Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biomedical SERS – the current state and future trends
Dana
Cialla-May
*ab,
Alois
Bonifacio
 c,
Thomas
Bocklitz
c,
Thomas
Bocklitz
 abd,
Alexey
Markin
abd,
Alexey
Markin
 e,
Natalia
Markina
e,
Natalia
Markina
 e,
Stefano
Fornasaro
e,
Stefano
Fornasaro
 f,
Aradhana
Dwivedi
ab,
Tony
Dib
ab,
Edoardo
Farnesi
f,
Aradhana
Dwivedi
ab,
Tony
Dib
ab,
Edoardo
Farnesi
 b,
Chen
Liu
ab,
Arna
Ghosh
ab and
Juergen
Popp
b,
Chen
Liu
ab,
Arna
Ghosh
ab and
Juergen
Popp
 ab
ab
aLeibniz Institute of Photonic Technology, Member of Leibniz Health Technologies, Member of the Leibniz Centre for Photonics in Infection Research (LPI), Albert-Einstein-Straße 9, 07745 Jena, Germany. E-mail: dana.cialla-may@leibniz-ipht.de
bInstitute of Physical Chemistry (IPC) and Abbe Center of Photonics (ACP), Friedrich Schiller University Jena, Member of the Leibniz Centre for Photonics in Infection Research (LPI), Helmholtzweg 4, 07743 Jena, Germany
cDepartment of Engineering and Architecture, University of Trieste, Via Alfonso Valerio 6, 34127 Trieste (TS), Italy
dFaculty of Mathematics, Physics and Computer Science, University of Bayreuth (UBT), Nürnberger Straße 38, 95440 Bayreuth, Germany
eInstitute of Chemistry, Saratov State University, Astrakhanskaya Street 83, 410012 Saratov, Russia
fDepartment of Chemical and Pharmaceutical Sciences, University of Trieste, Via Licio Giorgieri 1, 34127 Trieste (TS), Italy
First published on 7th August 2024
Abstract
Surface enhanced Raman spectroscopy (SERS) is meeting the requirements in biomedical science being a highly sensitive and specific analytical tool. By employing portable Raman systems in combination with customized sample pre-treatment, point-of-care-testing (POCT) becomes feasible. Powerful SERS-active sensing surfaces with high stability and modification layers if required are available for testing and application in complex biological matrices such as body fluids, cells or tissues. This review summarizes the current state in sample collection and pretreatment in SERS detection protocols, SERS detection schemes, i.e. direct and indirect SERS as well as targeted and non-targeted SERS, and SERS-active sensing surfaces. Moreover, the recent developments and advances of SERS in biomedical application scenarios, such as infectious diseases, cancer diagnostics and therapeutic drug monitoring is given, which enables the readers to identify the sample collection and preparation protocols, SERS substrates and detection strategies that are best-suited for their specific applications in biomedicine.
Alois Bonifacio studied Chemistry at the University of Trieste, and obtained his PhD at the Vrije Universteit Amsterdam on surface enhanced Raman spectroscopy of heme proteins. From 2007, he became post-doc and then assistant professor at the University of Trieste, where he is now associate professor of chemistry. His research interest is the application of Raman and SERS spectroscopy to complex biological samples, with the purpose of developing clinical applications. Stefano Fornasaro obtained a degree in pharmacy and then earned a PhD in nanotechnology from the University of Trieste. He is currently an assistant professor of analytical chemistry at the same university. His interests lie in chemometrics and SERS spectroscopy, with a specific focus on their applications in the biomedical and bioanalytical fields. |
Key learning points• The experimental parameters affecting a SERS measurement for a biological samples• The difference among different detection strategies and how to choose them evaluating pros and cons. • The possibilities and limits of SERS bioanalysis for various biological samples and diseases. • How SERS can tackle the issue of chemical complexity of biological samples and or matrix interference. |
1. Introduction
Biomedical science requires highly sensitive and specific analytical tools. Whereas gold standard methods (such as gas and high performance liquid chromatography combined with mass spectrometric detection) often require sending the samples to centralized labs, surface enhanced Raman spectroscopy (SERS), especially when combined with a streamlined sample pre-treatment and portable instrumentation, is an excellent candidate for point-of-care-testing (POCT). The attractiveness of SERS in analytical science is due to its molecular specificity combined with the sensitivity enabled by the signal-enhancing plasmonic nanostructures, allowing detection limits that in some cases can reach the single molecule regime.1–3 One of the first biomedical applications of SERS include (i) the investigation of the intercalation of antitumor drugs into DNA double strands to draw conclusions about the conformation within the DNA duplex, (ii) the analysis of sialic acid residues on living cells to shed light into the biological activity of inhibiting viruses, as well as (iii) the identification of drugs as complexed or free form within the cell interior.4 Since then, SERS was further developed by increasing the number and type of SERS-active substrates and probes, their functionalization with biocompatible layers, and their application towards in vivo investigations,5 POCT6,7 or clinical decision-making processes.For instance, tumor margins could be identified in vivo thanks to a specific enrichment in the tumor tissues of bright SERS-active metallic nanoparticles conjugated with Raman reporter molecules, in some cases even using an endoscope.5,7 Similar concepts are applied in drug delivery and photothermal therapy within mouse models, where the decrease in SERS signal intensity over time illustrates the release of the drug or the accumulation of nanoparticles at the target site allowing for treatment with thermal energy. However, before its application in humans, the assessment of the toxicity of these SERS probes and safe-by-design concepts are essential. The investigation of cells by applying SERS-based methods shed light into their microenvironment, and will be important to understand the development of diseases on a cellular level in future.7
Although a qualitative or semi-quantitative detection is enough for many biomedical SERS applications such as identifying of tumor margins or cancer sites, other applications require the use of quantitative methods, as in the case of therapeutic drug monitoring. Since a reliable quantification is strongly correlated with the signal-enhancing performance of the specific SERS substrate, the use of internal standards is very effective to address temporal or spatial substrate-related variability of SERS intensity, e.g. due to the aggregation state in colloids or variations in nanostructured sensor surfaces from spot-to-spot or batch-to-batch.2 Finally, the prediction of the target analyte's concentration in biomedical and clinical detection schemes needs to be validated to confirm the accuracy of the developed SERS-based detection scheme.8 In quantitative SERS, several figures of merits need to be estimated to allow for a final successful application in a clinical environment:8,9 (i) the accuracy of the detection schemes within the clinically relevant concentration range, e.g. the required drug dose or the expected biomarker concentration; (ii) the limit of detection (LOD), which should be lower than the actual required sensitivity for the envisioned application scenario; (iii) the apparent recovery rate, i.e. the ratio of estimated and real concentration, which is related with a validation against the gold standard; (iv) the accuracy of the prediction via its root mean square error. It needs to be mentioned that these figures of merits are dependent on the quality of the SERS-active substrates and might vary between labs when using different Raman systems. The more reproducible the SERS substrate is, the more precise the concentration prediction is. In qualitative or semi-quantitative detection for clinical applications, however, the requirements for the reproducibility of a SERS substrate are often less strict.
The necessity of a methodology to compare quantitative SERS results acquired by various Raman systems but using the same SERS substrate and sample preparation protocol was addressed in a European inter-laboratory study.9 An inter-laboratory comparison is of high importance towards standardization of SERS-based detection schemes, not only in the framework of biomedical and clinical application scenarios, but also for a wider acceptance of SERS as a reliable quantitative technique in analytical chemistry. Within the study, concentration-dependent SERS spectra of adenine were recorded employing colloidal and solid SERS substrates, which were distributed among the participants. As a result, it was shown that the root mean square error of prediction (RMSEP) was between 13 and 29%, depending on the specific method considered, i.e. SERS substrate and excitation wavelength, which is promising for future applications of SERS on a large scale. Despite the point marked by this collaborative effort, the biomedical SERS community still lacks widely accepted standardized protocols. Hopefully other inter-laboratory studies will follow, perhaps using the same or a similar methodology as the one used by that study to report results. The need for benchmarks in SERS have increasingly been recognized by various authors,10,11 setting this goal as one of the most important for the coming years.
To summarize, SERS is available for both qualitative as well as quantitative analysis, the latter requiring more effort. This review article aims at introducing recent developments mainly from the last 3 years and advances of SERS in biomedical application scenarios, and at guiding the readers to identify the sample collection and preparation protocols (Section 2), SERS substrates (Section 3) and detection strategies (Section 4) that are best-suited for their specific applications. Moreover, Section 5 provides work flow for data analysis of SERS data by means of artificial intelligence-based approaches, and Section 6 summarizes recent examples from the field of SERS-based detection in biomedicine.
2. Sample collection and pretreatment for a successful SERS detection
SERS has proven to be a versatile and valuable tool for the ex vivo and in vitro analysis of a wide variety of clinical and pre-clinical biological matrices, e.g. biofluids, extracts, or tissues, in various physiological and pathological states. In this section, we present an overview of the samples that can be successfully analyzed using SERS as well as a discussion of the distinctive benefits and difficulties associated with each type of matrix and specimen.Biological fluids, including blood, serum, plasma, urine, and saliva, were the most frequently analyzed samples. This is because of the ease with which these samples can be collected, as well as their high relevance in the clinical setting as a partly noninvasive and cost-effective route for obtaining bio-analytes and biomarkers for diagnosis and monitoring. SERS analysis of biofluids can be pursued by both direct and indirect detection (see Section 4), enabling the identification of a specific biochemical signature or biomarker that characterizes the pathobiology and response to therapy for infectious diseases, inflammation, metabolic disorders, and cancer. Moreover, SERS detection and quantitation in biofluids of exogenous agents, such as bacterial and viral loads, drugs, and related metabolites, are becoming increasingly important for assessing the severity of an active infection or for understanding the pharmacokinetics of selected drugs within the framework of personalized treatment. SERS detection is favorable in aqueous environments. Therefore, it is common practice to monitor the intra and extracellular microenvironment of cultured and collected cells by analyzing cell lysates or various culture media.12,13
Serum and plasma are liquid portions of blood and serve as the primary specimens of clinical interest. Blood collection is a commonly performed clinical procedure that is largely standardized, although some variations in collection protocols can occur. Blood samples are typically obtained by venipuncture, and subsequent centrifugation allows for the collection of separated serum and plasma samples. Serum offers an advantage over plasma in terms of the absence of spectral interference caused by platelets, clotting factors, and anticoagulants.14 Urine is easily obtained and is a rich source of information about kidney and liver function, metabolic disorders, and specific biomarkers related to several conditions. Saliva and crevicular fluids are other valuable biofluids that can be easily collected on paper substrates and do not require intrusive procedures or the involvement of specialized medical staff, making them ideal media for SERS detection of various internal metabolites in fields such as oral health monitoring, hormone profiling, and the detection of cancer and infectious diseases.15,16 Sweat is another biofluid that possesses diagnostic potential. An interesting approach using SERS is the direct analysis of sweat using a flexible plasmonic meta surface to capture distinctive spectral patterns.17 Cerebrospinal fluid (CSF) samples have been successfully subjected to SERS analysis to diagnose neurological disorders and central nervous system infections.18,19 However, a CSF sample can only be collected by a specially trained doctor performing a lumbar puncture, an invasive technique.
The interstitial fluid (ISF), which exists between blood vessels and cells, is similar to plasma, and numerous studies have explored its use in conjunction with SERS and microneedle arrays (MN), creating minimally invasive platforms for chemical sensing. An example of that is the continuous glucose monitoring carried out by skin-worn MN-based biosensors,20 where the ISF can be utilized as a rich biomarker source to monitor glucose levels in diabetes patients (Fig. 1 left).21 An MN-assisted paper-based sensing platform has been developed22 that combines rapid, sufficient extraction of ISF with good structural integrity and enables painless biofluid analysis with ultrasensitive molecular recognition capacity. Notably, the analyte molecules in the needles can be recovered into a moist cellulose paper through spontaneous diffusion. More importantly, the paper can be functionalized with enzymatic colorimetric reagents or a plasmonic array, enabling a desired detection capacity of a diverse set of molecules (cefazolin, nicotine, paraquat, methylene blue) whose levels could vary significantly in response to metabolism (Fig. 1 right).22 The minimally invasive sensing systems could have great benefit in precision medicine, especially for personal healthcare monitoring.
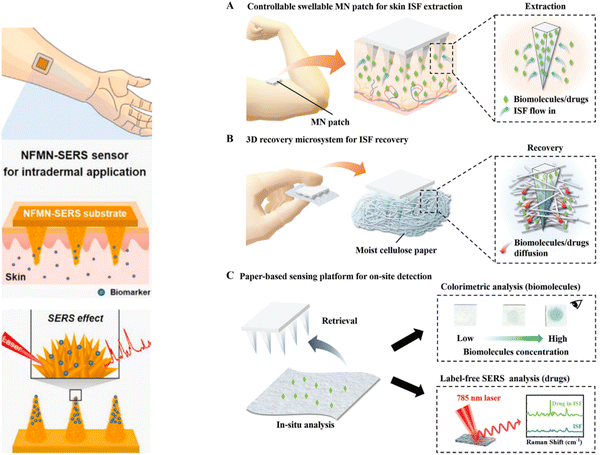 | ||
| Fig. 1 (Left) Microneedle patches for biopsy sampling and biosensing applications. Schematic of intradermal application of the MN-SERS sensor. Inset in (v) represents the flower-like nanostructure formed on the microneedle. Used with permission of [Elsevier Science & Technology Journals], from ref. 21; permission conveyed through Copyright Clearance Center, Inc. (Right) Schematic representation of a rapid, minimally invasive, ultrasensitive detection system for healthcare monitoring. (A) Extraction of ISF from skin tissue using the csMN patch. (B) Recovery of biomolecules and drugs from the ISF-extracted MN patch using a 3D recovery microsystem. (C) Optical analysis of biomolecules or drugs using paper-based sensing platforms. Used with permission of John Wiley & Sons – Books, from ref. 22; permission conveyed through Copyright Clearance Center, Inc. | ||
Lacrimal fluid—that makes up the liquid of tears—is another body fluid tested in health monitoring applications using SERS. For example, SERS contact lens materials (SERS-LM) have been developed for noninvasive selective glucose sensing in human tears.23 The tear's glucose levels measured by these contact lenses before and after a meal followed the corresponding blood glucose levels, suggesting their potential application as wearable biosensors.23 The synovial fluid (SF) is also worth mentioning in this context, which is produced by the connective tissue lining the joints. It functions as cartilage nourishment and joints lubricant for which lubricin, a glycocomposite binded to hyaluronan (HA), plays a major role. As a decrease in HA is associated with a loss of SF viscoelastic properties in the onset of osteoarthritis, it could constitute a potential biomarker for the early stages of the disease. Human SF derived from knee osteoarthritis patients has been examined using combined SERS and resonant Raman spectra to discriminate between low- and high-grade groups, and has achieved high classification accuracy.24
Moisoiu et al. identified three types of SERS protocols that are usually employed for the analysis of biofluids: (1) the mixing of biofluids with metal colloids, and the subsequent SERS analysis of the sample in its liquid form for the detection of purine metabolites, i.e. “liquid SERS”, (2) an analogous protocol but with the use of iodide-modified substrate, for the detection of proteins, (3) the SERS analysis of dried drops of biofluids–metal colloids mixtures for the detection of both purine metabolites and proteins, i.e. “dried SERS”. Many diseases cause perturbations to cellular turnover rate or inflammation, so that many experimental data observed in SERS liquid biopsy research make sense in this context. An altered purine metabolism, and in particular elevated uric acid levels, in addition to being a marker of cellular turnover, is linked to specific illnesses like gout or cardiovascular conditions, but it is also proposed as a protection against neurodegenerative disorders.14
Human exhaled breath carries a wealth of information about human health, with breath volatile organic compounds (VOCs) serving as excellent biomarkers for noninvasive screening of a variety of cancers (including stomach, lung, and oral cancer) as well as lung-related disorders such as COVID-19.25–28 The relative concentration of breath VOCs varies significantly with the progression of a disease and is distinct from the VOCs produced during a normal physiological activity. However, these changes are often as low as a sub-ppb (parts per billion). In this regard, SERS breath analysis has recently emerged as a potential tool for a routine VOC-based illness detection platform. The fundamental issue for SERS analysis of exhaled breath is the detection of gas molecules, which are difficult to enrich and capture in SERS “hotspot” regions of substrates. Many solutions have been proposed to address this issue, with the most successful being the combination of noble metal nanoparticles with porous materials (such as graphene oxide28 or metal–organic framework (MOF)27) or the construction of three-dimensional (3D) SERS substrates.26
In many biological specimens, a specific analyte of interest is typically found alongside hundreds or even thousands of other chemical compounds (referred to as the “matrix”), which can potentially interfere with its detection. Furthermore, analytes concentrations are often much lower than that of many other potentially interfering substances. This situation holds true for most analytical techniques, and it's not specific to SERS. To address this “matrix issue” effectively, a practical solution is to reduce the chemical complexity through sample pretreatment before conducting the analysis. In SERS, interfering species from the matrix can either compete for “hot spots” or disrupt the interaction between a recognition element and the analyte. Ideally, SERS has the advantage that sample pretreatment can be minimized if the analyte exhibits an exceptionally strong affinity for the metal substrate or when the recognition element in indirect detection is highly selective.29 In the former case, an analyte can be directly detected even in the presence of a complex matrix because its SERS signal dominates over most biomolecules. This is the case, for instance, of molecules having a sulfur atom capable of binding Au and Ag surface, such as thiopurine drugs or the amino acid ergothioneine, both of which can be easily detected with SERS even when found together with many other biomolecules in complex biological matrices (e.g. cell lysates, serum seminal plasma).30,31 Additionally, when metal colloids are used as substrates, the matrix can interfere with hot-spot formation, for example, by forming large protein coronas around nanoparticles. Generally, analytical approaches that heavily rely on sample pretreatment are less desirable, as they shift complexity from the actual SERS detection to a pre-analytical step. Although there is space for improving the efficiency of such pretreatments, there are currently time-consuming, and in many cases their development remains experimentally challenging. Simpler one- or two-step pretreatment methods, like de-proteinization (e.g. by centrifugation, filtration, or precipitation), dilution and solvent extraction, often provide a reasonable compromise to reduce matrix complexity to a level that permits SERS detection of desired targets. De-proteinization solves the hot-spots formation issues when using metal colloids, and prevents the protein fouling of the metal nanostructure used as substrate in the case of solid SERS substrates. Dilution, by lowering the concentration of proteins and other matrix component, can be an effective method to minimize matrix interferences. However, it works best with analytes who show a strong affinity for the SERS substrates, as their concentration is lowered as well. Target analytes can be concentrated with the help of solvent extraction and liquid–liquid extraction (LLE). By exploiting the difference in solubility or partition coefficient between the target analyte and an organic solvent, it is possible to selectively extract the molecules of interest into a smaller volume of a more suitable solvent.32 It is essential to recognize that sample pretreatment strategies may vary depending on the specific research or clinical goal. The key to the successful SERS analysis of biofluids lies in tailoring the sample preparation and pretreatment steps to overcome the inherent challenges while preserving the integrity of the biological specimen. This concept has been recently explored in a comprehensive review.32
3. SERS substrates in biomedical SERS
As in most SERS applications, the performance of SERS in biomedical analysis is highly dependent on the plasmonic properties of SERS substrates. Therefore, the design, exploration, and construction of high-performance SERS-active nanostructures remain crucial in developing the SERS technology. An ideal SERS substrate must be biocompatible, reproducible, and robust. Overcoming the challenges of biomedical SERS, such as limited functionality and target selectivity as well as mitigation of complex matrix interferences, is also critical.33 The development of SERS substrates with increasing complexity has gone through many stages. Various materials and structures, including single and multi-component metals, semiconductors, 2D materials, and composite nanostructures, have been explored as potential SERS substrates.34 In biomedical applications, especially for molecules whose fluorescence signal is in the visible range of wavelengths or which are prone to photodegradation, the SERS substrate should ideally be suitable for excitation by lasers with wavelengths in the red-near IR region (e.g. 633, 785 or 830 nm) to avoid the generation of an interfering fluorescence background and the destruction of the analyte.35 In addition, lasers at these wavelengths achieve high signal enhancements with all coinage metals used for SERS substrates preparation (see Fig. 2), as they match their localized surface plasmon resonances.36 This section provides a brief introduction to SERS substrates in biomedical applications, along with references to the appropriate literature for more in-depth information on each substrate category. An illustrative summary of the various approaches for SERS substrates is provided in Fig. 3.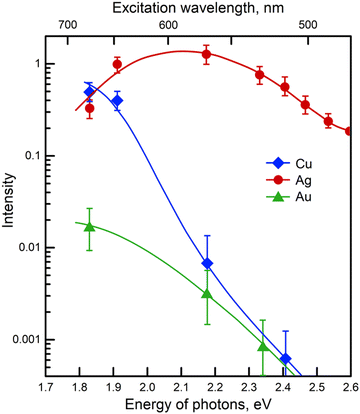 | ||
| Fig. 2 Excitation profiles from copper, silver, and gold films deposited under ultrahigh vacuum. Pyridine was used as the test analyte. Adapted with permission from ref. 36. | ||
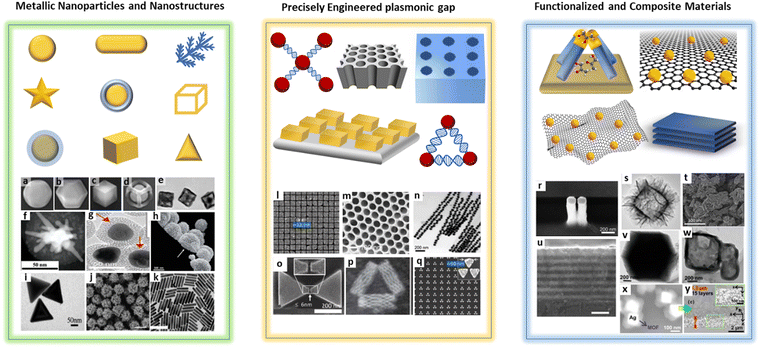 | ||
| Fig. 3 Schematic illustration and respective SEM images of different types of plasmonic SERS substrates, (a) gold nanoparticle, (b) gold@silver shell, (c) gold@silver cage, (d) gold@silver frame, (e) hollow gold nanocubes, (f) gold nanostar, reprinted from ref. 37, Copyright 2022, with permission from Elsevier, (g) gap separated core–satellite nanoparticle33 (h) mesoporous gold nanoparticle38 (i) gold nanotriangle (j) sea urchin-shaped gold particles (k) gold nanorods, used with permission of Royal Society of Chemistry, from ref. 39, permission conveyed through Copyright Clearance Center, Inc., (l) and (q) diverse gold structures with nanogaps, (o) bowtie nanoantenna, reprinted from ref. 40, Copyright 2023, with permission from Elsevier, (m) porous gold substrate and (p) triangular DNA origami38 (n) gap separated linear chain of gold nanoparticles33 (r) gold nanopillar on fused silica, reprinted from ref. 37, Copyright 2022, with permission from Elsevier, (s) amorphous ZnO nanocage (t) semiconductor–metal hybrid (Au@DFH-4T) nanostructure (u) atomic-layer WTe2 and MoTe2 substrate41 (v) metal–organic framework (ZIF-67) and (w) porous metal–organic framework, used with permission of Royal Society of Chemistry, from ref. 34, permission conveyed through Copyright Clearance Center, Inc., (x) and (y) silver with a metal–organic framework (Ag@MOF), reprinted from ref. 37, Copyright 2022, with permission from Elsevier. | ||
Metallic nanostructures made of gold and silver are widely used because of their adaptability and ease of synthesis. These nanostructures can be tailored into various shapes and sizes, allowing their properties to be modified. Moreover, they can be deposited on solid supports, using various bottom-up approaches, to prepare solid SERS substrates. Bottom-up synthesis involves the creation of larger systems from smaller building blocks such as atoms, molecules, polymers or nanoparticles.42 Chemical synthesis,43,44 template-based methods,45 optical printing46 and self-assembly47 are some of the techniques used for bottom-up approaches. This synthesis approach is cost effective and more accessible without the requirement of heavy instruments or cleanroom facilities. In addition, they offer precise control over morphology and composition to achieve denser hot spots relevant to SERS. Despite these advantages, they sometimes require multi-step synthesis protocols, have limited scalability and are subject to lab-to-lab variability.48 Spherical nanoparticles made of silver and gold have traditionally been the preferred choice in SERS-based applications. However, a wide range of structures have been developed, including anisotropic nanostructures such as nanorods, nanostars, nanocubes, and nanotriangles with sharp edges or corners. All these metal nanoparticles are able to generate intense localized electromagnetic fields upon illumination with a laser having an adequate wavelength, because of their plasmonic properties. When nanoparticles clusters are formed, particularly intense electromagnetic fields are generated at the nano-sized gaps between particles (“hot spots”), enhancing the intensity of the Raman scattering for those molecules located there. Their plasmonic profile can be tuned by adjusting their size and shape and various excitation wavelengths in the visible and NIR region are accessible to take advantage from additional resonance Raman enhancement or to avoid fluorescence background by using the best suited excitation wavelength.39 Complex three-dimensional plasmonic structures, such as nanocages, nanoframes, and bimetallic nanostructures, have the ability to enhance signals by trapping the analytes and exploiting the combined properties of different metals.37 Different aggregating agents can be utilized to increase the reproducibility and enhancement factors in the hot spots. The main factors impacting SERS sensitivity are nanoparticles size, shape, and choice of metal. New SERS substrates should always be compared with the most frequently used ones such as citrate-stabilized Ag or AuNPs.
Precisely engineered plasmonic nanostructures with hot spots-generating nanogaps, such as nanogratings and nanogrooves, offer explicit control over Raman signal enhancement. These structures are created by top–down techniques such as photolithography, electron-beam lithography, focused ion beam lithography, etc. ensuring a well-defined and ordered surface. One major challenge of these substrates is capturing and placing the analyte at the plasmonic nanogaps. The fabrication strategies for sub-10 nm gap-engineered substrates and techniques for placing analytes within these gaps were thoroughly discussed.40 Combined with DNA templates, metal-based nanostructures can create plasmonic gaps as small as 1 nm and can reach single molecule sensitivity. Utilizing specific DNA scaffold sequences, it is possible to create DNA origami-based substrates with programmable 2D or 3D nanostructures and controlled nanogaps.49 These techniques are sophisticated, more complex, and require expert facilities (e.g., cleanroom). Due to the hotspot localization, usually, these substrates offer better signal reproducibility and uniformity.
Inorganic materials such as MXene, graphene, and semiconductor materials like TiO2 and ZnO have attracted attention due to their large surface area and molecular adsorption capabilities, as well as enhancing the charge transfer between substrate and analyte.41 Plasmonic nanostructures with sophisticated optical properties were functionalized with these materials showing well-studied electronic properties to combine the tailored SERS applications with reproducibility and stability. Metal–organic frameworks (MOFs), hybrids of organic–inorganic materials, offer precise control over porosity, periodicity of metallic nodes, and functionalization, making them promising substrates.38 These new types of substrates have the ability to overcome the challenges of SERS such as reproducibility, reusability, longer shelf life, cost-effective fabrication, and increased adsorption of the analyte. They can excel in achieving both maximum charge transfer and maximum plasmonic enhancement.
To summarize, there is no one-size-fits-all SERS substrate for all envisioned applications in biomedicine and other detection schemes, but the SERS substrate must be designed, adapted, and modified to meet the needs of the user for the designated application field. Furthermore, material choices are largely dependent on the given application. To achieve maximum sensitivity and reliability in biomedical applications, it is recommended that a substrate has intra- and inter-batch reproducibility with a standard deviation of less than 10%, and that it is free of impurities that cause an interfering signal overlap with the target analyte. Ideally, SERS substrates should fulfill all the above-mentioned requirements, but in reality, one often needs to compromise on some features in accordance with their desired application. For example, quantitative assays in biomedical applications and therapeutic drug monitoring require a robust, uniform, and reproducible detection rather than the most sensitive detection that is required in forensic applications.
When evaluating the use of SERS in biomedical applications, various molecules such as drugs, nucleic acids, proteins, viruses, and bacteria are potential analytes of interest. The selection of SERS substrates and excitation wavelength should be determined based on the type of target analyte. For instance, a substrate with a nanogap is better suited to detect smaller molecules like metabolites or drugs, but cannot be used for the analysis of larger macromolecules or cells due to restricted access to the nanogap. For larger macro-molecules or cells and when using imaging-based methods such as tissue and cell imaging, colloidal SERS substrates are more desirable. However, colloidal substrates also present some drawbacks, including in the possibility of a non-uniform hot spot distribution, uncontrolled aggregation, and the generation of a signal with limited intensity reproducibility. On the other hand, hybrid methods incorporating plasmonic materials functionalized with inorganic substrates such as MXene and 2D materials have also shown some promise as viable options. In the future, multidisciplinary strategies will drive the design of substrates, focusing on careful design for precise and minimally invasive biomedical analysis.
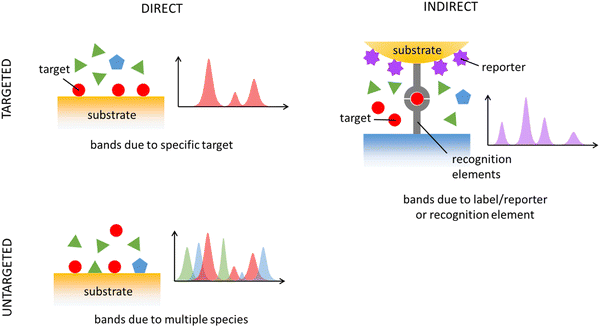
4. Detection strategies of SERS in bioanalytics
The detection and quantification of analytes with SERS can be achieved through two approaches: direct and indirect. In direct detection, analyte-specific bands are directly sought, while in indirect detection, the focus is on observing bands of a different, well-characterized molecule connected to the analyte, often referred to as a “reporter” or “label.” These bands of these reporters can either appear/disappear or shift when the analyte interacts with them. Direct detection is known as “label-free” as it doesn't rely on additional labels. However, since an interaction between the analyte and metal nanostructures is still needed, some authors prefer the term “direct” over “label-free” to avoid ambiguity. Both detection strategies have pros and cons. The indirect approach helps address the inherent complexity of SERS by using well-known reporters with predictable and strong SERS spectra, providing researchers with better control over the system. However, this control comes with a trade-off: typically, the reporter must be linked to the analyte, either through a direct covalent bond or by using a recognition element like an antibody or aptamer. An overview of direct/indirect and targeted/untargeted approaches is schematically depicted in Fig. 4. We will first consider relevant aspects of direct approaches (both targeted and untargeted), and then continue to those involving an indirect targeted strategy.Direct SERS detection relies on a favorable analyte–substrate interaction, typically influenced by the analyte's structural characteristics. Predicting whether a molecule will yield a strong SERS spectrum upon binding to metals like Ag and Au, the common SERS substrates, lacks precise rules. Certain molecule classes, such as thiols (e.g., mercaptopurines, glutathione, cysteine, ergothioneine), N-containing heterocyclic aromatic compounds (e.g., adenine, uric acid, xanthine, hypoxanthine), and substances with amine or carboxylate groups, especially when linked to an aromatic structure, usually yield intense SERS spectra. These molecules possess functional groups allowing chemisorption onto Ag and Au surfaces, resulting in a stable interaction and prolonged presence in “hot spots”. Conversely, carbohydrates and lipids tend to lack strong affinity for SERS metals, making them challenging to detect directly. Polypeptides, proteins, and nucleic acids generally have better chances of observation with a direct SERS detection.50–52 Overall, a direct detection strategy works well with a limited range of biomolecules on usual Ag and Au SERS substrates. For analytes with low metal affinity, the key challenge lies in promoting an efficient analyte–surface interaction by functionalizing the metal surface. Despite these limits, a direct SERS detection has been successfully applied to detect clinically relevant biomolecules and drugs in biofluids, exosomes, bacterial and eukaryotic cells, and tissues.
SERS applications typically aim to detect or quantify, both directly and indirectly, specific targets, such as metabolites, drugs, proteins, nucleic acid sequences, or specific cell types like circulating tumor cells (CTCs). This type of application is referred to as “targeted” analysis. In a targeted diagnostic SERS method, the goal may be to identify the presence of a specific biomarker, classifying the sample as either “disease” or “non-disease” (classification task). Alternatively, it may involve quantifying the concentration of a marker (regression task) and relating it to the likelihood of a pathological condition. Targeted direct SERS methods are often used for the quantification of particular drugs in biofluids, particularly for therapeutic drug monitoring (see Section 6.3). Direct, targeted SERS detection applied to tissue sections is uncommon, but a method for SERS visualization of polysulfides on tissue sections of ovarian carcinoma was reported, which could predict the prognosis of cisplatin-based chemotherapy.53
In many applications, specific and reliable markers are still unavailable, rendering targeted analysis unsuitable. In such cases, alternative approaches are necessary to discover potential markers. An “untargeted” detection strategy leverages on all available biochemical information in a sample, as conveyed by a SERS spectrum, for diagnostic or prognostic purposes. In untargeted approaches, the biochemical complexity of biological specimens becomes valuable rather than problematic. Untargeted approaches typically employ a direct SERS detection strategy, which can spontaneously detect multiple biomolecules simultaneously, depending on their affinity for the metal substrate. Since the spectral contribution of a biomolecule is driven more by its substrate affinity than its concentration, SERS spectra are often dominated by a few species, simplifying the representation of a biological sample. When certain SERS bands are identified as characteristic of a particular condition or cell type, further spectral analysis may reveal whether these bands originate from one or more molecular species, potentially leading to the discovery of new markers. Untargeted, direct SERS approaches have primarily been used for diagnostic purposes in biofluids,54 but applications extend to exosomes,55 tissue extracts,56 cell lysates,57 and even bacteria detection and identification.58
In SERS diagnostics, an untargeted direct detection strategy relies on the assumption that biochemical differences among sample groups, as revealed by SERS, may correlate with distinct medical conditions. If such a correlation is established and validated, spectral differences need to be translated into biochemical differences by associating each spectral feature with a specific metabolite. This interpretation of SERS spectra obtained through an untargeted direct detection strategy is critical and should involve a direct comparison of the specimen spectrum with SERS spectra of reference biomolecules obtained under the same experimental conditions (i.e., using the same substrate and excitation wavelength). It is important to avoid using normal Raman spectra as references because normal Raman and SERS spectra of the same species often exhibit significant differences.
Indirect detection, conversely, has seen a surge in interest in the context of SERS liquid biopsy, owing to the possibility to detect CTCs, cancer-related proteins, and nucleic acids such as DNA and microRNAs (miRNA). In such complex heterogeneous mixtures as biofluids, the SERS signal of the target analyte is most often overwhelmed in the strong background or has no chance to reach the “hot spots” region due to the competitive interference from the remaining components of the matrix. A typical way to overcome this issue is to combine specific recognition elements (MIPs, antibodies, enzymes, and aptamers) with SERS reporters, together with a pretreatment step in which interfering molecules are filtered out (see Section 2). SERS lateral flow assays (LFA) and immunoassay (LFIA) are two examples of popular POCT approaches based on an indirect detection because of their simplicity, affordability, cost-effectiveness, and versatility, as they can be used for different matrices and can be carried out by non-specialized personnel.59 Their combination with nanostructures providing SERS features has improved their sensitivity, rendering LFA-SERS and LFIA-SERS promising technologies.
Besides being used in LFA- and LFIA-SERS methods, SERS reporters have also proven to be excellent labels for cellular and tissue bioimaging and SERS mapping has also become a useful imaging technique for isolated living tumor cells, excised tumor tissues, and in vivo tumors. Indirect detection strategies coupled with the integration with endoscopes also enabled the identification of the presence and location of tumors, visualization of lesion sites and surgical navigation.60
5. Data analysis and modelling through artificial intelligence (AI)-based approaches
The standard way of applying Raman spectroscopy and a common way of applying SERS is to apply the techniques in a direct manner, as opposed to the use of fluorescent labels, SERS labels or Raman tags. The direct Raman and SERS measurements provide a characteristic molecular fingerprint of the sample. This fingerprint can be exploited by artificial intelligence (AI) methods such as chemometrics and machine learning and translated into high-level information such as disease markers. To exploit the full potential of SERS and Raman data, AI-based data analysis pipelines are being constructed to correct, standardize and cleanse the Raman data before it is translated into high-level information. As the same data analysis pipeline for direct Raman spectral data and direct SERS data is used, in the following this combined data pipeline is described.5.1. AI based data pipelines Raman spectroscopic data including SERS spectra
An AI-based data analysis pipeline for SERS and Raman spectra comprises a series of algorithms for correcting, standardizing, and refining measured Raman data;61 however, it is crucial to avoid errors. Fig. 5 illustrates the data pipeline for Raman-effect-based spectra such as SERS spectra. An appropriate experimental plan, including sample size planning,62 should be implemented prior to commencing the data pipeline. The sample size planning (SSP) technique calculates the quantity of measurements required to derive significant outcomes. Upon completion of planning and data collecting for SERS, the analysis process starts with the elimination of cosmic spikes arising from high-intensity cosmic particles or their secondary particles. Occasionally, additional artefacts like fixed pattern noise are also eliminated. The next stage is to perform calibration for wavelength, wavenumber, and intensity.63–65 The initial two calibration procedures endeavour to create a steady wavenumber axis in relation to the excitation wavelength. This is essential as relative measurement with respect to the excitation wavelength is carried out in SERS and Raman spectroscopy. The aim of intensity calibration is to produce setup-independent Raman spectra by correcting the spectral transfer function of optical components, including lenses, filters, dispersive elements, and the quantum efficiency of the detector. However, these procedures still require further research and refinement as a notable variation in Raman and SERS spectra has been observed among different laboratories and Raman devices.9,66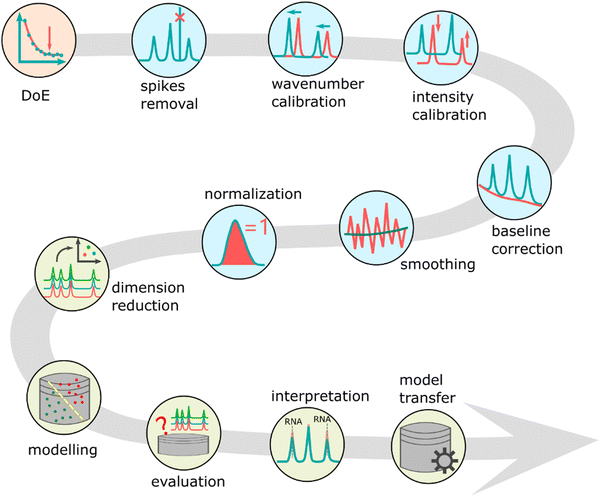 | ||
| Fig. 5 The data pipeline for Raman and SERS data. It contains sequential applied steps to allow a meaningful modelling of the spectral data and to extracting information and knowledge from it. The main steps are design of experiments (DoE) including SSP, pre-processing, data modelling, model evaluation and model interpretation. Adapted from ref. 67. | ||
After this stage, several preprocessing techniques are utilized to enhance data standardization. The preprocessing phase generally encompasses baseline correction due to the Raman effects energetic overlap with fluorescence. A denoising and normalization procedure is then performed to ensure that spectra from different measurement periods can be compared. Although a denoising may not always be executed, the algorithm's selection needs to factor in the mixed Poisson–Gaussian noise. Finally, feature extraction or dimension reduction techniques, such as principal component analysis (PCA), are employed.68 The identified features are then subjected to analysis using simple machine learning or chemometric models. These final two stages may be integrated with deep learning methods. Finally, the evaluation of machine learning or deep learning models included in the entire data analysis pipeline determines the models' generalization performance, such as their performance on a novel dataset.69 This evaluation step – if carried out correctly – reduces overfitting and the estimated performance of the model is a good estimate of the performance of the model on unseen (test) data. It should be mentioned here that overfitting is more severe in small sample size scenarios, especially when the independent data packages is limited to 3–5. Due the technical variability resulting from the measurement, SERS substrate generation, and intrinsic variability, including biological variability, another effect overlaying overfitting is overestimation, which occurs if information leakage between trainings and test data is happening. Finally, the interpretation of the models is undertaken using visualization methods.70
5.2. AI driven applications to analyze biomedical SERS spectra
The AI-driven data analysis pipelines for Raman and SERS data mentioned above have exciting potential applications, which we will review below. While this list is not comprehensive, the selected studies provide insight into how AI-based analysis techniques can be used for SERS spectra.A quantitative analysis of DNA methylations utilizing an AI-assisted label-free SERS biosensor known as iMeSERS was reported.71 In this research, the plasmonic pickering emulsions were used as a biosensing platform for direct SERS detection, enabling simultaneous quantification of both C5-methylcytosine (5mC) level and methylation ratio in DNA samples. Additionally, a deep learning algorithm was trained using SERS signatures to enable quantitative global DNA methylation analysis. This demonstrates the potential for utilizing direct SERS techniques combined with deep learning algorithms to accurately quantify epigenetic DNA modifications. SERS spectra of plasma exosomes were utilized and integrated with data modelling based on convolution neural network (CNN) to develop a new and accurate diagnostic approach for major depressive disorder (MDD).72 The authors applied a CNN architecture comprising of 5 convolutional layers, 1 flatten layer and 2 fully connected layers. This resulted in an area under the receiving operator curve (AUC) of 0.939, a sensitivity of 91.4%, and a specificity of 88.6% in the evaluation of a test set of 70 samples.
Similarly, SERS spectra of serum exosomes were investigated.73 This study aimed at a breast cancer subtype classification and the authors claimed that a 100% classification of breast cancer subtypes was possible. In addition, even the surgical outcome could be predicted. SERS spectra were employed to develop a diagnostic algorithm.74 The objective was to diagnose oral cancer (OC) using SERS spectra of exhaled breath, which was achieved through implementing an AI-based technology with plasmonic–metal organic framework (MOF) nanoparticles. The authors demonstrated that an artificial neural network (ANN) model could accurately classify both healthy and OC subjects with a 99% accuracy rate. Moreover, a tool for non-invasive secretome screening to utilise secretome composition as predictive biomarkers was developed.75 In this study, it was demonstrated that SERS spectra and cell secretome variations can trace relevant metabolites while extracting standard features for cell death classification. The microfluidic-based measurement of the SERS spectra was also demonstrated. Finally, a simultaneous detection system for six types of early-stage cancers (lung, breast, colon, liver, pancreas, and stomach) utilizing SERS spectra was developed.76 An AI-based analysis was applied to this dataset, resulting in an AUC of 0.945 for classifying the tumor organ type.
6. Application of SERS-based detection schemes in biomedicine
Within the previous chapters, information is provided on sampling and sample treatment of biomedical relevant objects, SERS substrates, SERS-based detection schemes as well as AI tools for data analysis. Thus, the interested reader is able to design a suited SERS workflow for the envisioned application scenario. In order to compare own ideas with state-of-the-art examples from literature, we briefly discuss recent developments in biomedicine, whereas most examples can be found in oncology and infectious disease related research work.6.1. Infectious diseases
Infectious diseases are a group of diseases caused by various pathogens, such as viruses, bacteria, parasites and fungi, transmitted between humans and animals. The identification of pathogens is becoming increasingly important in clinical diagnosis and customized treatment.77 Therefore, numerous detection assays have been developed. Among these, SERS is widely used in clinical analysis and bioanalysis.78 Using this method, various pathogenic microorganisms were directly detected and their type and number were determined. In addition, pathogens can be also detected indirectly by analyzing the metabolites or viral lysate and serological investigation from clinical samples. In order to translate the SERS-based detection related with infectious diseases into clinical practice, point-of-care systems were developed, which was summarized in a review article.79Due to bacterial cell structure, which is rich in proteins and amino acid residues and peptidoglycan, providing vibrational fingerprinting information, bacteria were considered as a common pathogenic microorganism suitable for SERS detection.80 The progress on SERS-based detection of bacteria in relation with food safety and public health was summarized emphasizing the necessity of point-of-care systems pursuing the identification bacterial infections.81 A classic SERS substrate, a silver nanorod array, was used to classify 22 different bacterial strains, including four types of biological warfare agent pathogens, one type of pathogen related to strong infectious diseases, and seven types of pathogens associated with food safety.82 By applying mathematical analysis methods, they revealed the unique spectral characteristics of each bacterial strain, allowing 20 of them to be clearly differentiated in a highly sensitive and specific manner.
Antibiotic susceptibility testing (AST) is another important direction of SERS-based bacterial analysis (AST-SERS) used to identify bacterial species with antimicrobial resistance (including multidrug resistant pathogens).83 AST-SERS involves addition of certain antibiotic to a target bacterial culture with further study of the cellular stress by measuring SERS spectra. SERS analysis before and after the addition of antibiotics enables to monitor (i) changes in the structure and composition of bacterial cell walls84 and (ii) changes in the concentration of stress biomarkers in the cell lysate85 or expressed in the extracellular space.86 Importantly, AST-SERS is usually performed using a direct SERS strategy. However, the metabolic response to the addition of antibiotics usually results in the expression of multiple signaling metabolites, which often have quite similar SERS spectra (e.g., purine bases). Thus, chemometric analysis of AST-SERS spectral data has been proposed to get accurate AST results.85 Finally, although SERS enables the analysis of micro amounts of samples, additional sample manipulations have been proposed to speed up SERS-AST analysis. For example, the use of bacteria cultivating in a microfluidic microwell device87 and electrokinetic extraction88 of bacterial cells enables to perform AST-SERS in less than 2 h, including bacteria incubation step.
Viruses as pathogenic microorganisms such as influenza A virus (H1N1), severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), and respiratory syncytial virus (RSV), are widely known for triggering pandemics. The capsid proteins of these viruses can be directly detected by SERS-based methods.89 To produce SERS-active hotspots, calcium ions were utilized as aggregating agents due to the generation of complexes with citrate molecules and moreover, to reduce citrate interference on the surface of silver nanoparticles for the detection of these capsid proteins. Acetonitrile was used to enhance the calcium-induced hot-spot effect and this developed method significantly improved the LOD of direct detection of SARS-CoV-2 as low as 2 × 103 copies per test in saliva. Alternatively, SERS sensors can be constructed to capture viruses and assess their concentration, in combination with techniques such as enzyme-linked immunosorbent assay (ELISA) and lateral flow assays (LFA), which are also applied in colorimetry detection of viruses. In preparation of ELISA-SERS or LFA-SERS sensors, the classical antigen–antibody–secondary antibody sandwich immunoassay, or antibody–antigen–antibody sandwich immunoassay are employed. Differently to classical tests, the enzyme-coupled protein or the colorimetric test marker are replaced by so-called SERS tags. A SERS-based LFA is provided for rapid and accurate detection of the HIV-1 dsDNA, which can be detected in serum with LOD as low as 0.3 fM.90 Moreover, a SERS-based LFA was developed for the detection of West Nile virus (WNV) non-structural protein 1 (NS1) and actual WNV samples with the portable system.91 The LOD of the developed method was 0.2 × 102 copies per μL, respectively, which was equal to that of the routinely diagnosis standard of virus clinic detection, i.e. quantitative fluorescence RT-PCR.92 Different to the above-mentioned methods, DNA-based sensors are also used to develop a more sensitive SERS method, such as sandwich-like assay or aptasensor locking amplifier to detect the targeted virus material.93,94 The amplification principle of the locking mechanism relies on the continuous growth through the complementary chain pairing of the DNA bases. This process effectively captures and amplifies the DNA sequences of the analyte, thereby further enhancing the signal. Here, instead of antibodies, DNA sequences are applied and dependent on the concentration of the target analyte, such as virus DNA sequences, the SERS signal varies due to the change of distance between the applied dye label and the SERS active sensing surface. Moreover, a homogeneous, i.e. reagent-less, SERS sensing mechanism was investigated, which is different from classical catalyzed hairpin assembly and other DNA self-assembly processes, to detect Plasmodium falciparum and SARS-CoV-2 B.1.1.529 in clinic samples.95 The optimized LOD of the sensor was observed to be down to the sub-pM level.
Viral lysates can also be used to trace respiratory viral infections, which is illustrated by the investigation of the lysates of SARS-CoV-2 lysate, influenza A subtype H1N1 lysate and respiratory syncytial virus lysate in human saliva as the real sample matrix.96 A SERS detection scheme combined with electrochemistry (EC-SERS) was utilized and the three viruses were classified based on their different proportions of DNA bases in the lysates. In addition to viral lysates, respiratory metabolites are targeted by indirect assays; here, breath volatile organic compounds (BVOCs) serve as COVID-19-specific biomarkers. Coronavirus-induced immune responses and metabolic changes can alter the concentration of BVOCs such as aldehydes, ketones and alcohols in breath.25 Based on this observation, the authors developed a method based on the functionalization of Ag nanocubes with 4-mercaptobenzoate (MBA), 4-mercaptopyridine (MPY) and 4-aminothiophenol (ATP), the schematics diagram of detection workflow was shown in the Fig. 6. The interaction between the metabolite of interest and the functionalization molecules leads to the incorporation of a hydrogen bonding structure, resulting in structural changes and causing a shift in the peak position of the complex compound molecule. Moreover, combined with machine learning, this metabolite detection is much suitable for rapid large-scale screening. Finally, serological investigation plays an important clinical characterization and further studies of changes of molecular composition of body fluids, such as serum, before and after a viral infection. The change in concentration of amino acids, carbohydrates, lipids and proteins, or its ratios, can be indicated as the result of viral infection in order to be able to categorize diseased patients from healthy controls.97 Within this study, SARS-CoV-2 was used as an example to perform statistical investigation utilizing direct SERS and incorporating machine learning to study serum metabolic signature differences between patients 4- and 16-weeks post infection in non-hospitalized adults.
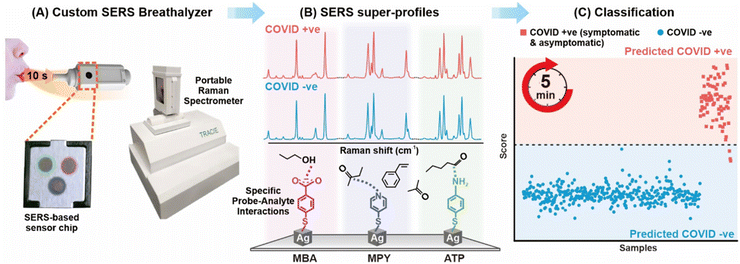 | ||
| Fig. 6 Workflow of SERS-based detection to identify COVID-positive individuals via using BVOCs. Reprinted with permission from ref. 25. Copyright 2023 American Chemical Society. | ||
6.2. Cancer diagnosis and treatment monitored by means of SERS
During and post-pandemic, cancer has remained one of the leading causes of death worldwide. Delay in cancer screenings and treatments due to the COVID-19 pandemic has had significant repercussions on patients' health outcomes. Early detection of cancer plays a crucial role in improving survival rates and reducing the need for aggressive treatments. With postponed screenings, there is a higher likelihood of cancers being diagnosed at later stages when they are more advanced and difficult to treat. Moreover, SARS-CoV-2, given its endemic future, may lead to increasing the risk of developing several types of cancer as the infection targets certain proteins involved in diseases, i.e. non-small cell lung cancer, breast carcinoma and multiple human cancers. On the other side, people who have been weakened by tumor and/or its treatments could show altered immune functions, resulting susceptible to serious illness if they get COVID-19. Based on these aforementioned issues, as announced by American Cancer Society's National Consortium for Cancer Screening and Care (ACS National Consortium), enhancing cancer test to pre-pandemic levels is strongly needed.98,99In this section, we consider critical to understanding and summarizing how could be improved the diagnosis and treatment of cancer by means of SERS, scanning which samples may be taken in account for the screening. It is considered as cancer biomarker any gene, protein, cell-derived small vesicle or other substance, produces by cancer or non-cancer cells, that can be tested for cancer diagnostics. The discovery of the relationship between malignant tumors and genetic changes has led the scientific community to consider cancer as genetic disease. Genomic biomarkers, originated from alterations in nucleic acids, include for example DNA sequence point mutations, increased expression levels of miRNAs and altered levels of DNA methylation. Tumor-related nucleic acids are present not only in tissues but also in human body-fluids, i.e. blood, saliva and urine, opening the route to using liquid biopsy to detect genomic biomarkers with high sensitivity.13 Gaining attention for sensing in cancer theranostics in the last years, indirect SERS, i.e. the application of SERS tags, allows ultrasensitive detection of cancer-derived nucleic acids for cancer diagnostics and staging. The possible strategies for SERS-based nucleic acids sensors design and both challenges and limitations for a future clinical translation were compared and critically discussed, underlining the importance in increasing accuracy and reproducibility of SERS detection in physiological conditions.13 Furthermore, SERS-based methods in genomic biomarkers detection evaluating their limit of detection and possible testing in liquid biopsy were reported.100 Five main groups of experimental protocols, i.e. capture-based, displacement-based, sandwich-based, enzyme-assisted and specialized protocols, have been identified and deeply described. Cancer-derived extracellular vesicles (EVs) and cancer-derived exosome, are particular subgroups of lipid bilayer vesicles secreted in the extracellular environment from malignancy cells. Circulating in the body-fluids and carrying own bio-components (DNA, RNA, proteins), they have recently garnered scientific interest as cancer biomarkers in liquid biopsy. Descriptive examples of integrated SERS-microfluidics platforms to isolate and characterize EVs have been reported, also providing their insight for the integrated setups application and discussing their limitations in clinical trials.101 An overview about the direct as well as labelled SERS-based detection of EVs is provided in Fig. 7. The above-described approaches, once the obstacles have been overcome, will be valuable in cancer diagnosis and precision medicine. Regarding exosomes, their detection and analysis by mean of SERS approaches has been summarized, pointing out limitations and advantages for cancer diagnostics, highlighting how the exploration of new high-performance SERS substrates and combination with microfluid chips would be the right direction for the future exosome-based cancer research.102 CTC in body fluids has been recently discovered as the most important stimuli of metastasis. A summary on the state-of-art of SERS microfluidic sensors for isolating and analyzing them, plus a future perspective in biosensing, has been provided.103 In particular, using e.g. carbon nanotubes and paper for microfluidic biosensors, will make them more user-friendly and low cost for the future commercialization. Finally, the advances and challenges of direct SERS in cancer screening, detection of cancer-related biomarkers, cancer prognosis and treatment monitoring, testing body-fluids (blood, urine, saliva, sweat) and breath is introduced.104 Here, given the fundamental role played by chemometrics in the development of SERS point-of-care sensors in clinical trials, an overview regarding the most performing machine learning algorithms for SERS data analysis has been also provided.
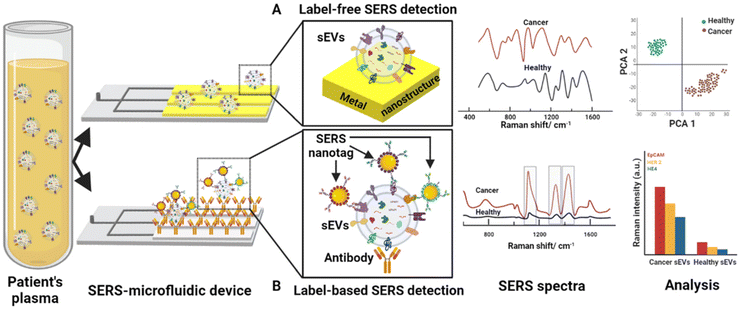 | ||
| Fig. 7 Workflow for the detection of extracellular vesicles (EVs) by integrated SERS-microfluidic device (A) in direct SERS approach, i.e. EVs are absorbed on the plasmonic surface which will enhance the Raman signals of EVs’ molecules and the spectra have been analysed by multivariate analysis (e.g. PCA), and (B) in indirect SERS-based detection, i.e. specific antibodies on the chip captured EVs and the SERS response is associated with the Raman reporter of the applied SERS nanotags. Used with permission of Royal Society of Chemistry, from ref. 101; permission conveyed through Copyright Clearance Center, Inc. | ||
SERS is largely applied for imaging (also in vivo) cancer cells and tissues especially aiming to identify tumor margins for improving decision-making processes during surgery and more it is receiving great attention in cancer therapy as therapeutic/drug delivery platform and for anti-cancer drugs monitoring. For in vivo tumor targeting, antibody-modified SERS nanoprobes are equipped with a variety of Raman reporters allowing for multiplexing in combination with hierarchical clustering for data analysis,105 and the potential of the multiplexed SERS immuno-imaging of tumor sites was demonstrated in mouse models.106,107 Recently optimistic outlooks for a soon to happen translation of SERS in clinical trial for in vivo imaging in cancer screening were reported, i.e. recent developments in using SERS nanoparticles as nanoprobes for multiplexed and multimodal in vivo imaging applications in oncology were summarized.108 Showing good extravasation and uptake in cancer sites, nanoparticles allow multiple targets, i.e. in organs, detection with promising specificity and spatial resolution. Moreover, SERS plays a role in in vivo imaging and biosensing applications related with in vivo intraoperative guidance, especially the SERS role in the identification and resection of microtumors and its possible use as therapeutic platform. Furthermore, paving again a possible future in oncology trials, the challenges associated with SERS active interfaces, setups and clinical translation were discussed and are currently explored by many research groups worldwide.33 Additionally, high cytotoxicity of anticancer drugs and appearance of severe side effects during the therapy lead to development of diverse assays (including SERS-based ones) for monitoring of blood circulating drugs and precise controlling treatment process. The use of SERS analysis for monitoring of therapeutic drugs (including anticancer) is overviewed in detail in the next section. More specific information about advances in the SERS-based assays and SERS-assisted drug-delivery systems for widely used anticancer drugs can be found in recent reviews.109,110
6.3. Therapeutic drug monitoring employing SERS detection schemes
Therapeutic drug monitoring (TDM) is one of the tools of personalized medicine aimed at improving medical treatment by monitoring changes in drug concentration in body fluids (blood serum and plasma, urine, saliva). TDM is particularly important for monitoring drugs with a narrow therapeutic window, such as anticancer (doxorubicin,111 methotrexate,112–115 5-fluorouracil,116etc.), antibacterial (cephalosporins,117 fluoroquinolones,118etc.), and psychotropic drugs (stimulants,119 opioids,120etc.), and the literature has traditionally been dominated by reports of SERS assays for TDM of these drugs. In this section we will focus only on discussing the most general and important aspects of the use of SERS in TDM, referring to the interesting reports published since 2020. More detailed information with a comprehensive analysis of the results achieved over the last 10 years can be found in the recent reviews.121–123The SERS-based TDM is mainly performed using SERS assays with direct detection strategy utilizing common colloidal gold or silver nanoparticles as SERS substrates. As discussed in previous sections, SERS signal has a multiband nature that is a great advantage for TDM because it allows increasing the accuracy of the analysis by eliminating false-positive results. However, as already stated in section 2, SERS is not a universal method because direct SERS has a very limited selectivity that is a critical limitation for the analysis of such complex objects as body fluids. Therefore, combination of the SERS detection with sample pretreatment and separation techniques is usually a mandatory practice for solving TDM tasks.122 Usually the analysis involves sample dilution with further separation of target analyte molecules from the matrix using, e.g., advanced filtration techniques,113,115 solid phase112,115 or liquid–liquid extraction.116,119,120 The extraction also significantly improves the analysis sensitivity (up to one order) that is especially critical for TDM of drugs in saliva samples.120 In the case of plasma and serum, the proteins have to be removed by filtration,113 centrifugation115 or precipitation with an organic solvent.118 The modification of the SERS-active surface also enables to maximize the interaction of the surface with the analyte molecules and to minimize one for the molecules of admixtures. For example, modification with an anti-fouling (L-cysteine111) or supramolecular compound (β-cyclodectrin114) enabled to simplify sample pretreatment by limiting it to only the dilution step.
Regarding Raman spectrometers we are pleased to report that modern SERS-based TDM is very often performed using portable and easy-to-use spectrometers, proving the applicability of the proposed assays to hospital laboratories. Indeed, all original studies cited in this section were performed using such spectrometers. In addition, stationary Raman spectrometers coupled with confocal optical microscopes typically used in scientific laboratories are not a good choice for the SERS-based TDM not only due to complexity of such equipment, but also because their sensitivity to spatial inhomogeneities in the analyzed samples is too high, which leads to a deterioration in the analysis precision.
Despite the difficulties with selectivity, the availability of information about consumed drugs drastically simplifies the use of SERS in TDM, making identification step unnecessary. Moreover, the purpose of TDM is to determine whether the concentration of a drug in a body fluid is within a certain range (therapeutic window of concentrations) or not.123 Therefore, the extreme sensitivity of SERS assays (e.g., fM or aM levels) is often not necessary since drug concentrations within therapeutic ranges are usually quite high (usually nM to μM levels). In addition, normal concentration ranges of the intrinsic body fluid components (i.e., potential interferences) are also usually well-known and can be effectively used in preliminary studies of the assay selectivity and optimization of detection conditions.112 Abnormal concentrations of the metabolites in the case of some diseases (e.g., glucose, creatinine, electrolytes, etc.) can also be taken into account to make SERS-based TDM even more universal and accurate. For example, the accuracy of the SERS assay was evaluated for antibiotic determination using real urine samples with artificially increased (2- and 4-fold) concentrations of the main urine metabolites.117,118 Importantly, the composition of urine and saliva significantly changes over daytime and it has to be taken into account because it influences background signal and, consequently, accuracy of SERS analysis.114,120 Therefore, the development of SERS assay for TDM should always include thorough analysis of all available information about the sample composition to minimize the effect of interferences on the analytical signal. Schematic representation of the SERS-based TDM with summing-up information is shown in Fig. 8.
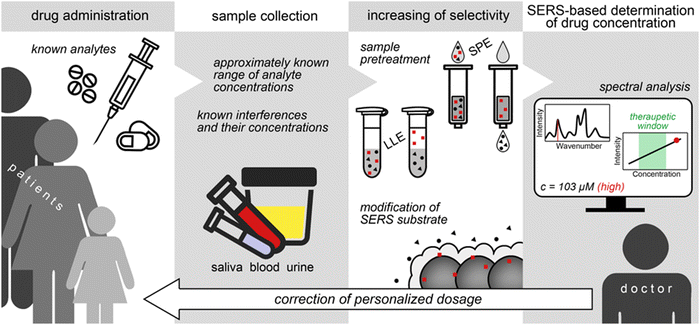 | ||
| Fig. 8 Application of SERS-based analysis for therapeutic drug monitoring and personalized medicine. SPE and LLE are solid phase and liquid–liquid extractions, respectively. | ||
In the following paragraphs we suggest several directions, which, in our opinion, will be the most promising for the SERS-based TDM.
Most of the published SERS assays (not just for TDM) have been currently proposed for the determination of only one analyte. However, any drug usually belongs to a certain class of drugs whose representatives have quite similar chemical structure and properties. Consequently, it is expected that drugs of the same class should have comparable conditions for (i) their separation from body fluids during sample pretreatment and (ii) collection of an intense SERS signal. From an analytical point of view, this fact should facilitate the development of more universal SERS assays. SERS assays with improved versatility have been recently proposed for determination of anticancer drug and its metabolite115 as well as representatives of psychotropic119 and antibacterial117,118 drugs. Moreover, if the representatives have similar molecular parts, which provide an intense SERS signal (e.g., fluoroquinolone antibiotics), these drugs will have comparable spectral profiles, additionally facilitating analysis of the results.118 Therefore, we believe that the development of a more universal analysis protocols suitable for the analysis of a specific class of drugs is a practically demanding direction due to the significant simplification of the TDM process.
The key difference between sensors and single-shot assays is the ability to monitor changes in analyte concentration over time (like digital pH electrodes). Unfortunately, strong analyte adsorption onto the SERS-active sites causes appearance of memory effects which significantly complicate development of SERS sensors. Therefore, most of the SERS assays are disposable ones. Despite significant progress in the development of continuous-flow analytical SERS systems based on microfluidics, there is still a lack of reports describing the application of SERS sensors in TDM. In this regard, we suggest the application of (i) EC-SERS analytical systems113 and (ii) planar SERS substrates modified with anti-fouling and molecular recognition compounds111,114 as two most promising directions because both approaches allow achieving reversible analyte adsorption, preserving SERS-active sites from degradation.
Another important aspect of pharmaceutical treatment is that it often involves administration of multiple drugs during, e.g., anticancer therapy or treatment of bacterial infections with antimicrobial resistance. Consequently, it would be useful to develop the assays suitable for simultaneous determination of two or more analytes, i.e. multiplex analysis. For example, Chen et al.119 combined SERS with liquid–liquid extraction to determine psychoactive compounds (methcathinone and 4-methylmethcathinone) in urine. However, although the authors were able to detect both analytes simultaneously, they showed that the signal intensities in the mixture were weaker compared to those of individual analytes. Therefore, to achieve the success in quantitative multiplex SERS analysis, it is necessary to take into account competitive adsorption on SERS-active sites that is a quite challenging task.
We believe that any progress in the listed directions will facilitate the penetration of SERS analysis into clinical laboratories due to improvement of cost efficiency and performance of the SERS-based TDM that consequently improves efficiency of the medical treatment. In addition, we also should note that the SERS assays suitable for TDM have strict requirements for high precision, accuracy, and selectivity. As a result, the approaches used in the development of such assays can also be successfully used to develop assays for other exogenous compounds (e.g., illicit drugs, anticoagulants, neuroleptics, etc.) and various low molecular weight metabolites.
6.4. SERS as detection scheme in further diseases – from neurological dysfunction to organ function assessment
The neurotransmitter dopamine was detected by a DNA-based approach including the detachment of the SERS probe in presence of dopamine with a sensitivity in the aM range.124 An array of SERS-active nanoparticles is modified with a specific DNA sequence, capturing a SERS probe equipped with a Raman reporter and an aptamer sequence specific for dopamine. Thus, nanogaps between the nanoparticles are generated, enhancing the Raman signal of the reporter molecule. Upon the release of dopamine, the target is bound specifically by the aptamer of the SERS probe resulting in its detachment which is accompanied with a decrease in SERS signal intensity. It is illustrated that within a complex serum matrix, the SERS approach shows a linear dynamic range between 1 pM and 10 nM. In the case of Alzheimer disease, the assessment of the secondary structure of 42-residue-long amyloid-β (Aβ42) proteins is of high importance, which is achieved by employing chiral plasmonic triangular nanorings.125 The direct SERS approach is employed to discriminate enantiomers based on a selective resonance coupling between the dipole modes which are related to the enantiomers and the chiral nanostructures, i.e. SERS-chiral anisotropy. As limit of detection, the values of 45 and 4 fM were estimated for the Aβ42 monomer and its fibrils, respectively. Finally, the developed test protocol was applied to cerebrospinal fluid of Alzheimer patients and being compared with results of control assay kits. The intensity of the Raman mode at 1245 cm−1 associated with the formation of the β-sheet increases as function of the estimated concentration of Aβ42 fibers, illustrating the potential of chiral-assisted direct SERS in the identification of diseases based on protein-misfolding. Magnetic SERS-active nanostructures were applied to allow the adsorption of periodontal pathogens and its separation from matrix components of human saliva matrices within a microfluidic environment.126 By investigating the SERS data with principal component analysis, Porphyromonas gingivalis can be identified and differentiate from other relevant bacterial strains such as Aggregatibacter actinomycetemcomitans and ubiquitous oral Streptococcus spp. The importance of this detection scheme is due to the development of periodontitis by P. gingivalis which are recently found also in the brain of Alzheimer disease patients, which might be a related co-morbidity with increasing importance in the future. Employing a SERS-based immunoassay approach, the biomarkers matrix metalloproteinase (MMP-9), soluble platelet-derived growth factor receptor, and major facilitator superfamily domain containing Mfsd2a were detected within blood achieving a detection limit of 1 fg mL−1.127 By employing 3 different SERS tags equipped with different Raman marker molecules as well as specific antibodies, barcoding was achieved as each binding event between antibody and its biomarker was recognized by unique SERS fingerprint information. The investigated biomarkers are described as markers to predict the progression of white matter lesions and its early detection in blood is important for fast treatment decisions.Addressing cardiovascular diseases, Raman and SERS-based techniques are discussed being a powerful alternative to the clinical gold standard methods with showing the great potential of SERS-based techniques in biomarker detection.128 Today, for the detection of cardiac troponin I (cTnI), a biomarker related with acute myocardial infarction, direct and label-based approaches are available heading towards the point-of-care diagnosis employing for example microfluidics or paper-based assays.129 As an example, cTnI was captured towards the surface of aptamer-modified magnetic beads equipped with silver nanoparticles.130 Thus, cTnI could be separated from matrix components and other proteins. Within the next step, bicinchoninic acid (BCA) and Cu2+ (BCA assay) are added to the solution and after the reduction to Cu+ by the protein under alkaline conditions, 2 BCA molecules forms a complex with Cu+. The estimated LOD employing the combined BCA-SERS assay was 0.23 pg mL−1, and the specificity of the assay was confirmed by investigating other proteins such as BSA or HSA. Multiplexing for the detection of two or more cardiac biomarker, which is relevant for an improved accuracy of the diagnosis, is achieved for example by applying SERS tags modified with different Raman reporter molecules and recognition elements being specific for different biomarkers.131
To determine the activity of N-acetyl-β-D-glucosaminidase (NAG) as tissue damage indicator in body fluid such as urine or serum, a SERS-based detection was developed which is based on the azo coupling of NAG-enzymatic products with diazonium dye in alkaline conditions.132 As function of the NAG concentration, the intensity of the SERS fingerprint of the reaction product increases proportional. By employing silver nanostructures as SERS sensing platform, LOD values for 3 matrices, i.e. urine, milk and plasma, were estimated with 0.07, 0.47 and 0.50 mU mL−1, respectively. Finally, the NAG activity level was analyzed in a sheep model, validated with a conventional assay. To predict the concentration of α1-acid glycoprotein (AGP), which is associated with inflammation and immunomodulation, in human serum, a SERS based approach was combined with CNN transfer learning.133 First, a SERS-active gold surface was modified with phenylboronic acid (–PhB(OH)2) chemical moieties to capture AGP from complex body fluids. A spectral data base was recorded to establish a CNN-based model for quantification of AGP within the medical relevant concentration range, i.e. 2–8 μg mL−1 with a detection limit around 1 μg mL−1. The authors were able to detect an increase of AGP in serum which is assigned as alarming.
As an example of identification of diseases in humans by targeting related biomarkers employing a portable SERS approach, a POCT detection scheme for jaundice in complex tear fluids is introduced.134 Here, bilirubin is the target molecule as its total amount is increased in patients with jaundice. To separate the target analyte from its complex matrix, i.e. tear fluid, and to allow for its specific and sensitive detection, a SERS-active surface with pores was developed. Thus, during the drying process of the tear fluid, un-bound matrix molecules such as proteins diffuse through pores below the SERS-active sensing surface and thus, the number of molecules contributing to the overall SERS spectrum is reduced. Employing this approach, patients with jaundice were successfully identified within a clinical setting. Moreover, a non-invasive detection scheme of non-alcoholic steatohepatitis was developed based on the acquiring SERS spectral data after mixing serum samples from more than 200 individuals with silver nanoparticles.135 Here, no specific modification of the nanostructure surface is applied allowing all present molecules in the matrix to interact with the SERS-active substrate. The SERS data were analyzed employing neural network approaches in relation with a liver biopsy dataset for development and validation of the routine to predict the disease under investigation. The potential of a CNN-assisted SERS detection scheme for the assessment of an organ function was illustrated employing the kidney function-related biomarkers serum creatinine and blood urea nitrogen.136 Various conditions were simulated by mixing both biomarkers in varying concentration in fetal bovine serum. A prediction of the concentration of both target analytes was achieved in spiked serum samples.
Fructose was detected in artificial urine by means of a deboronization reaction of 4-mercaptophenylboronic acid directly on the surface of silver nanoparticles initiated by the target analyte fructose.137 By doing so, thiophenol is generated on the SERS sensing surface and its SERS signal intensity increases as function of the concentration of fructose. As detection limit, the values of 0.084 μmol L−1 in water and 0.535 μmol L−1 in artificial urine were found. The potential for a sensitive and specific detection of fructose in human urine is illustrated which is of importance in screening of fructokinase deficiency as well as hereditary fructose intolerance.
7. Challenges and perspectives
Although SERS holds an enormous potential for biomedical analysis, it is also a complex technique, presenting several challenges to the non-expert. A careful choice of the detection strategy, sample pre-treatment, the right nanostructured substrate and adequate experimental parameters relies on a sound understanding of SERS basic principles. The main difficulties in biomedical direct SERS analysis are associated with the chemical complexity of biological samples. In the case of direct analysis, the SERS substrate will enhance the Raman signal of any molecule (analyte or interference) adsorbed onto its surface. Consequently, the most critical limitation of SERS-based analysis of complex mixtures is intrinsically low selectivity of SERS, i.e., the ability to enhance the Raman signal of only those analytes having a high affinity for the SERS substrates used. An excessive concentration of intrinsic body fluids components having a strong interaction with the SERS substrate surface can lead to competition with the analyte molecules for SERS-active sites. As a result, this competition can cause the appearance of a strong background signal or suppression of the analyte signal. For cells and tissues, a direct detection strategy also implies that the SERS substrate must penetrate through biological membranes and the compounds of interest diffuse to and adsorb onto the substrate surface. Finally, if the experimental conditions (e.g., pH, solvent properties, ionic strength, etc.) are not favorable for the analyte adsorption onto the SERS substrate, even an increase in the analyte concentration may not lead to the appearance of the practically useful SERS signal. For analytes with structures featuring lone electron pairs (e.g. –NH2, –COO−, heterocycles and –SH), intense SERS spectra are expected, as a consequence of a strong interaction with the SERS-active sites of metallic nanostructures. Less so for other classes of biological molecules, for which a direct detection strategy will be more challenging. In order to simplify the SERS response and to increase the specificity towards these molecules and other samples such as proteins, viruses or bacteria, an indirect detection strategy can be used. Thus, a specific recognition is achieved and the marker modes associated with the SERS label, i.e. Raman reporter, is dominating the SERS response or allows detection even in a complex biological matrix. Although SERS methods based on indirect detection strategies are more reliable for managing issues with selectivity, however, direct information about the Raman spectrum of the object is lost. As in other bioanalytical techniques, sample pre-treatment is also a key step when applying SERS to biological samples, and it usually aims at minimizing the interference from the matrix (e.g. by totally or partially separating the analyte from other components) and/or at enriching the analyte to make its detection easier. In the best case, separation and/or enrichment can also be performed directly on the SERS substrate surface upon functionalization with recognition compounds such as molecularly imprinted polymers, aptamers, or antibodies. Additionally, surface modification can improve the interaction of SERS substrates with hydrophobic analytes. The development of multifunctional surface-modified SERS substrates has been one of the main goals in SERS for decades, and significant progress in this field has been reached. In order to analyze the complex SERS spectra, data analysis approaches and/or multivariate data analysis gained an increasing importance within the last years. However, multivariate methods need to be applied to SERS data with care and expertise, as an unskilled use can easily lead to artifacts or biases. Particular attention should be paid to methods evaluation (e.g. validation) and interpretation of the results. If SERS is applied correctly and all the parameters are adequately selected, complex bioanalytical tasks can be achieved. Concentration profiles in body fluids or local distributions in cells or tissues of the targeted analytes can be visualized, so that conclusions for treatment and improvement of decision-making processes in clinics can be drawn.7.1. SERS substrates in biomedical research
The plasmonic characteristics, i.e., the surface plasmon resonance, need to be matched by the excitation wavelength. For practical use, laser excitations can be selected in the visible and NIR range, depending on the plasmonic material, aggregation state, size and shape of the nanostructures as well as surrounding medium. Silver and gold nanostructures are mostly applied for SERS substrates for biomedical tasks. Here, gold nanostructures are more stable and longer storable after fabrication. Although silver nanostructures are associated with more intense SERS spectra, it is very susceptible to oxidative degradation of the signal intensity. For silver nanostructures, excitation wavelengths from the visible to NIR spectral region are applicable depending on the shape and size of the nanostructures. In the case of Au nanostructures, typical excitation wavelengths are in the red and NIR spectral range. Ease of fabrication, stability, signal uniformity, and potential interaction with the analyte molecule are some of the most important criteria to consider for SERS substrates.Classical spherical silver and gold nanoparticles, as well as those with different shapes and sizes, are easy to synthesize, the LSPR can be precisely tuned over a wide excitation range by its shape, and comparable high laser power can be applied in aqueous solution. However, they are susceptible to degradation by aggregation and tend to form a more stable structure without a protective agent, so their batch-to-batch reproducibility can be low when measured over a period of a few days to weeks. When dried on the solid substrates, they can be very unevenly distributed from point to point and from sample to sample resulting in randomly distributed hotspots. Moreover, the surface of classical colloidal nanostructres is occupied by capping agents applied in their synthesis.
Well-defined plasmonic structures with nano-sized gaps give a uniform signal, and their hotspots can be tuned according to the size of the analyte. These substrates are manufactured in a very defined and sophisticated manner. They can, therefore, have large wafer sizes, a clean surface with no capping agents to interfere with the SERS signal, and a high reproducibility from batch to batch and sample to sample, making them easier to standardize and commercialize. However, they are more expensive and time-consuming in production due to the need for specialized facilities and expensive laboratory equipment.
Semiconducting and dielectric materials offer high stability and can be fabricated once and stored for months to years, but they mostly favor chemical enhancement and weaker SERS than plasmonic metals. Functionalized and composite materials have the potential to offer the best of both worlds with high sensitivity, hotspot tunability, reproducibility, and uniformity. However, they are not widely studied and still have more to discover regarding enhancement mechanism and versatility of use; for example, most semiconductors contain hydroxyl groups that can bind to proteins without destroying the secondary structure. However, more research is needed in this area.
Before selecting SERS substrates for biomedical application, it is advisable to analyze the size and chemical structure of the analyte and the complexity of the matrix and then select the most suitable substrates accordingly, considering which properties of the substrates are more critical for the given analytical task.
7.2. Targeted bio objects and its complex composition
In direct SERS, the Raman signal of any molecule (analyte or interference) adsorbed onto the SERS-active surface will get enhanced. Consequently, the application of SERS in biomedical research is limited by the intrinsically low selectivity of SERS, i.e., the ability to enhance the Raman signal of only target molecules (analyte molecules). In the case of excessive concentrations of the admixtures (e.g., intrinsic components of body fluids) a competition with the analyte molecules for free SERS-active sites is observed. As a result, this competition can cause the appearance of a strong background signal or suppression of the analyte signal. For cells and tissue samples, their internal composition can be studied only if the SERS substrate penetrates into the object and the compounds of interest diffuse to and adsorb onto the substrate surface. To summarize, if the experimental conditions (e.g., pH, solvent properties, ionic strength, etc.) are not favorable for the analyte adsorption onto the SERS substrate, even an increase in the analyte concentration may not lead to the appearance of the practically useful SERS signal. Analyte molecules with moieties with lone electron pairs (–NH2, –COO−, and –SH) provide strong interaction with the SERS-active sites of metallic nanostructures and can be enriched on the SERS active surface even in complex matrices. By applying SERS labels or tags, the selectivity is increased by employing recognition elements such as antibodies or aptamers and the SERS response is simplified as mostly the fingerprint modes of the Raman reporter molecule is dominating the SERS spectrum. Although indirect SERS is a more reliable tool for managing issues with selectivity, direct information about the Raman spectrum of the target analyte is lost.7.3. How to minimize the competition for free binding sites in biomedical SERS, i.e. sample preparation strategies
By adjusting pH, ionic strength, dielectric constant (changing the solvent) and/or dilution with pure water or buffer, the interference between target analyte and matrix molecules in the SERS response can be lowered and in the case of a high affinity of the target analyte towards the metallic surface, the SERS signal intensity can be even increased although its concentration is lowered by e.g. dilution. Simple treatment protocols are very attractive for POCT; however, they are not universal enough and its effectiveness decreases when the analyte concentration is too low compared to concentrations of interferences. In this case, it is necessary to separate the analyte molecules from the interferences before SERS analysis, and this can be done using extraction (solid phase or liquid–liquid), chromatography or protein precipitation from body fluids. The separation can also be performed directly on the SERS substrate surface after its modification with recognition compounds such as molecularly imprinted polymers, aptamers, or antibodies. Additionally, surface modification can improve the interaction of SERS substrates with hydrophobic analytes. To summarize, a detailed preliminary study of the effect of experimental conditions on the signal of the target analyte and the analyte-free object allows better control of the selectivity and, consequently, accuracy of the analysis.7.4. Analytical performance of biomedical SERS
From the point of view of analytical chemistry, the most reliable practice during a SERS study of a bio object/sample or development of a SERS assay for bioanalysis is to collect as many control SERS spectra as possible and under various registration conditions. This includes the signal of the analyte(s), pure SERS substrate, and pure bio object/sample without the analyte molecules. This practice facilitates spectral interpretation and identification of spurious/anomalous bands, e.g., signal of stabilizer of the SERS substrate; signal of elemental carbon appeared due to photothermal degradation of organic compounds onto SERS substrate surface; etc. Ideally, the signals of most important interferences should also be collected at experimental conditions intended for use during analyte detection. In quantification assays by using SERS, the accuracy (apparent recovery rate), root mean square error of prediction (RMSEP), limit of detection (LOD) as well as limit of quantification (LOQ) needs to be estimated and the validation should be done employing the gold standard technique. In cases with complex spectral profile, data analysis approaches/multivariate data analysis is required and should be implemented in the analysis chain.7.5. Typical interferences in biomedical SERS spectra
In SERS investigations, the main priority has to be given to the interferences with large concentration in the sample, strong/moderate affinity to the SERS substrate surface, and/or strong SERS signal. Creatinine, uric acid, and bilirubin can serve as an example of such interferences in serum and other body fluids and they can form quite strong background signal complicating the analysis. In contrast, the effect of common inorganic ions is usually less important because their SERS signal is very weak (complex anions) or absent (metal ions), and they mainly influence intensity of analytical signal through ionic strength by causing aggregation of colloidal SERS substrates. The effect of potential interferences should be studied at the concentration levels corresponding to those in the real objects, i.e., at realistic «analyte – interfering compound» ratios. As an example, the effect of serum albumins on the analyte signal has to be studied at 50–80 mg mL−1 of albumin, which is a huge, but realistic concentration. In the case of proteins, it is known that a corona is formed around the SERS-active nanoparticles, which impede the interaction of the target analytes with the sensing surface and illustrating the necessity of sample pretreatment.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Federal Ministry of Education and Research, Germany (Bundesministerium für Bildung und Forschung, BMBF) and PTJ (Projektträger Jülich) for supporting the project grant 03ZU1214EA (ThWIC: Vorort-Bestimmung von Mikroschadstoffen in Abwasserproben mit plasmonisches Multiplex-Assays und kombinierter Fingerprintanalyse (MIKA) – A). We thank BMBF for financial support of the EU-LAK project ABSarbo (01DN23003). The Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) supported this work under grant 465289819. Funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC 2051 – Project-ID 390713860. This work is supported by the BMBF, funding program Photonics Research Germany (13N15466 (LPI-BT1)) and is integrated into the Leibniz Center for Photonics in Infection Research (LPI). The LPI initiated by Leibniz-IPHT, Leibniz-HKI, Friedrich Schiller University Jena and Jena University Hospital is part of the BMBF national roadmap for research infrastructures. The authors acknowledge financial support from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 860185 (PHAST = Photonics for Healthcare: multiscale cancer diagnosis and Therapy).References
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. G. de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J. F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J. M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L. L. Tay, K. G. Thomas, Z. Q. Tian, R. P. V. Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- A. Pérez-Jiménez, D. Lyu, Z. Lu, G. Liu and B. Ren, Chem. Sci., 2020, 11, 4563–4577 RSC.
- D. Cialla, A. März, R. Böhme, F. Theil, K. Weber, M. Schmitt and J. Popp, Anal. Bioanal. Chem., 2012, 403, 27–54 CrossRef CAS PubMed.
- I. Nabiev, I. Chourpa and M. Manfait, J. Raman Spectrosc., 1994, 25, 13–23 CrossRef CAS.
- S. Laing, L. E. Jamieson, K. Faulds and D. Graham, Nat. Rev. Chem., 2017, 1, 0060 CrossRef CAS.
- J. Perumal, Y. Wang, A. B. E. Attia, U. Dinish and M. Olivo, Nanoscale, 2021, 13, 553–580 RSC.
- D. Cialla-May, X.-S. Zheng, K. Weber and J. Popp, Chem. Soc. Rev., 2017, 46, 3945–3961 RSC.
- Z. Ye, C. Li, Q. Chen, Y. Xu and S. E. Bell, Angew. Chem., Int. Ed., 2019, 58, 19054–19059 CrossRef CAS PubMed.
- S. Fornasaro, F. Alsamad, M. Baia, L. A. Batista de Carvalho, C. Beleites, H. J. Byrne, A. Chiadò, M. Chis, M. Chisanga, A. Daniel, J. Dybas, G. Eppe, G. Falgayrac, K. Faulds, H. Gebavi, F. Giorgis, R. Goodacre, D. Graham, P. L. Manna, S. Laing, L. Litti, F. M. Lyng, K. Malek, C. Malherbe, M. P. M. Marques, M. Meneghetti, E. Mitri, V. Mohaček-Grošev, C. Morasso, H. Muhamadali, P. Musto, C. Novara, M. Pannico, G. Penel, O. Piot, T. Rindzevicius, E. A. Rusu, M. S. Schmidt, V. Sergo, G. D. Sockalingum, V. Untereiner, R. Vanna, E. Wiercigroch and A. Bonifacio, Anal. Chem., 2020, 92, 4053–4064 CrossRef CAS PubMed.
- S. E. J. Bell, G. Charron, E. Cortés, J. Kneipp, M. L. de la Chapelle, J. Langer, M. Procházka, V. Tran and S. Schlücker, Angew. Chem., Int. Ed., 2020, 59, 5454–5462 CrossRef CAS PubMed.
- J.-F. Masson, ACS Sens., 2021, 6, 3822–3823 CrossRef CAS PubMed.
- L. Xiao, C. Wang, C. Dai, L. E. Littlepage, J. Li and Z. D. Schultz, Angew. Chem., 2020, 132, 3467–3471 CrossRef.
- L. Wu, A. Dias and L. Dieguez, Biosens. Bioelectron., 2022, 204, 114075 CrossRef CAS PubMed.
- V. Moisoiu, S. D. Iancu, A. Stefancu, T. Moisoiu, B. Pardini, M. P. Dragomir, N. Crisan, L. Avram, D. Crisan, I. Andras, D. Fodor, L. F. Leopold, C. Socaciu, Z. Bálint, C. Tomuleasa, F. Elec and N. Leopold, Colloids Surf., B, 2021, 208, 112064 CrossRef CAS PubMed.
- A. C. Hernández-Arteaga, A. C. S. Gómez, L. Godínez-Hernández, A. Hernández-Cedillo, M. M. Huerta, M. J. Yacamán and H. R. Navarro-Contreras, Placenta, 2022, 130, 12–16 CrossRef PubMed.
- S. Fornasaro, F. Berton, C. Stacchi, F. Farina, A. Esposito, V. Sergo, R. Di Lenarda and A. Bonifacio, Analyst, 2021, 146, 1464–1471 RSC.
- Y. Wang, C. Zhao, J. Wang, X. Luo, L. Xie, S. Zhan, J. Kim, X. Wang, X. Liu and Y. Ying, Sci. Adv., 2021, 7, eabe4553 CrossRef CAS PubMed.
- J. Liu, C. Cai, Y. Wang, Y. Liu, L. Huang, T. Tian, Y. Yao, J. Wei, R. Chen, K. Zhang, B. Liu and K. Qian, Adv. Sci., 2020, 7, 1903730 CrossRef CAS PubMed.
- Y. Lu, L. Lin and J. Ye, Mater. Today Bio, 2022, 13, 100205 CrossRef CAS PubMed.
- J. Ju, L. Li, S. Regmi, X. Zhang and S. Tang, Biosensors, 2022, 12, 606 CrossRef CAS PubMed.
- V. T. N. Linh, S.-G. Yim, C. Mun, J.-Y. Yang, S. Lee, Y. W. Yoo, D. K. Sung, Y.-I. Lee, D.-H. Kim, S.-G. Park, S. Y. Yang and H. S. Jung, Appl. Surf. Sci., 2021, 551, 149411 CrossRef CAS.
- Y. C. Hsieh, C. Y. Lin, H. Y. Lin, C. T. Kuo, S. Y. Yin, Y. H. Hsu, H. F. Yeh, J. Wang and D. Wan, Adv. Healthcare Mater., 2023, 12, 2300321 CrossRef CAS PubMed.
- W.-C. Lee, E. H. Koh, D.-H. Kim, S.-G. Park and H. S. Jung, Sens. Actuators, B, 2021, 344, 130297 CrossRef CAS.
- C. D. Bocsa, V. Moisoiu, A. Stefancu, L. F. Leopold, N. Leopold and D. Fodor, Nanomedicine, 2019, 20, 102012 CrossRef CAS PubMed.
- S. X. Leong, Y. X. Leong, E. X. Tan, H. Y. F. Sim, C. S. L. Koh, Y. H. Lee, C. Chong, L. S. Ng, J. R. T. Chen, D. W. C. Pang, L. B. T. Nguyen, S. K. Boong, X. Han, Y.-C. Kao, Y. H. Chua, G. C. Phan-Quang, I. Y. Phang, H. K. Lee, M. Y. Abdad, N. S. Tan and X. Y. Ling, ACS Nano, 2022, 16, 2629–2639 CrossRef CAS PubMed.
- L. Huang, Y. Zhu, C. Xu, Y. Cai, Y. Yi, K. Li, X. Ren, D. Jiang, Y. Ge, X. Liu, W. Sun, Q. Zhang and Y. Wang, ACS Sens., 2022, 7, 1439–1450 CrossRef CAS PubMed.
- Y. Huang, T. Xie, K. Zou, Y. Gu, G. Yang, F. Zhang, L.-L. Qu and S. Yang, Nanoscale, 2021, 13, 13344–13352 RSC.
- Y. Chen, Y. Zhang, F. Pan, J. Liu, K. Wang, C. Zhang, S. Cheng, L. Lu, W. Zhang, Z. Zhang, X. Zhi, Q. Zhang, G. Alfranca, J. M. de la Fuente, D. Chen and D. Cui, ACS Nano, 2016, 10, 8169–8179 CrossRef CAS PubMed.
- R. Xiao, L. Lu, Z. Rong, C. Wang, Y. Peng, F. Wang, J. Wang, M. Sun, J. Dong, D. Wang, L. Wang, N. Sun and S. Wang, Biosens. Bioelectron., 2020, 168, 112524 CrossRef CAS PubMed.
- S. Pagarin, A. Bolognese, S. Fornasaro, M. Franzin, U. Hofmann, M. Lucafò, R. Franca, M. Schwab, G. Stocco, G. Decorti and A. Bonifacio, Chem.-Biol. Interact., 2024, 387, 110792 CrossRef CAS PubMed.
- S. Fornasaro, V. Sergo and A. Bonifacio, FEBS Lett., 2022, 596, 1348–1355 CrossRef CAS PubMed.
- H. Lai, Z. Yu, G. Li and Z. Zhang, J. Chromatogr. A, 2022, 1675, 463181 CrossRef CAS PubMed.
- Q. Li, H. Huo, Y. Wu, L. Chen, L. Su, X. Zhang, J. Song and H. Yang, Adv. Sci., 2023, 10, 2202051 CrossRef CAS PubMed.
- Y. Ying, Z. Tang and Y. Liu, Nanoscale, 2023, 15, 10860–10881 RSC.
- R. A. Álvarez-Puebla, J. Phys. Chem. Lett., 2012, 3, 857–866 CrossRef PubMed.
- I. Pockrand, Chem. Phys. Lett., 1982, 85, 37–42 CrossRef CAS.
- S. Mitra and M. Basak, Mater. Today, 2022, 57, 225–261 CrossRef CAS.
- O. Guselnikova, H. Lim, H. J. Kim, S. H. Kim, A. Gorbunova, M. Eguchi, P. Postnikov, T. Nakanishi, T. Asahi, J. Na and Y. Yamauchi, Small, 2022, 18, 2107182 CrossRef CAS PubMed.
- X. Liu, J. Guo, Y. Li, B. Wang, S. Yang, W. Chen, X. Wu, J. Guo and X. Ma, J. Mater. Chem. B, 2021, 9, 8378–8388 RSC.
- P. P. P. Kumar, S. Kaushal and D.-K. Lim, TrAC, Trends Anal. Chem., 2023, 168, 117341 CrossRef CAS.
- X. Wang and L. Guo, Angew. Chem., Int. Ed., 2020, 59, 4231–4239 CrossRef CAS PubMed.
- I. Haidar, A. Day, U. Martino, A. Chevillot-Biraud, N. Felidj and L. Boubekeur-Lecaque, Appl. Mater. Today, 2019, 15, 462–471 CrossRef.
- N. G. Bastús, J. Comenge and V. Puntes, Langmuir, 2011, 27, 11098–11105 CrossRef PubMed.
- M. Grzelczak, J. Pérez-Juste, P. Mulvaney and L. M. Liz-Marzán, Colloidal Synth. Plasmonic Nanomet., 2020, 197–220 CAS.
- C. Matricardi, C. Hanske, J. L. Garcia-Pomar, J. Langer, A. Mihi and L. M. Liz-Marzán, ACS Nano, 2018, 12, 8531–8539 CrossRef CAS PubMed.
- J. Gargiulo, I. L. Violi, S. Cerrota, L. S. Chvátal, E. Cortés, E. M. Perassi, F. Diaz, P. Zemánek and F. D. Stefani, ACS Nano, 2017, 11, 9678–9688 CrossRef CAS PubMed.
- H. Zhang, J. Cadusch, C. Kinnear, T. James, A. Roberts and P. Mulvaney, ACS Nano, 2018, 12, 7529–7537 CrossRef CAS PubMed.
- G. A. Vinnacombe-Willson, Y. Conti, A. Stefancu, P. S. Weiss, E. Cortés and L. Scarabelli, Chem. Rev., 2023, 123, 8488–8529 CrossRef CAS PubMed.
- Y. Zhao and C. Xu, Adv. Mater., 2020, 32, 1907880 CrossRef CAS PubMed.
- A. Garcia-Leis and S. Sanchez-Cortes, ACS Appl. Nano Mater., 2021, 4, 3565–3575 CrossRef CAS.
- Y. Bao, Y. Li, L. Ling, X. Xiang, X. Han, B. Zhao and X. Guo, Anal. Chem., 2020, 92, 14325–14329 CrossRef CAS PubMed.
- L. Guerrini and R. A. Alvarez-Puebla, Frontiers of Nanoscience, Elsevier, 2020, 16, 9–23 Search PubMed.
- K. Honda, T. Hishiki, S. Yamamoto, T. Yamamoto, N. Miura, A. Kubo, M. Itoh, W.-Y. Chen, M. Takano, T. Yoshikawa, T. Kasamatsu, S. Sonoda, H. Yoshizawa, S. Nakamura, Y. Itai, M. Shiota, D. Koike, M. Naya, N. Hayakawa, Y. Naito, T. Matsuura, K. Iwaisako, T. Masui, S. Uemoto, K. Nagashima, Y. Hashimoto, T. Sakuma, M. Sakamoto, T. Kato, Y. Ino, H. Yoshida, H. Tsuda, N. Hiraoka, Y. Kabe and M. Suematsu, Redox Biol., 2021, 41, 101926 CrossRef CAS PubMed.
- E. Avci, H. Yilmaz, N. Sahiner, B. G. Tuna, M. B. Cicekdal, M. Eser, K. Basak, F. Altıntoprak, I. Zengin, S. Dogan and M. Culha, Cancers, 2022, 14, 5021 CrossRef CAS PubMed.
- L. Guerrini, E. Garcia-Rico, A. O’Loghlen, V. Giannini and R. A. Alvarez-Puebla, Cancers, 2021, 13, 2179 CrossRef CAS PubMed.
- M. Czaplicka, A. Kowalska, A. Nowicka, D. Kurzydłowski, Z. Gronkiewicz, A. Machulak, W. Kukwa and A. Kamińska, Anal. Chim. Acta, 2021, 1177, 338784 CrossRef CAS PubMed.
- S. Liu, H.-S. Su, Z. Yang and Y. Zhang, ACS Appl. Nano Mater., 2021, 5, 269–276 CrossRef.
- W. Ahmad, J. Wang, H. Li, T. Jiao and Q. Chen, TrAC, Trends Anal. Chem., 2021, 142, 116310 CrossRef CAS.
- S. Sloan-Dennison, E. O’Connor, J. W. Dear, D. Graham and K. Faulds, Anal. Bioanal. Chem., 2022, 414, 4541–4549 CrossRef CAS PubMed.
- Y. Hang, J. Boryczka and N. Wu, Chem. Soc. Rev., 2022, 51, 329–375 RSC.
- S. Guo, J. Popp and T. Bocklitz, Nat. Protoc., 2021, 16, 5426–5459 CrossRef CAS PubMed.
- N. Ali, S. Girnus, P. Rösch, J. R. Popp and T. Bocklitz, Anal. Chem., 2018, 90, 12485–12492 CrossRef CAS PubMed.
- S. Mostafapour, T. Dörfer, R. Heinke, P. Rösch, J. Popp and T. Bocklitz, Spectrochim. Acta, Part A, 2023, 302, 123100 CrossRef CAS PubMed.
- S. Guo, A. Kohler, B. Zimmermann, R. Heinke, S. Stöckel, P. Rösch, J. R. Popp and T. Bocklitz, Anal. Chem., 2018, 90, 9787–9795 CrossRef CAS PubMed.
- S. Guo, R. Heinke, S. Stöckel, P. Rösch, J. Popp and T. Bocklitz, J. Raman Spectrosc., 2018, 49, 627–637 CrossRef CAS.
- S. Guo, C. Beleites, U. Neugebauer, S. Abalde-Cela, N. K. Afseth, F. Alsamad, S. Anand, C. Araujo-Andrade, S. Askrabic, E. Avci, M. Baia, M. Baranska, E. Baria, L. A. E. B. de Carvalho, P. de Bettignies, A. Bonifacio, F. Bonnier, E. M. Brauchle, H. J. Byrne, I. Chourpa, R. Cicchi, F. Cuisinier, M. Culha, M. Dahms, C. David, L. Duponchel, S. Duraipandian, S. F. El-Mashtoly, D. I. Ellis, G. Eppe, G. Falgayrac, O. Gamulin, B. Gardner, P. Gardner, K. Gerwert, E. J. Giamarellos-Bourboulis, S. Gizurarson, M. Gnyba, R. Goodacre, P. Grysan, O. Guntinas-Lichius, H. Helgadottir, V. M. Grošev, C. Kendall, R. Kiselev, M. Kölbach, C. Krafft, S. Krishnamoorthy, P. Kubryck, B. Lendl, P. Loza-Alvarez, F. M. Lyng, S. Machill, C. Malherbe, M. Marro, M. P. Marques, E. Matuszyk, C. F. Morasso, M. Moreau, H. Muhamadali, V. Mussi, I. Notingher, M. Z. Pacia, F. S. Pavone, G. Penel, D. Petersen, O. Piot, J. V. Rau, M. Richter, M. K. Rybarczyk, H. Salehi, K. Schenke-Layland, S. Schlücker, M. Schosserer, K. Schütze, V. Sergo, F. Sinjab, J. Smulko, G. D. Sockalingum, C. Stiebing, N. Stone, V. Untereiner, R. Vanna, K. Wieland, J. Popp and T. Bocklitz, Anal. Chem., 2020, 92, 15745–15756 CrossRef CAS PubMed.
- S. Guo, J. Popp and T. Bocklitz, Spectroscopy, 2023, 38, 30–33 Search PubMed.
- S. Guo, P. Rösch, J. Popp and T. Bocklitz, J. Chemom., 2020, 34, e3202 CrossRef CAS.
- S. Guo, T. Bocklitz, U. Neugebauer and J. Popp, Anal. Methods, 2017, 9, 4410–4417 RSC.
- J. Contreras and T. Bocklitz, Biomed. Opt. Express, 2023, 14, 3259–3278 CrossRef PubMed.
- Y. Xie, M. Chen, X. Liu, X. Su and M. Li, Adv. Funct. Mater., 2023, 33, 2307091 CrossRef CAS.
- H. Shin, Y. Kang, K. W. Choi, S. Kim, B.-J. Ham and Y. Choi, Anal. Chem., 2023, 95, 6410–6416 CrossRef CAS PubMed.
- Y. Xie, X. Su, Y. Wen, C. Zheng and M. Li, Nano Lett., 2022, 22, 7910–7918 CrossRef CAS PubMed.
- X. Xie, W. Yu, Z. Chen, L. Wang, J. Yang, S. Liu, L. Li, Y. Li and Y. Huang, Nanoscale, 2023, 15, 13466–13472 RSC.
- J. Plou, P. S. Valera, I. García, D. Vila-Liarte, C. Renero-Lecuna, J. Ruiz-Cabello, A. Carracedo and L. M. Liz-Marzán, Small, 2023, 19, 2207658 CrossRef CAS PubMed.
- H. Shin, B. H. Choi, O. Shim, J. Kim, Y. Park, S. K. Cho, H. K. Kim and Y. Choi, Nat. Commun., 2023, 14, 1644 CrossRef CAS PubMed.
- A. Haleem, M. Javaid, R. P. Singh, R. Suman and S. Rab, Sens. Int., 2021, 2, 100100 CrossRef.
- X.-S. Zheng, I. J. Jahn, K. Weber, D. Cialla-May and J. Popp, Spectrochim. Acta, Part A, 2018, 197, 56–77 CrossRef CAS PubMed.
- H. Chen, A. Das, L. Bi, N. Choi, J.-I. Moon, Y. Wu, S. Park and J. Choo, Nanoscale, 2020, 12, 21560–21570 RSC.
- P. A. Mosier-Boss, Biosensors, 2017, 7, 51 CrossRef PubMed.
- X. Zhou, Z. Hu, D. Yang, S. Xie, Z. Jiang, R. Niessner, C. Haisch, H. Zhou and P. Sun, Adv. Sci., 2020, 7, 2001739 CrossRef CAS PubMed.
- S. Liu, Q. Hu, C. Li, F. Zhang, H. Gu, X. Wang, S. Li, L. Xue, T. Madl and Y. Zhang, ACS Sens., 2021, 6, 2911–2919 CrossRef CAS PubMed.
- N. E. Dina, M. A. Tahir, S. Z. Bajwa, I. Amin, V. K. Valev and L. Zhang, Biosens. Bioelectron., 2023, 219, 114843 CrossRef CAS PubMed.
- A. Pramanik, D. Davis, S. Patibandla, S. Begum, P. Ray, K. Gates, Y. Gao and P. C. Ray, Nanoscale Adv., 2020, 2, 2025–2033 RSC.
- W. J. Thrift, S. Ronaghi, M. Samad, H. Wei, D. G. Nguyen, A. S. Cabuslay, C. E. Groome, P. J. Santiago, P. Baldi and A. I. Hochbaum, ACS Nano, 2020, 14, 15336–15348 CrossRef CAS PubMed.
- J. C. Gukowsky and L. He, Spectrochim. Acta, Part A, 2022, 282, 121680 CrossRef CAS PubMed.
- H.-K. Huang, H.-W. Cheng, C.-C. Liao, S.-J. Lin, Y.-Z. Chen, J.-K. Wang, Y.-L. Wang and N.-T. Huang, Lab Chip, 2020, 20, 2520–2528 RSC.
- K.-H. Chen, S.-H. Lee, L.-C. Kok, T.-O. Ishdorj, H.-Y. Chang and F.-G. Tseng, Biosens. Bioelectron., 2022, 197, 113740 CrossRef CAS PubMed.
- Z. Zhang, D. Li, X. Wang, Y. Wang, J. Lin, S. Jiang, Z. Wu, Y. He, X. Gao, Z. Zhu, Y. Xiao, Z. Qu and Y. Li, Chem. Eng. J., 2022, 438, 135589 CrossRef CAS PubMed.
- Y. Pang, Q. Li, C. Wang, Z. Sun and R. Xiao, Chem. Eng. J., 2022, 429, 132109 CrossRef CAS.
- X. Jia, Z. Liu, Y. Peng, G. Hou, W. Chen and R. Xiao, Microchim. Acta, 2021, 188, 206 CrossRef CAS PubMed.
- M. Dramé, M. T. Teguo, E. Proye, F. Hequet, M. Hentzien, L. Kanagaratnam and L. Godaert, J. Med. Virol., 2020, 92, 2312 CrossRef PubMed.
- G. Zhdanov, E. Nyhrikova, N. Meshcheryakova, O. Kristavchuk, A. Akhmetova, E. Andreev, E. Rudakova, A. Gambaryan, I. Yaminsky, A. Aralov, V. Kukushkin and E. Zavyalova, Front. Chem., 2022, 10, 937180 CrossRef CAS PubMed.
- C. Tian, L. Zhao, G. Qi, J. Zhu and S. Zhang, Sens. Actuators, B, 2022, 371, 132445 CrossRef CAS PubMed.
- S. M. Quarin, A. C. Macke, L. N. Kissell, M. S. Kelly, A. Dayananda, J. Ungvary, G. Stan, R. I. Dima and P. Strobbia, ACS Sens., 2023, 8, 2000–2010 CrossRef CAS PubMed.
- S. H. Lee, I. B. Ansah, W.-C. Lee, J.-Y. Yang, C. Mun, H. Jang, S. Kim, S. Jung, M.-Y. Lee, H. S. Jung, T. Kang, S. Lee, D.-H. Kim and S.-G. Park, Chem. Eng. J., 2023, 454, 140066 CrossRef.
- M. Chisanga, H. Williams, D. Boudreau, J. N. Pelletier, S. Trottier and J.-F. Masson, Anal. Chem., 2023, 95, 3638–3646 CrossRef CAS PubMed.
- C. R. Wells and A. P. Galvani, Lancet Public Health, 2022, 7, e490–e491 CrossRef PubMed.
- M. B. Allen, J. Appl. Clin. Med. Phys., 2022, 23, e13628 CrossRef PubMed.
- A. Issatayeva, E. Farnesi, D. Cialla-May, M. Schmitt, F. M. A. Rizzi, D. Milanese, S. Selleri and A. Cucinotta, Talanta, 2023, 267, 125198 CrossRef PubMed.
- L. Ngo, A. Tukova, A. Hassanzadeh-Barforoushi, W. Zhang and Y. Wang, Lab Chip, 2023, 23, 2899–2921 RSC.
- J. Li, Y. Li, P. Li, Y. Zhang, L. Du, Y. Wang, C. Zhang and C. Wang, Acta Biomater., 2022, 144, 1–14 CrossRef CAS PubMed.
- F. Farshchi and M. Hasanzadeh, Biomed. Pharmacother., 2021, 134, 111153 CrossRef CAS PubMed.
- M. Constantinou, K. Hadjigeorgiou, S. Abalde-Cela and C. Andreou, ACS Appl. Nano Mater., 2022, 5, 12276–12299 CrossRef CAS PubMed.
- O. E. Eremina, A. T. Czaja, A. Fernando, A. Aron, D. B. Eremin and C. Zavaleta, ACS Nano, 2022, 16, 10341–10353 CrossRef CAS PubMed.
- C. Andreou, K. Plakas, N. Berisha, M. Gigoux, L. E. Rosch, R. Mirsafavi, A. Oseledchyk, S. Pal, D. Zamarin, T. Merghoub, M. R. Detty and M. F. Kircher, Nanoscale Horiz., 2022, 7, 1540–1552 RSC.
- J. H. Yu, I. Steinberg, R. M. Davis, A. V. Malkovskiy, A. Zlitni, R. K. Radzyminski, K. O. Jung, D. T. Chung, L. D. Curet, A. L. D’Souza, E. Chang, J. Rosenberg, J. Campbell, H. Frostig, S. Park, G. Pratx, C. Levin and S. S. Gambhir, ACS Nano, 2021, 15, 19956–19969 CrossRef CAS PubMed.
- Kenry, F. Nicolson, L. Clark, S. R. Panikkanvalappil, B. Andreiuk and C. Andreou, Nanotheranostics, 2022, 6, 31–49 CrossRef CAS PubMed.
- S. S. Panikar, D. Cialla-May, E. De la Rosa, P. Salas and J. Popp, TrAC, Trends Anal. Chem., 2021, 134, 116122 CrossRef CAS.
- O. A. Goryacheva, P. S. Pidenko, A. V. Markin, N. E. Markina, D. V. Tsupka, E. A. Mordovina, T. D. Ponomaryova, S. A. Meshcheryakova, D. A. Kornilov, P. D. Strokin, D. D. Drozd, Y. A. Podkolodnaya, A. A. Kovyrshina, I. V. Morozova, T. V. Shelekhova and I. Y. Goryacheva, TrAC, Trends Anal. Chem., 2023, 169, 117373 CrossRef CAS.
- S. S. Panikar, N. Banu, E.-R. Escobar, G.-R. García, J. Cervantes-Martínez, T.-C. Villegas, P. Salas and E. De la Rosa, Talanta, 2020, 218, 121138 CrossRef CAS PubMed.
- N. E. Markina, A. M. Zakharevich and A. V. Markin, Anal. Bioanal. Chem., 2020, 412, 7757–7766 CrossRef CAS PubMed.
- Y. Goksel, E. Dumont, R. Slipets, S. T. Rajendran, S. Sarikaya, L. H. Thamdrup, K. Schmiegelow, T. Rindzevicius, K. Zor and A. Boisen, ACS Sens., 2022, 7, 2358–2369 CrossRef CAS PubMed.
- N. E. Markina, I. Y. Goryacheva and A. V. Markin, Colloids Interfaces, 2023, 7, 42 CrossRef CAS.
- G. Soufi, E. Dumont, Y. Göksel, R. Slipets, R. A. Raja, K. Schmiegelow, H. Bagheri, A. Boisen and K. Zor, Biosens. Bioelectron.: X, 2023, 14, 100382 CAS.
- H. Liu, Y. Liu, T. Zhou, P. Zhou, J. Li and A. Deng, Molecules, 2022, 27, 4019 CrossRef CAS PubMed.
- N. E. Markina, S. N. Ustinov, A. M. Zakharevich and A. V. Markin, Anal. Chim. Acta, 2020, 1138, 9–17 CrossRef CAS PubMed.
- N. E. Markina, A. V. Markin and D. Cialla-May, Talanta, 2023, 254, 124083 CrossRef CAS PubMed.
- Y.-C. Chen, S.-W. Hong, H.-H. Wu, Y.-L. Wang and Y.-F. Chen, Nanomaterials, 2021, 11, 1789 CrossRef CAS PubMed.
- S. Farquharson, C. Shende, J. Newcomb, I. L. Petrakis and A. J. Arias, Molecules, 2023, 28, 2010 CrossRef CAS PubMed.
- C. Li, Z. Ye, Y. Xu and S. E. Bell, Analyst, 2020, 145, 6211–6221 RSC.
- C. Liu, S. Weber, R. Peng, L. Wu, W.-S. Zhang, P. B. Luppa, J. Popp and D. Cialla-May, TrAC, Trends Anal. Chem., 2023, 164, 117094 CrossRef CAS.
- S. Fornasaro, D. Cialla-May, V. Sergo and A. Bonifacio, Chemosensors, 2022, 10, 128 CrossRef CAS.
- L. Li, Y. Lu, Z. Qian, Z. Yang, K. Yang, S. Zong, Z. Wang and Y. Cui, Biosens. Bioelectron., 2021, 180, 113100 CrossRef CAS PubMed.
- G. Wang, C. Hao, W. Ma, A. Qu, C. Chen, J. Xu, C. Xu, H. Kuang and L. Xu, Adv. Mater., 2021, 33, 2102337 CrossRef CAS PubMed.
- E. Witkowska, A. M. Łasica, K. Niciński, J. Potempa and A. Kamińska, ACS Sens., 2021, 6, 1621–1635 CrossRef CAS PubMed.
- Q. Jiang, Z. Lin, M. Peng, B. Zhou, E. Liu, Z. Li, Y. Wei, H. Yang, F. Song and M. Yin, ACS Appl. Nano Mater., 2023, 6, 13524–13532 CrossRef.
- A. Chaichi, A. Prasad and M. R. Gartia, Biosensors, 2018, 8, 107 CrossRef CAS PubMed.
- A. I. Saviñon-Flores, F. Saviñon-Flores, G. Trejo, E. Méndez, Ş. Ţălu, M. A. González-Fuentes and A. Méndez-Albores, Front. Chem., 2022, 10, 1017305 CrossRef PubMed.
- C. Lin, X. Huang, J. Ning and D. He, IEEE Trans. Dependable Secure Computing, 2022, 20, 4536–4550 Search PubMed.
- V. Mani, C. Durmus, W. Khushaim, D. C. Ferreira, S. Timur, F. Arduini and K. N. Salama, Biosens. Bioelectron., 2022, 216, 114680 CrossRef CAS PubMed.
- N. R. Nirala, J. Asiku, H. Dvir and G. Shtenberg, Talanta, 2022, 239, 123087 CrossRef CAS PubMed.
- M. Erzina, A. Trelin, O. Guselnikova, A. Skvortsova, K. Strnadova, V. Svorcik and O. Lyutakov, Sens. Actuators, B, 2022, 367, 132057 CrossRef CAS.
- C. Y. Zhao, Y. Y. Liu, X. X. Zhang, G. C. He, H. Liu, D. W. Ji, Y. C. Hu and Q. A. Chen, Angew. Chem., 2022, 61, e202207202 CrossRef CAS PubMed.
- F. Gao, D.-C. Lu, T.-L. Zheng, S. Geng, J.-C. Sha, O.-Y. Huang, L.-J. Tang, P.-W. Zhu, Y.-Y. Li, L.-L. Chen, G. Targher, C. D. Byrne, Z.-F. Huang and M.-H. Zheng, Hepatology Int., 2023, 17, 339–349 CrossRef PubMed.
- P. Lu, D. Lin, N. Chen, L. Wang, X. Zhang, H. Chen and P. Ma, Anal. Methods, 2023, 15, 322–332 RSC.
- X. Wang, S. Wen, X. Du, Y. Zhang, X. Yang, R. Zou, B. Feng, X. Fu, F. Jiang and G. Zhou, Cell Death Dis., 2023, 14, 718 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |