Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
iSERS: from nanotag design to protein assays and ex vivo imaging
Namhyun
Choi
a,
Yuying
Zhang
 b,
Yuling
Wang
b,
Yuling
Wang
 *c and
Sebastian
Schlücker
*c and
Sebastian
Schlücker
 *a
*a
aDepartment of Chemistry and Center of Nanointegration Duisburg-Essen (CENIDE) & Center of Medical Biotechnology (ZMB), University of Duisburg-Essen, Essen, 45141, Germany. E-mail: sebastian.schluecker@uni-due.de
bSchool of Medicine, Nankai University, Tianjin, 300071, China
cSchool of Natural Sciences, Faculty of Science and Engineering, Macquarie University, Sydney, NSW 2109, Australia. E-mail: yuling.wang@mq.edu.au
First published on 3rd June 2024
Abstract
Proteins are an eminently important class of ubiquitous biomacromolecules with diverse biological functions, and numerous techniques for their detection, quantification, and localisation have been developed. Many of these methods exploit the selectivity arising from molecular recognition of proteins/antigens by immunoglobulins. The combination of surface-enhanced Raman scattering (SERS) with such “immuno”-techniques to immuno-SERS (iSERS) is the central topic of this review, which is focused on colloidal SERS nanotags, i.e., molecularly functionalised noble metal nanoparticles conjugated to antibodies, for their use in protein assays and ex vivo imaging. After contrasting the fundamental differences between label-free SERS and iSERS, including a balanced description of the advantages and drawbacks of the latter, we describe the usual workflow of iSERS experiments. Milestones in the development of the iSERS technology are summarised from a historical perspective. By highlighting selected examples from the literature, we illustrate the conceptual progress that has been achieved in the fields of iSERS-based protein assays and ex vivo imaging. Finally, we attempt to predict what is necessary to fully exploit the transformative potential of the iSERS technology by stimulating the transition from research in academic labs into applications for the benefit of our society.
1. Introduction
Undoubtedly, the field of surface-enhanced Raman scattering (SERS) has witnessed a rapid development in the past 50 years, and this is one reason why we, as the entire SERS community, celebrate it together. With its roots in fundamental research on spectroelectrochemistry, SERS has now become a widely used analytical tool with a broad range of diverse applications. The deeper one delves into the fundamentals of SERS,1 the more one recognises that it touches quite a number of different areas in chemistry and physics: plasmonics, colloidal chemistry, surface science, molecular spectroscopy, quantum chemistry and molecular dynamics, charge transfer processes, etc.In parallel, tremendous achievements in the field of physical and chemical nanotechnology have been made in the past decades. Examples of this progress are control over top-down nanofabrication of sophisticated nanoantennas as well as over the synthesis of complex colloids, including anisotropic nanoparticles and assemblies of particles. Experimental techniques for characterisation at the single-particle or single-antenna level, as well as computations of their optical properties by electromagnetic modelling, are nowadays routinely performed. Theory can certainly not only explain existing experimental findings but also a priori predict properties of structures that have not even yet been synthesised. Quantum chemical modelling of SERS spectra slowly catches up with what we observe experimentally. Applications of SERS in bioassays detect target molecules at ultra-low concentrations. Nanoparticles are used for SERS in vitro and in vivo, including animal studies. Overall, these are truly amazing developments with an impact on many other scientific fields.2,3
One of these fields is protein science. Proteins are an eminently important class of ubiquitous biomacromolecules with diverse biological functions; the latter range, for example, from their defence role in the immune system over control of gene expression to the conversion of chemicals into mechanical energy.4 Due to the outstanding role of proteins, numerous methods for their detection, quantification, and localisation have been developed. Many of these techniques rely on molecular recognition of proteins/antigens by immunoglobulins/antibodies via non-covalent forces; examples are immunoassays for protein detection in solution, immunocytology and immunohistochemistry for protein localisation in cells and on tissue, respectively. The combination of SERS with such “immuno”-techniques to immuno-SERS (iSERS) is the central topic of this review.
Our intention as authors is to put the subfield of iSERS into the bigger picture of the entire SERS field. We restrict the content of this review to colloidal SERS nanotags in combination with antibodies for protein detection in vitro and localisation ex vivo. Since applications in vivo require additional disjunct considerations, they are not covered here.
For the novice, this contribution may serve as a starting point for further reading prior to experimental work in this exciting field. For the iSERS expert, it may serve as a useful resource in research and/or teaching.
Our selection of highlighted examples from the literature is naturally subjective. We apologise in advance to all respected colleagues in the iSERS field whose work is not mentioned or cited here. The body of literature is simply overwhelming and forces us to focus on conceptual advances that, in our opinion, were important.
The outline of this review on iSERS is briefly summarised in this paragraph. Fundamental differences between conventional label-free SERS and iSERS are contrasted in Section 2. The workflow of iSERS experiments is described in Section 3. Selected examples from the literature that, in our opinion, represent conceptual milestones in the development of the iSERS field are highlighted in Section 4. Then, Section 5 introduces different technology platforms employed in iSERS assays, followed by a discussion of selected examples that, in our opinion, were important to the advancement of the field. The same concept also applies to Section 6, which is focused on iSERS bioimaging and, after introducing general areas of application, highlights the continuous progress with respect to the multiplexing capacity of iSERS. Finally, in Section 7, we attempt to predict what is necessary to fully exploit the transformative potential of the iSERS technology by stimulating the transition from research in academic labs into applications for the benefit of our society.
2. Two concepts: conventional label-free SERS vs. iSERS
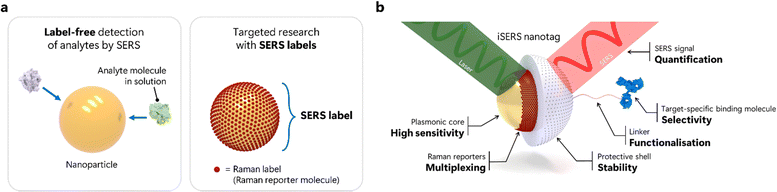
SERS substrates, i.e., plasmonic nanostructures supporting a localised surface plasmon resonance (LSPR), can be broadly classified as colloidal or solid SERS substrates. The majority of current iSERS applications are based on colloidal SERS particles that are usually prepared by wet nanochemistry with a high degree of control over size and shape. Most conventional SERS applications in analytical chemistry exploit the molecular fingerprinting capabilities of vibrational Raman spectroscopy for the direct label-free, i.e., non-targeted detection of analytes that adsorb onto the metal surface (Fig. 1a left). Albeit seemingly simple from an initial, superficial perspective, the large number of degrees of freedom makes standardisation in SERS anything but a trivial task.5The concept of iSERS is fundamentally different from that of label-free SERS. iSERS is inherently an indirect scheme for protein detection that exploits the target specificity of recognition elements, in particular antibodies, that are chemically bound to SERS labels/nanotags for optical identification; the latter comprise Raman labels/reporter molecules that are permanently chemi- or at least physisorbed on the metal surface and determine the characteristic Raman spectroscopic fingerprint of the labelling agent (Fig. 1a right).
iSERS nanotags are relatively large nanoparticle-based labels, typically 20 to 100 nm in size, for the selective and sensitive optical detection of protein target molecules. Integral components of an iSERS nanotag are (Fig. 1b):
(i) The plasmonic core for providing high sensitivity via efficient LSPR excitation. To achieve this, the laser excitation wavelength must be matched to a LSPR peak of the metal colloid. Red to near-infrared laser excitation is often preferred in a biological context, for example, to avoid interference with autofluorescence or to provide large penetration depths through biological tissues. Gold is often the preferred metal in bio-applications due to its high chemical inertness. In contrast, silver provides larger plasmonic enhancements; however, it is chemically less inert and prone to oxidation.
(ii) Raman reporters for identification and spectral multiplexing. The characteristic Raman spectrum of the Raman reporter molecules that are permanently adsorbed onto the surface of the plasmonic core, for example, via a strong gold–sulfur bond, is used for the unambiguous spectral identification of the entire iSERS nanotag – similar to the characteristic colour of fluorescence emitted by a fluorescent dye or a quantum dot. The spectral multiplexing capacity of SERS is enormous: due to the small linewidth of vibrational Raman bands, typically 2–20 cm−1, a large number of Raman reporter molecules can be spectrally distinguished,6 either by simple univariate analysis or more sophisticated multivariate approaches from chemometrics or machine learning algorithms. In contrast to fluorophores, either molecular or nanocrystal-based, for which the excitation must be tuned to the corresponding absorption maximum, only a single laser excitation wavelength is necessary to excite the Raman spectra of all SERS labels. The high photostability of SERS nanotags is another key advantage compared to fluorescence-based approaches that suffer from photobleaching. The fact that iSERS is a spectroscopic technique relying on spectra rather than just a single intensity value, together with the fact that the SERS signal is directly proportional to the number of SERS labels, makes it an inherently quantitative technique – an aspect that is routine in the physical sciences but becomes increasingly important in the life sciences.
(iii) A protective shell is optional but strongly recommended since it guarantees colloidal stability. The shell should prevent desorption of the Raman reporter molecules from the surface of the plasmonic core that is in chemical equilibrium with its environment, normally water or buffers as suspension medium. This guarantees constant SERS signal levels as the number of adsorbed Raman reporters remains constant. The shell also provides colloidal stability to the SERS nanotags, usually either by electrostatic or steric stabilisation. Raman reporter molecules and molecular components of the shell may be co-adsorbed on the metal surface of the plasmonic core, or they are preferably spatially separated from each other. Specifically, a full monolayer of Raman reporters, which is disjunct from the shell and avoids co-adsorption, represents maximum surface coverage and, therefore, maximal SERS intensities.7 Finally, the shell is also the chemical bridge for binding the actual SERS nanotag to the target-specific ligand for yielding the iSERS nanotag.
(iv) A linker may, for example, be a functional group on the surface of the shell or also a separate, often bifunctional molecule that at one side binds to the shell and at its other terminus binds to the target-specific ligand. Generally, covalent or non-covalent bioconjugation chemistry is employed. Covalent bioconjugation is usually recommended due to its higher robustness, for example, in situations involving harsh conditions such as high ionic strength of the suspension medium or multiple washing steps in assays or microscopy. Direct conjugation of the target-specific ligand to either the Raman reporter or the plasmonic core is not recommended since conjugation to the Raman reporters increases the likelihood of its desorption, while direct conjugation to the plasmonic core via co-adsorption, reduces the number of chemisorbed Raman reporters.
(v) The target-specific binding molecule, usually an antibody that selectively recognises its antigen, is key for selectivity in iSERS: it is the binding specificity of the antibody that determines the selective detection of proteins using iSERS nanotags. The term iSERS nanotag refers to SERS nanotag conjugated to the antibody or vice versa, a SERS nanotag-labelled antibody.
Overall, the rational design of SERS nanotags with defined physical and chemical properties is essential for successful iSERS applications. At all stages of their synthesis, analytical techniques must be employed to characterise their performance and uniformity.
Specificity is the central advantage of iSERS over conventional label-free Raman/SERS, and it is directly related to the quality of the target-specific ligand through its selective recognition capabilities and high binding strength. Generally, antibodies are outstanding ligands, famous for their high binding specificities and association constants. A disadvantage of SERS nanotags is certainly their relatively large size and weight compared with molecular fluorophores. This might be critical for live cell imaging, especially in intracellular studies, but is typically not critical for cell surface antigens, tissue sections and assays. Drawbacks of each targeted strategy, either with SERS nanotags or fluorophores as optical labelling agents, are that you need to know a priori which target you want to detect, that a target-specific ligand must be available and that the latter might be costly. Other aspects that should be considered are whether the intrinsic binding properties of the ligand are modified or even compromised upon bioconjugation to the SERS nanotag and whether cross-reactions to other targets occur. In any case, negative control experiments must be performed in every reliable iSERS study. One important negative control is the use of isotype antibodies that do not bind to the target.
3. The workflow of iSERS experiments
The workflow of iSERS experiments is shown in Fig. 2. It starts with the choice and chemical synthesis of the plasmonic core of the SERS nanotag, usually via the reduction chemistry of noble metal salts. A variety of plasmonic nanoparticles with different sizes, shapes and compositions is available. Standard quasi-spherical gold nanoparticles generally suffer from modest signal enhancements at the monomer level; their LSPR peak, in combination with green laser excitation, is unfavourable for various biological and biomedical applications due to intense autofluorescence; this can be circumnavigated, for example, by hollow gold/silver spheres that exhibit tuneable resonances across the visible spectrum.8 Anisotropic particles such as rods also exhibit a tuneable longitudinal LSPR depending on the aspect ratio. Plasmonic coupling in superstructures such as core/satellite particles is a second efficient strategy to control the LSPR energy and additionally generates very high enhancement factors (EFs) in the narrow gap between the central large core and the smaller satellites. Their detection at the single-particle level was demonstrated in correlative optical/electron-microscopic experiments.9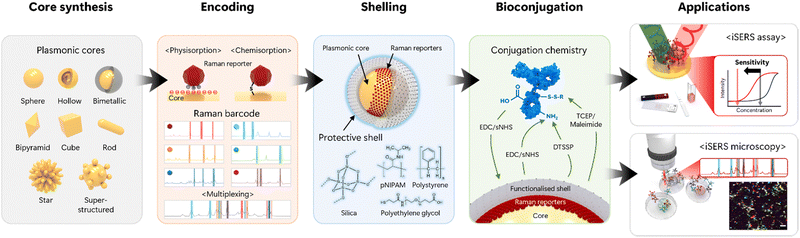 | ||
| Fig. 2 Overview of the workflow in iSERS experiments: from core synthesis to encoding, shelling and bioconjugation to applications in assays and microscopy. | ||
The second step is encoding the plasmonic core with Raman reporter molecules. Each type of reporter has a characteristic “molecular fingerprint”; its corresponding Raman barcode is a simplified and intuitive representation, with peak positions and intensities encoded in the horizontal positions and widths of the bars, respectively. While the plasmonic core must support a LSPR for optically resonant excitation that leads to the generation of plasmon-assisted Raman scattering, the laser excitation may also overlap with the electronic absorption spectrum of the Raman reporter. The choice of either electronically resonant or non-resonant Raman reporters has both specific pros and cons. A key advantage of resonant Raman dyes is their additional signal enhancement via surface-enhanced resonance Raman scattering (SERRS). Key disadvantages are, however, photobleaching and a reduced spectral multiplexing capacity compared to the smaller non-resonant Raman reporters that, due to the smaller number of atoms N, exhibit fewer vibrational degrees of freedom (3N − 6 for a non-linear molecule).7
Encapsulation of the plasmonic core with a shell is an optional but highly recommended step, as colloidal stability is key to successful iSERS experiments. Numerous shell materials, in particular inorganic polymers such as silica, organic polymers such as poly(ethylene glycol), and biopolymers such as proteins, are available. The shell does not only provide stability to the SERS nanotags by electrostatic or sterical stabilisation but is also central to subsequent bioconjugation; the shell either already contains suitable functional groups or needs to be functionalised in an additional step. Once the shell is functionalised, one has to decide on either covalent or non-covalent bioconjugation; the latter is, for example, routinely done in the preparation of immunogold conjugates in conventional lateral flow assays via electrostatic binding of antibodies to citrate-capped gold nanoparticles. However, for iSERS – based on many years of our own experience – we strongly recommend employing covalent conjugation chemistry because of the significantly higher strength and robustness of covalent chemical bonds compared to non-covalent interactions.
Multiple functional groups can be addressed in covalent bioconjugation, for example, carboxyl (COOH), amino (NH2) and thiol (SH)/disulfide (S–S) groups. In many cases, the functional group must first be activated for bioconjugation, for example, in the case of unreactive carboxyl groups via the activated esters by using carbodiimides. The quality of the iSERS nanotags, i.e., SERS nanotag-labelled antibodies, is key for successful iSERS applications. Central properties that must be checked are the target-specific binding capabilities of the antibody as well as the colloidal stability and the brightness of the iSERS nanotags. Once the integrity of the iSERS nanotags is confirmed, the stage is set for either iSERS assays or microscopy. Again, we stress the importance of negative control experiments using isotype antibodies to unambiguously demonstrate binding specificity.
4. Historical perspective: milestones in the development of iSERS
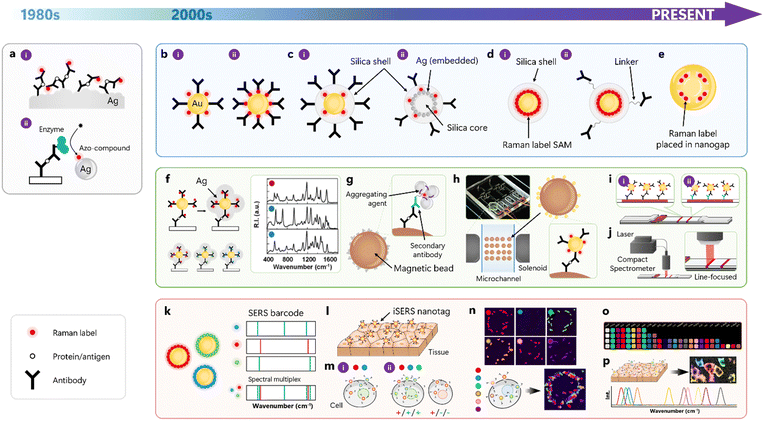
This section highlights milestones in the development of iSERS from a historical perspective (Table 1). We selected conceptually innovative and original work that, in our opinion, stimulated the field and, therefore, is an important contribution to the development of the iSERS field. This selection can be broadly classified into iSERS assay development with a focus on different technology platforms, the rational design and synthesis of SERS nanotags, and multiplexed iSERS microscopy on cells and tissues. (Fig. 3).| Year | Development | Ref. |
|---|---|---|
| 1989 | First SERS-based sandwich immunoassay on Ag substrate | 10 |
| 1997 | ELISA-SERS sandwich with azo-compounds as Raman label (Ra) | 11 |
| 1999 | First conception of iSERS nanotag: gold nanoparticles bioconjugated to antibodies | 12 |
| 2003 | Synthesis of iSERS nanotags using direct conjugation of antibodies to Ra | 13 |
| 2003 | Conception of iSERS multiplex assay and improvement of sensitivity by silver deposition | 14 |
| 2003 | Syntheses of silica-encapsulated SERS nanotags | 15,16 |
| 2006 | Bi-silica iSERS nanotags: silica-encapsulated silver-embedded silica particle | 17 |
| 2006 | Proof-of-concept for iSERS microscopy on tissue section | 18 |
| 2007 | Multiplexed immunoassay with SERS-encoded beads | 19 |
| 2008 | Magnetic bead-based iSERS assay | 20 |
| 2009 | Silica-encapsulated iSERS nanotags with a full monolayer of Ra for red laser excitation | 7 |
| 2009 | Introduction of the SERS barcode concept | 21 |
| 2010 | First conception of on-chip iSERS assay | 22 |
| 2011 | iSERS ex vivo on fresh tissue | 23 |
| 2011 | Gap-enhanced SERS nanotags | 24 |
| 2012 | 2-colour iSERS microscopy | 25 |
| 2014 | First SERS-LFA/ICA | 26 |
| 2015 | 3-colour iSERS microscopy | 27 |
| 2016 | Quantitative 6-plex with SERS nanotags | 28 |
| 2018 | Correlative SERS/SEM/LSPR of single SERS nanotags | 9 |
| 2019 | First compact and fast iSERS-LFA reader | 29 |
| 2020 | 6-colour iSERS microscopy on single cells | 30 |
| 2022 | Demonstration of 26-plex iSERS microscopy | 31 |
| 2023 | 10-plex iSERS tissue sectioning | 32 |
A sandwich immunoassay based on SERS was pioneered by Rohr, Cotton, and co-workers in 1989 (Fig. 3a-i).10 A roughened silver film, coated with the capture antibody and blocked with bovine serum albumin, served as SERS substrate. After the capture of the antigen, in this case, thyroid-stimulating hormone, sandwich complexes were formed upon the addition of the Raman-active immunoconjugate. In this pioneering study, the authors already explicitly stated that the SERRS signal generation from the Raman reporter molecule p-dimethylamino azobenzene was not limited to silver films and electrodes, but could be extended to silver (or other metal) colloids, too. The enzyme-linked immunosorbent assay (ELISA), utilising enzyme-labelled detection antibodies, is very popular in clinical diagnostics due to its high-throughput capabilities. In conventional ELISA, the enzyme catalyses the conversion of a colourless substrate into a coloured product that can be quantitatively determined by UV/Vis absorption spectroscopy. ELISA based on SERS was introduced by Dou, Ozaki, and co-workers in 1997 (Fig. 3a-ii).11 A peroxidase that was covalently bound to the detection antibody oxidised o-phenylenediamine to azo-aniline, which then served as Raman reporter dye for SERRS signal generation after adsorption to the surface of silver nanoparticles. The limit of detection was ca. one order of magnitude better compared to that of the first SERRS study.10
4.1. Design of SERS nanotags
Porter and co-workers published a SERS nanotag-based iSERS immunoassay in 1999.12 An obvious advantage of this strategy was that in these iSERS nanotags, the capabilities for SERS signal generation (SERS nanotag) and specific binding (antibody) were both incorporated into an iSERS nanotag as a single bifunctional unit (Fig. 3b-i). In contrast to the preceding configurations, aromatic thiols as Raman reporter molecules were directly attached to the surface of 30 nm colloidal gold via strong Au–S bonds. The sensitivity was in the pM regime. Drawbacks of this configuration were co-adsorption of (i) antibodies and Raman reporters, which avoided the formation of a complete self-assembled monolayer (SAM) of Raman reporter molecule (Ra) and thus reduced sensitivity and of (ii) other (bio)molecules onto the gold surface, increasing the risk of unspecific binding and the appearance of unwanted additional SERS signals. In 2003, they presented a significant improvement in sensitivity down to the fM regime (1 pg mL−1) for the detection of prostate-specific antigen (PSA).13 In their modified iSERS nanotag design shown in Fig. 3b-ii, the detection antibody was covalently bound to the Raman reporter molecule rather than to the gold surface as in their earlier approach, ensuring a high surface coverage with the Raman reporter 4-nitro-2-mercaptobenzoic acid.Silica encapsulation for stabilisation of SERS nanotags was introduced independently by the groups of Natan and Nie in 2003, respectively (Fig. 3c-i). Mulvaney, Natan, and co-workers introduced the concept of glass encapsulation to single, Raman reporter-labelled silver or gold nanoparticle cores to provide both mechanical and chemical stability to the core while essentially maintaining its optical properties.15 Co-adsorption of an amino silane and the Raman reporter results in a vitreophilic SERS label onto which a silica shell is grown by the addition of active silica, followed by silica growth with the famous Stöber method known from sol–gel chemistry. The resulting glass shell also allows further functionalisation, employing a variety of established conjugation protocols. In the same year, Doering and Nie presented a very similar approach to glass-coated nanoparticles, in which they exploited Raman reporter molecules carrying an isothiocyanate group or multiple sulfur atoms.16 The synthesis protocol comprised the addition of Raman reporter onto gold nanoparticle surface, the addition of a mercaptosilane, silica deposition by using sodium silicate and finally, condensation of remaining silicate onto the existing shell. In addition to the encapsulation of single noble metal nanoparticles, also core/satellite assemblies of nanoparticles, which are attractive due to their high SERS signal levels arising from plasmonic coupling between the satellites, were encapsulated by silica (Fig. 3c-ii).17 The multi-step synthesis comprises the preparation of spherical silica nanoparticles as core particles that were functionalised with a mercaptosilane for binding of silver nanoparticles onto the thiolated core, followed by co-adsorption of Raman reporter for SERS and mercaptosilane for silica encapsulation of the entire assembly. In both approaches (Fig. 3c-i and ii), only submonolayer coverage with Raman reporter molecules was achieved due to the co-adsorption of silanes as precursors for subsequent sol–gel chemistry.
Schlücker and co-workers resolved this drawback by separating the formation of a full monolayer of Raman reporters from the formation of a silica shell around it in a layer-by-layer approach followed by a modified Stöber synthesis (Fig. 3d).7 The complete SAM yielded 22 times higher SERS compared to sub-monolayer coverage; together with the use of Au/Ag nanoshells optimised for red laser excitation rather than conventional quasi-spherical Au nanoparticles, 180 times stronger SERS was detected. In addition to this 2-step procedure involving the addition of two polyelectrolytes, the same group also developed a direct route to silica encapsulation by integrating the Raman reporter and a silane anchor into a single molecular entity via covalent chemistry33 or, in a non-covalent approach, by using noncovalently bound silane precursors.34
In addition to maximising the surface coverage with Raman reporters, maximising the plasmonic enhancement is a second important aspect in the rational design of SERS nanotags. The key to obtaining a maximal enhancement factor in SERS is to use either the tips of gold nanostars35 as SERS nanotags or by employing coupled plasmonic nanostructures with very small gaps (<2 nm). Nam, Suh, and co-workers employed a DNA-modified gold core as a seed for obtaining gold nano-bridged nanogap particles with a well-defined, hollow, ca. 1 nm interior gap (Fig. 3e).24 The use of DNA not only facilitated the formation of the nano-bridged nanogap but also enabled the precise positioning of Raman dyes inside the gap, providing strong and highly stable SERS.
A fair direct comparison of different SERS nanotag designs is challenging for several reasons. For example, cuvette experiments at the ensemble level are difficult to interpret since usually not only monomers, but also a small fraction of clusters and even larger aggregates may be present; the latter yield significantly higher SERS intensities than the monomers due to hot spots resulting from plasmonic coupling.36 In other words, the overall SERS signal of the mixture is largely dominated by contributions from minority species and, therefore, this is not representative for the entire colloid. Single-particle studies circumnavigate this problem. To this end, the presence and identity of single nanoparticles must be checked. Correlative optical and electron-microscopic studies are best suited to provide this type of information. Schlücker and co-workers, for example, identified in correlative LSPR/SERS/SEM experiments on four different types of isotropic and anisotropic nanoparticles that Au/Au/core/satellite nanoparticles and Au nanostars can be detected at the single-particle level, while hollow Au/Ag nanoshells and normal quasi-spherical Ag nanoparticles cannot.9
4.2. iSERS assays
Mirkin and co-workers introduced Raman dye-labelled nanoparticles for protein detection by modifying probes that were originally designed for DNA and RNA detection.14 Seeded growth of silver on small 13 nm gold nanoparticles was performed via electroless silver deposition for producing SERS-active nanostructures (Fig. 3f left). Nanoparticle probes were based on either protein-small molecule (type I) or protein–protein (type II) interactions and employed the dyes Cy3, Cy3.5 and Cy5 as Ra for SERS detection in the corresponding 3-plex assays (Fig. 3f right).In real-world samples, proteins are usually abundant in complex mixtures involving disturbing matrices, necessitating the use of purification steps for isolating a particular target protein. Micron-sized magnetic beads coated with suitable capture antibodies are an elegant approach for extraction and enrichment since they can be mixed with the corresponding complex sample in solution for target binding and then rapidly pulled down using an external magnet. The supernatant can be removed, and the conjugate comprising magnetic bead, capture antibody and target antigen can be redispersed and washed several times. Smith, Graham and co-workers demonstrated the first combination of a magnetic bead-based pull-down assay and iSERRS (Fig. 3g).20 Specifically, they employed a dye-labelled secondary antibody for recognising the primary detection antibody. Silver nanoparticles and aggregating agents were added for SERRS-based detection of the dye in silver colloids aggregated on the surface of the magnetic bead. Note that in this example, the SERRS signal is generated in situ and is not provided by an extrinsic SERS nanotag.
A second important technology platform is microfluidics as a key component for realising the idea of the “lab on a chip”. The underlying central idea is to perform all chemical reactions in small volumes on a chip as a mini-lab in an automated fashion, ideally without the need for manual handling by users. Choo and co-workers pioneered the combination of microfluidic sample handling and processing with iSERS.22 Specifically, they developed a gradient optofluidic sensor comprising three compartments for (i) serial dilution of the target protein in a gradient channel, (ii) injection and mixing of the sample with both capture antibody-coated magnetic beads and SERS nanotags comprising Raman dye-labelled antibodies adsorbed on the surface of hollow gold nanoparticles, and finally (iii) the trapping area for capture of the immunocomplexes (Fig. 3h).
A third important technology platform is a rapid test that is user-friendly and easy to perform anytime and anywhere. Lateral flow immunoassays (LFAs or LFIAs), also called immunochromatographic assays (ICAs), are widely used portable strips for point-of-care testing (POCT). While the term LFA highlights the flow aspect along the membrane, typically nitrocellulose with defined pore sizes, the term ICA stresses the role of both liquid and membrane acting as the mobile and stationary phase, respectively. Deng and co-workers demonstrated the first SERS-based ICA/LFA (Fig. 3i).26 Conventional gold conjugates used in standard LFAs were replaced by iSERS nanotags comprising detection antibodies labelled with gold/silver core/shell particles with Raman reporters adsorbed on the gold core as SERS nanotags. The corresponding limit of detection was improved by 1–3 orders of magnitude to conventional LFA based on naked-eye detection, depending on the choice of a particular target.
A major drawback of all initial SERS-based LFAs was the very long readout time until the test result was available for the user. Often, research-grade confocal Raman microscopes were used for point-mapping of either the entire TL, with typical dimensions of 3–7 mm in width and 1–2 mm in length, or at least a large portion of it. This is obviously a time-consuming procedure due to the larger number of scanning steps along both x- and y-dimensions employing point illumination with a laser spot of usually one to few microns. Schlücker and co-workers circumnavigated this long acquisition time and the use of expensive research-grade Raman instrumentation by designing and building a compact SERS reader for rapid scanning of the test strip (Fig. 3j).29 Line illumination along the entire width of the TL was achieved by a custom-designed fibre optical probe with line focus illumination in combination with a powerful yet compact 785 nm diode laser for providing a sufficiently high laser power density. The test strip is scanned along the flow direction, i.e., orthogonal to the line focus, using a motorised stage, reducing the overall acquisition time by several orders of magnitude from many hours down to a few seconds. Overall, this proof-of-concept study demonstrated the feasibility (fast and easy-to-use) and affordability (compact reader vs. expensive confocal Raman microscopes) of the SERS-LFA technology for end users of Raman-based POCT in and especially outside academic laboratories.
4.3. iSERS microscopy
Early multiplexing studies on protein assays by iSERS employed a colour legend as a simplified representation of the Raman spectrum.14 While this is an obvious analogy to fluorescence, this representation marginalises the complexity of a vibrational spectrum with its 3N − 6 vibrational modes/peaks for a non-linear molecule, peak intensities and widths, and even peak shapes/line profiles. Additionally, only single-color cases and no mixtures were presented. Schlücker and co-workers introduced the barcode concept as an alternative simplified representation that reduces the complexity of a vibrational Raman spectrum to a 1D barcode in which peak positions/wavenumbers are encoded in the horizontal positions of the bars and peak intensities in their corresponding widths (Fig. 3k).21 Later, the same group demonstrated a quantitative 6-plex in colloidal suspension employing silica-encapsulated SERS nanotags and a conventional least-square algorithm for signal decomposition into the six spectrally distinct contributions of each tag.28Sequential analysis and spatial multiplexing are common strategies when sufficient sample material is available, and multiple samples must be analysed in a high-throughput format. Prominent examples include microarrays and 96-well plates. Spectral multiplexing is a powerful extension of the capabilities of spatial multiplexing that employs only one or few “colours”, e.g., in colourimetry (ELISA) or fluorescence. The spectral multiplexing capacity of iSERS is especially attractive and advantageous in situations where the sample itself is a limiting factor. In assays, for example, this is the case when the sample volume is limited – e.g., the lysate from a single cell. In microscopy, for example, this is the case when the sample number is limited – e.g., analysis of the same single cell or tissue section. Formalin-fixed and paraffin-embedded (FFPE) tissue is the workhorse in conventional pathology for subsequent haematoxylin & eosin (H&E) and immunohistochemical staining after antigen retrieval. Schlücker and co-workers presented the proof of concept for iSERS microscopy on tissue ex vivo in early 2006.18 Hollow gold/silver nanoshells optimised for red laser excitation, conjugated to an anti-PSA antibody, were employed as iSERS nanotags for localising PSA in the epithelium of prostate tissue section (Fig. 3l).7 In 2011, Ghambir and co-workers demonstrated the application of iSERS microscopy to fresh tissue.23 Since 2006, the number of different spectral barcodes (“colours”) employed in iSERS microscopy on cells and tissue specimens has constantly been raised and has now reached a level that is multiple times higher than that of conventional fluorescence. Two- and three-colour iSERS microscopy with different target proteins was demonstrated by the group of Choo in 201225 and the groups of Pérez-Juste and Liz-Marzán in 2015,27 respectively (Fig. 3m). Four-color iSERS microscopy on tissue was demonstrated by Liu and co-workers in 2018.37 Yet, four “colours” (N = 4) were still in the range of what conventional fluorescence microscopy with fluorescent dyes can achieve.
As mentioned above, spectral multiplexing is particularly advantageous in situations where the sample itself is a limiting factor, for example, in single-cell studies. To this end, and in order to also highlight the superior multiplexing capacity of iSERS compared to conventional fluorescence (N > 4), Schlücker and co-workers demonstrated 6-colour iSERS microscopy on a single cancer cell (Fig. 3n).30 Single cancer cells from the human breast cancer line SkBr-3 that overexpress human epidermal growth factor receptor 2 (HER2), the most prominent breast cancer marker that is localised on the cell membrane, were analysed. Each type of six spectrally distinct SERS nanotags was conjugated to anti-HER2 antibodies for localising HER2 on the cell membrane of the same single cancer cell. This work paved the way for future multi-colour/multi-target imaging for characterising heterogeneous protein expression at the single-cell level.
In 2022, Zavaleta and co-workers presented an impressive expanded library of 26 spectrally distinct, silica-encapsulated SERS nanotags (Fig. 3o).31 Raman reporters were selected in a theory-guided approach using vibrational Raman spectra computed by density functional theory (DFT) in order to ensure a high degree of spectral uniqueness. Highly multiplexed optical imaging was demonstrated in the context of targeting cultured cells and profiling cancerous human tissue sections.
In 2023, Ye and co-workers presented 32 spectrally distinct SERS nanotags with characteristic peaks from stretching vibrations of C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C and C
C and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N triple bonds in the so-called Raman-silent region (1900–2300 cm−1) (Fig. 3p).32 Ten-colour iSERS imaging with high spectral resolution in clinical biopsies of breast cancer tissues was demonstrated. Both numbers (N = 32 and N = 10) represent the highest values reported for multiplexing in iSERS to date, i.e., at the end of 2023.
N triple bonds in the so-called Raman-silent region (1900–2300 cm−1) (Fig. 3p).32 Ten-colour iSERS imaging with high spectral resolution in clinical biopsies of breast cancer tissues was demonstrated. Both numbers (N = 32 and N = 10) represent the highest values reported for multiplexing in iSERS to date, i.e., at the end of 2023.
5. iSERS assays
Besides iSERS, various immunoassay platforms based on colourimetry38 and fluorescence39 as well as electrochemical40 and field-effect transistor (FET)41,42 sensors have been developed. These have been effectively deployed in the diagnostic field. Colourimetric methods are cost-effective, simple, and intuitive without the need for readout devices, but limited sensitivity poses a challenge. In contrast, fluorescence is based on direct reading of emission signals, enabling highly sensitive quantification, and limited multiplexing is possible using a variety of fluorophores. However, signal degradation caused by photobleaching of the fluorophore and background noise resulting from autofluorescence interference are critical since they ultimately diminish the sensitivity of the assay. Electrochemical assays have gained significant attention for their ability to facilitate highly sensitive real-time monitoring. However, their sensitivity is critically impacted by electrode fouling and the composition of the sample matrix. The presence of contaminants on the electrode leads not only to a decrease in sensitivity but also to inaccuracies due to reduced selectivity, while the matrix composition of the sample also creates interferences; both negatively affect the overall assay performance. FET has recently emerged as a label-free assay platform with significant potential for miniaturisation, for example, enabling the integration into wearable sensors. However, it has been observed that the sensitivity of FET-based sensors is susceptible to environmental factors. Moreover, multiplexed detection with FET is limited. Overall, it is noteworthy that FET is being considered as the next generation of sensors, owing to their unique properties and potential applications in in vitro diagnostics (IVD). Nonetheless, it is essential to be aware of the potential limitations of such FET sensors, including their susceptibility to environmental factors and limited capabilities for multiplexing.The distinctive features of iSERS – highly sensitive detection, multiplexing capabilities, and stable as well as reproducible outcomes – present a promising solution to the challenges encountered by other assay platforms. iSERS can detect low concentrations of analytes, particularly in samples containing trace amounts of targets, which makes it an advantageous technique for comprehensive immunoassays. The second most significant benefit of iSERS is its capability to detect multiple analytes in a single readout due to the use of Raman reporter molecules with spectrally narrow and well-defined peaks. In addition, the spectral specificity of iSERS is particularly crucial for distinguishing closely related analytes and minimising interference from background signals, which improves the signal-to-noise ratio and detection accuracy for complex sample matrices. iSERS is intrinsically capable to generate reliable and reproducible signals in immunoassays. This reliability is critical for obtaining accurate and consistent results when real-life samples (e.g., blood) are employed. Furthermore, iSERS can be customised for specific applications by adjusting the nanotag's size, shape, and configuration. Therefore, the flexibility of iSERS allows it to be optimised to meet the needs of a variety of assays. Overall, iSERS strongly benefits from the unique combination of superior sensitivity with multiplexing, spectral specificity, reliability, and flexibility, making it an excellent choice for immunoassays. The integration of iSERS into existing platforms is straightforward: substitution of (i) the label by a SERS nanotag and (ii) the readout by a SERS reader.
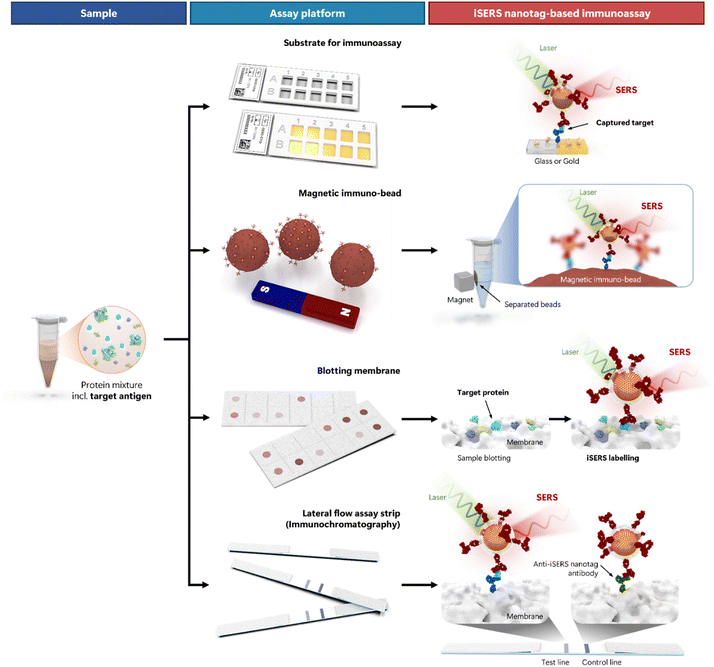
Fig. 4 depicts the most widely used formats of iSERS assays (from top to bottom). The workflow is schematically shown (from left to right) and begins with the sample (left column). Real-world samples are usually complex mixtures that, among many other molecular components, may also contain the target protein. A matched pair of antibodies directed against two different epitopes of this antigen (= target protein) is employed: capture and detection antibody. The role of the capture antibody is to immobilise the target protein onto a substrate to separate it from all other components of the mixture. All four depicted iSERS assay types essentially differ only in the type of substrate that is used in a particular assay platform (middle column). The capture antibody is immobilised to this substrate (from top to bottom): a glass or gold substrate, a magnetic immuno-bead, a blotting membrane, or a lateral flow assay strip.
The role of the detection antibody is to enable the detection of the immobilised target proteins. In iSERS nanotag-based immunoassays (right column), the detection antibody is conjugated to a SERS nanotag (= iSERS nanotag). Molecular recognition of the target protein by the iSERS nanotags leads to the formation of a sandwich immuno-complex: the target protein is sandwiched between capture and detection antibodies. Unbound iSERS nanotags must be washed away since they give rise to false-positive results. The SERS signal of the SERS nanotag in the sandwich immuno-complexes is directly proportional to their number and thereby enables quantitative detection of proteins. Since iSERS nanotags can be detected even at the single-particle level (see Section 1 and 3), iSERS assays are exquisitely sensitive.
Validity and reliability are two key parameters of each test. Validity means that you actually measure what is specified and intended to be measured. Reliability means that you measure something in a reproducible way with both accuracy and precision. Both of these central aspects, validity and reliability, certainly also apply to iSERS assays.
Validity in the context of iSERS assays is synonymous with specificity. Since iSERS – as every “immuno”-technique – relies on molecular recognition of proteins by antibodies, the validity of an iSERS assay critically depends on the functional integrity of the corresponding capture and target antibodies. Cross-reactions with proteins other than the target antigen should be minimal and tested in control experiments. Non-specific binding due to electrostatic or non-covalent interactions is a limitation to specificity. Attempts to increase binding affinity and to reduce non-specific binding are therefore important. Porter and co-workers, for example, presented a strategy employing a rotating disk for increasing binding affinity (Fig. 5a-i).43 Shiddiky, Trau, and co-workers developed molecular nano shearing for reducing non-specific adsorption of non-target protein molecules from solid/liquid interfaces by applying an alternating electric field to the electrode surface (Fig. 5a-ii).44
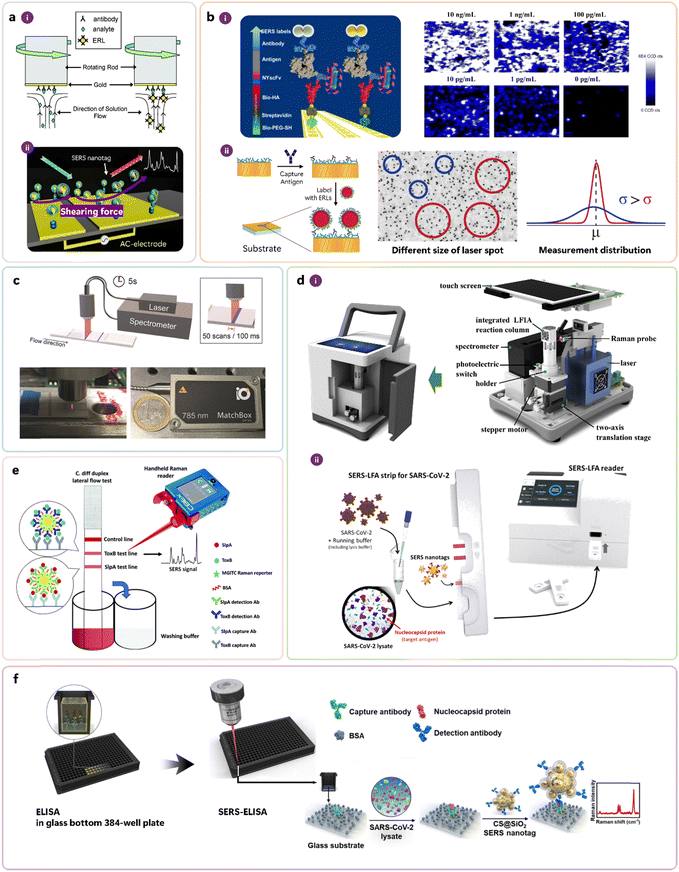 | ||
| Fig. 5 Conceptually important work for the development of iSERS assays, addressing the following key aspects: (a) improving binding affinity (i) and minimising non-specific binding (ii), (b) increasing precision via mapping and sampling, (c) reducing acquisition time and instrumentation costs by orders of magnitude, (d) portable instrumentation, (e) handheld instrumentation, and (f) high-troughput screening in SERS-ELISA. (a) Reprinted with permission from ref. 43. Copyright 2007 American Chemical Society. Reprinted with permission from ref. 44. Copyright 2015 American Chemical Society. (b) Reprinted with permission from ref. 47. Copyright 2014 American Chemical Society. Reprinted with permission from ref. 48. Copyright 2016 American Chemical Society. (c) Reprinted with permission from ref. 29. Copyright 2019 Wiley-VCH. (d) Reprinted with permission from ref. 50. Copyright 2020 Elsevier. Reprinted with permission from ref. 51. Copyright 2022 American Chemical Society. (e) Reproduced from ref. 52 with permission from the Royal Society of Chemistry. (f) Reprinted with permission from ref. 53. Copyright 2023 Elsevier. | ||
Reliability in the context of iSERS assay means that the standard deviation of the measured values is small (precision), and their mean value is close to the true value (accuracy). The addition of a defined amount (= spiking) of an isotope-edited specimen as an internal standard to a sample is an elegant and commonly employed approach in analytical chemistry, for example, in mass spectrometry, for improving accuracy. Zhang, Ben-Amotz, and co-workers introduced the isotope-edited internal standard method to the SERS field.45 Sampling errors are a common error source in SERS measurements since the diameter of the laser spot, typically on the order of only a few microns, is much smaller than the large dimensions of the macroscopic sample that is typically in the mm to cm range. In order to reduce the sampling error and to increase the precision of iSERS assays, Wang, Schlücker, and co-workers employed a point-mapping strategy in which the solid substrate was raster-scanned in both x- and y-direction; the inhomogeneous spatial distribution of the immunocomplexes at low target concentrations was visualised in the corresponding SERS false-colour images (Fig. 5b-i).46,47 Porter and co-workers employed a different strategy to address the sampling error: they increased the laser spot size and thereby covered larger areas of the sample within an individual measurement (Fig. 5b-ii).48 On LFA strips (cf.Fig. 4 bottom), the immunocomplexes are immobilised on a test line with dimensions on the order of a few mm, typically 3–4 mm in width and 1–2 in length. Also in this case, the sampling problem does apply when research-grade confocal Raman microscopes with diffraction-limited laser spots are employed for raster scanning with point illumination. Long acquisition times of several hours prevented applications in point-of-care testing (POCT). Additionally, the large price of research-grade Raman instrumentation was an obvious hurdle to applying this powerful technology outside academic laboratories. Schlücker and co-workers addressed both issues – the drastic reduction of both time and prize by 2–3 orders of magnitude, respectively – by developing a home-built compact SERS reader (Fig. 5c) comprising a powerful small laser, a custom-made fibre optic probe with line focus illumination, a motor-driven linear stage for moving the LFA strip along the flow direction, and a portable spectrometer with CCD.29,49 Portable instrumentation has additional technical requirements, such as battery-driven devices that make the user independent from power grid connections. Towards this end, Wang and co-workers, as well as Kang, Lee, Chen, Choo and co-workers, presented portable SERS readers (Fig. 5d-i and ii).50,51 Faulds, Keegan, Graham and co-workers demonstrated even a handheld approach: instead of using a conventional benchtop Raman instrument, a handheld Raman spectrometer was employed for optical excitation and readout (Fig. 5e).52 Finally, some applications demand the detection of many samples in a high-throughput format. Microwell plates, often with 96 or even 384 wells, are an ideal platform for analysing multiple samples. Kang, Chen, Yoon, Choo and co-workers integrated ELISA (not shown in Fig. 4, cf. Section 3) using a 384-well plate (glass substrate) with an iSERS assay (Fig. 5f).53 A 10-fold improvement compared to conventional ELISA and a 100-fold improvement compared to LFA detection was shown.
6. iSERS microscopy
Fluorescence imaging is an extensively utilised optical technology that employs fluorophores as labels to gather diverse information on biomolecules, cell organelles, etc. One of the unique facets of this technology is that various colours can be employed to track different biomolecules. Multicolour fluorescence microscopy enables researchers to observe colocalisation and, in turn, to elucidate potential interactions between different types of biomolecules. Notwithstanding the benefits, there are critical inherent limitations associated with this technology. For instance, photobleaching is a ubiquitous phenomenon where exposure to continuous excitation light can alter the molecular structure, causing non-fluorescent behavior. Consequently, this technology is not ideally suited for continuous imaging observations. In addition, organic fluorophores usually exhibit poor solubility in aqueous solutions, which causes the compounds themselves to aggregate or cause protein aggregation.54 Although multiple detection channels are possible, the number of available colours is restricted by the relatively broad spectral emission profiles of molecular fluorophores that are prone to spectral overlap. Additionally, the technology is vulnerable to autofluorescence, leading to image distortion and blurring.55The ideal labels/tags for bioimaging possess specific characteristics such as photostability, high contrast, capability of spectral multiplexing, and can be applied in aqueous environment. iSERS microscopy is a suitable candidate as it meets all of these requirements. The utilisation of iSERS nanotags for bioimaging has emerged as a powerful multi-colour technique, e.g., for identifying colocalised targets. Additionally, the spectral intensity directly provides quantitative information about the tagged protein.56 Furthermore, the optical stability of iSERS offers advantages for continuous imaging. The flexibility of choosing the laser excitation wavelength matching the nanotag's plasmonic properties enables distortion-free and blur-free observations through bioimaging in the ‘biological window’,57 which is immune to influences such as autofluorescence. However, one notable drawback of iSERS is the relatively large size of the nanotag, which should be considered in the experimental design for bioimaging. Overall, iSERS microscopy is a powerful method for multi-colour quantitative bioimaging in situations where the target proteins are readily accessible.
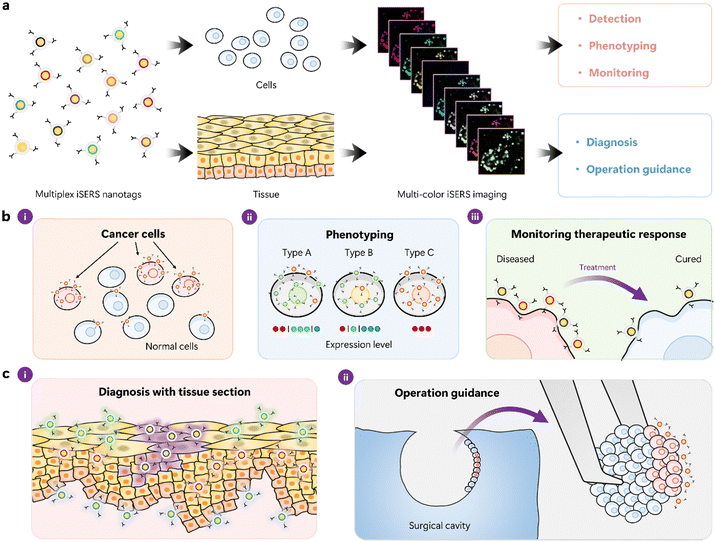
Fig. 6 depicts the concept of multi-colour cell- and tissue-based iSERS microscopy (Fig. 6a) together with selected applications in cell (Fig. 6b) and tissue (Fig. 6c) research. The different Raman reporter molecules on the surface of the plasmonic core are colour-encoded in (Fig. 6a): each colour represents the spectral barcode of the iSERS nanotag. Hyperspectral data sets in iSERS microscopy are usually obtained by raster scanning the sample in two dimensions. At each pixel, a Raman spectrum is detected and subsequently spectrally decomposed into the contributions from each iSERS nanotag by using their known individual spectral signatures. Finally, a set of false-colour Raman images is obtained, where each image shows the localisation of the corresponding antigen (Fig. 6a right).
Common to all applications of multiplexed iSERS microscopy in cell – (Fig. 6b) and tissue-based (Fig. 6c) research is the ability to simultaneously probe a set of protein biomarkers in a quantitative approach. This includes the localisation of each target protein (spatial information) as well as the degree of its over- or underexpression (quantitative information). For example, cancer cells usually differ from normal cells in the expression of so-called cancer biomarkers (Fig. 6b-i). Even cells from the same cell line, i.e., with the “same” genetic information, may be quite heterogeneous in terms of their phenotype. Quantification of the expression levels of each biomarker, therefore, allows phenotyping (Fig. 6b-ii). Also, the response to therapy can be monitored at the molecular level, for example, the reduction in the expression of a particular cancer biomarker after drug treatment (Fig. 6b-iii). The latter is of importance, for example, in the context of personalised medicine. Tissue-based applications of multiplexed iSERS microscopy also benefit from the simultaneous, quantitative localisation of a set of protein biomarkers (Fig. 6c). Multiplexed immunohistochemistry by iSERS microscopy, for example, offers more detailed insights compared to conventional immunohistochemistry, addressing only single biomarkers. This type of “molecular profiling” can also be exploited in or near the operating room when decisions on tumour margins must be made by differentiating between tissue material that is kept (healthy) vs what needs to be removed (cancerous).
Fig. 7 highlights selected examples of iSERS microscopy on single cells. HER2 is an important biomarker in breast cancer. Jeong, Cho, and Lee were the first to demonstrate the selective localisation of HER2 and CD10 on cellular membranes by iSERS microscopy in late 2006 by using relatively large (>100 nm) SERS nanotags (“SERS dots”): a comparison of the bright-field image and the signals recorded in the corresponding Raman mapping is shown in Fig. 7a.17
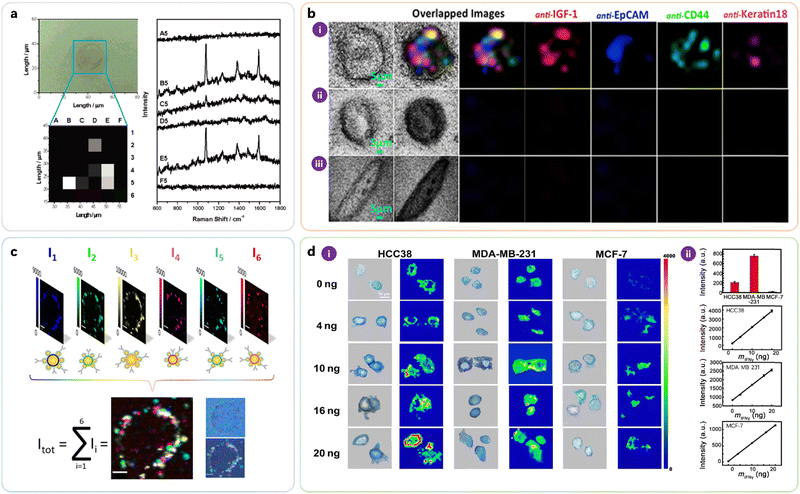 | ||
| Fig. 7 Selected examples for cell-based iSERS microscopy. (a) Localisation of the breast cancer marker HER2 on a single MCF-7 cell. Reprinted with permission from ref. 17. Copyright 2006 American Chemical Society. (b) Four-colour iSERS microscopy on a single MCF-7 cell in the presence of an excess of fibroblasts. Reprinted with permission from ref. 58. Copyright 2014 Springer Nature. (c) 6-Colour iSERS localisation of HER2 on the same single SKBR3 cell. Reprinted with permission from ref. 30. Copyright 2020 American Chemical Society. (d) Localisation of PD-L1 on single triple-negative breast cancer cells and monitoring of its expression variation during drug treatment. Reprinted with permission from ref. 59. Copyright 2018 American Association for the Advancement of Science. | ||
Biris, Zharov, and co-workers demonstrated the identification of circulating tumour cells (CTC) by four-colour iSERS microscopy (Fig. 7b). Results from the transmission and multiplex iSERS imaging of a single MCF-7 cell among 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fibroblast cells have demonstrated that iSERS nanotags, due to their high sensitivity and high multiplexing capacity, are extremely attractive to be modified with recognition molecules against different biomarkers for CTC profiling.58
000 fibroblast cells have demonstrated that iSERS nanotags, due to their high sensitivity and high multiplexing capacity, are extremely attractive to be modified with recognition molecules against different biomarkers for CTC profiling.58
In order to further promote multiplexed imaging applications of single-cell iSERS microscopy, Schlücker, Kasimir-Bauer and co-workers demonstrated 6-color iSERS imaging of HER2 on the same single SkBr-3 cell (Fig. 7c). The six false-colour Raman images show the spectral intensities I1 to I6 for each individual iSERS nanotag. Their overlap Itot is the sum of contributions from all iSERS nanotags; the selective localisation of HER2 on the cell membrane is clearly visible and is additionally confirmed by comparison with the corresponding white light image on the right.30
Quantification of important marker proteins is essential for the clinical classification of tumour stages and the evaluation of therapeutic effects. For example, quantitative analysis of programmed cell death receptor ligand 1 (PD-L1), which is an important immune checkpoint molecule, may guide PD-L1-based personalised therapy. Tian, Zheng and co-workers localised PD-L1 on single triple-negative breast cancer cells and also monitored its expression variation during drug treatment (Fig. 7d). The Raman intensity of the peak at 1530 cm−1 reflected that the PD-L1 expression levels of HCC38, MDA-MB-231, and MCF-7 cells progressively increased after stimulation with different concentrations of IFN-γ.59 Tsao and coworkers monitored cellular phenotypic changes of melanoma cell lines harbouring BRAF mutations in response to BRAF inhibitor (PLX4720), showing that distinct subpopulations were formed upon PLX4720 treatment. They further collected blood samples serially from 10 stage-IV melanoma patients at different time points during the treatment course and monitored the phenotypic changes of the CTC. CTC populations were found to shift after treatment with dabrafenib and trametinib for 40 days, demonstrating the capability of iSERS imaging in characterising CTC evolution in clinical applications.60
Fig. 8 highlights selected applications of tissue-based iSERS microscopy. Schlücker and co-workers demonstrated the proof of concept for tissue-based iSERS microscopy in early 2006 (Fig. 8a) by using SERS-labeled primary antibodies to localise PSA on FFPE prostate tissue sections.18 The numbers (1) to (5) in the white light image (i) indicate different regions: (1)–(3) epithelium, (4) stroma, and (5) lumen. The corresponding spectra recorded at these locations demonstrate the specific presence of PSA in the epithelium.
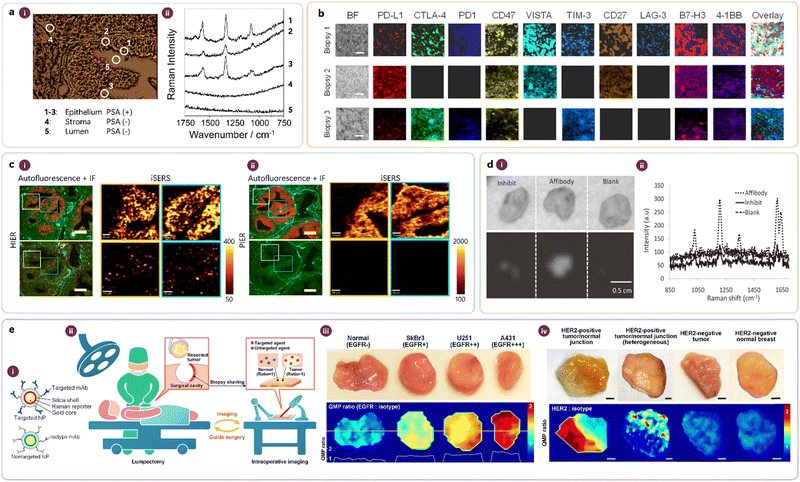 | ||
| Fig. 8 (a) First demonstration of iSERS microscopy on tissue specimen: (a-i) White light images and (a-ii) corresponding Raman spectra. Reprinted with permission from ref. 18. Copyright 2006 Wiley-VCH. (b) Multiplexed iSERS imaging of immune checkpoint inhibitors from three patients. Reprinted with permission from ref. 32. Copyright 2023 Elsevier. (c) Heat- vs. protein-induced epitope retrieval (HIER vs. PIER) of FFPE tissue. The bottom false-colour Raman images are obtained with isotope antibodies as negative controls. Reprinted with permission from ref. 61. Copyright 2018 American Chemical Society. (d) Ex vivo iSERS microscopy on fresh tissue: staining with affibody-based iSERS nanotags vs. two negative controls (inhibit, blank). Reprinted with permission from ref. 23. Copyright 2011 Wiley-VCH. (e) Ratiometric iSERS imaging for reliable molecular profiling during surgery for intraoperative guidance. Reprinted with permission from ref. 62. Copyright 2016 Springer Nature. | ||
In the context of personalised medicine, a cocktail of immune checkpoint (ICP) inhibitors is expected to maximise the immune response rates of patients compared to the conventional single-drug treatment strategy in cancer immunotherapy. The prediction of such optimal combinations requires multiplexed imaging techniques such as iSERS microscopy and suitable data analysis. Ye and co-workers applied 10-colour iSERS microscopy for probing a panel of 10 ICP inhibitors (Fig. 8b) on breast cancer biopsies; the results obtained from three patients are shown together with the corresponding bright-field image on the left.32
FFPE tissue sections, the workhorse in traditional pathology, require antigen retrieval before immunostaining. Zhang, Schlücker, and co-workers studied the role of antigen retrieval on the quality of the resulting Raman false-colour images obtained in iSERS microscopy for localising PSA on FFPE prostate tissue (Fig. 8c).61 Specifically, they compared heat-induced epitope retrieval (HIER) (Fig. 8c-i) and protease-induced epitope retrieval (PIER) (Fig. 8c-ii). PIER clearly produced better results than HIER, as evidenced by the staining quality, including negative controls with isotype antibodies.
Surgery is one of the most effective and widely used procedures in treating human cancers, but the major problem is that the surgeon often fails to remove the entire tumour, leaving behind tumour-positive margins, metastatic lymph nodes, and/or satellite tumour nodules. If postoperative pathology identifies residual tumours at the surgical margins, re-excision surgeries are often necessary, which comes along with additional expenses and risks for the patient. Therefore, intraoperative methods that can identify residual tumours and guide their complete removal during tumour-resection procedures are highly desired in the clinic. In 2011, Gambhir and coworkers demonstrated the feasibility of iSERS microscopy for detecting the tumour marker epidermal growth factor receptor (EGFR) ex vivo in explanted xenograft tumours from mice (Fig. 8d).23Fig. 8d-i shows bright-field (top) and SERS false-colour (bottom) images of tissue incubated with iSERS nanotags directed against EGFR by using an EGFR affibody (Affibody) as well as two negative controls: tissue pre-incubated with free EGFR affibody for competitive inhibition (Inhibit) and untargeted SERS nanotags (Blank); the corresponding Raman spectra are shown in Fig. 8d-ii.
Non-specific binding limits the reliability of iSERS imaging and it is difficult to eliminate completely. Based on the assumption that nontargeted SERS nanotags reflect the non-specific binding of iSERS nanotags, Liu et al. developed a ratiometric approach: an equimolar mixture of targeted NPs (iSERS nanotags) and nontargeted NP (SERS nanotags) are topically applied (Fig. 8e-i) ex vivo to fresh tissue specimens resected during surgery for intraoperative guidance (Fig. 8e-ii). Ratiometric SERS images, constructed using the ratio of the Raman signals from both NP types, should more reliably reflect the molecular expression of tumour markers. Fig. 8e-iii depicts results on EGFR expression in tumour xenografts (SkBr3, U251, and A431) that express various levels of EGFR, while Fig. 8e-iv depicts results on HER2 expression in human breast cancer specimens.62 In a subsequent clinical study, multiplexed iSERS imaging of four biomarkers (HER2, ER, EGFR, and CD44) in 57 freshly excised breast specimens enabled highly sensitive and specific discrimination of carcinoma, demonstrating the potential of iSERS for intraoperative guidance to reduce the rate of re-excision in cancer patients.63
7. Outlook and perspectives
iSERS nanotags are an attractive and powerful optical labelling technique for use in both protein bioassays and ex vivo imaging because they offer a unique combination of features such as quantification, sensitivity, photostability, and dense multiplexing for simultaneous target detection/localisation. iSERS has also already shown enormous potential in biomedical applications, in particular for the development of clinical diagnostic assays and imaging tools. Since both iSERS bioassays and imaging rely on iSERS nanotags, substantial efforts should be undertaken to control the monodispersity and homogeneity of iSERS nanotags to achieve more reliable qualitative and, in particular, quantitative results.To this end, significant advances in the rational design and synthesis of bright noble metal nanoparticles as the core of SERS nanotags have been accomplished, including the control over size and shape as well as over nano assemblies with multiple hot spots.64 We feel that more concerted efforts from both material scientists and colloidal/physical chemists are still necessary to increase reproducibility by producing highly uniform and bright SERS nanotags that can be detected at the single-particle level. Such SERS nanotags with uniform brightness across many single particles require both uniform plasmonic enhancement and Raman reporters with uniform Raman signal contributions; the latter is the result of two effects: surface coverage and Raman scattering cross-section. Accurate quantification, especially at low or even ultra-low concentrations, requires uniform SERS nanotags. Uniform iSERS nanotags, in addition to uniform brightness, require uniform binding properties. Important aspects in this respect are the number of antibodies as well as their orientation and accessibility. Therefore, optimal bio-conjugation strategies need to be applied to ensure maximum binding of the iSERS nanotags to the target protein. Overall, the workflow of producing iSERS nanotags is complicated since it requires multiple, time- and labour-consuming steps. In the future, the production of uniform iSERS nanotags or at least uniform SERS nanotags might be achieved by automated synthesis using a robot.65 If the synthesis yields are not close to 100%, then purification strategies for obtaining purified colloids with a well-defined composition (no mixtures) are recommended.66 Currently, iSERS nanotags are usually freshly prepared prior to iSERS experiments. To reduce the workload for sample preparation, large batches of particles should be produced and stored. Storage of iSERS nanotags requires improvement of their stability by a protective coating (e.g., polymer, silica). For example, for POCT in tropical and subtropical locations, iSERS nanotags in LFA strips must tolerate high temperatures.67 Also, other applications may involve harsh conditions such as high ionic strength or cryogenic temperatures.
Most publications of iSERS assays employed purified systems, such as proteins in water or buffer, and obtained very low limits of detection (LOD). In contrast, clinical specimens are much more complex mixtures involving unknown matrix effects. A few studies have shown that the LOD is much higher when using patient samples compared to pure proteins in the buffer.49 A substantial effort should be invested in sample preparation, not only to purify or isolate the target proteins from the complex matrix but also for the design and optimisation of the entire iSERS workflow. Only then, in our opinion, we will be able to fully exploit the so far unexplored area of biologically highly active species that are abundant at very low concentrations in complex clinical specimens.
The field of immunoassays continues to face challenges, such as specificity, that require further improvements.68,69 This applies to all receptor–ligand interactions, including that between antigens and antibodies.70 A major obstacle is non-specific binding including cross-reactivity. Obviously, improving the specificity of iSERS assays also faces these challenges. The advent of SERS readout has paved the way for highly sensitive detection, enabling studies in the low concentration regime that were hitherto unexplored. However, this development also poses a new challenge in the form of assay errors caused by non-specific binding, which so far has been overlooked but became observable by highly sensitive SERS.53 An illustrative example71 that highlights this aspect in a quantitative fashion is the discrepancy between the detection limits of protein targets in buffer vs. serum: at high concentrations, there is almost no difference in the detection limits, while they differ by three orders of magnitude at low concentrations in that particular case. This observation suggests an association with non-specific binding to components of the complex sample matrix and/or platform materials such as assay substrates.72
Various solutions to this debate have been proposed in the clinical assay field for a long time, and researchers have implemented designs and strategies that take them into account. Representative solutions for minimising interference with non-specific binding and cross-reactions include separation or chemical destruction of non-specific reactants such as ligands,73 selection and optimisation of assay methods,74 optimisation of incubation time based on thermodynamics and reaction kinetics,75 selection of the most specific antibodies76,77 and the most effective blocking.78 The extended surface area of the colloidal nanoparticles within the iSERS platform requires careful considerations to impede non-specific binding. In addition to BSA, which is commonly used as a blocking reagent, other types of blocking strategies can be applied.79 Alternative strategies may be inspired from colocalization in two-colour single-molecule fluorescence microscopy for identifying accurately formed sandwich immuno-complexes.80 Another approach to minimise non-specific binding, in addition to those mentioned in Section 5, is to improve the specificity of iSERS nanotags by optimising the bioconjugation of the antibodies with the nanotags including their orientation on the particle surface.81,82
Quantification of target proteins is still a challenge of practical iSERS applications on real-world samples. There are two potential ways to improve quantification by iSERS. One way is to adopt the method of the internal Raman standard and integrate it into the workflow of iSERS assays with iSERS nanotags.83 The second way is based on digital reading where, in contrast to a single intensity detected in ensemble measurements, the distribution function of the molecules is measured by counting the signal.84 Validation is a further important aspect of obtaining accurate quantification. We strongly recommend that results from iSERS assays should be validated with a gold standard (e.g., ELISA, ECL) or, if not possible, at least cross-validated with other methods.85,86 This is especially important in the context of clinical samples.
Compact and affordable instrumentation for iSERS assays is necessary for applications outside academic laboratories, especially for POCT. Ideally, portable or handheld readers are available. This suggests the engagement of industry, especially Raman instrument manufacturers. In this respect, the requirements of iSERS microscopy differ from those of iSERS assays. Since the first demonstration of iSERS microscopy by combining Raman microscopy with iSERS nanotags for tissue imaging,18 significant advances have been made. Future developments should aim to further develop iSERS imaging to make it an important diagnostic tool that complements other imaging modalities such as magnetic resonance imaging (MRI), fluorescence imaging, and computed tomography. Developing multi-modal approaches is quite promising since multiple independent imaging techniques provide more comprehensive data (morphology, location, biodistribution) than a single technique and open a new window for more accurate diagnosis.
The imaging speed in iSERS is still a major concern, especially in scenarios that require fast decision-making. The challenge here is still the development of rapid confocal Raman microscopes that could be used for large-area tissue imaging. Although wide-field Raman imaging for a few colours has been developed, more effort is necessary to develop imaging tools for multiplexed imaging in combination with fast data acquisition (e.g., using line-focus imaging rather than point-by-point mapping).87,88 For improving image contrast for accurate diagnosis, minimisation of autofluorescence using time-gated Raman techniques89 might be considered for use in iSERS imaging.
Translation to a clinical environment and POCT not only requires suitable instrumentation but also has additional constraints. For example, end users typically do not have expertise in nanotechnology, molecular spectroscopy, or data analysis. In other words, sample handling, instrumentation, and software must be intuitive, user-friendly, and easily integrated into the clinical workflow. The latter includes not only the actual test but also sample preparation, including pre-processing. For example, current whole blood testing in traditional clinical laboratories necessitates sample pre-processing by centrifugation, which is time-consuming and requires a centrifuge as an instrument. This can be circumnavigated by elegant pre-processing directly on the LFA strip using separation pads, which is important for the implementation of POCT for reducing assay time and cost.90 Only in collaboration with clinicians, comprehensive clinical studies involving many patient samples will be feasible. This will then require standardisation efforts and finally certification according to medical device and IVD regulations.
Overall, we already have most of the technology in our hands; it is now the time to show the added value of iSERS to users outside our iSERS community for really generating a much broader impact! In the future, multiplexed ultrasensitive iSERS may even contribute to identify panels of biologically highly active biomarkers for next-generation molecular profiling. The future of iSERS, indeed, is bright and colourful!
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from the University of Duisburg-Essen (UDE), the Center for Nanointegration Duisburg-Essen (CENIDE), and the Center of Medical Biotechnology (ZMB) is acknowledged. This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (grant number 2021R1A6A3A14038507). Y. Zhang acknowledges that this work was supported by Natural Science Foundation of Tianjin City (No. 22JCZDJC00320). Y. Wang acknowledges funding support from Australian Research Council (ARC) Future Fellowship (FT210100737). Y. Wang and S. Schlücker acknowledge funding support from the Alexander von Humboldt Foundation and the German Research Foundation (DFG).References
- E. C. Le Ru and P. G. Etchegoin, Principles of Surface-Enhanced Raman Spectroscopy, Elsevier, Amsterdam, 2009 Search PubMed.
- S. Schlücker, Angew. Chem., Int. Ed., 2014, 53, 4756–4795 CrossRef PubMed.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. García de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J.-F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J.-M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L.-L. Tay, K. G. Thomas, Z.-Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- T. E. Creighton, Proteins: Structures and Molecular Properties, ed. W. H. Freeman, New York, 1993 Search PubMed.
- S. E. J. Bell, G. Charron, E. Cortés, J. Kneipp, M. L. de la Chapelle, J. Langer, M. Procházka, V. Tran and S. Schlücker, Angew. Chem., Int. Ed., 2020, 59, 5454–5462 CrossRef CAS PubMed.
- S. Schlücker, ChemPhysChem, 2009, 10, 1344–1354 CrossRef PubMed.
- B. Küstner, M. Gellner, M. Schütz, F. Schöppler, A. Marx, P. Ströbel, P. Adam, C. Schmuck and S. Schlücker, Angew. Chem., Int. Ed., 2009, 48, 1950–1953 CrossRef PubMed.
- M. Gellner, B. Küstner and S. Schlücker, Vib. Spectrosc., 2009, 50, 43–47 CrossRef CAS.
- V. Tran, C. Thiel, J. T. Svejda, M. Jalali, B. Walkenfort, D. Erni and S. Schlücker, Nanoscale, 2018, 10, 21721–21731 RSC.
- T. E. Rohr, T. Cotton, N. Fan and P. J. Tarcha, Anal. Biochem., 1989, 182, 388–398 CrossRef CAS PubMed.
- X. Dou, T. Takama, Y. Yamaguchi, H. Yamamoto and Y. Ozaki, Anal. Chem., 1997, 69, 1492–1495 CrossRef CAS.
- J. Ni, R. J. Lipert, G. B. Dawson and M. D. Porter, Anal. Chem., 1999, 71, 4903–4908 CrossRef CAS PubMed.
- D. S. Grubisha, R. J. Lipert, H.-Y. Park, J. Driskell and M. D. Porter, Anal. Chem., 2003, 75, 5936–5943 CrossRef CAS PubMed.
- Y. C. Cao, R. Jin, J.-M. Nam, C. S. Thaxton and C. A. Mirkin, J. Am. Chem. Soc., 2003, 125, 14676–14677 CrossRef CAS PubMed.
- S. P. Mulvaney, M. D. Musick, C. D. Keating and M. J. Natan, Langmuir, 2003, 19, 4784–4790 CrossRef CAS.
- W. E. Doering and S. Nie, Anal. Chem., 2003, 75, 6171–6176 CrossRef CAS PubMed.
- J.-H. Kim, J.-S. Kim, H. Choi, S.-M. Lee, B.-H. Jun, K.-N. Yu, E. Kuk, Y.-K. Kim, D. H. Jeong, M.-H. Cho and Y.-S. Lee, Anal. Chem., 2006, 78, 6967–6973 CrossRef CAS PubMed.
- S. Schlücker, B. Küstner, A. Punge, R. Bonfig, A. Marx and P. Ströbel, J. Raman Spectrosc., 2006, 37, 719–721 CrossRef.
- B.-H. Jun, J.-H. Kim, H. Park, J.-S. Kim, K.-N. Yu, S.-M. Lee, H. Choi, S.-Y. Kwak, Y.-K. Kim, D. H. Jeong, M.-H. Cho and Y.-S. Lee, J. Comb. Chem., 2007, 9, 237–244 CrossRef CAS PubMed.
- P. Douglas, R. J. Stokes, D. Graham and W. E. Smith, Analyst, 2008, 133, 791–796 RSC.
- M. Gellner, K. Kömpe and S. Schlücker, Anal. Bioanal. Chem., 2009, 394, 1839–1844 CrossRef CAS PubMed.
- H. Chon, C. Lim, S.-M. Ha, Y. Ahn, E. K. Lee, S.-I. Chang, G. H. Seong and J. Choo, Anal. Chem., 2010, 82, 5290–5295 CrossRef CAS PubMed.
- J. V. Jokerst, Z. Miao, C. Zavaleta, Z. Cheng and S. S. Gambhir, Small, 2011, 7, 625–633 CrossRef CAS PubMed.
- D.-K. Lim, K.-S. Jeon, J.-H. Hwang, H. Kim, S. Kwon, Y. D. Suh and J.-M. Nam, Nat. Nanotechnol., 2011, 6, 452–460 CrossRef CAS PubMed.
- S. Lee, H. Chon, S.-Y. Yoon, E. K. Lee, S.-I. Chang, D. W. Lim and J. Choo, Nanoscale, 2012, 4, 124–129 RSC.
- M. Li, H. Yang, S. Li, K. Zhao, J. Li, D. Jiang, L. Sun and A. Deng, J. Agric. Food Chem., 2014, 62, 10896–10902 CrossRef CAS PubMed.
- G. Bodelón, V. Montes-García, C. Fernández-López, I. Pastoriza-Santos, J. Pérez-Juste and L. M. Liz-Marzán, Small, 2015, 11, 4149–4157 CrossRef PubMed.
- M. Schütz and S. Schlücker, J. Raman Spectrosc., 2016, 47, 1012–1016 CrossRef.
- V. Tran, B. Walkenfort, M. König, M. Salehi and S. Schlücker, Angew. Chem., Int. Ed., 2019, 58, 442–446 CrossRef CAS PubMed.
- E. Stepula, X.-P. Wang, S. Srivastav, M. König, J. Levermann, S. Kasimir-Bauer and S. Schlücker, ACS Appl. Mater. Interfaces, 2020, 12, 32321–32327 CrossRef CAS PubMed.
- O. E. Eremina, A. T. Czaja, A. Fernando, A. Aron, D. B. Eremin and C. Zavaleta, ACS Nano, 2022, 16, 10341–10353 CrossRef CAS PubMed.
- J. Li, F. Liu, X. Bi and J. Ye, Biomaterials, 2023, 302, 122327 CrossRef CAS PubMed.
- M. Schütz, B. Küstner, M. Bauer, C. Schmuck and S. Schlücker, Small, 2010, 6, 733–737 CrossRef PubMed.
- M. Schütz, M. Salehi and S. Schlücker, Chem. – Asian J., 2014, 9, 2219–2224 CrossRef PubMed.
- M. Schütz, D. Steinigeweg, M. Salehi, K. Kömpe and S. Schlücker, Chem. Commun., 2011, 47, 4216–4218 RSC.
- Y. Zhang, B. Walkenfort, J. H. Yoon, S. Schlücker and W. Xie, Phys. Chem. Chem. Phys., 2015, 17, 21120–21126 RSC.
- J. Li, H. Liu, P. Rong, W. Zhou, X. Gao and D. Liu, Nanoscale, 2018, 10, 8292–8297 RSC.
- A. Mohamad, H. Teo, N. A. Keasberry and M. U. Ahmed, Crit. Rev. Biotechnol., 2019, 39, 50–66 CrossRef CAS PubMed.
- X. Fang, Y. Zheng, Y. Duan, Y. Liu and W. Zhong, Anal. Chem., 2019, 91, 482–504 CrossRef CAS PubMed.
- K. R. Wehmeyer, R. J. White, P. T. Kissinger and W. R. Heineman, Annu. Rev. Anal. Chem., 2021, 14, 109–131 CrossRef CAS PubMed.
- J. Rexha, N. Perta, A. Roscioni, S. Motta, A. La Teana, L. Maragliano, A. Romagnoli and D. Di Marino, Adv. Sensor Res., 2023, 2, 2300053 CrossRef CAS.
- T. T.-H. Nguyen, C. M. Nguyen, M. A. Huynh, H. H. Vu, T.-K. Nguyen and N.-T. Nguyen, J. Nanobiotechnol., 2023, 21, 411 CrossRef CAS PubMed.
- J. D. Driskell, J. M. Uhlenkamp, R. J. Lipert and M. D. Porter, Anal. Chem., 2007, 79, 4141–4148 CrossRef CAS PubMed.
- Y. Wang, R. Vaidyanathan, M. J. A. Shiddiky and M. Trau, ACS Nano, 2015, 9, 6354–6362 CrossRef CAS PubMed.
- D. Zhang, Y. Xie, S. K. Deb, V. J. Davison and D. Ben-Amotz, Anal. Chem., 2005, 77, 3563–3569 CrossRef CAS PubMed.
- Y. Wang, M. Salehi, M. Schütz, K. Rudi and S. Schlücker, Analyst, 2013, 138, 1764–1771 RSC.
- Y. Wang, S. Rauf, Y. S. Grewal, L. J. Spadafora, M. J. A. Shiddiky, G. A. Cangelosi, S. Schlücker and M. Trau, Anal. Chem., 2014, 86, 9930–9938 CrossRef CAS PubMed.
- A. C. Crawford, A. Skuratovsky and M. D. Porter, Anal. Chem., 2016, 88, 6515–6522 CrossRef CAS PubMed.
- S. Srivastav, A. Dankov, M. Adanalic, R. Grzeschik, V. Tran, S. Pagel-Wieder, F. Gessler, I. Spreitzer, T. Scholz, B. Schnierle, O. E. Anastasiou, U. Dittmer and S. Schlücker, Anal. Chem., 2021, 93, 12391–12399 CrossRef CAS PubMed.
- R. Xiao, L. Lu, Z. Rong, C. Wang, Y. Peng, F. Wang, J. Wang, M. Sun, J. Dong, D. Wang, L. Wang, N. Sun and S. Wang, Biosens. Bioelectron., 2020, 168, 112524 CrossRef CAS PubMed.
- Y. Joung, K. Kim, S. Lee, B.-S. Chun, S. Lee, J. Hwang, S. Choi, T. Kang, M.-K. Lee, L. Chen and J. Choo, ACS Sens., 2022, 7, 3470–3480 CrossRef CAS PubMed.
- W. A. Hassanain, J. Spoors, C. L. Johnson, K. Faulds, N. Keegan and D. Graham, Analyst, 2021, 146, 4495–4505 RSC.
- Q. Yu, H. D. Trinh, Y. Lee, T. Kang, L. Chen, S. Yoon and J. Choo, Sens. Actuators, B, 2023, 382, 133521 CrossRef CAS PubMed.
- J. Ou-Yang, C.-Y. Li, Y.-F. Li, J. Fei, F. Xu, S.-J. Li and S.-X. Nie, Sens. Actuators, B, 2017, 240, 1165–1173 CrossRef CAS.
- M. Malak, J. James, J. Grantham and M. B. Ericson, Sci. Rep., 2022, 12, 16584 CrossRef CAS PubMed.
- S. Lee, H. Chon, J. Lee, J. Ko, B. H. Chung, D. W. Lim and J. Choo, Biosens. Bioelectron., 2014, 51, 238–243 CrossRef CAS PubMed.
- S. Diao, G. Hong, A. L. Antaris, J. L. Blackburn, K. Cheng, Z. Cheng and H. Dai, Nano Res., 2015, 8, 3027–3034 CrossRef CAS.
- Z. A. Nima, M. Mahmood, Y. Xu, T. Mustafa, F. Watanabe, D. A. Nedosekin, M. A. Juratli, T. Fahmi, E. I. Galanzha, J. P. Nolan, A. G. Basnakian, V. P. Zharov and A. S. Biris, Sci. Rep., 2014, 4, 4752 CrossRef CAS PubMed.
- E. Feng, T. Zheng, X. He, J. Chen and Y. Tian, Sci. Adv., 2018, 4, eaau3494 CrossRef CAS PubMed.
- S. C.-H. Tsao, J. Wang, Y. Wang, A. Behren, J. Cebon and M. Trau, Nat. Commun., 2018, 9, 1482 CrossRef PubMed.
- Y. Zhang, X.-P. Wang, S. Perner, A. Bankfalvi and S. Schlücker, Anal. Chem., 2018, 90, 760–768 CrossRef CAS PubMed.
- Y. Wang, S. Kang, A. Khan, G. Ruttner, S. Y. Leigh, M. Murray, S. Abeytunge, G. Peterson, M. Rajadhyaksha, S. Dintzis, S. Javid and J. T. C. Liu, Sci. Rep., 2016, 6, 21242 CrossRef CAS PubMed.
- Y. W. Wang, N. P. Reder, S. Kang, A. K. Glaser, Q. Yang, M. A. Wall, S. H. Javid, S. M. Dintzis and J. T. C. Liu, Cancer Res., 2017, 77, 4506–4516 CrossRef CAS PubMed.
- M. Gellner, D. Steinigeweg, S. Ichilmann, M. Salehi, M. Schütz, K. Kömpe, M. Haase and S. Schlücker, Small, 2011, 7, 3445–3451 CrossRef CAS PubMed.
- A. Dankov, L. Hensen, M. Aydin, N. Choi, H. Giesler, D. Toker, S. Schlücker and R. Grzeschik, ChemRxiv, 2023, preprint DOI:10.26434/chemrxiv-2023-rpg77.
- D. Steinigeweg, M. Schütz, M. Salehi and S. Schlücker, Small, 2011, 7, 2443–2448 CrossRef CAS PubMed.
- J. Jeon, S. H. Lee, Y. Joung, K. Kim, N. Choi and J. Choo, Sens. Actuators, B, 2020, 321, 128521 CrossRef CAS.
- J. J. Miller and R. Valdes, Jr., Clin. Chem., 1991, 37, 144–153 CrossRef CAS.
- E. Güven, K. Duus, M. C. Lydolph, C. S. Jørgensen, I. Laursen and G. Houen, J. Immunol. Methods, 2014, 403, 26–36 CrossRef PubMed.
- I. Buchwalow, V. Samoilova, W. Boecker and M. Tiemann, Sci. Rep., 2011, 1, 28 CrossRef PubMed.
- X. Gao, P. Zheng, S. Kasani, S. Wu, F. Yang, S. Lewis, S. Nayeem, E. B. Engler-Chiurazzi, J. G. Wigginton, J. W. Simpkins and N. Wu, Anal. Chem., 2017, 89, 10104–10110 CrossRef CAS PubMed.
- C. Stavnsbjerg, J. S. Jørgensen, T. B. Engel, A. Brus, L. Ringgaard, A. E. Hansen, A. Kjaer and T. L. Andresen, J. Immunol. Methods, 2022, 500, 113177 CrossRef CAS PubMed.
- S. W. Graves, K. Sharma and A. B. Chandler, Clin. Chem., 1986, 32, 1506–1509 CrossRef CAS.
- A. R. Mire-Sluis, Y. C. Barrett, V. Devanarayan, E. Koren, H. Liu, M. Maia, T. Parish, G. Scott, G. Shankar, E. Shores, S. J. Swanson, G. Taniguchi, D. Wierda and L. A. Zuckerman, J. Immunol. Methods, 2004, 289, 1–16 CrossRef CAS PubMed.
- R. F. Vining, P. Compton and R. McGinley, Clin. Chem., 1981, 27, 910–913 CrossRef CAS.
- P. B. Williams, J. H. Barnes, S. L. Szeinbach and T. J. Sullivan, J. Allergy Clin. Immunol., 2000, 105, 1221–1230 CrossRef CAS PubMed.
- W. Hao, Y.-X. Pan, Y.-Q. Ding, S. Xiao, K. Yin, Y.-D. Wang, L.-W. Qiu, Q.-L. Zhang, C. Y. Woo Patrick, K. P. Lau Susanna, K.-Y. Yuen and X.-Y. Che, Clin. Vaccine Immunol., 2008, 15, 194–202 CrossRef CAS PubMed.
- R. Ahirwar, S. Bariar, A. Balakrishnan and P. Nahar, RSC Adv., 2015, 5, 100077–100083 RSC.
- F. A. Dourbash, A. A. Shestopalov and L. J. Rothberg, Sens. Actuators, B, 2022, 360, 131657 CrossRef CAS.
- A. A. Hariri, S. S. Newman, S. Tan, D. Mamerow, A. M. Adams, N. Maganzini, B. L. Zhong, M. Eisenstein, A. R. Dunn and H. T. Soh, Nat. Commun., 2022, 13, 5359 CrossRef CAS PubMed.
- A. K. Trilling, J. Beekwilder and H. Zuilhof, Analyst, 2013, 138, 1619–1627 RSC.
- S. Gao, J. M. Guisán and J. Rocha-Martin, Anal. Chim. Acta, 2022, 1189, 338907 CrossRef CAS PubMed.
- H. Chen, S.-G. Park, N. Choi, H.-J. Kwon, T. Kang, M.-K. Lee and J. Choo, ACS Sens., 2021, 6, 2378–2385 CrossRef CAS PubMed.
- J. Li, A. Wuethrich, A. A. I. Sina, H.-H. Cheng, Y. Wang, A. Behren, P. N. Mainwaring and M. Trau, Nat. Commun., 2021, 12, 1087 CrossRef CAS PubMed.
- Z. Cheng, N. Choi, R. Wang, S. Lee, K. C. Moon, S.-Y. Yoon, L. Chen and J. Choo, ACS Nano, 2017, 11, 4926–4933 CrossRef CAS PubMed.
- Y. Wang, S. Zong, L. Wu, Y. Zhang, Z. Wang, Z. Wang, B. Chen and Y. Cui, ACS Appl. Mater. Interfaces, 2018, 10, 24999–25005 CrossRef CAS PubMed.
- S. Schlücker, M. D. Schaeberle, S. W. Huffman and I. W. Levin, Anal. Chem., 2003, 75, 4312–4318 CrossRef PubMed.
- S. E. Bohndiek, A. Wagadarikar, C. L. Zavaleta, D. Van de Sompel, E. Garai, J. V. Jokerst, S. Yazdanfar and S. S. Gambhir, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12408–12413 CrossRef CAS PubMed.
- M. Kögler and B. Heilala, Meas. Sci. Technol., 2020, 32, 012002 CrossRef.
- X. Gao, J. Boryczka, S. Kasani and N. Wu, Anal. Chem., 2021, 93, 1326–1332 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |