Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Event-driven data management with cloud computing for extensible materials acceleration platforms
Michael J.
Statt
*a,
Brian A.
Rohr
*a,
Dan
Guevarra
bc,
Santosh K.
Suram
 d and
John M.
Gregoire
d and
John M.
Gregoire
 *bc
*bc
aModelyst LLC, Palo Alto, CA 94306, USA. E-mail: brian.rohr@modelyst.io; michael.statt@modelyst.io
bDivision of Engineering and Applied Science, California Institute of Technology, Pasadena, CA 91125, USA. E-mail: gregoire@caltech.edu
cLiquid Sunlight Alliance, California Institute of Technology, Pasadena, CA 91125, USA
dToyota Research Institute, Los Altos, CA 94022, USA
First published on 21st December 2023
Abstract
The materials research community is increasingly using automation and artificial intelligence (AI) to accelerate research and development. A materials acceleration platform (MAP) typically encompasses several experimental techniques or instruments to establish a synthesis-characterization-evaluation workflow. With the advancement of workflow orchestration software and AI experiment design, the scope and complexity of MAPs are increasing, however each MAP typically operates as a standalone entity with dedicated experiment, compute, and database resources. The data from each MAP is thus siloed until subsequent efforts to integrate data into complex schema such as knowledge graphs. To lower the latency of data integration and establish an extensible community of MAPs, we must expand our automation efforts to include data handling that is decoupled from the resources of each MAP. Event-driven pipelines are well established in the computational community for building decoupled data processing systems. Such pipelines can be difficult to implement de novo due to their distributed nature and complex error handling. Fortunately, the broader computational science community has established a suite of cloud services that are well suited for this task. By leveraging cloud computing resources to establish event-driven data management, the MAP community can better realize the ideals of extensibility and interoperability in materials chemistry research.
1 Introduction
Materials Acceleration Platforms (MAPs) comprise the integration of automation and computation in experimental workflows to accelerate the discovery of materials as well as the underlying scientific knowledge.1,2 Critical analysis within the MAP community has led to the identification of a portfolio of remaining challenges,3–5 which can be broadly explained as furthering the extensibility and interoperability of MAPs as well as establishing universal data management protocols. Meanwhile, the successes of individual autonomous and self-driving laboratories has inspired and clarified the vision of global, interconnected MAPs.6–9 This vision may realize a million-fold increase in knowledge generation by accelerating scientific learning cycles from the traditional year-long cadence set by publications and conferences to sub-1 minute learning cycles via deep integration of artificial intelligence (AI). At a scientific level, realizing this vision requires development of AI that comprehends and reasons about scientific data so that the automated learning cycles better emulate those of human scientists.2,10–12 At a practical level, the greatest obstacle is the development of extensible and scalable management of MAPs and the data they produce. Recent progress in the design of ontologies13,14 and their integration with complex data schema such as knowledge graphs,14–18 provide a vision for the encoding of knowledge from a community of MAPs. For example, MatKG17 represents the knowledge from published abstracts and figure captions as relationships (edges) among materials properties, descriptors, applications, etc. (nodes).17 Such approaches provide new opportunities for human and machine learning from diverse data. The machinery for real-time interaction among MAPs and databases is relatively underdeveloped in the materials chemistry community.While the present discussion focuses on data management for MAPs, these challenges of constructing data pipelines mirror the broader challenges in management of materials and chemistry experiment data. Beyond the throughput of data generation, the experimental methods themselves are constantly evolving, requiring the data pipelines to be equally dynamic. The experimental data are produced by a wide variety of data acquisition software and instruments, which may be located across multiple labs. Metadata may be within the scope of the software or may be exclusively known by human researchers, requiring manual data entry and linking to the primary data. The workflows and associated data management requirements are often unique to a lab, and consequently the researchers typically construct bespoke data management pipelines, which dilutes effort toward their primary domain of expertise. These challenges are exacerbated by the standard researcher turnover in academic groups and have ultimately resulted in a lack of community-wide data pipeline infrastructure. Improved data engineering methodologies are needed to handle these problems and make scientific data pipelines more flexible, transparent, maintainable, and interoperable.
A key strategy to depart from the status quo is to decouple data management from the resources of experiment execution. The scope of automation within an instantiation of a MAP is at the level of a workflow, which includes all physical and computational resources to design and execute a series of experiment processes, typically spanning synthesis, characterization, and performance evaluation.19–21 Traditionally, a MAP has a dedicated database that may serve as the source data for AI-based experiment design as well as the destination of data produced by the experimental and/or computational workflow. The deep integration of data handling and workflow orchestration is the most straightforward way of creating a fast learning loop within a single workflow, but implementing fast learning cycles across interconnected MAPs requires data to be managed by an independent arbiter that is robust to the continual addition and removal of operational MAPs. Cloud-based workflow orchestration has been demonstrated,22,23 and extending the use of cloud services to include event-driven pipelines will enable the next generation of data management.
We have recently presented an event-based schema for data management,24 a schema for the resting state of data, for which a knowledge graph is a complementary schema.18 While event-driven pipelines and schema are natural partners, the pipeline can interface with any number of databases and a variety of schema.25 On the data generation side, the HELAO-async workflow orchestration software19 and the Globus platform23 explicitly represent workflow execution as a series of events, which also facilitates interfacing with event-driven data management. We assert that any computational or experimental workflow is naturally represented as a sequence of events, where each event comprises the set of actions and settings that produce a new piece of data. To help introduce the concepts and tools for implementing event-driven pipelines in materials chemistry, we herein summarize challenges, opportunities, and tools established in the broader field of computational science.26–29
2 Event-driven pipelines: advantages and key concepts
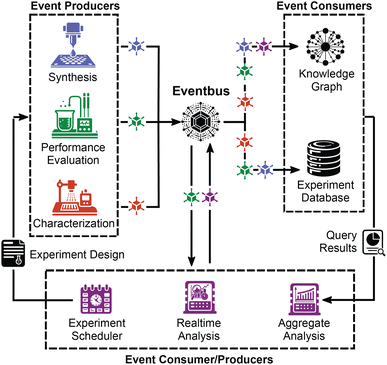
Fig. 1 illustrates an event-driven system for management of materials and chemistry data, where any synthesis, characterization, performance evaluation, etc. experiment constitutes an “event”. Events also include raw data being recorded by an instrument, a human entering metadata, or data analysis being performed. While the data flow in Fig. 1 is well suited for coupling to automated experiments, manually-performed experiments or analyses may also generate events, for example through a web form where manual data entry comprises an event producer whose published events enter the event bus alongside automatically-published events. Workflows that currently employ a laboratory information management system (LIMS) may seek to incorporate the LIMS into an event-based pipeline. Provided that the LIMS has an application programming interface (API) for accessing data, this API could be configured to send events to the event bus. Regular polling of the API may be necessary to detect new data, potentially causing delays in the system. Additionally, developing software to monitor for new data would create an obstacle in integrating data streams into a unified system. Therefore, it is advantageous to host user interfaces for data input on infrastructure that can directly interact with the event bus.Fig. 1 illustrates that events from any number of (manual or automated) producers are recorded, alongside their source and the time that they occurred, in a central “event bus”. Then, any number of functions can listen to the event bus and execute code when certain types of events occur. For example, this code could perform analysis of raw data, make insertions into a database, or trigger the execution of an active learning acquisition function that ultimately triggers the execution of new experiments.
The centralization of the event bus in lab operations has a variety of benefits. It offers lab-wide transparency of the experiment and computation events being executed. Since events can trigger execution of analysis functions, incorporating real-time data processing is straightforward, where completion of any analysis is modelled as a new event. Real-time data processing is critical to a future-proof data management system as researchers increasingly use AI-based decision-making in the lab.
An event-driven data pipeline allows downstream consumers to use events without needing to understand how they were produced. For example, if one researcher has an idea for a new way to analyze a stream of raw data being captured in the lab, they can implement a listener that runs the analysis without needing to interact with the data acquisition code or resources of the event producer. This independence bolsters the interoperability and maintainability of the system. If the researcher finds a bug in their analysis code or changes their database schema, they can use the event system to replay the historical event stream against the new code.
The event replay capability also eliminates the need for writing a translator to upgrade data to a new or additional database. The same code that ingests new data can also ingest legacy data by replaying the event stream. Legacy data and new data can even be sent to separate consumers based upon version identifiers within the event, enabling specificity in the responsibility of each consumer by reducing the scope of data any given consumer needs to consider. When there are multiple types of instruments, especially commercial instruments with different native data formats, the translation layer that unifies the data format for database ingestion and analysis can be developed asynchronously from the instrument control software. The event replay functionality thus enables new instruments (data producers) to be brought online without waiting for full development of the data consumers. Perhaps most foundationally, the event store serves as a ledger of what occurred in the lab, establishing a ground truth that is the cornerstone of traceability and reproducibility efforts.
3 Cloud computing solutions to challenges in implementing event-driven pipelines
Although event-based systems are very powerful, they can be difficult to implement from scratch. Regarding data security, creating an event bus that only certain users can interact with means that an identity management and permissions system needs to be in place. Per the design principle of unifying data management over many experiment workflows, an event-driven system uses distributed computing to aggregate events from many producers, requiring the event bus to robustly handle concurrent requests, which is not trivial to implement. Furthermore, error handling and debugging become increasingly difficult as more decoupled systems are chained together, motivating incorporation of robust logging systems. Altogether, it would take an experienced team of programmers a significant amount of time to implement such a system from scratch.Cloud computing addresses the majority of the challenges with implementing such systems. As detailed in Table 1, modern cloud service providers offer an event system, identity management, web security features, permissions system, compute platform to run custom code, managed database services, and robust logging systems. These services may be deployed to bring various aspects of lab automation into the cloud,22,23,30 and we believe that event-based data management comprises the most universally useful implementation of cloud computing for MAPs.
| Service | AWS | GCP | Azure |
|---|---|---|---|
| Executing functions | Lambda | Cloud functions | Azure functions |
| Event bus | Event bridge | Pub/sub | Event grid |
| Queues | SQS | Cloud tasks | Queue storage |
| Logging | Cloud watch | Cloud logging | Monitor |
| Permissions | IAM | IAM | Azure AD/RBAC |
While we recognize that learning to use these tools creates an activation barrier to widespread adoption of event-driven data management, we believe that this barrier is less significant than that faced by the ongoing transformation of experimental science via custom programming of automated workflows. Among materials and chemistry experimentalists, programming skills went from a rarity to an expectation in a matter of years. A similar evolution of skill set will occur as the value of cloud computing is increasingly recognized.
Cloud-based event-driven pipelines streamline complexity by using configuration files that are easily shared, in stark contrast to the extensive and intricate codebases typical of traditional git repositories. As the community establishes and shares performant configurations, the modular and intuitive nature of event-driven data management via cloud services will foster widespread deployment. In this manner, the general cloud computing tools summarized in Fig. 1 will be implemented into broadly-applicable materials chemistry data management systems within the next 1–2 years.
4 Conclusion
Materials and chemistry research inherently presents challenges for realizing flexible, maintainable, interoperable, and transparent data pipelines. The decoupled nature of event-based systems helps address these key challenges. Although event-based systems were once very difficult to implement, cloud computing has greatly reduced the barrier to using them, and it is now realistic for materials and chemistry data pipelines to take advantage of event-based architectures. The event-replay feature of the event bus enables experimental platforms to continue generating event-managed data while the community refines the ontologies and schema that enable global integration of materials chemistry data. To enhance data consistency and foster collaboration, it is crucial for the community to embrace standardized formats and bolster data standardization efforts, which pave the way for a streamlined integration across MAPs.31,32Conflicts of interest
Modelyst LLC implements custom data management systems in a professional context. J. M. G. is a consultant for companies that aim to accelerate materials discovery.Acknowledgements
This work was primarily funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Award DE-SC0023139 and the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Fuels from Sunlight Hub under Award DE-SC0021266. Additional support was provided by the Toyota Research Institute through their Accelerated Materials Design and Discovery program and the Resnick Sustainability Institute through an RSI Impact Grant.Notes and references
- M. M. Flores-Leonar, L. M. Mejía-Mendoza, A. Aguilar-Granda, B. Sanchez-Lengeling, H. Tribukait, C. Amador-Bedolla and A. Aspuru-Guzik, Curr. Opin. Green Sustainable Chem., 2020, 25, 100370 CrossRef.
- C. P. Gomes, B. Selman and J. M. Gregoire, MRS Bull., 2019, 44, 538–544 CrossRef.
- E. Stach, B. DeCost, A. G. Kusne, J. Hattrick-Simpers, K. A. Brown, K. G. Reyes, J. Schrier, S. Billinge, T. Buonassisi, I. Foster, C. P. Gomes, J. M. Gregoire, A. Mehta, J. Montoya, E. Olivetti, C. Park, E. Rotenberg, S. K. Saikin, S. Smullin, V. Stanev and B. Maruyama, Matter, 2022, 4, 2702–2726 CrossRef.
- J. Yano, K. J. Gaffney, J. Gregoire, L. Hung, A. Ourmazd, J. Schrier, J. A. Sethian and F. M. Toma, Nat. Rev. Chem, 2022, 6, 357–370 CrossRef.
- P. M. Maffettone, P. Friederich, S. G. Baird, B. Blaiszik, K. A. Brown, S. I. Campbell, O. A. Cohen, R. L. Davis, I. T. Foster, N. Haghmoradi, M. Hereld, H. Joress, N. Jung, H.-K. Kwon, G. Pizzuto, J. Rintamaki, C. Steinmann, L. Torresi and S. Sun, Digital Discovery, 2023, 1644–1659 RSC.
- J. Bai, L. Cao, S. Mosbach, J. Akroyd, A. A. Lapkin and M. Kraft, JACS Au, 2022, 2, 292–309 CrossRef CAS PubMed.
- M. Vogler, J. Busk, H. Hajiyani, P. B. Jørgensen, N. Safaei, I. E. Castelli, F. F. Ramirez, J. Carlsson, G. Pizzi, S. Clark, F. Hanke, A. Bhowmik and H. S. Stein, Matter, 2023, 6(9), 2647–2665 CrossRef.
- F. Strieth-Kalthoff, H. Hao, V. Rathore, J. Derasp, T. Gaudin, N. H. Angello, M. Seifrid, E. Trushina, M. Guy, J. Liu, X. Tang, M. Mamada, W. Wang, T. Tsagaantsooj, C. Lavigne, R. Pollice, T. C. Wu, K. Hotta, L. Bodo, S. Li, M. Haddadnia, A. Wolos, R. Roszak, C.-T. Ser, C. Bozal-Ginesta, R. J. Hickman, J. Vestfrid, A. Aguilar-Gránda, E. L. Klimareva, R. C. Sigerson, W. Hou, D. Gahler, S. Lach, A. Warzybok, O. Borodin, S. Rohrbach, B. Sanchez-Lengeling, C. Adachi, B. A. Grzybowski, L. Cronin, J. E. Hein, M. D. Burke and A. Aspuru-Guzik, Delocalized, Asynchronous, Closed-Loop Discovery of Organic Laser Emitters, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-wqp0d.
- Z. Ren, Z. Ren, Z. Zhang, T. Buonassisi and J. Li, Nat. Rev. Mater., 2023, 1–2 Search PubMed.
- A. Ourmazd, Nat. Rev. Phys., 2020, 2, 342–343 CrossRef.
- M. Ziatdinov, Y. Liu, K. Kelley, R. Vasudevan and S. V. Kalinin, ACS Nano, 2022, 16, 13492–13512 CrossRef CAS PubMed.
- H. Wang, T. Fu, Y. Du, W. Gao, K. Huang, Z. Liu, P. Chandak, S. Liu, P. Van Katwyk, A. Deac, A. Anandkumar, K. Bergen, C. P. Gomes, S. Ho, P. Kohli, J. Lasenby, J. Leskovec, T.-Y. Liu, A. Manrai, D. Marks, B. Ramsundar, L. Song, J. Sun, J. Tang, P. Veličković, M. Welling, L. Zhang, C. W. Coley, Y. Bengio and M. Zitnik, Nature, 2023, 620, 47–60 CrossRef CAS PubMed.
- J. Morbach, A. Yang and W. Marquardt, Eng. Appl. Artif. Intell., 2007, 20, 147–161 CrossRef.
- M. Kraft, J. Bai, S. Mosbach, C. Taylor, D. Karan, K. F. Lee, S. Rihm, J. Akroyd and A. Lapkin, Research Square, 2023 DOI:10.21203/rs.3.rs-3141873/v1.
- R. Choudhury, M. Aykol, S. Gratzl, J. Montoya and J. Hummelshøj, J. Open Source Softw., 2020, 5, 2105 CrossRef.
- K. S. Aggour, A. Detor, A. Gabaldon, V. Mulwad, A. Moitra, P. Cuddihy and V. S. Kumar, Integr. Mater. Manuf. Innov., 2022, 11, 467–478 CrossRef.
- V. Venugopal, S. Pai and E. Olivetti, arXiv, 2022, preprint, arXiv:2210.17340, DOI:10.48550/arXiv.2210.17340.
- M. J. Statt, B. A. Rohr, D. Guevarra, S. K. Suram, T. E. Morrell and J. M. Gregoire, Sci. Data, 2023, 10, 184 CrossRef.
- D. Guevarra, K. Kan, Y. Lai, R. J. R. Jones, L. Zhou, P. Donnelly, M. Richter, H. S. Stein and J. M. Gregoire, Digital Discovery, 2023, 2, 1806–1812 RSC.
- T. Konstantinova, P. M. Maffettone, B. Ravel, S. I. Campbell, A. M. Barbour and D. Olds, Digital Discovery, 2022, 1, 413–426 RSC.
- M. Sim, M. Ghazi Vakili, F. Strieth-Kalthoff, H. Hao, R. Hickman, S. Miret, S. Pablo-García and A. Aspuru-Guzik, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-v2khf.
- J. Li, J. Li, R. Liu, Y. Tu, Y. Li, J. Cheng, T. He and X. Zhu, Nat. Commun., 2020, 11, 2046 CrossRef CAS.
- R. Chard, J. Pruyne, K. McKee, J. Bryan, B. Raumann, R. Ananthakrishnan, K. Chard and I. T. Foster, Future Generat. Comput. Syst., 2023, 142, 393–409 CrossRef.
- M. J. Statt, B. A. Rohr, K. Brown, D. Guevarra, J. Hummelshøj, L. Hung, A. Anapolsky, J. M. Gregoire and S. K. Suram, Digital Discovery, 2023, 2, 1078–1088 RSC.
- M. Kleppmann, Designing data-intensive applications: The big ideas behind reliable, scalable, and maintainable systems, O'Reilly Media, Inc., 2017 Search PubMed.
- L. Chanussot, A. Das, S. Goyal, T. Lavril, M. Shuaibi, M. Riviere, K. Tran, J. Heras-Domingo, C. Ho and W. Hu, et al. , ACS Catal., 2021, 11, 6059–6072 CrossRef CAS.
- M. Uhrin, S. P. Huber, J. Yu, N. Marzari and G. Pizzi, Comput. Mater. Sci., 2021, 187, 110086 CrossRef.
- L. Talirz, S. Kumbhar, E. Passaro, A. V. Yakutovich, V. Granata, F. Gargiulo, M. Borelli, M. Uhrin, S. P. Huber and S. Zoupanos, et al. , Sci. Data, 2020, 7, 299 CrossRef.
- M. W. Gaultois, A. O. Oliynyk, A. Mar, T. D. Sparks, G. J. Mulholland and B. Meredig, APL Mater., 2016, 4, 053213 CrossRef.
- M. Segal, Nature, 2019, 573, S112–S113 CrossRef CAS.
- L. Bromig, D. Leiter, A.-V. Mardale, N. von den Eichen, E. Bieringer and D. Weuster-Botz, SoftwareX, 2022, 17, 100991 CrossRef.
- E. Huerta, B. Blaiszik, L. C. Brinson, K. E. Bouchard, D. Diaz, C. Doglioni, J. M. Duarte, M. Emani, I. Foster and G. Fox, et al. , Sci. Data, 2023, 10, 487 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |