-Luciferin to Produce Light, Biochem. J., 1996, 317(1), 273–277, DOI:10.1042/bj3170273.
- A. Dragulescu-Andrasi, G. Liang and J. Rao, In Vivo Bioluminescence Imaging of Furin Activity in Breast Cancer Cells Using Bioluminogenic Substrates, Bioconjugate Chem., 2009, 20(8), 1660–1666, DOI:10.1021/bc9002508.
- T. Monsees, W. Miska and R. Geiger, Synthesis and Characterization of a Bioluminogenic Substrate for α-Chymotrypsin, Anal. Biochem., 1994, 221(2), 329–334, DOI:10.1006/abio.1994.1421.
- T. A. Su, K. J. Bruemmer and C. J. Chang, Caged Luciferins for Bioluminescent Activity-Based Sensing, Curr. Opin. Biotechnol, 2019, 60, 198–204, DOI:10.1016/j.copbio.2019.05.002.
- M. A. O’Brien, W. J. Daily, P. E. Hesselberth, R. A. Moravec, M. A. Scurria, D. H. Klaubert, R. F. Bulleit and K. V. Wood, Homogeneous, Bioluminescent Protease Assays: Caspase-3 as a Model, SLAS Discov., 2005, 10(2), 137–148, DOI:10.1177/1087057104271865.
- B. L. Zerfas and D. J. Trader, Synthesis and Application of an Activity-Based Peptide-Peptoid Hybrid Probe for the Immunoproteasome, Curr. Protoc. Chem. Biol., 2019, 11(4) DOI:10.1002/cpch.76.
- G. L. Beutner, I. S. Young, M. L. Davies, M. R. Hickey, H. Park, J. M. Stevens and Q. Ye, TCFH–NMI: Direct Access to N -Acyl Imidazoliums for Challenging Amide Bond Formations, Org. Lett., 2018, 20(14), 4218–4222, DOI:10.1021/acs.orglett.8b01591.
- B. A. Pulaski and S. Ostrand-Rosenberg, Mouse 4T1 Breast Tumor Model, Curr. Protoc. Immunol., 2000, 39, 20.2.1–20.2.16, DOI:10.1002/0471142735.im2002s39.
- KZR-616, a First-in-Class Selective Inhibitor of the Immunoproteasome, Ameliorates Polymyositis in a Murine Model. ACR Meeting Abstracts, https://acrabstracts.org/abstract/kzr-616-a-first-in-class-selective-inhibitor-of-the-immunoproteasome-ameliorates-polymyositis-in-a-murine-model/, (accessed 2022-11-18).
- J. Lickliter, D. Bomba, J. Anderl, A. Fan, C. J. Kirk and J. Wang, AB0509 Kzr-616, a Selective Inhibitor of the Immunoproteasome, Shows a Promising Safety and Target Inhibition Profile in a Phase i, Double-Blind, Single (SAD) and Multiple Ascending Dose (MAD) Study in Healthy Volunteers, SLE, Sjögren's and APS – treatment, BMJ Publishing Group Ltd and European League Against Rheumatism, 2018, pp. 1413.3–1414 DOI:10.1136/annrheumdis-2018-eular.3344.
- T. Muchamuel, J. L. Anderl, R. A. Fan, H. W. B. Johnson, C. J. Kirk and E. Lowe, FRI0296 Kzr-616, a Selective Inhibitor of the Immunoproteasome, Blocks the Disease Progression in Multiple Models of Systemic Lupus Erythematosus (SLE). In Friday, 15 June 2018, BMJ Publishing Group Ltd and European League Against Rheumatism, 2018, pp. 685.1–685 DOI:10.1136/annrheumdis-2018-eular.1100.
- R. Furie, S. Parikh, J. Wang, D. Bomba, R. Leff, C. Kirk and N. Henig, POS0695 KZR-616, A Selective Immunoproteasome Inhibitor for the Treatment of Systemic Lupus Erythematosus: Results from the Completed Dose Escalation Phase 1b Portion of the Mission Study, Ann. Rheum. Dis., 2021, 80(Suppl. 1), 595.2–596, DOI:10.1136/annrheumdis-2021-eular.2158.
- R. Sheyi, B. G. de la Torre and F. Albericio, Linkers: An Assurance for Controlled Delivery of Antibody-Drug Conjugate, Pharmaceutics, 2022, 14(2), 396, DOI:10.3390/pharmaceutics14020396.
- Y. Anami, C. M. Yamazaki, W. Xiong, X. Gui, N. Zhang, Z. An and K. Tsuchikama, Glutamic Acid–Valine–Citrulline Linkers Ensure Stability and Efficacy of Antibody–Drug Conjugates in Mice, Nat. Commun., 2018, 9(1), 2512, DOI:10.1038/s41467-018-04982-3.
- M. R. Sorkin, J. A. Walker, F. Ledesma, N. P. Torosian and C. A. Alabi, Design of Protein-Based “Turn on” Molecular Probes for Intracellular Bond Cleavage, Mol. Syst. Des. Eng., 2020, 5(1), 385–391, 10.1039/C9ME00147F.
- S. Y. Y. Ha, Y. Anami, C. M. Yamazaki, W. Xiong, C. M. Haase, S. D. Olson, J. Lee, N. T. Ueno, N. Zhang, Z. An and K. Tsuchikama, An Enzymatically Cleavable Tripeptide Linker for Maximizing the Therapeutic Index of Antibody-Drug Conjugates, Mol. Cancer Ther., 2022, 21(9), 1449–1461, DOI:10.1158/1535-7163.MCT-22-0362.
- N. Jain, S. W. Smith, S. Ghone and B. Tomczuk, Current ADC Linker Chemistry, Pharm. Res., 2015, 32(11), 3526–3540, DOI:10.1007/s11095-015-1657-7.
- A. C. Love, D. R. Caldwell, B. Kolbaba-Kartchner, K. M. Townsend, L. P. Halbers, Z. Yao, C. K. Brennan, J. Ivanic, T. Hadjian, J. H. Mills, M. J. Schnermann and J. A. Prescher, Red-Shifted Coumarin Luciferins for Improved Bioluminescence Imaging, J. Am. Chem. Soc., 2023, 145(6), 3335–3345, DOI:10.1021/jacs.2c07220.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00148f |
| This journal is © The Royal Society of Chemistry 2024 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cell per well) incubated with iCP inhibitor KZR-616 (500 nM) followed by incubation with
000 cell per well) incubated with iCP inhibitor KZR-616 (500 nM) followed by incubation with