Open Access Article
Open Access ArticleTransglutaminase-mediated proximity labeling of a specific Lys residue in a native IgG antibody†
Riko
Nishioka
 a,
Ryuya
Iida
a,
Kosuke
Minamihata
a,
Ryo
Sato
a,
Michio
Kimura
a and
Noriho
Kamiya
a,
Ryuya
Iida
a,
Kosuke
Minamihata
a,
Ryo
Sato
a,
Michio
Kimura
a and
Noriho
Kamiya
 *ab
*ab
aDepartment of Applied Chemistry, Graduate School of Engineering, Kyushu University, 744 Motooka, Fukuoka 819-0395, Japan. E-mail: kamiya.noriho.367@m.kyushu-u.ac.jp
bDivision of Biotechnology, Center for Future Chemistry, Kyushu University, 744 Motooka, Fukuoka 819-0395, Japan
First published on 17th July 2024
Abstract
The fusion protein of an engineered zymogen of microbial transglutaminase (EzMTG) with a protein G variant, EzMTG-pG, enabled the proximity-based, tag-free labeling of Lys65 in the heavy chain of a native IgG antibody (trastuzumab) with a Gln-donor peptidyl substrate functionalized with a fluorescent molecule.
Site-specific conjugation of a chemical payload to a protein-based therapeutic has attracted attention to achieve the desired properties for optimal function.1,2 In particular, antibody–drug conjugates (ADCs) are an important class of cancer drugs that combine an antibody with a small molecule drug via a linker, enabling both target specificity and pharmacological efficacy.3,4 Conventionally, chemical methods targeting Lys and Cys residues on the antibody surface have been employed in ADC synthesis. However, the presence of numerous Lys and Cys residues on the antibody surface complicates the control of the drug-to-antibody ratio (DAR) and the binding positions. For effectiveness and safety, achieving more homogeneous modification is desirable, which requires site-specific antibody modification.5
It has been previously suggested that enhancing the reactivity and specificity of chemical modification processes can be achieved by targeting relatively less reactive amino acid residues on the surface of antibodies using proximity-based strategies.6 Proximity-based strategies are used not only in chemical methods but also in enzymatic methods to enhance the specificity. Recently, the fusion of an antibody-binding peptide7,8 or protein9 has been used to facilitate the proximity labeling of native antibodies with target molecules. Yu et al. have successfully achieved the site-specific modification of human IgG antibodies using mutant variants that could fuse antibody-binding proteins, such as protein Z (pZ) and protein G (pG), with the enzyme sortase A. In this approach, the residues of both heavy chains (K5, K123, K135, K292, and K441) and light chains (K207) were modified, resulting in a reported DAR of 2-3. Hence, enzymatic bioconjugation that exhibits high specificity and react under mild conditions are attracting attention.2
One of the main enzymes used in the production of ADCs is microbial transglutaminase (MTG). Unlike mammalian-derived transglutaminase, MTG exhibits stable catalytic activity across a broad range of pH and temperature conditions without dependency on cofactors, such as Ca2+.10 MTG catalyzes acyl transfer reactions between the γ-carboxamide group of Gln residues and the ε-amino group of Lys residues within proteins and primary amines.11 However, IgG antibodies possess numerous Gln and Lys residues, and these amino acids on the antibody surface exhibit low enzymatic reactivity. Therefore, the pretreatment of IgG via enzymatic deglycosylation12,13 or genetic mutagenesis14 is necessary for the site-specific modification of native antibodies. These pre-treatment procedures may potentially adversely affect the inherent functionality and the production yield of the antibodies.15 Therefore, site-specific, tag-free modification methods for native antibodies have garnered considerable attention.16
To date, enzymatic modification methods for native antibodies have included the use of MTG, which was designed to facilitate the substitution of Gly residues near the active site loop with Ser residues for modification at Q295 of IgG.17 Additionally, the modification of an exposed Tyr residue (Y57) on the surface of IgG antibodies with an N-methylated luminol derivative has been reported using horseradish peroxidase.18 A new method for introducing an octanoic acid derivative to Lys residues on IgG antibodies has been developed using lipoyl ligase A, and the successful formulation of an ADC with the anti-HER2 antibody trastuzumab has also been recently demonstrated.19
Herein, we investigated the site-specific modification of a native IgG antibody by integrating the proximity effect with MTG-catalyzed crosslinking reactions. The molecular design used a recently developed engineered zymogen of MTG (EzMTG), which exhibits crosslinking activity even in the zymogen state.20 EzMTG is expressed as a soluble protein in Escherichia coli and does not require an additional activation process for the proteolytic cleavage of the propeptide, which expands the scope of the molecular design of new MTG mutants. In this study, we designed a pG-fused EzMTG (EzMTG-pG) by introducing pG at the C-terminus of EzMTG, which can be easily obtained in E. coli (Fig. 1a and Table S1, ESI†).
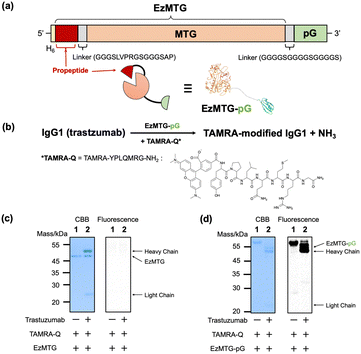 | ||
| Fig. 1 (a) Schematic illustration of DNA constructs encoding the EzMTG20 and pG. A model of the three-dimensional structure of EzMTG-pG (the PDB entry used for EzMTG is 3IU0.pdb and for pG is 1IGC.pdb). (b) Reaction scheme of EzMTG-pG-mediated antibody modification with TAMRA-Q. Evaluation of the cross-linking activity of (c) EzMTG and (d) EzMTG-pG by SDS-PAGE gel analysis and fluorescent imaging. The reaction was conducted with trastuzumab (1.3 μM), EzMTG or EzMTG-pG (2.6 μM), and TAMRA-Q (100 μM) in 40 mM Tris–HCl (pH 8.0) at 37 °C for 60 min. Raw images of electrophoretic gels (CBB staining, left) and their fluorescence images (derived from TAMRA, right) are shown in Fig. S2 (ESI†). | ||
As an initial step to demonstrate the concept, we conducted an evaluation of the crosslinking activity of EzMTG-pG using the Gln substrate peptide (TAMRA-YPLQMRG-NH2, TAMRA-Q) as a drug model against an IgG antibody (trastuzumab, Herceptin) (Fig. 1b). The labeling of the heavy chain of the antibody, which was not observed with EzMTG alone (Fig. 1c), was observed with TAMARA-Q (Fig. 1d, lane 2). This result indicated that the proximity of EzMTG to IgG enabled by the pG domain facilitated the generation of crosslinking activity. Because the labeling of EzMTG-pG itself with TAMARA-Q was also observed (Fig. 1d, lane 1), the self-labeling reaction was also promoted by the proximity-based intramolecular crosslinking reaction.
We also observed that the reaction solution became turbid when the concentrations of EzMTG-pG and IgG were increased, suggesting the formation of aggregates in the reaction mixture.
To solve these problems, we selected pG variants21,22 that specifically bind to the Fc or Fab region to yield EzMTG-pG(Fc) and EzMTG-pG(Fab) (Fig. 2a and Fig. S1, Table S1, ESI†). To circumvent the self-labeling of EzMTG-pG, the Lys and Gln residues in pG were replaced with Arg and Asn residues, respectively. To create Fc-specific pG that diminish the self-labeling reaction, five Lys residues (K3, K9, K12, K18, and K49) that did not appear to contribute to the interaction with the Fc domain were substituted with Arg residues (Fig. 2a(i)). For Fab-specific pG all the Lys and Gln residues located on the surface (K3, K9, K12, K18, K41, and Q31) were replaced with Arg and Asn residues, respectively (Fig. 2a(ii)). Both EzMTG-pG(Fc) and EzMTG-pG(Fab) expressed successfully in E. coli and catalyzed the labeling of the heavy chain of the IgG antibody. For EzMTG-pG(Fc), in which unmodified Lys residues (K27, K30) were present, the self-labeling with TAMRA-Q was still observed (Fig. 2b, lane 1). In contrast, EzMTG-pG(Fab), in which all the Lys residues were substituted, showed little self-labeling (Fig. 2c, lane 1). These results indicated that a Lys residue in the original pG domain caused the self-labeling.
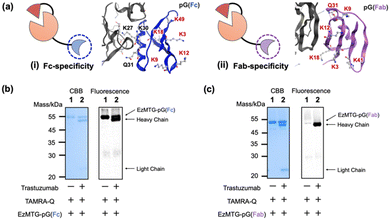 | ||
| Fig. 2 (a) Schematic illustration of (i) EzMTG-pG(Fc), in which a Fc-specific pG with five Lys residue substitutions is fused to EzMTG, and model of the three-dimensional structure of antibody (Fc) and Fc-specific pG. (ii) EzMTG-pG(Fab), in which a Fab-specific pG with all the Lys and Gln residues substituted to Arg and Asn, respectively, is fused to EzMTG, and model of the three-dimensional structure of antibody (Fab) and Fab-specific pG. Unsubstituted amino acids are shown in black and substituted amino acids are shown in red. The PDB entry for Fc and pG is 1FCC.pdb and the entry for Fab and pG is 1IGC.pdb. Evaluation of the cross-linking activity of (b) EzMTG-pG(Fc) and (c) EzMTG-pG(Fab) by SDS-PAGE gel analysis and fluorescent imaging. The reaction was conducted with trastuzumab (1.3 μM), each EzMTG mutant (2.6 μM) and TAMRA-Q (100 μM) in 40 mM Tris–HCl (pH 8.0) at 37 °C for 60 min. Raw images of electrophoretic gels (CBB staining, left) and their fluorescence images (derived from TAMRA, right) are shown in Fig. S2 (ESI†). | ||
The formation of aggregates was also suppressed by the substitution of pG domain (Fig. S3, ESI†), and therefore the observed aggregation was caused by the intrinsic characteristics of pG, which binds to distinct sites of IgG, Fab,23 and Fc24 regions. The fact that aggregation was sufficiently suppressed with both the newly designed EzMTG-pG(Fc) and EzMTG-pG(Fab) variants indicated the validity of increasing the specificity of pG to the Fab or Fc regions to achieve better handling of the EzMTG-pG variants.
Next, we conducted peptide mapping of the antibody modified with TAMRA-Q by EzMTG-pG(Fab) to identify the modification sites. The results revealed that Lys65 of the heavy chain was almost exclusively modified by TAMRA-Q (Fig. 3a and Fig. S4, S5, ESI†). To understand this highly selective modification of Lys65, though another lysine residue such as Lys76 is present in the vicinity (Fig. 3b), we characterized all the lysine residues in the heavy chain of the antibody using the molecular operating environment (MOE). The positive residue patch area (PRPA) of each residue determined by MOE revealed that the PRPA value of Lys65 was higher than those of other lysine residues (Table S2, ESI†). Judging from the solvent-exposure parameters such as ASA25 and Exp,26 both Lys65 and Lys76 were similarly exposed to the solvent, but their PRPA values were markedly different (Table S2, ESI†). The PRPA value represents the contribution of residues to positively charged regions on the protein surface. In the reaction mechanism of MTG, the Gln residue of a substrate is initially recognized at the hydrophobic pocket, followed by the recognition of the Lys residue formation at the negatively charged active site.11 Therefore, Lys65, surrounded by a positively charged region, was selectively modified because of favorable electrostatic interactions with the active site compared with the other Lys residues such as Lys76.
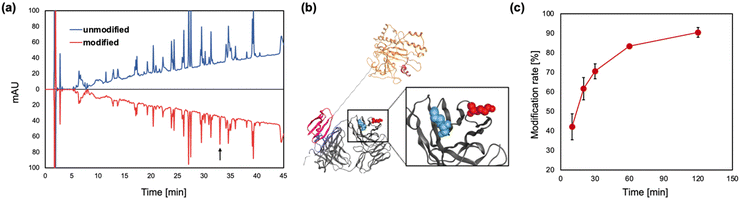 | ||
| Fig. 3 (a) Identification of Lys residues modified with TAMRA-Q by EzMTG-pG(Fab). RP-HPLC chromatographs of antibodies modified by EzMTG-pG(Fab). (b) Model of the three-dimensional structure of Fab and EzMTG-pG(Fab). Fab, MTG domain, pG(Fab) are shown in gray, orange, and purple, respectively. Lys65 and Lys76 of trastuzumab represented as the space-filling model are shown in red and blue, respectively. The PDB entries used for Fab, EzMTG, and Fab-specific pG are 1N8Z.pdb, 3IU0.pdb, and 1IGC.pdb, respectively. (c) Time course of modification of Lys65 in trastuzumab (1.3 μM) with TAMRA-Q (100 μM) by EzMTG-pG(Fab) (2.6 μM) in 40 mM Tris–HCl (pH 8.0) at 37 °C. N = 3; mean ± SD. | ||
The modification rate of IgG by EzMTG-pG(Fab) was 90.4 ± 2.5%, while EzMTG-pG(Fc) had a low modification rate of ca. 13% (Fig. 3c and Fig. S6a–d, ESI†). Using bio-layer interferometry, the binding affinity of EzMTG-pG(Fc) and EzMTG-pG(Fab) to trastuzumab was evaluated by the dissociation constant (KD). The KD values of EzMTG-pG(Fc) and EzMTG-pG(Fab) were 7.2 nM and 138 nM, respectively (Fig. S7, ESI†). Because both mutants showed sufficient affinity for the antibody under the experimental conditions and the modification site was found to be Lys65 in both cases (Fig. S4 and S6e and f, ESI†), the decrease in the modification rate was likely because of the low reactivity of EzMTG-pG(Fc) with Lys65. The structural predictions using MOE clearly showed differences in the distances between EzMTG and Lys65 when pG was bound to the Fab region compared with when bound to the Fc region. When pG was bound to Fab, MTG and Lys65 were in close proximity, leading to the high modification rate of EzMTG-pG(Fab) (Fig. S6g, ESI†). In contrast, when bound to Fc, EzMTG and Lys65 were physically separated, resulting in a considerable lowering of the reactivity, as it was only possible for these units to approach when the linker between EzMTG and pG was sufficiently extended (Fig. S6h, ESI†).
In addition, we evaluated the effect of the EzMTG domain on the IgG labeling by cleaving the propeptide by thrombin (Fig. 1a) to yield ΔproMTG-pG(Fab). The results showed that the initial reaction rate of ΔproMTG-pG(Fab) for labeling Lys65 was faster than that of EzMTG-pG(Fab), whereas the degree of final modification was saturated at 84.1% for ΔproMTG-pG(Fab) (Fig. S8, ESI†). The higher product yield can be ascribed by the suppression of competing hydrolysis of Gln of TAMRA-Q to Glu, implying the unique substrate-dependent reactivity of EzMTG.20
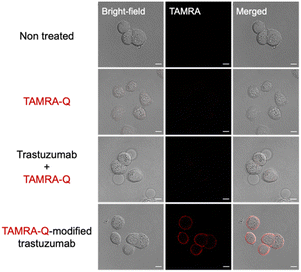
The Lys65 residue of trastuzumab is located in the framework region (FR3) but close to the edge of the complementarity-determining region (CDR) of the antibody (Fig. S9, ESI†). The CDR is the region directly involved in binding with antigens, and thus the CDR potentially affects the antigen binding affinity. Therefore, we evaluated the antigen binding capability of the K65-modified antibody (TAMRA-Q-modified trastuzumab) by examining the binding to HER2, the target macromolecular antigen of trastuzumab. TAMRA-Q-modified trastuzumab was purified by separating EzMTG-pG(Fab) from the cross-linked products. The dissociation of EzMTG-pG(Fab) was achieved by cation exchange chromatography under acidic conditions at pH 3.0, yielding TAMRA-Q-modified trastuzumab with a purity of approximately 57% (see ESI† for details, Fig. S10). The binding affinity of purified TAMRA-Q-modified trastuzumab to HER2 was evaluated. The apparent KD value of the purified sample was determined to be 3.8 nM, indicating that TAMRA-Q-modified trastuzumab maintained a binding affinity comparable with that of the unmodified antibody (KD value of 6.3 nM) (Fig. S11, ESI†). With the HER2 binding confirmed, we proceeded to investigate the binding of purified TAMRA-Q-modified trastuzumab to the HER2-positive cell line, SK-BR-3. TAMRA-Q-modified trastuzumab was applied to SK-BR-3 cells for 1 h, and the cells were then observed using confocal laser scanning microscopy (CLSM) after the cells had been washed and fixed by paraformaldehyde. The results clearly showed fluorescence from the cell membrane region, indicating the binding of TAMRA-Q-modified trastuzumab to the SK-BR-3 cells (Fig. 4). Red fluorescence was not observed from the cell membrane region of the HER2-negative cell line, MDA-MB-231 (Fig. S12, ESI†), which indicated that the binding of TAMRA-Q-modified trastuzumab was specific to the membrane antigen.
To test our concept on other IgGs, we applied EzMTG-pG(Fab) to the modification of rituximab and ramucirumab, and found that their modification rates seemed to saturate at about 28% and 12%, respectively (Fig. S13a, b and c, ESI†). Although the microenvironment around Lys65 of rituximab is similar to that of trastuzumab, those amino acid sequences are different (Table S3 and Fig. S13d, ESI†). By contrast, different microenvironment around Lys65 should lead to the low labeling efficiency because ramucirumab has the same surrounding amino acid sequence (Table S3 and Fig. S13e and f, ESI†). These results illustrate the high substrate specificity of EzMTG-pG toward Lys65 of trastuzumab, and in turn, suggest that further molecular design is required to ensure the general utility.
In conclusion, by using the proximity effect by fusing an engineered protein G to an active form of a zymogen of MTG (EzMTG), we achieved the site-specific labeling of Lys65 in the heavy chain of a native IgG antibody (trastuzumab). Through the optimization of the pG domain, EzMTG-pG(Fab) was developed, which enabled an approximately 90% modification rate of the IgG heavy chain, corresponding to a dye-antibody ratio of 1.8. It was confirmed that the antibody retained antigen-binding affinity even after the modification. Although the applicability of the current format of EzMTG fusion protein may be limited to trastuzumab, our results will pave the way for the site-specific and tag-free labeling of native IgGs by altering the substrate preferences of EzMTG, e.g. by introducing mutations or changing the composition of the Gln substrate.20
R. N.: writing the original draft, investigation, and methodology. R. I., R. S., M. K.: investigation, and methodology.; K. M.: conceptualization, methodology, validation, editing.; N. K.: conceptualization, methodology, supervision, validation, writing/reviewing, and editing. All authors contributed to the discussion of the paper and approved the manuscript.
This study was supported by AMED under Grant Number JP21ae0121004 and JSPS KAKENHI Grant number JP23H00247 (to N. K.). We thank Victoria Muir, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- S. B. Ebrahimi and D. Samanta, Nat. Commun., 2023, 14, 2411 CrossRef CAS PubMed.
- A. Debon, E. Siirola and R. Snajdrova, JACS Au, 2023, 3, 1267–1283 CrossRef CAS PubMed.
- A. Beck, L. Goetsch, C. Dumontet and N. Corvaïa, Nat. Rev. Drug Discovery, 2017, 16, 315–337 CrossRef CAS PubMed.
- J. Z. Drago, S. Modi and S. Chandarlapaty, Nat. Rev. Clin. Oncol., 2021, 18, 327–344 CrossRef.
- S. J. Walsh, J. D. Bargh, F. M. Dannheim, A. R. Hanby, H. Seki, A. J. Counsell, X. Ou, E. Fowler, N. Ashman, Y. Takada, A. Isidro-Llobet, J. S. Parker, J. S. Carroll and D. R. Spring, Chem. Soc. Rev., 2021, 50, 1305–1353 RSC.
- E. von Witting, S. Hober and S. Kanje, Bioconjugate Chem., 2021, 32, 1515–1524 CrossRef CAS PubMed.
- S. Kishimoto, Y. Nakashimada, R. Yokota, T. Hatanaka, M. Adachi and Y. Ito, Bioconjugate Chem., 2019, 30, 698–702 CrossRef CAS PubMed.
- K. Yamada, N. Shikida, K. Shimbo, Y. Ito, Z. Khedri, Y. Matsuda and B. A. Mendelsohn, Angew. Chem., Int. Ed., 2019, 58, 5592–5597 CrossRef CAS PubMed.
- W. Yu, K. P. Gillespie, B. Chhay, A. Svensson, P. Nygren, I. A. Blair, F. Yu and A. Tsourkas, Bioconjugate Chem., 2021, 32, 1058–1066 CrossRef CAS.
- H. Ando, M. Adachi, K. Umeda, A. Matsuura, M. Nonaka, R. Uchio, H. Tanaka and M. Motoki, J. Agric. Food Chem., 1989, 53, 2613–2617 CAS.
- T. Kashiwagi, K. Yokoyama, K. Ishikawa, K. Ono, D. Ejima, H. Matsui and E. Suzuki, J. Biol. Chem., 2002, 277, 44252–44260 CrossRef CAS.
- P. Dennler, A. Chiotellis, E. Fischer, D. Bregeon, C. Belmant, L. Gauthier, F. Lhospice, F. Romagne and R. Schibli, Bioconjugate Chem., 2014, 25, 569–578 CrossRef CAS.
- J. J. Bruins, J. A. M. Damen, M. A. Wijdeven, L. P. W. M. Lelieveldt, F. L. van Delft and B. Albada, Bioconjugate Chem., 2021, 32, 2167–2172 CrossRef CAS PubMed.
- A. Hadjabdelhafid-Parisien, S. Bitsch, A. M. Palacios, L. Deweid, H. Kolmar and J. N. Pelletier, RSC Adv., 2022, 12, 33510–33515 RSC.
- S. Boune, P. Hu, A. L. Epstein and L. A. Khawli, Antibodies, 2020, 9, 22 CrossRef CAS.
- K. Wu, C. Yu, C. Lee, C. Zuo, Z. T. Ball and H. Xiao, Bioconjugate Chem., 2021, 32, 1947–1959 CrossRef CAS PubMed.
- S. Dickgiesser, M. Rieker, D. Mueller-Pompalla, C. Schröter, J. Tonillo, S. Warszawski, S. Raab-Westphal, S. Kühn, T. Knehans, D. Könning, J. Dotterweich, U. A. K. Betz, J. Anderl, S. Hecht and N. Rasche, Bioconjugate Chem., 2020, 31, 1070–1076 CrossRef CAS PubMed.
- S. Sato, M. Matsumura, T. Kadonosono, S. Abe, T. Ueno, H. Ueda and H. Nakamura, Bioconjugate Chem., 2020, 31, 1417–1424 CrossRef CAS PubMed.
- S. Yamazaki, K. Ito, T. Aoki, N. Arashida, T. Watanabe, T. Fujii and Y. Matsuda, Bioconjugate Chem., 2024, 63, 644–650 CAS.
- R. Ariyoshi, T. Matsuzaki, R. Sato, K. Minamihata, K. Hayashi, T. Koga, K. Orita, R. Nishioka, R. Wakabayashi, M. Goto and N. Kamiya, Bioconjugate Chem., 2024, 35, 340–350 CrossRef CAS PubMed.
- Y. Jung, J. M. Lee, J. Kim, J. Yoon, H. Cho and B. H. Chung, Anal. Chem., 2009, 81, 936–942 CrossRef CAS PubMed.
- F. Unverdorben, M. Hutt, O. Seifert and R. E. Kontermann, PLoS One, 2015, 10, e0139838 CrossRef PubMed.
- J. Derrick and D. Wigley, Nature, 1992, 359, 752–754 CrossRef CAS.
- A. E. S. Eriksson, G. J. Kleywegt, M. Uhlén and T. A. Jones, Structure, 1995, 3, 265–278 CrossRef.
- B. Lee and F. M. Richards, J. Mol. Biol., 1971, 55, 379–400 CrossRef CAS PubMed.
- S. Miller, J. Janin, A. M. Lesk and C. Chothia, J. Mol. Biol., 1987, 196, 641–656 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, amino acid sequence of recombinant proteins, raw images of electrophoretic gels, evaluation of aggregation formation, identification of the modified peptide fragment, evaluation of the antigen-binding ability of the modified antibody. See DOI: https://doi.org/10.1039/d4cc01728e |
| This journal is © The Royal Society of Chemistry 2024 |