Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multivalent dendritic DNA aptamer molecules for the enhancement of therapeutic effects†
Yusuke
Kawamoto
 *a,
You
Wu
a,
Soyoung
Park
*a,
You
Wu
a,
Soyoung
Park
 b,
Kumi
Hidaka
c,
Hiroshi
Sugiyama
b,
Kumi
Hidaka
c,
Hiroshi
Sugiyama
 cd,
Yuki
Takahashi
cd,
Yuki
Takahashi
 a and
Yoshinobu
Takakura
*a
a and
Yoshinobu
Takakura
*a
aDepartment of Biopharmaceutics and Drug Metabolism, Graduate School of Pharmaceutical Sciences, Kyoto University, 46-29 Yoshida-shimoadachicho, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: y.kawamoto@pharm.kyoto-u.ac.jp; takakura@pharm.kyoto-u.ac.jp
bImmunology Frontier Research Center, Osaka University, Yamadaoka, Suita, Osaka 565-0871, Japan
cDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa-oiwakecho, Sakyo-ku, Kyoto 606-8502, Japan
dInstitute for Integrated Cell-Material Sciences (iCeMS), Kyoto University, Yoshida-ushinomiyacho, Sakyo-ku, Kyoto 606-8501, Japan
First published on 13th May 2024
Abstract
Dendritic DNA molecules, referred to as DNA dendrons, consist of multiple covalently linked strands and are expected to improve the cellular uptake and potency of therapeutic oligonucleotides because of their multivalency. In this study, we developed an efficient synthetic method for producing DNA dendrons using strain-promoted azide–alkyne cycloaddition. Integration of the antitumor aptamer AS1411 into DNA dendrons enhanced cellular uptake and antiproliferative activity in cancer cells. These findings demonstrate that the incorporation of multivalent aptamers into DNA dendrons can effectively boost their therapeutic effects.
Synthetic oligonucleotides have emerged as a therapeutic modality for regulating disease-related genes and proteins.1 In addition to antisense oligonucleotides and small interfering RNAs that interact with nucleic acids based on sequence-specific base pairing, aptamers are therapeutic oligonucleotides that fold into three-dimensional (3D) structures and bind to target biomolecules with high specificity and affinity by recognizing their 3D structure, which is similar to that of an antibody.2 Extensive research has been conducted to explore the therapeutic potential of aptamers. As of December 2023, the Food and Drug Administration has clinically approved two aptamer medicines that can bind and inhibit target proteins.3,4 Aptamers have also been explored as targeting agents for drug delivery.5 To improve their functionality, a multivalent aptamer system has been developed to mimic the multivalency of natural ligand–receptor interactions.6 For example, polymeric scaffolds and nanoparticles have been utilized to fabricate multivalent aptamers;7,8 however, achieving precise control over aptamer valency in these constructs remains challenging. DNA nanostructures composed of self-assembly of complementary base pairing enable precise control of the size of DNA scaffolds and the number of incorporated aptamers;9 however, nanostructures dependent on inherent base pairing and noncovalent interactions are susceptible to degradation in biological environments,10 thereby limiting their clinical applicability.
The development of organic chemistry-based technology has enabled the synthesis of well-defined molecules that can be linked to a precise number of oligonucleotide ligands via covalent bonds. We focused on covalently branched DNA molecules, termed DNA dendrons, which represent a novel structure of therapeutic oligonucleotides.11,12 DNA dendrons are composed of multiple clustered DNA branches linked via covalent bonds. Their high-density structure not only resembles that of multiple oligonucleotide-conjugated nanoparticles, but also allows for controllable valency through molecular structure tuning. Multivalent DNA dendrons have demonstrated improved uptake by immune cells and increased immunostimulatory activity of conjugated agonists compared with linear DNA.12–14 This dendritic structure applies to oligonucleotide-based therapeutics based on their multivalency and is a promising alternative for improving the potency of aptamers. However, studies on the valency of aptamer moieties at the single-molecular level using DNA dendrons and similar clustered molecules have been limited,15–17 probably because of the lack of an efficient synthetic method.
In this study, we developed a facile synthetic method for DNA dendrons to improve their synthetic yield by employing a strain-promoted azide–alkyne cycloaddition for coupling two DNA fragments: the stem and branch regions (Fig. 1A and Fig. S1A, ESI†).18 As a model for clustered functional oligonucleotide ligands, we opted for the antiproliferative aptamer AS1411, which specifically targets nucleolin, which is highly expressed on the tumor cell membrane. This aptamer has been investigated for its potential in antitumor medicine and tumor-targeting drug delivery systems.19–22 Using this new synthetic approach, three-, six-, and nine-branched aptamers were successfully synthesized. We characterized the properties of the G-quadruplex-forming aptamer within the DNA dendrons and evaluated the effect of different valences at the molecular level on the biological activity of the aptamers. We demonstrated that DNA dendrons, which make oligonucleotides multivalent at the single molecular level, can improve the therapeutic effects of aptamers and the improvement is dependent on valency.
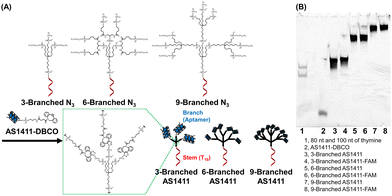 | ||
| Fig. 1 (A) Structures and synthesis of DNA dendrons composed of the multivalent aptamer via SPAAC. (B) Denaturing polyacrylamide gel electrophoresis analysis of the synthesized DNA dendrons. | ||
Whole structures of DNA dendrons have been prepared by solid-phase synthesis from the 3′- to 5′-end using branching phosphoramidites;11 however, this method suffers from low isolated yield.12 To address this issue, Mirkin et al. optimized the reaction conditions for solid-phase synthesis.13 However, we attributed the low yield to the low coupling efficiency of solid-phase synthesis after the branching region. Thus, we adopted a solution-based approach for coupling two fragments: the stem and branch regions (Fig. 1A), employing strain-promoted azide–alkyne cycloaddition (SPAAC).18 The stem region composed of a decamer of thymine (T10), branching moieties, and three, six, or nine azide groups at the 5′-end (referred to as 3-Branched N3, 6-Branched N3 or 9-Branched N3, respectively, Table S1, ESI†) was synthesized by solid-phase synthesis using trebler phosphoramidite 4 (Scheme S1, ESI†), doubler phosphoramidite 5 (Fig. S2, ESI†), and 6-bromohexyl phosphoramidite 6 (Scheme S2, ESI†). This was followed by the substitution of bromide groups at the 5′-end with azido groups and cleavage from the solid support.23,24 3′-Amine modified AS1411 DNA was synthesized through solid-phase synthesis, which was then conjugated with dibenzocyclooctyne (DBCO) via amide bond formation in solution to obtain AS1411-DBCO. Subsequently, the stem region modified with azide groups at the 5′-end was coupled with branched DNA modified with DBCO via SPAAC, resulting in trivalent 3-Branched AS1411, hexavalent 6-Branched AS1411, nonavalent 9-Branched AS1411, and their FAM-labeled counterparts (3-Branched AS1411-FAM, 6-Branched AS1411-FAM, and 9-Branched AS1411-FAM, Table S1, ESI†). The overall DNA dendron yields from the solid support of the stem regions were 19%, 9.7%, and 7.5% for 3-Branched AS1411, 6-Branched AS1411, and 9-Branched AS1411, respectively. These yields represented an improvement compared with our previously reported solid-phase synthesis of whole dendron molecules, ∼3.5-fold for 3-branch and ∼17-fold for 9-branch.12 The obtained DNA dendrons were characterized by denaturation PAGE (Fig. 1B and Fig. S1B, ESI†) and AFM imaging (Fig. S3, ESI†), and 3- and 6-Branched DNA dendrons were analyzed using MALDI-TOF MS (Table S3, ESI†). We observed the results corresponding to the calculated values, showing the successful synthesis of 3- and 6-Branched DNA dendrons composed of three and six branch moieties, respectively. Denaturing PAGE analysis of the reaction mixture of 9-Branched N3 and AS1411-DBCO suggested that 9-Branched AS1411 is composed of nine AS1411 branches (Fig. S4, ESI†).
The guanine-rich aptamer AS1411 can form four-stranded secondary G-quadruplex structures through the stacking of successive G-quartet planes composed of Hoogsteen base pairing, in which cations are coordinated.21 The structural features of the G-quadruplex are considered crucial for the antiproliferative activity of AS1411.25 We investigated the characteristics of the G-quadruplex of DNA dendrons composed of multiple AS1411 aptamers. In the presence of potassium ions, the circular dichroism (CD) spectra of 3-Branched AS1411, 6-Branched AS1411, and 9-Branched AS1411 exhibited a profile similar to that of AS1411, showing positive peaks at ∼260 and ∼210 nm, along with a negative band at ∼240 nm (Fig. 2A). This result implies that, similar to their intact structure, the aptamers within DNA dendrons could form a G-quadruplex structure, demonstrating a typical CD spectrum for the parallel motif.26 Next, we evaluated the thermal stability of the G-quadruplex within the DNA dendrons to examine the biophysical properties of the G-quadruplex of AS1411 aptamer moieties that could be utilized to estimate their biological activity. Thermal denaturation analysis of the G-quadruplex27 demonstrated sigmoidal melting curves for all aptamers (Fig. S5, ESI†). Meanwhile, clear sigmoid curves were not observed for stem DNAs (Fig. S6, ESI†), suggesting that they do not form a G-quadruplex. The melting temperatures of dendritic aptamers closely matched those of the intact aptamer (66.8 °C, 67.4 °C, 65.6 °C and 66.3 °C for AS1411, 3-Branched AS1411, 6-Branched AS1411 and 9-Branched AS1411, respectively) (Fig. 2B). This indicated that the clustered dendritic structure did not affect the thermal stability of the incorporated G-quadruplex. These results suggest that the properties of the G-quadruplex can be retained within the clustered structure of the DNA dendrons, allowing us to estimate that AS1411 aptamer moieties within DNA dendrons could also retain their biological activity.
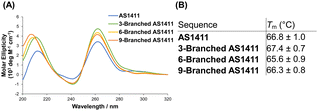 | ||
| Fig. 2 Characterization of the G-quadruplex of dendritic aptamers. (A) CD spectra, and (B) melting temperature (Tm). | ||
Nuclease resistance of oligonucleotides is an important factor affecting their therapeutic potency, and the G-quadruplex can contribute to the stability of oligonucleotides.28 To investigate the nuclease resistance of the G-quadruplex-forming aptamers within DNA dendrons, we treated linear AS1411 and dendritic 3-Branched AS1411, 6-Branched AS1411, and 9-Branched AS1411 with 50% FBS at 37 °C, followed by evaluating the degradation of DNA using PAGE. DNA degradation proceeded over time (Fig. S7, ESI†); however, bands of full-length strands were observed even after 24 h of treatment for all aptamers. The degradation rates of all dendritic aptamers were comparable to those of linear AS1411, indicating that incorporation of the G-quadruplex within DNA dendrons did not significantly affect nuclease resistance.
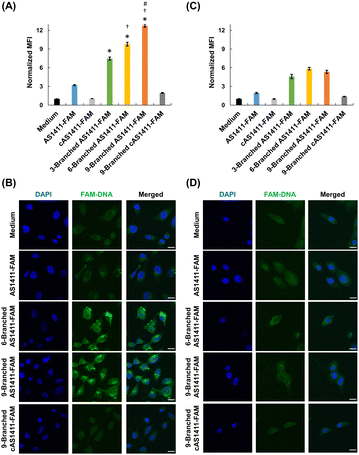
We investigated the effect of the multivalency of dendritic aptamers on cellular uptake and cancer-targeting capabilities. We selected HeLa cells that exhibited high expression levels of nucleolin and normal MCF-10A cells expressing low levels of nucleolin.8 We used flow cytometry and confocal microscopy to evaluate the cellular uptake efficiency of the linear and dendritic aptamers whose 3′-ends were modified with FAM. Linear cAS1411-FAM and dendritic 9-Branched cAS1411-FAM, including the nonactive sequence,22 were used as negative controls (Table S1, ESI†). AS1411-FAM and the three dendritic aptamers exhibited comparable fluorescence intensity at the same concentration (Fig. S8, ESI†). The three dendritic aptamers exhibited higher relative mean fluorescence intensity (MFI) values than the linear aptamer for HeLa cells (Fig. 3A). This result may be attributed to the enhancement of the cellular uptake by increase of an interaction between the aptamer moiety and nucleolin due to multivalency. In addition, as the number of aptamer moieties within the DNA dendrons increased, the relative MFI values increased. Furthermore, stronger fluorescent signals of dendritic aptamers 6-Branched AS1411-FAM and 9-Branched AS1411-FAM were observed in HeLa cells compared with cells treated with AS1411-FAM and 3-Branched AS1411-FAM (Fig. 3B and Fig. S9, ESI†). This result may be attributed to the enhanced cellular interaction of DNA molecules facilitated by the multivalence of aptamers within DNA dendrons. In contrast, the uptake efficiency of the two negative control DNAs was lower than that of all aptamers, indicating the potential contribution of aptamer sequences to uptake induction. Notably, when MCF-10A cells were treated, the relative MFI values were lower than those of HeLa cells (Fig. 3C), and the fluorescent signals of dendritic aptamers in MCF-10A cells were considerably weaker (Fig. 3D and Fig. S10, ESI†). This suggests that the aptamer within DNA dendrons retains its targeting ability, specifically toward HeLa cells. The results of competition assays suggested that the uptake of AS1411-FAM and 9-Branched AS1411-FAM to HeLa cells was mediated by nucleolin (Fig. S11, ESI†). The endocytosis pathway study suggested that the dendritic aptamer was mainly internalized through actin-dependent endocytosis including macropinocytosis (Fig. S12, ESI†). Macropinocytosis is known to be the main endocytosis pathway of AS1411.29 Lysosomal colocalization images implied that 9-Branched AS1411-FAM enters cells via endocytosis and then it is transported to the lysosome (Fig. S13, ESI†).
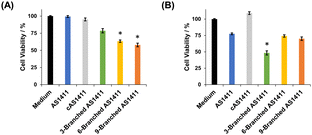
We examined the effect of the multivalency of aptamer AS1411 incorporated in DNA dendrons on its antiproliferative activity. We treated HeLa and MCF-10A cells with aptamers for 72 h and then evaluated cell viability. First, the concentration of DNA dendron molecules was kept constant. When treating HeLa cells with 3 μM of DNA dendrons, dendritic aptamers exhibited significantly higher potency than AS1411 (Fig. 4A). The trend in changes in antiproliferative effect based on the number of aptamer moieties correlated well with the observed cellular uptake (Fig. 3A). This result can be attributed to the induction of an interaction between DNA dendrons and nucleolin with an increase in valence. Stem DNAs did not exhibit antiproliferative effects on HeLa cells (Fig. S14, ESI†), demonstrating that these stem DNAs do not interfere with the evaluation of cell viability. In contrast, MCF-10A cells showed a lower sensitivity to the dendritic aptamers than HeLa cells and stem DNAs did not influence the proliferation of MCF-10A cells (Fig. S15, ESI†). To further investigate the antiproliferative activity, HeLa cells were treated with a constant aptamer concentration, ensuring consistency in the amount of aptamer delivered to cells across the examined compounds. Notably, the trend in antiproliferative effects was observed to be different when the amount of aptamer delivered to cells was constant from that when the dendron concentration was kept constant. At an aptamer concentration of 18 μM, 3-Branched AS1411 exhibited stronger antiproliferative activity in HeLa cells than AS1411, whereas the antiproliferative activity of 6-Branched AS1411 and 9-Branched AS1411 was comparable to that of AS1411 (Fig. 4B). In other words, at the same aptamer concentration, an enhancement in antiproliferative activity was observed with an increase in valency from one to three, indicating that the 3-Branched AS1411 structure has beneficial interactions with nucleolin compared with the equivalent amount of the monovalent aptamer. However, when the valency increased from three to six and nine, the increase in the aptamer's valency on DNA dendrons did not induce a significant increase in antiproliferative activity. We consider that this result could be due to the following reason: 3-Branched AS1411 could interact with multiple nucleolin molecules leading to a stronger antiproliferative effect than the same amount of the monovalent aptamer,30 however, this interaction could be hindered by the steric hindrance resulting from a higher density of aptamer moieties, as previously reported in DNA dendrons incorporating TLR9 ligands.14 Further studies are underway to improve the antiproliferative activity considering the steric hindrance derived from the higher density of aptamer moieties in 6-Branched AS1411 and 9-Branched AS1411.
In conclusion, we developed a useful synthetic method for DNA dendrons comprising multiple aptamer moieties, in which ligation of a stem region and multiple branch strands via SPAAC was employed. This methodology facilitates the efficient synthesis of aptamers with multivalency at the molecular level. The multivalent anti-nucleolin aptamers within DNA dendrons formed G-quadruplexes, with properties comparable to those of the intact aptamer. However, the multivalent aptamers demonstrated enhanced cellular uptake and antiproliferative activity compared with intact aptamers. The cellular uptake and antiproliferative potency of the aptamer within DNA dendrons were dependent on valency. We contend that our study provides valuable insights into the structural design of therapeutic aptamers and aptamer-based drug delivery systems.
We would like to thank Dr S. Hirashima and T. Kumagai (Kyoto University) for their technical assistance of solid-phase synthesis and helpful discussion. This work was supported by Japan Society for the Promotion of Science (21H02616, 20H04533, 21H04705, 23H04076, 21K15225 and 23H04072), National Key R&D Program of China (2022YFE0111600), Takeda Science Foundation and China Scholarship Council.
Conflicts of interest
There are no conflicts to declare.Notes and references
- Y. Kawamoto, Y. Wu, Y. Takahashi and Y. Takakura, Adv. Drug Delivery Rev., 2023, 199, 114872 CrossRef CAS PubMed.
- H. G. Sun, X. Zhu, P. Y. Lu, R. R. Rosato, W. Tan and Y. L. Zu, Mol. Ther.--Nucleic Acids, 2014, 3, e182 CrossRef CAS PubMed.
- E. W. M. Ng, D. T. Shima, P. Calias, E. T. Cunningham, D. R. Guyer and A. P. Adamis, Nat. Rev. Drug Discovery, 2006, 5, 123–132 CrossRef CAS PubMed.
- A. Mullard, Nat. Rev. Drug Discovery, 2023, 22, 774 Search PubMed.
- S. Kruspe, F. Mittelberger, K. Szameit and U. Hahn, ChemMedChem, 2014, 9, 1998–2011 CrossRef CAS PubMed.
- M. H. Lin, J. Zhang, H. Wan, C. Y. Yan and F. Xia, ACS Appl. Mater. Interfaces, 2021, 13, 9369–9389 CrossRef CAS PubMed.
- L. Yang, L. Meng, X. B. Zhang, Y. Chen, G. Z. Zhu, H. P. Liu, X. L. Xiong, K. Sefah and W. Tan, J. Am. Chem. Soc., 2011, 133, 13380–13386 CrossRef CAS PubMed.
- D. H. M. Dam, K. S. B. Culver and T. W. Odom, Mol. Pharmacol., 2014, 11, 580–587 CrossRef CAS PubMed.
- P. Charoenphol and H. Bermudez, Mol. Pharmacol., 2014, 11, 1721–1725 CrossRef CAS PubMed.
- H. Bila, E. E. Kurisinkal and M. M. C. Bastings, Biomater. Sci., 2019, 7, 532–541 RSC.
- M. S. Shchepinov, I. A. Udalova, A. J. Bridgman and E. M. Southern, Nucleic Acids Res., 1997, 25, 4447–4454 CrossRef CAS PubMed.
- Y. Kawamoto, W. Liu, J. H. Yum, S. Park, H. Sugiyama, Y. Takahashi and Y. Takakura, ChemBioChem, 2022, 23, e202100583 CrossRef CAS PubMed.
- M. E. Distler, M. H. Teplensky, K. E. Bujold, C. D. Kusmierz, M. Evangelopoulos and C. A. Mirkin, J. Am. Chem. Soc., 2021, 143, 13513–13518 CrossRef CAS PubMed.
- M. E. Distler, J. P. Cavaliere, M. H. Teplensky, M. Evangelopoulos and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2215091120 CrossRef CAS PubMed.
- Y. Kim, D. M. Dennis, T. Morey, L. Yang and W. H. Tan, Chem. – Asian J., 2010, 5, 56–59 CrossRef CAS PubMed.
- R. Z. Li, X. H. Wu, J. Li, X. H. Lu, R. C. Zhao, J. B. Liu and B. Q. Ding, Nanoscale, 2022, 14, 9369–9378 RSC.
- J. X. Li, Z. J. Zhang, J. Gu, R. Amini, A. G. Mansfield, J. R. Xia, D. White, H. D. Stacey, J. C. Ang, G. Panesar, A. Capretta, C. D. M. Filipe, K. Mossman, B. J. Salena, J. B. Gubbay, C. Balion, L. Soleymani, M. S. Miller, D. Yamamura, J. D. Brennan and Y. F. Li, J. Am. Chem. Soc., 2022, 144, 23465–23473 CrossRef CAS PubMed.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046–15047 CrossRef CAS PubMed.
- P. J. Bates, E. M. Reyes-Reyes, M. T. Malik, E. M. Murphy, M. G. O'Toole and J. O. Trent, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 1414–1428 CrossRef CAS PubMed.
- X. Tong, L. Ga, J. Ai and Y. Wang, J. Nanobiotechnol., 2022, 20, 57 CrossRef CAS PubMed.
- P. J. Bates, J. B. Kahlon, S. D. Thomas, J. O. Trent and D. M. Miller, J. Biol. Chem., 1999, 274, 26369–26377 CrossRef CAS PubMed.
- S. Soundararajan, W. W. Chen, E. K. Spicer, N. Courtenay-Luck and D. J. Fernandes, Cancer Res., 2008, 68, 2358–2365 CrossRef CAS PubMed.
- A. D. Shaller, W. Wan, B. M. Zhao and A. D. Q. Li, Chem. - Eur. J., 2014, 20, 12165–12171 CrossRef CAS PubMed.
- J. Lietard, A. Meyer, J. J. Vasseur and F. Morvan, Tetrahedron Lett., 2007, 48, 8795–8798 CrossRef CAS.
- V. Dapic, V. Abdomerovic, R. Marrington, J. Peberdy, A. Rodger, J. O. Trent and P. J. Bates, Nucleic Acids Res., 2003, 31, 2097–2107 CrossRef CAS PubMed.
- J. Kypr, I. Kejnovská, D. Renciuk and M. Vorlícková, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS PubMed.
- J. L. Mergny, A. T. Phan and L. Lacroix, FEBS Lett., 1998, 435, 74–78 CrossRef CAS PubMed.
- V. Dapic, P. J. Bates, J. O. Trent, A. Rodger, S. D. Thomas and D. M. Miller, Biochemistry, 2002, 41, 3676–3685 CrossRef CAS PubMed.
- E. M. Reyes-Reyes, Y. Teng and P. J. Bates, Cancer Res., 2010, 70, 8617–8629 CrossRef CAS PubMed.
- F. Cheng, Y. J. Jiang, B. Kong, H. R. Lin, X. J. Shuai, P. P. Hu, P. F. Gao, L. Zhan, C. Z. Huang and C. M. Li, Adv. Healthcare Mater., 2023, 12, 2300102 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc00578c |
| This journal is © The Royal Society of Chemistry 2024 |