Open Access Article
Open Access ArticleRapid formation of Nε-(carboxymethyl)lysine (CML) from ribose depends on glyoxal production by oxidation†
Hikari
Sugawa
a,
Tsuyoshi
Ikeda
b,
Yuki
Tominaga
a,
Nana
Katsuta
c and
Ryoji
Nagai
 *a
*a
aDepartment of Food and Life Sciences, School of Agriculture, Tokai University, Japan. E-mail: nagai-883@umin.ac.jp
bFaculty of Pharmaceutical Sciences, Sojo University, Japan
cResearch Institute of Agriculture, Tokai University, Japan
First published on 18th September 2024
Abstract
N ε-(Carboxymethyl)lysine (CML) is a major advanced glycation end-product (AGE) involved in protein dysfunction and inflammation in vivo. Its accumulation increases with age and is enhanced with the pathogenesis of diabetic complications. Therefore, the pathways involved in CML formation should be elucidated to understand the pathological conditions involved in CML. Ribose is widely used in glycation research because it shows a high reactivity with proteins to form AGEs. We previously demonstrated that ribose generates CML more rapidly than other reducing sugars, such as glucose; however, the underlying mechanism remains unclear. In this study, we focused on the pathway of CML formation from ribose. As a result, glyoxal (GO) was the most abundant product generated from ribose among the tested reducing sugars and was significantly correlated with CML formation from ribose-modified protein. The coefficient of determination (R2) for CML formation between the ribose-modified protein and Amadori products or the ribose degradation product (RDP)-modified protein was higher for the RDP-modified protein. CML formation from ribose degradation products (RDP) incubated with protein significantly correlated with CML formation from GO-modified protein (rs = 0.95, p = 0.0000000869). GO and CML formation were inhibited by diethylenetriaminepentaacetic acid (DTPA) and enhanced by iron chloride. Additionally, flavonoid compounds such as isoquercetin, which are known to inhibit CML, also inhibited GO formation from ribose and CML formation. In conclusion, ribose undergoes auto-oxidation and oxidative cleavage between C-2 and C-3 to generate GO and enhance CML accumulation.
Introduction
Post-translational modifications (PTMs) are biochemical reactions that involve one or more amino acid residues in proteins. These reactions alter protein function through covalent modification by adding functional groups to proteins. Phosphorylation, glycosylation, methylation, and acetylation are enzymatic PTMs that play important roles in regulating biological systems, such as enzymatic activity and the three-dimensional structure of proteins. In contrast, glycation is a non-enzymatic PTM that occurs between the amino acid residues of proteins and the carbonyl group of reducing sugars, such as glucose and ribose. This leads to the formation of advanced glycation end-products (AGEs), resulting in the induction of protein dysfunction via conformational changes in proteins.1N ε-(Carboxymethyl)lysine (CML), a major antigenic AGE,2 accumulates in the body with aging3 and is involved in the pathogenesis of diabetic complications, such as neuropathy4 and vascular disease.5 Zhang et al. reported that plasma CML levels in patients with type 2 diabetes mellitus (DM) are inversely correlated with cognitive function.6 Southern et al. reported that CML accumulation in cortical neurons and cerebral vessels is related to cognitive impairment or anamnesis in patients with DM, suggesting that its accumulation may contribute to dementia in individuals with cerebrovascular disease.7 Ribose is a component of ATP, which is used as an energy source in organisms. Ribose is widely used in supplements, sports drinks, and medicine as it promotes ATP synthesis and contributes to the treatment of congestive heart failure.8 Moreover, ribose has been used in glycation research as it shows high reactivity with proteins to form AGEs.9 Ribose levels in the serum are highly correlated with glycated serum protein levels, indicating that ribose contributes to the glycation of proteins in vivo.10 Furthermore, a recent study reported that ribose levels in older adult urine were inversely correlated with cognitive function, suggesting that abnormal ribose metabolism was involved in the early stages of cognitive impairment.11 Yu et al. suggested that serum ribose levels are increased in type 1 DM cases and may be involved in cognitive impairment.12 These findings indicate that glycation by ribose may contribute to decreased cognitive function.
Ribose-modified proteins generate AGEs more rapidly than glucose-modified proteins do. Ribose readily forms an open-chain structure compared with that of glucose because the reactivity of reducing sugars depends on the presence of a carbonyl group in the structure.13 Nevertheless, little is known about the mechanism by which ribose generates more CML than glucose.
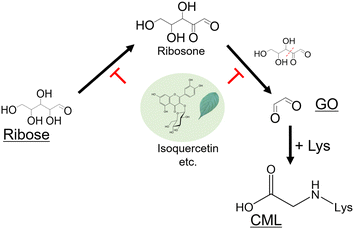
Glyoxal (GO), a key intermediate in AGE formation, demonstrates higher reactivity with proteins than with reduced sugars. This reactivity is significant because CML is formed from reducing sugars and dicarbonyl compounds, such as GO.14 Wells-Knecht et al. and Thornalley et al. reported that GO is formed by glucose autoxidation.15,16 This understanding led us to hypothesize that ribose is prone to form GO because it degrades more rapidly than reducing sugars, such as glucose. Here, we aimed to delineate the pathway by which ribose contributes to CML formation.
Results and discussion
CML formation is derived from reducing sugars
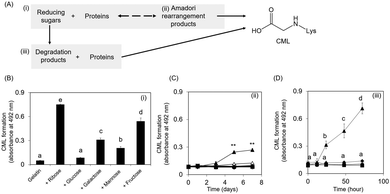
Collagen is the most abundant structural protein, constituting approximately 30% of the total protein mass in humans.17 CML accumulates with age in human skin.3 Therefore, the underlying mechanism of CML formation on collagen is important; however, investigating this mechanism has proven difficult because of the water-insolubility of collagen. This study focused on gelatin because it is a water-soluble protein obtained from collagen via the destruction of cross-links between polypeptide chains along with partial degradation of the polypeptide bonds.17CML formation was evaluated in three steps: (i) mixture of reducing sugars and proteins, (ii) Amadori rearrangement products, and (iii) mixture of reducing sugar degradation products and proteins (Fig. 1A). CML formation on ribose-modified proteins was the highest among the reducing sugar-modified proteins (Fig. 1B). Ribose tends to be more open-chain than glucose;13 therefore, it is more likely to react with proteins to form CML. Consistent with this observation, a non-competitive enzyme-linked immunosorbent assay (ELISA) revealed that ribose-derived Amadori rearrangement products had the highest CML levels among the reducing sugar-derived products (Fig. 1C). CML formation was observed when the ribose degradation product was incubated with gelatin. In contrast, when proteins were incubated with other reducing sugar degradation products, the CML level remained unchanged under the same conditions (Fig. 1D). These results suggest that ribose generates CML not only from Amadori rearrangement products but also from sugar degradation products.
Comparison contents of Amadori rearrangement product by protein modification of glucose and ribose
Evaluation of CML formation in Amadori products by oxidation shows that ribose-derived Amadori products generate more CML than other reducing sugar-derived products, including glucose (Fig. 1C). In the next step, the protein modification rate was evaluated by the TNBS method to clarify the formation of the Amadori product by ribose. As a result, the protein modification rate of 30 mM glucose and ribose increased in an incubation time-dependent manner. However, the rate of ribose modification was not significantly different from that of glucose (Table 1). Nevertheless, CML formation in ribose-derived Amadori rearrangement products by oxidation was higher than that in glucose-derived products (Fig. 1C). These results indicate that ribose-derived Amadori is more likely to generate CML than glucose-derived Amadori.| Incubation (day) | 0 | 1 | 3 | 5 | 7 | |
|---|---|---|---|---|---|---|
| Lys (nmol) | Glucose–gelatin | 52.8 (±0.9) | 46.7 (±1.0) | 37.7 (±0.8) | 35.9 (±1.5) | 29.1 (±2.9) |
| Ribose–gelatin | 57.4 (±1.2) | 50.3 (±0.4) | 44.0 (±0.7) | 39.9 (±2.0) | 32.2 (±1.2) | |
| Modification (%) | Glucose–gelatin | 0 (±1.6) | 11.6 (±1.8) | 28.6 (±1.5) | 32.0 (±2.8) | 45.0 (±5.5) |
| Ribose–gelatin | 0 (±2.2) | 12.3 (±0.7) | 23.4 (±1.1) | 30.4 (±3.4) | 44.0 (±2.1) | |
The Amadori product contents in the modified gelatin preparations (20 μg protein) were determined using TNBS, as described in the Experimental section. Amadori products were prepared as described in the Experimental section using reducing sugar-modified proteins (n = 3).
Correlation between ribose-derived CML and GO formation
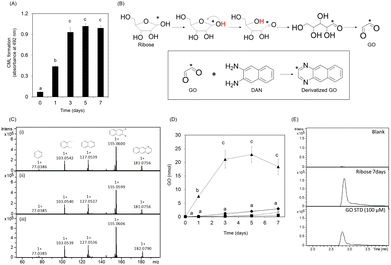
CML formation in the ribose-modified protein plateaued for only three days (Fig. 2A). Because CML is rapidly formed from GO,14 we investigated the formation of GO from ribose. The predicted structure of derivatized GO is illustrated in Fig. 2B. The derivatized GO in ribose solution or standard was analyzed in MS/MS analysis (derivatized GO: m/z 181.0756 ± 0.1, derivatized GO derived from 13C1 ribose: m/z 182.0790 ± 0.1) using liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry (LC-ESI-QTOF; Bruker Daltonics, Germany) as described in the Materials and methods section. The fragmentation pattern of GO derived from ribose and 13C1 ribose is consistent with that of the GO standard (Fig. 2C). In addition, the retention times of GO in the ribose solution are consistent with those of GO in the standard solution (Fig. 2E)These results demonstrate that GO is formed from ribose, suggesting that it is composed of C-1 and C-2 of ribose.
In addition, the intermediate (ribosone) was also detected from ribose and 13C1 ribose (Fig. S1A and B, ESI†).
Furthermore, the GO content in the ribose solution was the highest among the tested reducing sugars and was approximately 100-fold higher than that in the glucose solution (Fig. 2D). Ribosone formation also increased in a time-dependent manner (Fig. S1C, ESI†). In contrast, GO formation did not increase significantly compared to the other reducing sugars (Fig. 2D).
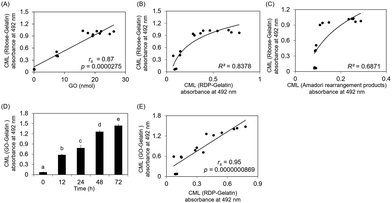
GO formation in the ribose solution also plateaued for only three days, similar to CML formation on ribose-modified gelatin. Fig. 3A shows that CML formation on ribose-modified proteins was significantly correlated with GO formation in ribose solution (rs = 0.87, p = 0.0000275). The determination coefficient (R2) between CML formation on ribose- and ribose degradation product (RDP)-modified proteins (R2 = 0.8378) was higher than that between CML formation on ribose-modified proteins and Amadori rearrangement products (R2 = 0.6871) (Fig. 3B and C). CML formation on GO- and RDP-modified proteins showed a significant correlation (rs = 0.95, p = 0.0000000869) (Fig. 3D and E).
These results suggest that CML on ribose-modified proteins is not only formed by the oxidation of Amadori rearrangement products, which is generated by the direct modification of proteins by ribose, but also by the modification of proteins via GO generated from ribose. Although the ribose concentration in the physiological environment is lower than that of other reducing sugars such as glucose,10 it could still significantly contribute to CML formation in vivo because it shows higher reactivity with proteins and generates GO more rapidly than other reducing sugars.
Ribose-derived CML formation pathway
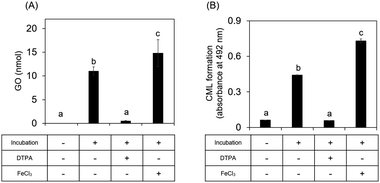
We next investigated whether ribose-derived GO formation is mediated by an oxidation reaction, as observed for GO derived from glucose.15,16 We found that GO and CML formation was inhibited by a metal chelator (antioxidative condition) and enhanced in the presence of iron chloride (enhanced oxidative condition) (Fig. 4). Ribosone formation was regulated by the presence or absence of oxidation conditions as well as by GO (Fig. S1D, ESI†).Our previous study revealed that flavonoid compounds in Eucommia ulmoides Oliver (E. ulmoides) leaf extract (ELE) inhibited ribose-derived CML formation.18 However, the mechanism underlying this inhibition remains unclear. Flavonoids exhibit antioxidant activity via chelate metals, such as free iron.19
Therefore, the inhibitory effects of flavonoid compounds from ELE on ribose-derived GO and CML formation were evaluated to determine whether the oxidation reaction led to their generation. As shown in Table 2, compounds 1–6 were found in ELE, whereas 7 and 8 were the aglycones of these compounds. Compounds 1–3 were quercetin glycosides and compounds 4–6 were kaempferol glycosides.
| ID | Chemical name |
|---|---|
| Compounds 1–6 contained quercetin glycosides and kaempferol glycosides in E. ulmoides leaf extract (ELE). Compounds 7 and 8 were the aglycones of these compounds. | |
| 1 | Isoquercetin |
| 2 | Rutin |
| 3 | Quercetin-3-O-sambubioside |
| 4 | Astragalin |
| 5 | Kaempferol-3-O-rutinoside |
| 6 | Kaempferol-3-O-(6-acetyl)-glucoside |
| 7 | Quercetin |
| 8 | Kaempferol |
Compounds in ELE, such as isoquercetin, inhibited ribose-derived GO and CML formation (Fig. 5B and C). These results indicate that CML rapidly forms from ribose-modified proteins because ribose generates GO via oxidation.
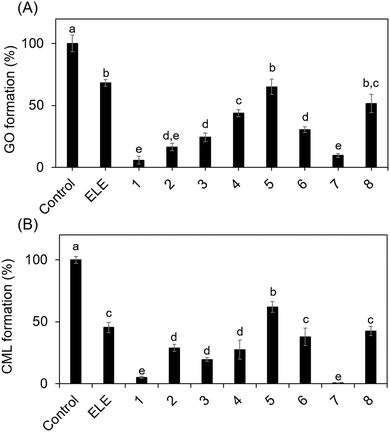 | ||
| Fig. 5 Inhibitory effect of ELE-related compounds on GO and CML formation. Ribose was incubated with/without (control) the ELE-related compounds. (A) Inhibitory effect on GO formation was evaluated using LC-ESI-QTOF. Gelatin and ribose were incubated in the presence or absence (control) of the ELE-related compounds. (B) The inhibitory effect on CML formation was then evaluated using ELISA (absorbance 492 nm). Data are expressed as formation (%) relative to that of the control (without ELE-related compounds). The ELE related compounds used are shown in Table 1: 1, isoquercetin; 2, rutin; 3, quercetin-3-O-sambubioside; 4, astragalin; 5, kaempferol-3-O-rutinoside; 6, kaempferol-3-O-(6′′-acetyl)-glucoside; 7, quercetin; 8, kaempferol. Data are presented as mean ± SD (n = 3). Different superscripts indicate statistically significant differences between groups (p < 0.05). | ||
Previous studies have reported the benefits of inhibiting CML formation. Bautista-Pérez et al. reported that administering methanol extract of spinach to diabetic rats prevented retinal degeneration by inhibiting CML–RAGE interaction and reducing CML accumulation.20 We previously reported that co-administration of Trapa bispinosa Roxb. extract and lutein in diabetic rats decreased serum CML levels and prevented the pathogenesis of diabetic cataract.21 Considering these findings, we believe that research on inhibiting CML formation is important from the perspective of preventive medicine. However, few studies have clarified the mechanisms underlying AGE formation. In this study, we identified a new pathway of AGE formation and found that compounds in E. ulmoides, such as isoquercetin, suppress CML formation by inhibiting this pathway. Therefore, our findings provide a reference for novel therapeutic strategies for CML-related diseases.
Quercetin glycosides had a greater inhibitory effect on GO and CML formation than kaempferol glycosides, suggesting that the inhibitory activity of E. ulmoides depends on their skeletal structure. However, it is important to note that glycosides are converted to aglycones by hydrolysis during intestinal absorption.22 Thus, we also evaluated the aglycones of these compounds. Quercetin inhibited the formation of GO and CML more potently than kaempferol did, which could be attributed to the number of hydroxyl groups in the compounds. Quercetin is used for the treatment of lifestyle diseases, such as diabetes, owing to its antioxidant and anti-inflammatory activities.23,24 Although we elucidated the mechanism by which natural compounds such as quercetin inhibit CML formation in vitro in this study, this should be verified in vivo in future studies.
Experimental
Preparation of reducing sugar-modified proteins
D-Ribose (Fuji Film Wako Pure Chemical, Japan), D-glucose (Kanto Chemical, Japan), D-galactose (Fuji Film Wako Pure Chemical, Japan), D-mannose (Kanto Chemical, Japan), and D-fructose (Kanto Chemical, Japan) were incubated with 2 mg mL−1 gelatin (Sigma-Aldrich, USA) in 200 mM phosphate buffer (pH 7.2) at 37 °C for up to 7 days. After incubation, unreacted sugars were removed by dialysis. Protein concentrations were determined using the Pierce™ bicinchoninic acid (BCA) Protein Assay Kit (Thermo Fischer Scientific, USA).Preparation of reducing sugar degradation product-modified proteins
Reducing sugars (glucose, mannose, galactose, fructose, or ribose; all 60 mM) were incubated in 200 mM phosphate buffer (pH 7.2) at 37 °C for 5 days. In this study, reducing sugar degradation products were defined as reducing sugar solutions that were incubated alone. Then, 30 mM reducing sugar degradation products were mixed with 2 mg mL−1 gelatin in 10 mM sodium phosphate buffer (pH 7.2) in the presence of 1 mM diethylenetriaminepentaacetic acid (DTPA) at 37 °C for 3 days, followed by the removal of unreacted sugars by dialysis. To prevent the production of reactive oxygen species that catalyze oxidation reactions, the proteins and reducing sugar degradation products were incubated with DTPA, a metal-chelating agent. The protein concentrations of the samples were determined using a BCA Protein Assay Kit.Preparation of reducing sugar-modified proteins
Reducing sugars (ribose, glucose, galactose, mannose, and fructose, all 30 mM) were incubated with 2 mg mL−1 gelatin solution in 10 mM sodium phosphate buffer (pH 7.2) in the presence of 1 mM DTPA and 0.1 mM aminoguanidine, and the solution was subsequently dialyzed against PBS at 4 °C for 24 h. Additionally, aminoguanidine was incorporated during the incubation to specifically trap any carbonyl compounds produced by sugar degradation, thus preventing their potential conversion into CML during the assay process. The protein concentrations of the samples were determined using a BCA protein assay kit. Amadori protein oxidation was performed as described previously.25 Briefly, the samples (0.1 mg) were incubated at 37 °C for 1 h with 0.4 mM anhydrous FeCl2 in the absence or presence of 5 mM H2O2, and CML formation was measured by ELISA.Measurement of glyoxal content in sugar solution
Sugar solutions (50 mM ribose, glucose, galactose, mannose, and fructose) were incubated with 200 mM phosphate buffer (pH 7.2) at 37 °C for up to 7 days. Subsequently, 50 μL of sugar solutions or GO standards were incubated with 200 μM 2,3-diaminonaphthalene (DAN; Tokyo Chemical Industry, Japan) at 37 °C for 2 h. As described previously, the GO content in the sugar solutions was measured using LC-ESI-QTOF.26 Briefly, the samples were injected into a Strata-X-C column and eluted with 2 mL of 7% ammonia in 75% acetonitrile (ACN). The pooled elution fractions were dried and resuspended in 80% ACN (0.5 mL) containing 0.1% formic acid. The samples (5 μL) were subjected to LC-ESI-QTOF analysis. 13C1D-ribose was purchased from Cambridge Isotope Laboratories (Cambridge, UK) and incubated in a manner similar to that described above. Fragment patterns were analyzed using LC-MS/MS to confirm GO formation.Evaluation of ribose-derived CML formation pathway
To evaluate CML formation, 30 mM ribose was incubated with 2 mg mL−1 gelatin in 200 mM sodium phosphate buffer (pH 7.2), in the presence of 2 mM DTPA or 1 mM FeCl3, at 37 °C for 24 h. Subsequently, unreacted sugars were removed by dialysis. The protein concentrations of the samples were determined using a BCA protein assay kit. CML formation was measured by ELISA. To evaluate GO formation, 50 mM ribose was incubated in 200 mM sodium phosphate buffer (pH 7.2) in the presence of 2 mM DTPA or 1 mM FeCl3 at 37 °C for 24 h, followed by mixing of incubated samples with 200 μM DAN at 37 °C for 2 h. The GO content was determined using LC-ESI-QTOF-MS.Evaluation of the inhibitory effect of flavonoid compounds on GO and CML formation
Natural compounds were isolated as described previously.18 To evaluate GO formation, 50 mM ribose was incubated in 200 mM sodium phosphate buffer (pH 7.2) in the presence of purified compounds (100 μM) at 37 °C for 24 h. The incubated samples were mixed with 200 μM DAN at 37 °C for 2 h and the GO content was determined using LC-ESI-QTOF. To evaluate CML formation, 30 mM ribose was incubated with 2 mg mL−1 gelatin in the presence of purified compounds (100 μM) at 37 °C for seven days. Subsequently, unreacted sugars were removed by dialysis, and CML formation was measured using ELISA.Measurement of CML formation by ELISA
ELISA was performed as described previously.18 Briefly, each well of a 96-well immuno-plate (Thermo Fischer Scientific, USA) was coated with 100 μL of 1 μg mL−1 modified gelatin in PBS at 25 °C for 2 h. After incubation, the wells were washed thrice with PBS containing 0.05% Tween 20 (washing buffer), blocked with blocking solution (0.5% gelatin hydrolysate in PBS), and incubated at 25 °C for 1 h. The wells were washed and incubated at 25 °C for 1 h with 100 μL of 0.1 μg mL−1 anti-CML monoclonal antibody (6D12)27 dissolved in washing buffer at 1 μg mL−1. This antibody is confirmed a stoichiometric correlation for CML content in BSA as determined by HPLC analysis.27The wells were washed and incubated at 25 °C for 1 h with 100 μL of 1 μg mL−1 HRP-conjugated goat anti-mouse IgG (H+L) antibody (KPL, USA) diluted in the washing buffer. Following this incubation period, the wells were rewashed and treated with 100 μL of 500 μg mL−1O-phenylenediamine dihydrochloride (Fuji Film Wako Pure Chemical, Japan) in citrate-phosphate buffer (pH 5.0) containing 5.9 mM hydrogen peroxide for 5 min. The reaction was stopped with 100 μL of 1.0 M sulfuric acid, and the absorbance was measured at 492 nm using a microplate reader (TECAN, Switzerland).
We previously demonstrated that glycated human serum albumin generates CML when incubated at temperatures above 70 °C for 40 minutes, while CML formation is negligible at room temperature.28
TNBS method
The modification rate of gelatin with glucose or ribose was determined using 2,4,6-trinitrobenzenesulfonic acid (TNBS),29 with Nα-acetyl-L-lysine (Tokyo Chemical Industry Co., Ltd, Japan) as the calibration standard. Briefly, 50 μL of 0.2 mmol L−1 borate (pH 9.0) and 20 μL of 15 mmol L−1 TNBS were added to 50 μL of each sample (20 μg protein). After incubation at 37 °C for 15 minutes, the reaction was terminated by adding 200 μL of 90% formic acid, and the absorbance was measured at 420 nm. The modification ratio for each sample is expressed as a percentage of modification relative to modified gelatin on day 0, which serves as the control.LC-MS system
LC was performed using ZIC-HILIC columns packed with 5 μm particles (2.1 × 150 mm, Merck Millipore, Billerica, MA, USA). The column oven temperature was maintained at 40 °C. The mobile phases were prepared using distilled water containing 0.1% formic acid (solvent A) and ACN containing 0.1% formic acid (solvent B). The flow rate was set to 0.2 mL min−1. MS was performed in the positive ion mode with an ionization source temperature of 200 °C. The capillary voltage was operated at 4.5 kV. Collision-induced dissociation was performed using nitrogen. The nebulizer pressure was set to 1.6 bar. Data were acquired with a stored mass ranging from 50 to 1000 (m/z). The collision energy in MS/MS analysis was set to 40 eV.Statistical analysis
All data are expressed as the mean ± standard deviation (SD), and statistical significance was determined using one-way analysis of variance with Bonferroni's post-hoc test. The correlation between GO and CML formation was examined for statistical significance using the Spearman's rank correlation coefficient (rs). Statistical analyses were performed using the EZR software.30Conclusions
CML, the most widely studied structure among AGEs, is associated not only with diabetes but also with various other conditions. The present study provides the first evidence of a novel CML formation pathway from ribose and clarifies the mechanism by which E. ulmoides-related flavonoids inhibit CML formation (Fig. 6). These findings are essential for understanding the Maillard chemistry.Author contributions
Hikari Sugawa: conceptualization, data curation, formal analysis, investigation, methodology, software, validation, visualization, writing – original draft. Tsuyoshi Ikeda: resources, validation, writing – review & editing. Yuki Tominaga: investigation. Nana Katsuta: investigation, methodology. Ryoji Nagai: resources, supervision, funding acquisition, writing – review & editing.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Adaptable and Seamless Technology Transfer Program Through Target-driven Research and Development of the Japan Science and Technology Agency (JST; grant number AS3015118U). We would like to thank Editage (https://www.editage.jp) for English language editing.Notes and references
- R. Nagai, J. Shirakawa, Y. Fujiwara, R. Ohno, N. Moroishi, N. Sakata and M. Nagai, J. Clin. Biochem. Nutr., 2014, 55, 1–6, DOI:10.3164/jcbn.13-112.
- S. Reddy, J. Bichler, K. J. Wells-Knecht, S. R. Thorpe and J. W. Baynes, Biochemistry, 1995, 34, 10872–10878, DOI:10.1021/bi00034a021.
- J. A. Dunn, D. R. McCance, S. R. Thorpe, T. J. Lyons and J. W. Baynes, Biochemistry, 1991, 30, 1205–1210, DOI:10.1021/bi00219a007.
- K. A. Sveen, B. Karimé, E. Jørum, S. I. Mellgren, M. W. Fagerland, V. M. Monnier, K. Dahl-Jørgensen and K. F. Hanssen, Diabetes Care, 2013, 36, 3712–3717, DOI:10.2337/dc13-0788.
- S. Kralev, E. Zimmerer, M. Brueckmann, S. Lang, T. Kälsch, A. Rippert, J. Lin, M. Borggrefe, H. P. Hammes and T. Süselbeck, Clin. Chem. Lab. Med., 2009, 47, 446–451, DOI:10.1515/CCLM.2009.100.
- J. H. Zhang, H. Z. Xu, Q. F. Shen, Y. Z. Lin, C. K. Sun, L. Sha, Y. S. Ge, Y. Liu and C. Wang, Can. J. Neurol. Sci., 2016, 43, 518–522, DOI:10.1017/cjn.2015.398.
- L. Southern, J. Williams and M. M. Esiri, BMC Neurol., 2007, 7, 35, DOI:10.1186/1471-2377-7-35.
- S. Wagner, J. Herrick and L. M. Shecterle, Prog Cardiovasc Nurs., 2009, 24, 59–60, DOI:10.1111/j.1751-7117.2009.00033.x.
- Y. Wei, C. S. Han, J. Zhou, Y. Liu, L. Chen and R. Q. He, Biochem. Biophys. Acta, 2012, 1820, 488–494, DOI:10.1016/j.bbagen.2012.01.005.
- Y. Chen, L. Yu, Y. Wang, Y. Wei, Y. Xu, T. He and R. He, Biochim. Biophys. Acta, Mol. Basis Dis., 2019, 1865, 2285–2292, DOI:10.1016/j.bbadis.2019.05.005.
- X. Zhu, Y. Wei, Y. He, R. He and J. Li, BMC Geriatr., 2022, 22, 693, DOI:10.1186/s12877-022-03288-w.
- L. Yu, Y. Chen, Y. Xu, T. He, Y. Wei and R. He, Aging, 2019, 11, 4943–4969 CrossRef CAS.
- H. F. Bunn and P. J. Higgins, Science, 1981, 213, 222–224, DOI:10.1126/science.12192669.
- M. A. Glomb and V. M. Monnier, J. Biol. Chem., 1995, 270, 10017–10026, DOI:10.1074/jbc.270.17.10017.
- K. J. Wells-Knecht, D. V. Zyzak, J. E. Litchfield, S. R. Thorpe and J. W. Baynes, Biochemistry, 1995, 34, 3702–3709, DOI:10.1021/bi00011a027.
- P. J. Thornalley, A. Langborg and H. S. Minhas, Biochem. J., 1999, 344, 109–116 CrossRef CAS.
- D. Liu, M. Nikoo, G. Boran, P. Zhou and J. M. Regenstein, Annu. Rev. Food Sci. Technol., 2015, 6, 527–557, DOI:10.1146/annurev-food-031414-111800.
- H. Sugawa, R. Ohno, J. Shirakawa, A. Nakajima, A. Kanagawa, T. Hirata, T. Ikeda, N. Moroishi, M. Nagai and R. Nagai, Food Funct., 2016, 7, 2566–2573, 10.1039/c5fo01563d.
- P. Sestili, G. Diamantini, A. Bedini, L. Cerioni, I. Tommasini, G. Tarzia and O. Cantoni, Biochem. J., 2002, 364, 121–128, DOI:10.1042/bj3640121.
- R. Bautista-Pérez, A. Cano-Martínez, E. Gutiérrez-Velázquez, M. Martínez-Rosas, R. M. Pérez-Gutiérrez, F. Jiménez-Gómez and J. Flores-Estrada, Antioxidants, 2021, 10, 717, DOI:10.3390/antiox10050717.
- S. Kinoshita, H. Sugawa, T. Nanri, R. I. Ohno, J. I. Shirakawa, H. Sato, N. Katsuta, S. Sakake and R. Nagai, J. Clin. Biochem. Nutr., 2020, 66, 8–14, DOI:10.3164/jcbn.19-34.
- G. D. Orfali, A. C. Duarte, V. Bonadio, N. P. Martinez, M. E. de Araújo, F. B. Priviero, P. O. Carvalho and D. G. Priolli, World J. Clin. Oncol., 2016, 7, 189–199, DOI:10.5306/wjco.v7.i2.189.
- G. Zu, K. Sun, L. Li, X. Zu, T. Han and H. Huang, Sci. Rep., 2021, 11, 22959, DOI:10.1038/s41598-021-02248-5.
- S. C. Bischoff, Curr. Opin. Clin. Nutr. Metab. Care, 2008, 11, 733–740, DOI:10.1097/MCO.0b013e32831394b8.
- R. Nagai, K. Ikeda, T. Higashi, H. Sano, Y. Jinnouchi, T. Araki and S. Horiuchi, Biochem. Biophys. Res. Commun., 1997, 234, 167–172, DOI:10.1006/bbrc.1997.6608.
- H. Sugawa, A. Yachi, Y. Fujimoto and R. Nagai, J. Biochem., 2021, 170, 587–592, DOI:10.1093/jb/mvab079.
- K. Ikeda, T. Higashi, H. Sano, Y. Jinnouchi, M. Yoshida, T. Araki, S. Ueda and S. N. Horiuchi, Biochemistry, 1996, 35, 8075–8083, DOI:10.1021/bi9530550.
- M. C. Hayashi, R. Nagai, K. Miyazaki, F. Hayase, T. Araki, T. Ono and S. Horiuchi, Lab. Invest., 2002, 82, 795–808, DOI:10.1097/01.lab.0000018826.59648.07.
- R. Fields, Biochem. J., 1971, 124, 581–590, DOI:10.1042/bj1240581.
- Y. Kanda, Bone Marrow Transplant, 2013, 48, 452–458, DOI:10.1038/bmt.2012.244.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00183d |
| This journal is © The Royal Society of Chemistry 2024 |