Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Reproducibility in materials informatics: lessons from ‘A general-purpose machine learning framework for predicting properties of inorganic materials’†
Daniel
Persaud
 a,
Logan
Ward
a,
Logan
Ward
 c and
Jason
Hattrick-Simpers
c and
Jason
Hattrick-Simpers
 *ab
*ab
aDepartment of Materials Science and Engineering, University of Toronto, Canada. E-mail: jason.hattrick.simpers@utoronto.ca
bAcceleration Consortium, University of Toronto, Canada
cData Science and Learning Division, Argonne National Laboratory, USA
First published on 17th January 2024
Abstract
The integration of machine learning techniques in materials discovery has become prominent in materials science research and has been accompanied by an increasing trend towards open data and open-source tools to propel the field. Despite the increasing usefulness and capabilities of these tools, developers neglecting to follow reproducible practices presents a significant barrier for other researchers looking to use or build upon their work. In this study, we investigate the challenges encountered while attempting to reproduce a section of the results presented in “A general-purpose machine learning framework for predicting properties of inorganic materials.” Our analysis identifies four major categories of challenges: (1) reporting software dependencies, (2) recording and sharing version logs, (3) sequential code organization, and (4) clarifying code references within the manuscript. The result is a proposed set of tangible action items for those aiming to make material informatics tools accessible to, and useful for the community.
1 Introduction
Ensuring reproducibility when applying machine learning (ML) is an important, yet overlooked aspect of research, leading some to consider it in a state of “crisis”.1–3 Computational reproducibility means to obtain consistent results with the original work using the same methods, code, and data.4 These three components play a fundamental role in material informatics (MI), a field where statistical methods, ML, and materials data are used to understand processing–structure–property–performance (PSPP) relationships.5 MI represents a paradigm shift in materials science research, from the expert-driven empirical approach, towards data-driven techniques to understand the PSPP relationship.6–13 As MI continues to grow, so too does the development of novel tools and platforms to support it's implementation.14–17 However, a gap persists in developers adopting reproducible practices when distributing their MI tools, which hinders the efforts of other researchers looking to implement them. Despite concerns about a developer's embedded bias, known or unknown, propagating to users,18 promoting reproducibility remains crucial for instilling trust and facilitating widespread adoption.An early seminal contribution to the field of MI was presented by Ward et al. in 2016,14 introducing a unique ML framework to predict properties of inorganic materials. Despite the significance of the framework, which has subsequently been extended and further developed, we found the version of the code provided with in the supplementary information (SI) of ref. 14 to be incomplete. Specifically for the bandgap study, SI of ref. 14 only included a text interface script to build the proposed framework (referred to as the model building script in this study) was provided, while any script necessary for model training, predictions or analysis (referred to as the model extensibility analysis in this study) were entirely missing. Since these scripts that are unavailable, in this work we focus on replicating (obtain consistent results using new data or methods4) the associated analysis presented in Ward's original work, for which replication efforts have been unsuccessful in the past.19 We provide an account of the methodology from the original work, describe difficulties encountered during the replication, and explain how we address these challenges, the result of which is a set of recommendations based on the findings during the replication process. This case study stands as an illustrative instance within the broader context of reproducibility issues in MI, with the intent to enhance the usability of future open-source tools.
2 Summary of “A general-purpose machine learning framework for predicting properties of inorganic materials”
The intention of Ward et al. was to create a framework for building ML models which could be applied to a variety of inorganic material problems. The central outcome was the Material Agnostic Platform for Informatics and Exploration (Magpie) descriptors, a method of obtaining quantitative and chemically meaningful quantitative descriptors for inorganic compounds based on its composition. In addition to the Magpie descriptors, their proposed framework incorporated a hierarchical modeling approach to group data into chemically similar sections, then train a model on each subset, reducing the breadth of the descriptors per model. The effectiveness of the framework was demonstrated by applying it to case studies.The SI of ref. 14 that was provided with the Ward et al. paper contained the source code and compiled version of the Magpie software; which is written in Java,20 a set of input scripts related to the main parts of the paper, data needed to train the ML models, and documentation from the author. The scripts describe how to use Magpie to solve a specific problem (e.g., build a machine learning model with specific parameters) and are written in a text interface language which is interpreted by Magpie. In theory, these are sufficient to recreate the paper in its entirety because they contain the same ingredients (software, data, inputs) used by the original authors. However, the SI of ref. 14 is missing the scripts used for the extensibility analysis presented in the paper.
A first key claim in Ward et al. involved predicting bandgap energies of crystalline compounds from an ensemble of Reduced Pruning Error Trees (REPTrees) then comparing it to a single REPTree and random selection. The novel hierarchical strategy made 67% accurate predictions, outperforming the single REPTree and random selection, which yielded 46% and 12% respectively. The performance difference between the hierarchical REPTrees and the single REPTree legitimized the concept, and the model building script was made available in the SI of ref. 14
The framework was then extended to address a practical material design problem: identifying new materials with bandgaps suitable for solar cells from a set of unexplored ternary compounds. The outcome of this task was a list of five candidate compounds and their predicted bandgap (Table 1). The model to make these predictions are available in the model building script, but instructions on model training and making predictions were not. Since the original scripts are unavailable and the predictions are unverified, we focused on replicating this extensibility analysis.
| Composition | Predicted bandgap (eV) |
|---|---|
| ScHg4Cl7 | 1.26 |
| V2Hg3Cl7 | 1.16 |
| Mn6CCl8 | 1.28 |
| Hf4S11Cl2 | 1.11 |
| VCu5Cl9 | 1.19 |
3 Replication of search for new solar cell materials and gaps in reproducibility
Ward et al. made significant efforts to promote reproducibility by providing raw data, a script to build the proposed hierarchical model, as well as extensive documentation, but even still it is challenging. In the following sections we detail the challenges encountered at each stage of replicated the results.3.1 Installation and recreating scripts
Our first problem was installing the dependencies needed to run Magpie. The requirements are not just buried several pages deep in documentation but are also misleading. The documentation states that Magpie requires “Java Runtime Environment (JRE) Version 7 (v7) or greater”, but it is incompatible with the current Java version (v20). Moreover, JREv7 is no longer supported by Oracle, lacks security patches, and is only available for developers. While this is a slight challenge, it is overcome by installing the still-supported Java v8, but the misleading requirements hinder reproducibility.As noted previously, we also had to resort to recreating extensibility analysis scripts that were not made available on publication. Learning the input language for Magpie presents a first barrier to reproducing the results from Ward et al. Building these scripts also requires relying on imprecise, human language descriptions from the manuscript. There are many points for deviation between original study and software, for example ambiguous references to ML models (“our model” vs. “the hierarchical model”), and data (OQMD vs. the ICSD entries in OQMD) without clarity on the specific entities being referenced. Disparity between documented methodologies and their practical implementation in code underscores a notable gap. We recreated the scripts as closely as possible using both the paper and, as needed, consulting with an author of Ward et al.
3.2 Raw data
The next step was identifying the data which are used for training and the search space. The training set is version 1.0 of Open Quantum Materials Database (OQMD),21,22 contains 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 crystalline compounds and their properties (energy, bandgap, etc.) computed via density functional theory, and was provided in the original work. The original test dataset, composed of around 4500 yet-undiscovered ternary compounds predicted to be stable by Meredig et al.,23 was neither openly accessible, nor made available via Ward et al. Additionally, the original predictions are unavailable for reference in our study, because they were not archived by Ward and have since been lost. Therefore, our test set is limited to the predictions of the five most promising compounds published in the original work (Table 1). This unavailability of the original predictions underscores the importance of maintaining version control systems in working directories to track and document the evolution of a project. The absence of such a system can lead to the loss of critical information, hindering reproducibility.
000 crystalline compounds and their properties (energy, bandgap, etc.) computed via density functional theory, and was provided in the original work. The original test dataset, composed of around 4500 yet-undiscovered ternary compounds predicted to be stable by Meredig et al.,23 was neither openly accessible, nor made available via Ward et al. Additionally, the original predictions are unavailable for reference in our study, because they were not archived by Ward and have since been lost. Therefore, our test set is limited to the predictions of the five most promising compounds published in the original work (Table 1). This unavailability of the original predictions underscores the importance of maintaining version control systems in working directories to track and document the evolution of a project. The absence of such a system can lead to the loss of critical information, hindering reproducibility.
3.3 Replication with available scripts
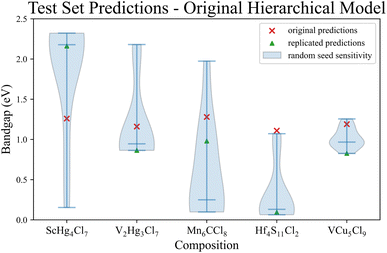
Following the resolution of computational dependency issues, we created models based on the model building script provided in the SI of ref. 14 Our replicated extensibility analysis is separated into two parts, the first script calculates and exports the descriptors for the provided training set and our test set. All the descriptors are created from the provided raw data, theoretically cloning the descriptors used in the original work. However, without a record to compare with directly or mention in the manuscript of the number of entries after data cleaning, validation of the replicated descriptors is not possible. The second script simply retrains a model from the model building script using the training dataset and subsequently generate predictions for the test dataset. Despite our best efforts, the bandgaps predicted by our model deviate significantly from the reported values (Fig. 1 green triangles) One potential explanation of the deviation is that our script is correct, and the differences are due to pseudo-randomness in the underlying algorithms. There is no record of the pseudo-random seed in the SI of ref. 14 so we performed a pseudo-random seed stability test with ten models, each with a different seed, ensuring to include the default pseudo-random seed of the underlying machine learning library, Weka,24 to determine whether the differences are due to different pseudo-random initialization.It is evident that none of the predictions from the original work can be replicated, based on the green triangles in Fig. 1. Most of the original predictions (Fig. 1 red x's) deviate toward the tail end of the pseudo-random seed sensitivity results (Fig. 1 blue violins), with the original prediction for Hf4S11Cl2 falling outside the spread of all replicated predictions. Additionally, the extreme sensitivity in pseudo-random seed variations in the replicated predictions raises concerns and has prompted further investigation. Utilizing the same descriptors, a SciKit-Learn Random Forest Regression (RF)25 model and an XGBoost Regression (XGB)26 model were trained. While all three models exhibit similar performance in 10-fold random cross-validation (ESI A),† the RF model proves considerably less sensitive to variations in the random seed, and the XGB model displays no sensitivity at all (ESI B).†
The contrast in the sensitivity to pseudo-random seed between the original hierarchical models and the modern models (RF and XGB) highlighting the potential susceptibility of the original hierarchical model to overfitting or other issues regarding generalization capabilities. Notably, the pseudo-random seed sensitivity test employed here was not a standard practice at the time of the original publication, emphasizing the evolving standards in model validation. Further exploration and validation with alternative models contribute to a nuanced understanding of the original hierarchical model's predictive performance and limitations. While the spread in the pseudo-random seed sensitivity test is an issue, not being able to reproduce the original predictions is of major concern within the scope of this work. Our primary hypothesis is that the Magpie codebase was being continually revised at the time of publishing, so it is not clear whether the version in the SI of ref. 14 is the same used to obtain the original results. The versioning issue was noted by an author as early as 2017.19 Without any documentation regarding the difference in Magpie versions or any of the intermediate results (descriptors, models, etc.), we cannot resolve this reproducibility challenge, as older versions are missing or have been overwritten.
4 Discussion
The framework proposed by Ward et al. achieved the objective of developing a generalized implementation of MI to aid materials research, while also demonstrating an effort to observe open science principles. Since the initial publication, Magpie has evolved to incorporate structural descriptors and has been integrated into Matminer, where it is now considered a standard MI baseline.16 However, the findings presented in this study indicate a crucial lesson: reproducibility demands deliberate effort, and without it, replication becomes very difficult. As a result, the following sections detail suggestions based on our replication effort, for developers of MI tools to aid reproducibility and implicitly, the ease of use for other researchers.4.1 Disseminate software dependencies
A significant hurdle to reproducibility emerges when developers presume that dependent software packages will remain compatible and readily accessible for years to come. However, the challenges posed by evolving, superseding, or abandoned dependencies are a well-recognized issues in computational reproducibility27,28 with many potential solutions, each accompanied by drawbacks. Docker, for instance, has gained popularity as an open platform for sharing and running tools within a loosely isolated environment to circumvent issues related to dependency management.29 However, it requires root access privileges, posing potential security concerns for large institutions or high-performance clusters. Singularity, an alternative solution that can be used without root access, but it is limited to Linux systems.30 Neither of these options are complete on their own, but either of them would have made our replication attempt easier. Going forward, developers must adopt a proactive stance in addressing dependency related issue, ensuring robust reproducibility when disseminating new MI tools, possibly by the containerization strategies we note above.4.2 Maintain and track versions
In 2017, Ward published a GitHub repository19 with an attempt to replicate the results of the original paper, noting that Magpie was in development at the same time published results were being gathered. Since there is no record of the Magpie version numbers at that time, nor a log of the machine learning operations (MLOps) for the results, it is very difficult to identify and address the root cause of our replicated prediction failing to align with the original. Employing a version control system, such as Git to track changes and the evolving state of a working directory over time aids reproducibility because changes can be easily identified and reverted.31 A step beyond Git tracking alone would be to record all vital aspects of an experiment systematically and meticulously on a run-to-run basis. MLFlow is a popular MLOps package which contains a tracking component to easily log numerous different aspects (start time, parameters, code version, metrics, etc.) of a project, ensuring the traceability of model lineage and fostering reproducible experiments.32 Publishing these logs to a digital library like Zenodo33 provides a method of long term data archival, in-line with Findable, Accessible, Interoperable and Reusable (FAIR) data management principles34 to aid in the reproducibility of materials informatics research in the long term.4.3 Utilize sequential coding practices
Most of the example scripts Ward et al. provided were in the form of a single, extensive input script that executed many tasks and demands a large learning-curve for new users, even with the inline comments. Implementing sequential coding practices that break the study into smaller subparts is an alternative approach that increases clarity, allowing new users to appreciate how new tools work more easily. The use of coding notebooks, for instance Jupyter notebooks,35 allow developers to separate code into task-specific cells with markdown blocks in-between for comprehensive subtask descriptions. This allows a new user to identify, understand, and implement a section of interest quickly. Moreover, we propose that examples should be formed by short and concise notebooks, each representing a fundamental step, allowing for each section to be executed independently, with the outputs exported for user validation. Although implementing these practices may require more effort from developers, they are valuable to users of varying skill levels, thereby fostering quicker and better implementation.4.4 Code clarity
Reproducibility inherently requires access to all relevant code and data; the code for the extensibility analysis was not provided by Ward et al., and one of the challenges with replication is the ambiguities in the original manuscript's procedures (as described in Section 3). To address this issue, we suggest incorporating clickable pointers in the manuscript that direct readers to specific files or lines of code, similar to how reference numbers are linked to citations in the reference section. Although this requires additional effort from the authors and publishers, the functionality for clickable references already exists and can greatly enhance the ability of researchers to understand the connection between the written rationale and the practical implementation.5 Conclusion
In this study we attempted to reproduce task presented in “A general-purpose machine learning framework for predicting properties of inorganic materials” by Ward et al., where they demonstrated the application of their framework to identify materials suitable for solar cell applications. Reproduction was not possible because of incomplete code, prompting us to replicate the results while highlighting challenges encountered and their resolutions. Facilitating a straightforward way to recreate computational environment alleviates the complexities of “dependency hell,” making it easier for users to reproduce results consistently. The use of version control system or dedicated tracking packages serves as a powerful mechanism for establishing a transparent and trustworthy record of the development process, instilling confidence in the reliability of published results. The division of a comprehensive script into discrete, self-contained segments, such as data cleaning, enhances user comprehension across various skill levels, enabling verification at distinct checkpoints. Lastly, incorporating clickable pointers from the manuscript to relevant files enhances clarity and reduces uncertainties inherent in concise writing. By adopting these practices, we anticipate that new tools will be more readily adopted and deployed, fostering further advancements in computational materials research.Data availability
The code and datasets used in this study are publicly available on GitHub and can be accessed at (https://github.com/dpersaud/ward2016_replication.git). The specific version of the code utilized for this research is version 1.0. Detailed information on data and code access, including versions and dates, is provided in the README.md file in the GitHub repository. All supporting data and code associated with this paper are included on GitHub.Author contributions
DP: data curation, formal analysis, visualization, writing – original draft, writing – review and editing. LW: supervision, writing – review and editing. JHS: conceptualization, supervision, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge funding provided by Natural Sciences and Engineering Research Council of Canada (NSERC), grant number RGPIN-2023-04843. LW was supported by the Exascale Computing Project (17-SC-20-SC) of the U.S. Department of Energy (DOE) and by DOE’s Advanced Scientific Research Office (ASCR) under contract DE-AC02-06CH11357.References
- National Academies of Sciences, Engineering, and Medicine, Open Science by Design: Realizing a Vision for 21st Century Research, The National Academies Press, Washington, DC, 2018 Search PubMed.
- M. B. A. McDermott, S. Wang, N. Marinsek, R. Ranganath, L. Foschini and M. Ghassemi, Sci. Transl. Med., 2021, 13, eabb1655 CrossRef PubMed.
- O. E. Gundersen and S. Kjensmo, Proceedings of the AAAI Conference on Artificial Intelligence, 2018, vol. 32, pp. 1644–1651 Search PubMed.
- Committee, Reproducibility and Replicability in Science, National Academies Press, Washington, D.C., 2019, p. 25303 Search PubMed.
- A. Agrawal and A. Choudhary, APL Mater., 2016, 4, 053208 CrossRef.
- R. K. Vasudevan, K. Choudhary, A. Mehta, R. Smith, G. Kusne, F. Tavazza, L. Vlcek, M. Ziatdinov, S. V. Kalinin and J. Hattrick-Simpers, MRS Commun., 2019, 9, 821–838 CrossRef CAS PubMed.
- R. Ramprasad, R. Batra, G. Pilania, A. Mannodi-Kanakkithodi and C. Kim, npj Comput. Mater., 2017, 3, 54 CrossRef.
- K. Rajan, Annu. Rev. Mater. Res., 2015, 45, 153–169 CrossRef CAS.
- J. Schmidt, M. R. G. Marques, S. Botti and M. A. L. Marques, npj Comput. Mater., 2019, 5, 83 CrossRef.
- T. Mueller, A. G. Kusne and R. Ramprasad, Reviews in Computational Chemistry, John Wiley & Sons, Inc, Hoboken, NJ, 2016, pp. 186–273 Search PubMed.
- Y. Liu, C. Niu, Z. Wang, Y. Gan, Y. Zhu, S. Sun and T. Shen, J. Mater. Sci. Technol., 2020, 57, 113–122 CrossRef.
- C. Chen, Y. Zuo, W. Ye, X. Li, Z. Deng and S. P. Ong, Adv. Energy Mater., 2020, 10, 1903242 CrossRef CAS.
- K. Choudhary, B. DeCost, C. Chen, A. Jain, F. Tavazza, R. Cohn, C. W. Park, A. Choudhary, A. Agrawal, S. J. L. Billinge, E. Holm, S. P. Ong and C. Wolverton, npj Comput. Mater., 2022, 8, 59 CrossRef.
- L. Ward, A. Agrawal, A. Choudhary and C. Wolverton, npj Comput. Mater., 2016, 2, 16028 CrossRef.
- K. Choudhary, K. F. Garrity, A. C. E. Reid, B. DeCost, A. J. Biacchi, A. R. Hight Walker, Z. Trautt, J. Hattrick-Simpers, A. G. Kusne, A. Centrone, A. Davydov, J. Jiang, R. Pachter, G. Cheon, E. Reed, A. Agrawal, X. Qian, V. Sharma, H. Zhuang, S. V. Kalinin, B. G. Sumpter, G. Pilania, P. Acar, S. Mandal, K. Haule, D. Vanderbilt, K. Rabe and F. Tavazza, npj Comput. Mater., 2020, 6, 173 CrossRef.
- A. Dunn, Q. Wang, A. Ganose, D. Dopp and A. Jain, npj Comput. Mater., 2020, 6, 138 CrossRef.
- L. Ward, A. Dunn, A. Faghaninia, N. E. Zimmermann, S. Bajaj, Q. Wang, J. Montoya, J. Chen, K. Bystrom, M. Dylla, K. Chard, M. Asta, K. A. Persson, G. J. Snyder, I. Foster and A. Jain, Comput. Mater. Sci., 2018, 152, 60–69 CrossRef.
- D. Danks and A. J. London, 26th International Joint Conference on Artificial Intelligence, (IJCAI 2017) Forthcoming, 2017, pp. 4691–4697 Search PubMed.
- L. Ward, Identify-Solar-Cell-Materials, Ipynb, 2017 Search PubMed.
- K. Arnold, J. Gosling and D. Holmes, The Java Programming Language, Addison-Wesley, Upper Saddle River, NJ, 4th edn, 2006 Search PubMed.
- S. Kirklin, J. E. Saal, B. Meredig, A. Thompson, J. W. Doak, M. Aykol, S. Rühl and C. Wolverton, npj Comput. Mater., 2015, 1, 15010 CrossRef CAS.
- J. E. Saal, S. Kirklin, M. Aykol, B. Meredig and C. Wolverton, JOM, 2013, 65, 1501–1509 CrossRef CAS.
- B. Meredig, A. Agrawal, S. Kirklin, J. E. Saal, J. W. Doak, A. Thompson, K. Zhang, A. Choudhary and C. Wolverton, Phys. Rev. B: Condens. Matter Mater. Phys., 2014, 89, 094104 CrossRef.
- E. Frank, M. A. Hall and I. H. Witten, The WEKA Workbench, Morgan Kaufhann, 4th edn, 2016 Search PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and É. Duchesnay, J. Mach. Learn. Res., 2011, 12, 5 Search PubMed.
- T. Chen and C. Guestrin, Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco California USA, 2016, pp. 785–794 Search PubMed.
- S. L. Sawchuk and S. Khair, JeSLIB, 2021, 10, 1206 Search PubMed.
- B. Grüning, J. Chilton, J. Köster, R. Dale, N. Soranzo, M. Van Den Beek, J. Goecks, R. Backofen, A. Nekrutenko and J. Taylor, Cell Syst., 2018, 6, 631–635 CrossRef PubMed.
- D. Merkel, Linux J, 2014, 239, 2 Search PubMed.
- G. M. Kurtzer, V. Sochat and M. W. Bauer, PLoS One, 2017, 12, e0177459 CrossRef PubMed.
- S. Chacon, Pro Git, Apress, New York, NY, 2nd edn, 2014 Search PubMed.
- A. Chen, A. Chow, A. Davidson, A. DCunha, A. Ghodsi, S. A. Hong, A. Konwinski, C. Mewald, S. Murching, T. Nykodym, P. Ogilvie, M. Parkhe, A. Singh, F. Xie, M. Zaharia, R. Zang, J. Zheng and C. Zumar, Proceedings of the Fourth International Workshop on Data Management for End-to-End Machine Learning, Portland OR USA, 2020, pp. 1–4 Search PubMed.
- European Organization For Nuclear Research, OpenAIRE, 2013 Search PubMed.
- M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. Da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. ’T Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. Van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. Van Der Lei, E. Van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, Sci. Data, 2016, 3, 160018 CrossRef PubMed.
- T. Kluyver, B. Ragan-Kelley, F. Pérez, B. Granger, M. Bussonnier, J. Frederic, K. Kelley, J. Hamrick, J. Grout, S. Corlay, P. Ivanov, D. Avila, S. Abdalla, C. Willing and J. D. Team, Jupyter Notebooks – a publishing format for reproducible computational workflows, Elpub, 2016, pp. 87–90 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00199g |
| This journal is © The Royal Society of Chemistry 2024 |