Open Access Article
Open Access ArticleARCaDia: single-round screening of a DNA-type targeted covalent binder possessing a latent warhead†
Masumi
Taki
 *ab,
Masayasu
Kuwahara
*ab,
Masayasu
Kuwahara
 c,
Chaohui
Li
a,
Naoko
Tomoda
d,
Naoyuki
Miyashita
af,
Tetsuo
Kan
a and
Jay
Yang
*ade
c,
Chaohui
Li
a,
Naoko
Tomoda
d,
Naoyuki
Miyashita
af,
Tetsuo
Kan
a and
Jay
Yang
*ade
aDepartment of Engineering Science, The Graduate School of Informatics and Engineering, The University of Electro-Communications (UEC), 1-5-1 Chofugaoka, Chofu, Tokyo 182-8585, Japan. E-mail: taki@pc.uec.ac.jp; Tel: +81-42-443-5980
bInstitute for Advanced Science, UEC, Chofu 182-8585, Japan
cGraduate School of Integrated Basic Sciences, Nihon University, Setagaya-ku, Tokyo 156-8550, Japan
dDepartment of GI Surgery II, Hokkaido University Graduate School of Medicine, Sapporo, Hokkaido, Japan
eSchool of Medicine and Public Health, University of Wisconsin, Madison, Wisconsin 53706, USA. E-mail: jyang75@wisc.edu
fDepartment of Biological Systems Engineering, Graduate School of Biology-Oriented Science and Technology, KINDAI University, 930 Nishimitani, Kinokawa, Wakayama 649-6493, Japan
First published on 8th November 2024
Abstract
A covalent binder for a target protein was obtained by a direct single-round screening of a latent-warhead-modified DNA library via affinity/reactivity-based co-selection of aptameric deoxyribonucleic acid (ARCaDia), followed by a top k-mer analysis. The optimal position of the conjugated warhead on the selected aptamer was simultaneously identified.
Covalent drugs, also known as targeted covalent inhibitors (TCIs), form permanent bonds to target proteins and show a prolonged duration of drug action.1,2 The half-life extension should result in less frequent drug dosing at lower amounts and improve the quality-of-life of patients.3 The major TCI developments have focused on small molecules,4 however, the interaction between a small molecule and a target protein is strongly influenced by hydrophobic interactions,5 which often induce irreversible side-effects from off-target covalent binding.3 To avoid this potential risk, the TCI modality in fundamental research is broadening from the conventional small molecules to larger biomolecules possessing more stringent target specificity.6 Such middle-to-macro biomolecular TCIs (bioTCIs) can be sub-categorized into peptidic, proteinic, and nucleotidic (i.e., aptameric) TCIs. Among them, reversible bioTCIs, especially nucleotidic TCIs where the semi-permanent drug effect can be reversed on demand by a selective complementary-strand (CS) antidote, would mitigate the major concern of a long-lasting drug side effect.7
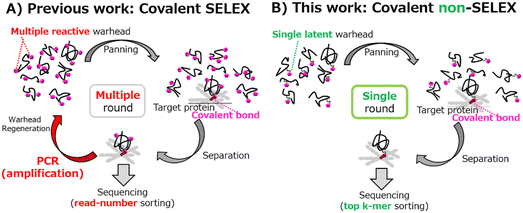
The aptameric covalent binders can be created by both indirect rational design, such as introducing a warhead8–10 into a pre-existing aptamer,11,12 and by the direct combinatorial screening method. In the latter, repetitive bio-panning of an aptamer library against the target by a process termed systematic evolution of ligands by exponential enrichment (SELEX)13 has been exclusively used. Historically, Smith and colleagues were the first to report covalently binding RNA and DNA aptamers targeting neutrophil elastase.14,15 The optimized binder was obtained by a screening denoted the blended SELEX using a splint-DNA comprising a small-molecule-TCI as the elastase-specific warhead, a spacer, and a 3′ overlap complimentary to the forward primer region of the nucleotide library. This method has a major advantage in that an unmodified conventional nucleotide library is used as the input. Other covalent SELEX methods using a generalized warhead reactable with any target protein, in theory, instead of the neutrophil elastase-specific warhead, have been reported16–18 (ESI† for details), but all require multiple cycle selection and regeneration of the multiple-warhead conjugated library every cycle (Fig. 1A).
To avoid the labor and time-consuming process of the traditional SELEX-based covalent aptamer selection, here we report a single-round selection of a covalent binder possessing a latent fosylate-type warhead19,20 and a linker between the warhead and the nucleotide base (Fig. 1B). Our library design differed from the previous attempts at combinatorial screening for covalent aptamers. In fact, our library design was previously reported as being potentially problematic for screening because of (1) linker flexibility,21 (2) low reactivity of the fosylate warhead,18 and (3) placement of only a single warhead within the aptamer.18 We chose this approach since placing a long linker between a single nucleotide base and the warhead likely positioned the covalent attachment site outside the aptamer-target docking domain, enabling on-demand reversal using the CS antidote.12 The fosylate warhead, although less reactive, shows high selectivity and reacts with the target protein in a microenvironment-dependent manner, theoretically limiting nonspecific conjugation and enabling a direct single-round screening of a latent-warhead-modified library via affinity/reactivity-based co-selection22 of aptameric deoxyribonucleic acid (ARCaDia). Using the non-SELEX method, we successfully identified a covalent-binder against thrombin through high throughput sequencing (HTS) after the selection, followed by a k-mer-based bioinformatics analysis.
We started with an N25-randomized DNA library where a single 5-octadinyl-dU is placed in the middle at nucleotide 13, with the primer sequences flanking the randomized region (Fig. 2A) to attempt the non-SELEX single round selection of aptamers covalently binding to the thrombin target protein. A relatively high-reactive warhead (benzene-sulfonyl fluoride; BSF, 1), the latent fosylate (aryl-fluorosulfate; AFS, 2), and a control non-reactive one (4) were conjugated at the central alkyne, respectively (Fig. 2A, right). After pre-selection with BSA to remove non-specific binders, the remaining library reacted with the thrombin target bound to magnetic nano-beads. The aptamer-target conjugate was subjected to a harsh wash using urea/SDS-containing buffer to remove non-covalent binders while retaining the covalently bound aptamers on the beads. That the warhead-conjugated library before conjugation with the target protein was adequately amplified by PCR was confirmed by a qPCR assay (Fig. S2, ESI†), but the aptamers after selection now covalently bound to the large target protein required digestion with proteinase K to serve as a template for PCR amplification (Fig. S3, ESI†). Comparison of the N25 libraries endowed with different warheads demonstrated a robust expected 64bp PCR product band for the AFS library (2) and a weak band for the BSF library (1) suggesting the potential presence of a selected aptamer surviving the strong wash (Fig. 2B, lanes 1 and 2). Other control libraries which presumably do not form a covalent conjugation with the thrombin target only gave a negligible PCR product band under the same condition (lanes 3–5). We focused on the robust PCR product resulting from the AFS library screening and further analyzed for potential covalent binders by HTS and bioinformatics analysis.
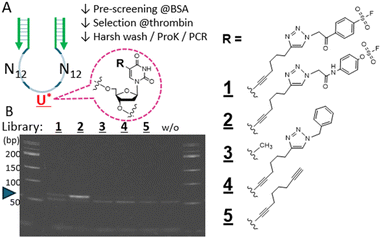 | ||
| Fig. 2 The DNA library and warheads used for the combinatorial screening. (A) The library comprises tandem N12 randomized sequences with a single linker (octadinyl-dU) with a terminal alkyne (U*) inserted at position 13. The flanking primer sequences are denoted in green. The R substitution on the uracil was benzenesulfonyl fluoride (BSF; 1), fosylate (AFS; 2), CH3 with no linker (3; natural T), benzyl (4), or unconjugated alkyne (5). The single-round selection comprised pre-screening of the library with BSA-beads, reaction with thrombin-beads, harsh (stringent) wash, proteinase K digestion to free the covalently bound aptamers, and PCR amplification. For details, see ESI,† Section S3.2. (B) PAGE separation of the PCR products corresponding to the respective starting libraries (1–5), without template (w/o), 50 bp markers on both edges. The blue arrow points to the expected 64 bp PCR-amplified aptamers. | ||
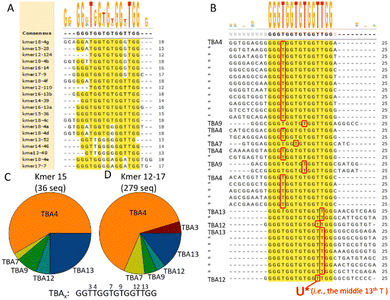
The traditional SELEX relied on the identification of binding aptamers by focusing on the “most abundant” sequences observed among the sequences obtained at the end of multiple rounds of selection.18 However, incorporation of HTS in the workflow with a dramatic increase in the observed sequences suggested that the sequence abundance at the end of multiple rounds of selection may not be the best measure to identify the strongest binding aptamers.23 Rather, the emergence of a motif or a critical subsequence of nucleotides during the selection cycles may better identify the best binders. Such a focus on subsequences of k-nucleotides or k-mers of non-SELEX HTS data was reported by Hoon,24 and we extend this approach to arrive at a systematic but a simple procedure for identifying potential covalent binders after a single-round selection and PCR amplification.
The bidirectional raw HTS reads were cleaned up (AptaSuite) and a FASTA file of 25 nucleotide long reads passed onto Tallymer for k-mer analysis (see ESI† for details and instructions on implementation of the freeware). The overall non-unique k-mer distribution of the HTS data shows an abundance of small k-mers with a rapid decrease in the number of k-mers with increasing k-mer length (Fig. S4, ESI†). The most and second abundant reads for each k-mer length are noted as seeds, since we use these sequences for the alignment in the next step. The ad hoc analysis employed by Hoon, namely the ASKAS method, applied to our data showed that the total counts of the top five k-mer reads jumped between k-mers 15 and 16 in agreement with their report.24 However, the objective definition of a significant decrease in the counts was not defined.
Without defining a priori, the critical k-mer length, as with Hoon,24 we took two approaches for further analysis of the HTS data. In the first approach, we took the top seed sequences for each k-mer and aligned the 20 seeds themselves (Fig. 3A). A loose consensus sequence of 15 nucleotides-long with a G-quad-like structure resembling the well-known thrombin-binding aptamer (TBA) emerged. The ambiguity, for example at the thymidine residues not forming the G-quad structure, is well tolerated with little impact on binding to the target (reviewed in Ricardi et al.).25 Inclusion of the second-best seeds in the alignment had little effect on the identified consensus sequence (Fig. S5, ESI†). Thus, a simple alignment of the top seed sequences appeared sufficient to identify the critical nucleotides and the length of the aptamers that presumably covalently bound the target thrombin in the single-round selection. We next took the respective seed sequences for the k-mer length and sought the HTS reads that contained the seeds. Fig. 3B shows the alignment of 36 HTS reads that contained the top seed sequence GGGTGGTGTGGTTGG for k-mer length 15. The alignments of actual HTS reads identified by other k-mer seeds are shown in Fig. S6 (ESI†). Since the reactive AFS warhead (U*) was placed at nucleotide 13 in the original library (see Fig. 2A) and read as ‘T’ by HTS, we can identify the precise position of the warhead from the alignment. The position of the warhead is denoted as TBAx, where the x indicates the position of the warhead (i.e., 3, 4, 7, 9, 12, or 13) in the original TBA sequence. The HTS reads identified from the 15mer seed showed a predominance of TBA4 followed by TBA13 (Fig. 3C). The distribution of TBAx-like sequence from 279 HTS reads identified by 12–17mer top seeds showed a similar overall distribution with predominance of TBA4 (Fig. 3D). It was previously noted that the position of warhead conjugation in the TBA aptamer was critical in determining the ability to form a covalent bond with the thrombin target.11,12 Thus far, the determination of the optimal position of warhead conjugation required a trial-and-error approach experimentally testing each potential position, but the asymmetric distribution of TBAx obtained during the covalent-aptamer screening starting from the warhead-endowed library suggests that the preferred position may be identified during the selection itself.
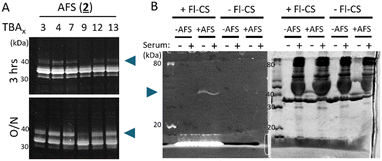
We tested the ability of AFS-conjugated TBAx to interact covalently with the thrombin target by a reducing SDS-PAGE band-shift assay where the covalently modified thrombin target appears as an upward mobility shifted band. AFS reacts slowly compared to the previously studied BSF warhead and only a faint mobility-shifted band can be observed at 3 hours reaction duration and a robust mobility-shifted band only observed after the overnight incubation (Fig. 4A). TBAx conjugated to a nonreactive benzyl mock warhead (4) or left unmodified as an alkyne (5) did not produce a band-shifted thrombin even after an overnight incubation. Of note is the observation that the mobility-shifted band is weak for TBA3 and TBA9, and stronger for TBA4 and TBA13 in agreement with the TBAx distribution abundance. The exception is TBA7, where a robust band shift is seen but less represented in the TBAx distribution.
The target-specificity of AFS warhead endowed-TBA4 was examined by a non-reducing SDS-PAGE mobility shift assay.12 The covalent conjugation of the aptamer was visualized by the addition of a fluorescent-label tagged complimentary strand against TBA (Fl-CS; Fig. 4B). The fluorescent signal (blue arrow) was observed only for AFS-TBA4, and a gross excess of human serum albumin revealed no non-specific binding supporting the high specificity of the AFS-endowed aptamer, as was shown previously for BSF.12 Control lanes with no Fl-CS showed no fluorescent signal.
Our combinatorial screening for covalent aptamers differs from the prior efforts in several respects. First, the starting library contained a single warhead in distinction to placing multiple warheads replacing all adenine18 or uracil16,17 nucleoside residues in the library like click-SELEX.21 Our approach enabled randomization of all four nucleotides, enhancing the starting library diversity. Although the single warhead in our library was tethered to the aptamer via an elongated octadinyl-dU linker, this modified unnatural nucleotide-containing library served well as a PCR substrate required for the single round of selection unlike the covalently binding library created by modification of the phosphodiester linkage.18 Second, we chose the AFS warhead with high specificity previously shown to be an optimal latent-warhead with little reactivity until placed in an optimal microenvironment created by the target protein and the warhead.22 This enabled a “slow-but-accurate” conjugation of the warhead-enabled aptamer to thrombin. Once a covalent aptamer is identified by selection, the slow AFS warhead could be altered to a more reactive warhead26 such as BSF12 to increase the rate of covalent reaction with the target important to potential utility as a drug. Third, the use of nano-beads to anchor the target protein allowed high stringency wash during the aptamer selection and recovery of only the covalently bound aptamers, reducing background noise in the HTS readout from non-covalent interactors or non-specific binders. This together with optimization of the overall protocol including proteinase K digestion to enable polymerase read-through of the target-bound aptamers, preselection of the library with BSA-nano beads to reduce non-specific interactors, and k-mer-based bioinformatics analysis enabled identification of covalent binders from the HTS readout after only one round of selection.
Despite the initial excitement over aptamers, fundamental limitations of rapid elimination from renal clearance and nuclease sensitivity have precluded the translational application of this promising drug modality. Recent exploration of covalent aptamers where the warhead-endowed single stranded nucleotide forms a covalent bond with the target protein suggests that covalent aptamers may circumvent these fundamental limitations.7 A significant remaining barrier to further acceleration of exploration of covalent aptamers as a drug modality has been the cumbersome trial-and-error approach required for determining the optimal position of warhead incorporation. The single round non-SELEX covalent aptamer screening that easily allows simultaneous identification of the aptamer sequence and the probable position of warhead incorporation described in this work, if reproducible for other targets, is likely to accelerate the resurgence of interest in aptamer drugs.
This work was supported by a JSPS KAKENHI grant (#21K05287, 21K12123, 20K20533), and JKA and its promotion funds from KEIRIN RACE. We thank Hokkaido Univ. Dept GI Surgery for the free use of the laboratory.
Data availability
Data supporting the findings of this study have been deposited in the ESI† of this article, including raw data of HTS results as three independent files of fastq format, and their summary on the basis of the top-k-mer analysis as an excel file. The details, including their manipulation methods using several web server, are written in the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- D. S. Johnson, E. Weerapana and B. F. Cravatt, Future Med. Chem., 2010, 2, 949–964 CrossRef CAS PubMed.
- J. Singh, R. C. Petter, T. A. Baillie and A. Whitty, Nat. Rev. Drug Discovery, 2011, 10, 307–317 CrossRef CAS PubMed.
- R. A. Bauer, Drug Discovery Today, 2015, 20, 1061–1073 CrossRef CAS PubMed.
- F. Sutanto, M. Konstantinidou and A. Domling, RSC Med. Chem., 2020, 11, 876–884 RSC.
- M. S. Rao, R. Gupta, M. J. Liguori, M. Hu, X. Huang, S. R. Mantena, S. W. Mittelstadt, E. A. G. Blomme and T. R. Van Vleet, Front. Big Data, 2019, 2, 25 CrossRef PubMed.
- Q. Li, Q. Chen, P. C. Klauser, M. Li, F. Zheng, N. Wang, X. Li, Q. Zhang, X. Fu, Q. Wang, Y. Xu and L. Wang, Cell, 2020, 182, 85–97e16 CrossRef CAS PubMed.
- J. Yang, Y. Tabuchi, R. Katsuki and M. Taki, Int. J. Mol. Sci., 2023, 24, 3525 CrossRef CAS PubMed.
- N. V. Mehta and M. S. Degani, Drug Discovery Today, 2023, 28, 103799 CrossRef CAS PubMed.
- N. Shindo and A. Ojida, Bioorg. Med. Chem., 2021, 47, 116386 CrossRef CAS PubMed.
- L. Hillebrand, X. J. Liang, R. A. M. Serafim and M. Gehringer, J. Med. Chem., 2024, 67, 7668–7758 CrossRef CAS PubMed.
- Y. Tivon, G. Falcone and A. Deiters, Angew. Chem., Int. Ed., 2021, 60, 15899–15904 CrossRef CAS PubMed.
- Y. Tabuchi, J. Yang and M. Taki, Chem. Commun., 2021, 57, 2483–2486 RSC.
- J. H. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS PubMed.
- D. Smith, G. P. Kirschenheuter, J. Charlton, D. M. Guidot and J. E. Repine, Chem. Biol., 1995, 2, 741–750 CrossRef CAS PubMed.
- J. Charlton, G. P. Kirschenheuter and D. Smith, Biochemistry, 1997, 36, 3018–3026 CrossRef CAS PubMed.
- K. B. Jensen, B. L. Atkinson, M. C. Willis, T. H. Koch and L. Gold, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 12220–12224 CrossRef CAS PubMed.
- I. S. MacPherson and T. Liedl, Aptamers, 2018, 2, 74–81 Search PubMed.
- Z. Qin, K. Zhang, P. He, X. Zhang, M. Xie, Y. Fu, C. Gu, Y. Zhu, A. Tong, H. Wei, C. Zhang and Y. Xiang, Nat. Chem., 2023, 15, 1705–1714 CrossRef CAS PubMed.
- Q. Zheng, J. L. Woehl, S. Kitamura, D. Santos-Martins, C. J. Smedley, G. Li, S. Forli, J. E. Moses, D. W. Wolan and K. B. Sharpless, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 18808–18814 CrossRef CAS PubMed.
- S. N. Carneiro, S. R. Khasnavis, J. S. Lee, T. W. Butler, J. D. Majmudar, C. A. W. Ende and N. D. Ball, Org. Biomol. Chem., 2023, 21, 1356–1372 RSC.
- F. Pfeiffer, F. Tolle, M. Rosenthal, G. M. Brandle, J. Ewers and G. Mayer, Nat. Protoc., 2018, 13, 1153–1180 CrossRef CAS PubMed.
- Y. Tabuchi, T. Watanabe, R. Katsuki, Y. Ito and M. Taki, Chem. Commun., 2021, 57, 5378–5381 RSC.
- T. Schütze, B. Wilhelm, N. Greiner, H. Braun, F. Peter, M. Mörl, V. A. Erdmann, H. Lehrach, Z. Konthur, M. Menger, P. F. Arndt and J. Glökler, PLoS One, 2011, 6, e29604 CrossRef PubMed.
- S. Hoon, B. Zhou, K. D. Janda, S. Brenner and J. Scolnick, Biotechniques, 2011, 51, 413–416 CrossRef CAS.
- C. Riccardi, E. Napolitano, C. Platella, D. Musumeci and D. Montesarchio, Pharmacol. Ther., 2021, 217, 107649 CrossRef CAS PubMed.
- S. Uematsu, Y. Tabuchi, Y. Ito and M. Taki, Bioconjugate Chem., 2018, 29, 1866–1871 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc04594g |
| This journal is © The Royal Society of Chemistry 2024 |