Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bioprocessing 4.0: a pragmatic review and future perspectives
Kesler
Isoko
a,
Joan L.
Cordiner
b,
Zoltan
Kis
 bc and
Peyman Z.
Moghadam
bc and
Peyman Z.
Moghadam
 *a
*a
aDepartment of Chemical Engineering, University College London, Torrington Place, London WC1E 7JE, UK. E-mail: p.moghadam@ucl.ac.uk
bSchool of Chemical, Materials and Biological Engineering, University of Sheffield, Mappin Street, Sheffield S1 3JD, UK
cDepartment of Chemical Engineering, Imperial College London, South Kensington Campus, London SW7 2AZ, UK
First published on 30th July 2024
Abstract
In the dynamic landscape of industrial evolution, Industry 4.0 (I4.0) presents opportunities to revolutionise products, processes, and production. It is now clear that enabling technologies of this paradigm, such as the industrial internet of things (IIoT), artificial intelligence (AI), and Digital Twins (DTs), have reached an adequate level of technical maturity in the decade that followed the inception of I4.0. These technologies enable more agile, modular, and efficient operations, which are desirable business outcomes for particularly biomanufacturing companies seeking to deliver on a heterogeneous pipeline of treatments and drug product portfolios. Despite the widespread interest in the field, the level of adoption of I4.0 technologies in the biomanufacturing industry is scarce, often reserved to the big pharmaceutical manufacturers that can invest the capital in experimenting with new operating models, even though by now AI and IIoT have been democratised. This shift in approach to digitalisation is hampered by the lack of common standards and know-how describing ways I4.0 technologies should come together. As such, for the first time, this work provides a pragmatic review of the field, key patterns, trends, and potential standard operating models for smart biopharmaceutical manufacturing. This analysis aims to describe how the Quality by Design framework can evolve to become more profitable under I4.0, the recent advancements in digital twin development and how the expansion of the Process Analytical Technology (PAT) toolbox could lead to smart manufacturing. Ultimately, we aim to summarise guiding principles for executing a digital transformation strategy and outline operating models to encourage future adoption of I4.0 technologies in the biopharmaceutical industry.
1 Introduction
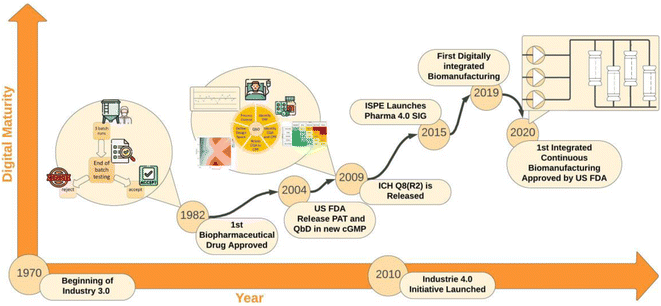
The adoption of Industry 4.0 (I4.0) technologies in the biomanufacturing industry is often referred to as Bioprocessing 4.0, a term that is rapidly growing in popularity. The term comes from Industrie 4.0,1 which was an initiative that began in Germany around 2010, where the interconnection of business models, supply chain,2 and processes through the Industrial Internet of Things (IIoT) was strategically used to drive manufacturing forward.3 The final objective of the initiative was to develop intelligent adaptive factories capable of delivering a more diverse and personalised product portfolio while running autonomously.3 However, since this manufacturing initiative did not involve bio-manufacturing,4 at the time of writing, biomanufacturing is still undergoing its third industrial revolution.3Fig. 1 shows the evolution in digital maturity of the biopharmaceutical manufacturing industry since its inception. In its early years biomanufacturing was predominantly performed using manual batch processes reliant on a paper-based manufacturing execution system (MES). Consequently, the revisions published by the US Food and Drugs Administrations (FDA) and subsequently consolidated by the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH), have led to an increased level of automation and digitalisation that marked a move towards Industry 3.0. Interestingly, although the third industrial revolution led to large-scale operations in other manufacturing industries such as oil and gas, where higher throughput is more desirable, in biomanufacturing, it began to foster a shift towards more efficient and smaller-scale operations. This is characterised by bioprocess intensification efforts aiming to “do more with less” by developing smaller, more modular factories that can produce various pharmaceuticals and require less resources to operate and build, while maintaining comparable levels of productivity.5
Technologies like single-use, plug-and-play, and advancements in cell-line development have unlocked transformative productivity gains in the previous decades. For example, using single-use bioreactors and modern cell lines, an expression level of 5 g L−1 can be achieved, making a 2000 L bioreactor as productive as a 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 L bioreactor from 20 years ago.6 Given the smaller operating volumes upstream, downstream processing is facilitated, thereby making the modern single-use bioreactors more attractive. Moreover, the development of end-to-end integrated continuous biomanufacturing (ICB) platforms comprising N-1 bioreactors, perfusion in production, multicolumn chromatography, simulated moving beds, true moving beds and single pass tangential flow filtration is advancing this trend further. The innovation in continuous bioprocessing culminated in 2019, when BiosanaPharma produced the first monoclonal antibody (mAb) using a fully integrated continuous biomanufacturing process.6 BiosanaPharma claimed that this continuous process significantly reduced production time, increased yield, and reduced costs compared to traditional batch manufacturing.7
000 L bioreactor from 20 years ago.6 Given the smaller operating volumes upstream, downstream processing is facilitated, thereby making the modern single-use bioreactors more attractive. Moreover, the development of end-to-end integrated continuous biomanufacturing (ICB) platforms comprising N-1 bioreactors, perfusion in production, multicolumn chromatography, simulated moving beds, true moving beds and single pass tangential flow filtration is advancing this trend further. The innovation in continuous bioprocessing culminated in 2019, when BiosanaPharma produced the first monoclonal antibody (mAb) using a fully integrated continuous biomanufacturing process.6 BiosanaPharma claimed that this continuous process significantly reduced production time, increased yield, and reduced costs compared to traditional batch manufacturing.7
More recently, these developments have received regulatory support with the release of Q13 (2023),8 which aims to provide guidelines to support continuous pharmaceutical manufacturing. As such, despite the relatively recent origins of this industry, the speed of technology transfer has been considerably faster compared to other manufacturing industries, as shown in Fig. 1. To this end, digital biomanufacturing represents one of the most recent attempts to enhance productivity. On October 15th, 2019, Sanofi launched a facility in Framingham, Massachusetts,9 that can be considered the first example of digital biomanufacturing. With a substantial investment of 4 billion dollars over five years, Sanofi established a digitally integrated biomanufacturing facility and was awarded the Facility of The Year Award in 2020.9 This facility featured paperless and data-driven manufacturing, continuous, intensified biologics production, connected and integrated processes, digital twins, as well as augmented reality.
1.1 The reality of bioprocessing 4.0
There have been many Bioprocessing 4.0 initiatives in Centres of Excellence (COE),10 expositions and studies in the literature.11–16 This has led to a fragmentation in the Bioprocessing 4.0 landscape with many small technical islands. Various companies and research institutes around the world are starting to implement Digital Twins, Artificial intelligence (AI), Computer Vision,17 augmented reality (AR)18 and additive manufacturing.19 There are even examples of robots performing automated aseptic sampling20 and implementations of blockchain technology to collect data from supply chains as proposed during the COVID-19 pandemic. However, there seems to be a lack of an overarching goal and an operating model for Bioprocessing 4.0 which clearly outlines how these technologies should come together.21 This could be attributed to a misconception of I4.0 as it is often described as being the application of machine learning (ML), Cyber Physical Systems (CPS), Internet of Things (IoT) and other technologies in a manufacturing setting.22 However, applications of AI in biomanufacturing go back to the early 1990s,23 thus merely applying these in manufacturing is neither novel nor will lead to tangible productivity gains as experienced in Industry 3.0. The misconception/misalignment hypothesis is supported by a survey carried out by the University College Cork in Ireland in 2020 where 78 respondents, representatives of the Irish multinational sector and thus global manufacturing scene were surveyed and they found that “Across all survey respondents, only 42% indicated any knowledge of 4.0”.24As such, characterising the I4.0 strategy by the technologies it leverages misses the core idea that has been successfully applied in other digitally transformed industries. The core idea is to connect Information Technologies (IT) and Operations Technologies (OT)14,17 such as Downstream Processing (DSP) and Upstream Processing (UPS) unit operations. In the original Industrie 4.0 vision, the composition of asset and administration shell was referred to as the I4.0 component,25 and the exchange of data and services within an I4.0 architecture forms a service oriented architecture (SOA) where every component along the industrial automation stack such as Enterprise Resource Planning (ERP), MES and Supervisory Control and Data Acquisition (SCADA) are connected. In this paradigm, data comes from various sources, including anything from raw materials distribution to market performance forming what was referred to as the value chain.2
Consequently, technologies can be considered tools rather than characteristics of the strategy, and should be deployed to solve the technical issues that arise from the desire to connect IT and OT. As a result, the convergence of IT and OT can lead to the development of CPS which help to monitor and automate parts of the manufacturing process.2,26 Therefore, one could argue that the culmination of a digital transformation journey is the development of high-fidelity digital representations which are a replica of the manufacturing process. These are often referred to as Digital Twins (DT). Using the analogy mentioned earlier, DTs are the most effective tool to tackle complex business objectives where optimal handling of materials, maintenance scheduling, and automation can present significant Return on Investment (ROI). The development of DTs comprises of data integration and process modelling. These two parts of DT development, should be explored in a complementary way. However, there has not been enough discourse around the data framework and the technologies available from other digitally transformed industries that can be readily applied to the biomanufacturing industry. As such, DTs are the key technology to transform industrial automation facilities from automated, as seen in Bioprocessing 3.0, to autonomous and achieve “The Smart Factory of the Future” capable of making intelligent decisions on its own.14
However, without understanding the underlying principles and problems that DTs and I4.0 technologies attempt to solve, those solutions may be seen as a black box by biomanufacturers that will not be able to effectively troubleshoot, customise, upgrade, or review their digital tools required by ICH, EMA, and FDA guidelines.27 Additionally, I4.0 opens up new forms of communications and automation that can disrupt the current operating models in the biopharmaceutical industry. This can unlock numerous opportunities for IT automation and advanced control strategies which need to be explored. In 2015, the International Society for Pharmaceutical Engineering (ISPE) trademarked the Pharma 4.0 initiative and organised Special Interest Groups (SIG) to come up with the Pharma 4.0 Operating Model, which aims to combine the original Industrie 4.0 model for organising manufacturing with the Pharmaceutical Quality System (PQS) outlined by ICH Q10.27 At the core of the Pharma 4.0 vision, there is the holistic control strategy which aims to interconnect and automate processes across the pharmaceutical value chain from supply through to patient feedback. Thanks to the structured nature of the PQS which present opportunities for automation, and with the advent of IIoT technologies, these objectives are now achievable. The work of ISPE serves as a guiding framework that can reduce the technical risk of moving some of the I4.0 technologies from COE and process development to full GMP manufacturing.27 Furthermore, in the literature, many roadmaps and frameworks can be found to start adopting Process Analytical Technologies (PAT)28 and DTs.29 In addition, the BioPhorum IT Digital Plant team has developed a maturity assessment tool to be used alongside the Digital Plant Maturity Model (DPMM).30
Another key enabler of Bioprocessing 4.0 is the Industrial Internet of Things (IIoT). However, contrary to popular belief, the technology and problems that IIoT is trying to solve have been partly addressed by SCADA systems, since the late 1990s. The implementation of SCADA enabled the monitoring and control of various components of large industrial infrastructures such as oil pipelines, from a remote location via streaming data often referred to as “telemetry”. As such, SCADA can be considered a pre-cursor of modern IIoT. Nevertheless, IIoT represents a philosophical shift from point-to-point integration to interoperability and interconnection across the stack, to accommodate the increasingly event-driven nature of industrial operations. This is made possible by technologies such as, Message Queuing Telemetry Transport (MQTT) for efficient and scalable data transmission and Open Platform Communication Unified Architecture (OPC UA) for machine-to-machine communication across SCADA, MES, and Process Control Systems (PCS). Additionally, thanks to gateway technologies provided by OT connectivity platforms such as Cogent DataHub and KEPserverEX that can convert traditional communication protocols i.e. Modbus into OPC UA, biomanufacturers can now leverage IIoT architectures. Nevertheless, the interconnection of IT and OT leads to data management challenges rising from the inconsistent identification of “tags”, assets, process variables and status codes across disparate MES, DCS and SCADA systems. A framework that is gaining popularity in industrial circles is the Unified Namespace, popularised by Walker Reynolds. This leverages common standards such as ISA-95 which provide a hierarchical structure that can be used to represent the plant digitally.31 This framework allows the coherent contextualisation of all the manufacturing events that can be used to automate resource planning and manufacturing execution. Complementing IIoT, the democratisation of ML, AI and cloud computing provides transformative opportunities for the biomanufacturing industry. Deploying ML models to production, where they can communicate with other manufacturing equipment and data collection systems, nowadays is as easy as configuring a Dockerfile. In only 20 lines of code, state-of-the-art (SOTA) ML models can be used to uncover patterns in large data sets which are not easy to find using traditional methods.
Another technology that can take advantage of IIoT connectivity are AR (Augmented Reality) and VR (Virtual Reality) technologies. These technologies are predominantly used for troubleshooting and training personnel or assisting in executing complex workflows. They offer significant benefits when connected to a centralized data hub containing Standard Operating Procedures (SOPs), real-time data collection and feedback. In bioprocessing, AR can overlay real-time data onto equipment, guiding operators through diagnostic procedures, thereby improving quality control and efficiency. For example, factory employees using smart glasses can control virtual equipment with hand gestures and receive real-time updates, such as maintenance reminders or alerts for worn-out parts. VR can simulate real-world scenarios, providing hands-on experience without the risks associated with actual operations. This immersive experience makes complex models easier to interact with and understand, thereby enhancing the effectiveness of DTs. These technologies have shown promising results in areas requiring complex, safer, scalable training, such as scientific research and laboratory procedures as exemplified by Lab 427 developed by Dr Stephen Hilton,32 and in industrial settings as shown by Körber Pharma's products.33
Overall, the technical maturity of I4.0 technologies, the guiding frameworks from the ISPE and the growing number of success stories in digital manufacturing, provide the right foundations to enable the fourth industrial revolution in the biomanufacturing industry. As such, in the present paper, we aim to synthesise the ongoing dialogues and initiatives in literature, regulatory environments, industry, and global institutions focused on the digital transformation of biomanufacturing adopting the Bioprocessing 4.0 strategy. We particularly focus on biopharmaceutical drug manufacturing, often termed red biotechnology, primarily on post-identification and profiling of risk phases of the target drug product within the Quality-by-Design framework. The objective is to identify key patterns, trends, and potential standard operating models for smart factories. This analysis will assist pharmaceutical companies embarking on a digital transformation journey, offering insights into the Bioprocessing 4.0 ecosystem. Additionally, the review aims to assist digital twin developers by outlining a pragmatic methodology to iteratively improve, connect and deploy digital twins in biomanufacturing. Finally, the perspective aims to highlight feasible future directions for the biomanufacturing industry and the Quality-by-Design framework in the context of Industry 4.0.
2 The evolution of the quality by design (QbD) framework towards bioprocessing 4.0
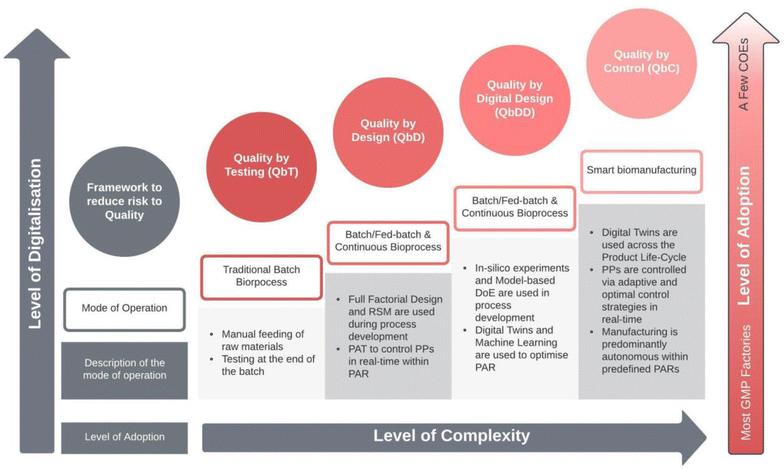
To discuss the evolution of biomanufacturing and the role of technology in shaping process development and compliance with current Good Manufacturing Practices (cGMP), it is useful to focus on how these advances have transformed process validation and quality assurance. While the methods of ensuring product quality in the biomanufacturing industry have evolved over time, the fundamental goals of these processes have remained consistent, reflecting the unique requirements and challenges of this sector. As such, in this section, an account of the evolution in process development and validation is provided through the lenses of four quality frameworks, Quality by Testing (QbT), Quality by Design (QbD), Quality by Digital Design (QbDD), and Quality by Control (QbC). In Fig. 2 the quality frameworks are laid out according to their digital maturity, level of complexity and the level of adoption as indicated by the intensity of the colour. In what follows, we will describe how the development and operation of biomanufacturing processes have evolved through regulatory and technological advances. Additionally, a technical perspective of how bioprocess development and operation will evolve under Bioprocessing 4.0 is provided.2.1 Quality by testing (QbT)
As of now, the biopharmaceutical manufacturing industry is still considered a relatively new field. Although, bioprocessing of products such as beer and wine has been around for centuries, the first biopharmaceutical drug was approved by the FDA and commercialised around 40 years ago. At that time, biopharmaceuticals were produced in batches.5 These batches were produced at specific operating conditions approved by regulatory bodies, and their quality was tested at the end of the process.34,35 This practice was called Quality by Testing (QbT)34 and required significant regulatory oversight. Adequate monitoring and analytical tools capable of capturing the multivariate state of bioprocesses were not available, making it difficult for biomanufacturers to know the quality of the batch until the end of the process. Additionally, process validation was seen as an activity on top of process development as multiple Process Performance Qualification (PPQ) runs would be required, often leading to sub-optimal biomanufacturing operations36 which could not be changed without regulatory notice. Furthermore, the quality control of drug products relied heavily on multiple univariate specifications. This approach, while useful, had its limitations as it often failed to detect anomalies that would only become apparent when multiple factors were analysed in conjunction, thereby overlooking complex interactions between variables. Consequently, this left potential outliers and critical variances in the production process undetected.Around the same time, the field of multivariate data analysis (MVDA) started to gain popularity. MVDA techniques such as Principal Component Analysis (PCA)37 have been used in fault detection and review of historical batch records since the early 90s, which enabled the rapid identification of outliers in the multivariate space. Further developments of this technique saw its applications in batch monitoring in real-time.38,39 These developments were accompanied by the emergence of chemometrics and by the use of Projection To Latent Structures (PLS) as a “calibration” technique which enabled the use of soft sensors that can estimate physical properties from spectral measurements. The growing popularity of MVDA techniques led to the development of software packages such as CAMO Analytics® and subsequently Umetrics SIMCA®, a package developed by the pioneers of Multivariate Statistical Process Control (MSPC), and chemometrics. At the time of writing, the software package has been acquired by Sartorius and is now part of the Umetrics® Software Suite.40 Additionally, MSPC capabilities have now been integrated in other products such as PharmaMV® from Perceptive Engineering and can be developed using open-source programming languages such as R (with plotly, shiny, PCAtools and chemometrics libraries) and python (with Dash and pyphi).
2.2 Quality by design (QbD)
In 2004, the US FDA published a revision of cGMP with a “risk-based approach”.41 The idea of this revision was that “quality cannot be tested into products, but rather should be built-in by design”, in other words, by designing biomanufacturing processes which can consistently make products with the predefined quality and specification; this was referred to as the Quality by Design (QbD). QbD starts with the end in mind; the first stage involves understanding the product by identifying the Quality Target Product Profile (QTPP),42 which is a summary of the quality characteristics of the drug product to ensure safety and efficacy.The second phase involves understanding the process by carrying out preliminary experiments and by using risk assessment tools such as Fishbone Diagrams (Ishikawa) and Failure-Mode-Effect Analysis (FMEA) to identify critical quality attributes (CQA),43 which are quality attributes with critical impact to the efficacy of the drug. This is accompanied by the use of statistical techniques such as Full Factorial Design of Experiments (DoE) and Response Surface Methodology (RSM), where the multivariate relationship between CQAs and process parameters (PP) is explored to find any statistically significant correlation. The PPs that correlate the most with the CQAs can be considered critical process parameters (CPPs) which are important to map out in the design space. This leads to the third phase, the process design, which involves finding the right combination of CPPs that can consistently produce the desired drug product. Finally, a control strategy needs to be developed with the goal of identifying process acceptance ranges (PAR) within the design space where the process can be controlled to produce optimal results. As such, the FDA provided important regulatory guidance by identifying a class of tools that can be used to monitor, analyse, and control processes referred to as Process Analytical Technologies (PAT).44 This led to a philosophical shift from QbT to real-time release testing (RTRT),34 a more responsive approach that can reduce the variability of the final product by adjusting the process in real time within the predefined PAR.
Additionally, the QbD framework outlined by the FDA was also subsequently promoted by the ICH, which published various quality guidelines to highlight the importance of pharmaceutical development Q8 (2005),42 quality risk management Q9 (2005) and the Pharmaceutical Quality System (PQS) Q10 (2008)45 thus introducing PQS elements for a more holistic approach. The PQS included management participation, corrective and preventive action (CAPA), change management systems enabled by robust knowledge management and quality risk management. At the time of writing, these developments constitute the standard operating model in the manufacturing of pharmaceutical medicines.
2.3 Quality by digital design (QbDD)
Issues with the QbD framework have been raised by a few reports.29,46 One of the most notable problems of the framework is the lack of use of existing and acquired process knowledge to speed up process development and reduce its costs. A more detailed business case for knowledge driven process development is provided by von Stosch et al.47 It is widely accepted that QbD can only be cost effective if knowledge is transferred from one product to the next, iteratively decreasing the number of experiments required to generate process understanding.47 However, given the extensive use of statistical tools such as Full Factorial DoE, which assumes no knowledge of the current bioprocess under development, and provides almost a brute-force approach to explore its design space, QbD remains resource intensive. Even considering a Fractional Factorial Design approach, that can reduce experimental efforts, without supplementary experiments to study the centre points of the design space, this may miss higher-order interactions and often provide inadequate levels of resolution. Furthermore, although RSM provides a robust methodology for identifying statistically significant relationships in the experimental data, it gives unsatisfactory performance when modeling highly non-linear systems.48The quality by digital design (QbDD) is a framework that is gaining popularity in the literature.36,49–52 This framework aims to leverage digital tools and process models to shift a significant amount of experimental design planning and execution, design space exploration and what-if-analysis from in vitro or in vivo to in silico, thereby enabling knowledge-based process development53 and computer aided QbD.36 By doing so, experimental effort has been reported to be reduced from 67% (ref. 54) up to 75%.53 This can make the QbD framework more profitable by reducing the amount of time and raw materials used in process development thereby increasing the lifetime of the drug in the market.29
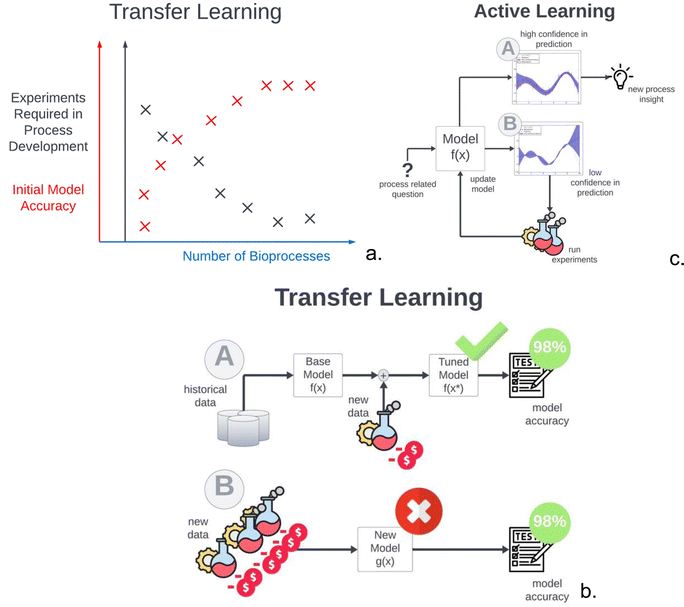
Fig. 3a provides a semi-quantitative illustration of how the number of experiments required in process development reduce exponentially as the initial model accuracy is improved by transferring knowledge from one bioprocess to the next. For this exercise, data driven models can be used to take advantage of historical data. A base model can be trained on existing data coming from other bioprocesses with the same cell-line with similar CPPs and CQAs, or the same bioprocess at different scales.55 Subsequently, the data-driven model parameters can be fine-tuned using data coming from the new process. This technique is illustrated in Fig. 3b and is often referred to as transfer learning. This allows the development of new models using historical data and a small amount of new data, while maintaining accuracy comparable to models trained solely on new data.56 Additionally, by exclusively conducting experiments in areas where the model exhibits significant uncertainty, this approach effectively minimises the number of necessary experiments. This method is displayed in Fig. 3c and is often referred to as active learning, which can drastically improve the sampling efficiency of the experimental campaign.
In a pivotal study by Hutter et al.,56 knowledge from one product to the other was efficiently transferred by modifying a Gaussian Process (GP) model. Here a novel method to represent different products (such as therapeutic proteins or vaccines) by embedding vectors, which are learned from bioprocess data using a GP model, has been proposed. The embedding vectors capture the similarities and differences between products and enable knowledge transfer across products. This novel method shows that the GP can reach comparable levels of accuracy to the model trained solely on new data, after only four experimental runs.
Nevertheless, in biomanufacturing, process knowledge often outweighs the amount of data available, especially during the early stages of process development. Therefore, first principle, mechanistic models have received a lot of attention in the literature57 due to their ability to capture expert knowledge. One of the most common process models is the macroscopic material balance with Monod-type kinetics, since it can satisfactorily describe cell growth and transport phenomena. However, when more data is available, hybrid semi-parametric models where machine learning and other statistical techniques have been used to describe the kinetics of the cell cultures with better performance. A more detailed account of these modelling techniques is provided in Section 3.1. However, the application of mechanistic process models in QbD under a QbDD framework has been presented in various scenarios.51,54,58,59 Two main scenarios where QbDD can be applied have been identified: (i) there are multiple competing hypothesis of bioprocess model structure. (ii) There are similar models available a priori from the literature or from similar bioprocesses.
In the first case, QbDD is applied via a model-based Design of Experiments (mbDoE) approach. This is a two-part exercise; initially competing model structures are discriminated systematically and then the selected model parameters are calibrated using information rich optimal experimental design.60 A list of optimal experimental designs has been provided by Herwig et al.55 However, one of the most common schemes for the mbDoE framework is the D-optimal design where the determinant of the Fisher Information Matrix61 of the model is calculated to obtain the best candidate experiments and reduce the variance in the model parameters.46 Once an accurate model is developed, this can be used for in silico design space exploration and identification of CPPs. For example, van de Berg et al.51 used variance based Global Sensitivity Analysis (GSA) by calculating Sobol Indices for all the inputs of the process model and ranking them in order of their level of contribution to the change in output. This narrowed down the number of PPs to investigate for design space optimisation.
It should be noted that the application of mbDoE has found more challenges in its adoption within the literature.55 However, this technique allows process developers to predict the information content of the next set of experiments which can consist of sequential or parallel experiments or can be a combination of both,46 thereby providing flexibility in the execution of those experiments. As such, with the commercialisation of the Ambr® 250, Sartorius, high-throughput experiments (HTE) have been made more accessible enabling process developers to perform parallel experiments which will facilitate mbDoE campaigns with parallel designs which makes the model discrimination phase less resource intensive.62 Additionally, open-source libraries such as AlgDesign and DoE.wrapper in R or pyDOE2 and statsmodels in python provide support for calculating D-optimal experimental designs and other crucial components of mbDoE. JMP® by SAS is also a popular option as it provides an interactive low-code, no-code graphical user interface that does not require programming experience.
In the second case, the QbDD applications show the most promising results. In this scenario, it is possible to use model assisted Design of Experiments (mDoE) where a Latin Hypercube Sampling (LHS) or similar screening experiments as shown by Herwig et al.55 can be performed to fit the initial model for calibration. The model is then used to explore in silico (via simulations) various experimental designs55 and boundary conditions providing insights as to what experiments are the most promising. The simulated data is then analysed using Analysis of Variance (ANOVA) and RSM to find the boundary process conditions where the response surface peaks and the PPs that explain the most variance in the response, thereby reducing the experimental space for the successive set of experiments. These set of experiments can be run and simulated using the model to then update the model with the obtained experimental observations.53 It is important to clarify that the main differences between mbDoE and mDoE are in the use of the model. In model assisted DoE, the model is used to assist the experimental campaign and different designs are simulated and analysed in silico in order to find optimal process conditions, whereas in mbDoE, the models are used to suggest optimal experiments which can improve its structure or parameters.55
If no initial process models are available, data-driven models can be used to develop adaptive sampling strategies which can reduce the experimental burden. Most notably, Bayesian Optimisation (BO) workflows, have been instrumental in enabling self-driving experiments in the small-molecule pharmaceutical industry and is now starting to see adoption in the bioprocess industry. In a study conducted by Rosa et al.,59 mRNA production was maximised using a surrogate model in a BO workflow. The surrogate model was interpreted using SHAP (SHapley Additive exPlanations) which provides a ranking of the process parameters level of impact on the model output. The surrogate model was also used to optimise the bioprocess and suggest optimal experiments.
Finally, during the IFAC Workshop Series on Control Systems and Data Science Toward Industry 4.0, Professor Richard Braatz discussed the progressive shift towards fully digitised process development in Bioprocessing 4.0, driven by the increasing reliance on in silico experiments and digital tools.63 He highlighted a case where his team successfully designed a continuous pharmaceutical manufacturing plant entirely in silico, which, upon actual construction, produced pharmaceuticals meeting all purity standards, while halving the total costs by eliminating experimental efforts.64 This demonstrates the benefits of the extensive use of computer aided QbD or QbDD. Additionally, his group at MIT, developed Algebraic Learning Via Elastic Net (ALVEN) a smart analytics algorithm which can be used to automate the data-driven model selection process in the absence of process knowledge.65 According to these studies, Automated Machine Learning (AutoML) tools, which select the optimal data driven model for a given data problem, combined with automated micro-scale technologies, which can perform experiments in parallel at a fraction of the cost, will become pillars of advanced manufacturing.
2.4 Quality by control (QbC)
So far, the use of digital tools to optimise process development has been discussed. However, the emerging trend of using these tools to optimise bioprocesses in operation could potentially disrupt the biomanufacturing industry in the coming years. This is because digital tools can enable autonomous UPS operation, which could lead to more profitable, sustainable, and efficient operations.Quality by control (QbC) is characterised by controlling the quality of the biopharmaceutical drug product via advanced process control (APC) strategies in real-time using the process knowledge and technologies developed during process development.35,66 As noted by Sommeregger et al.,46 the PAT guidance provided by the FDA “reads like a guide to realise APC” even though this is not explicitly addressed. Arguably, the models developed to relate CPPs and CQAs and the PARs can be used as dynamic process models and set points in APC.46
However, in the biomanufacturing industry, an adequate control system needs to be able to cope with both uncertainty and the nonlinearities of bioprocesses which make the use of PID controllers redundant.67 Bioprocesses are stochastic and complex in nature, and to predict their dynamics accurately and systematically, a number of advanced control strategies have been proposed in the literature.67 Model based control methods have shown promising results in the control of mammalian bioprocesses.67 Nonlinear Model Predictive Control (NMPC) can achieve more satisfactory controls than PID as it can handle nonlinear dynamics, suitable for Multi-Input Multi-Output (MIMO) systems where various process control loops interact. NMPC represent the SOTA in APC46 and can leverage the information gained through process development according to the QbD strategy and capture the complex dynamics and nonlinearities in mammalian cell culture processes to optimise the process inputs. NMPC can be manipulated, such that contradicting objectives i.e. minimal deviation of selected process states from the set-points and minimal control effort, are met in a suitable compromise.46
However, in the biomanufacturing industry measuring CQAs and Key Performance Indicators (KPIs) such as yield in real-time is significantly challenging. Therefore, given that NMPC excels when coupled with real-time CQA and CPP data, soft sensors capable of predicting or estimating NMPC inputs have become increasingly important. One such example are chemometric models used to predict the composition of the broth from Raman spectroscopy data. This technique gained popularity in the bioprocess industry from around 2011.68 Around that time, another pivotal study demonstrated the successful integration of Raman spectroscopy with a NMPC in a 15 litre bioreactor cultivating CHO cells.34 In this setup, Raman spectra were collected every 6 minutes to predict the concentrations of key metabolites such as glucose, glutamine, lactate, and ammonia using a PLS model. The NMPC then used these predictions to adjust the feeding rates, effectively maintaining a consistent glucose concentration in the bioreactor. This integration not only demonstrated enhanced bioprocess control but also showed a strong correlation with offline reference measurements. Controlling UPS in continuous biomanufacturing with the use of perfusion bioreactors is crucial, as the medium can often be expensive. In the literature, there are also successful implementations of MPC towards ICBs in perfusion bioreactors.69 On the other hand, DSP presents different control challenges that can be tackled by right sizing the downstream operation to handle the load generated upstream.
For DSP the common control objectives are to ensure that the control can be adjusted to changes in load coming from UPS and that uncertain perturbations are rejected.67 Nevertheless, just like in UPS, the most promising control strategies for DSP unit operations are spectroscopy instruments together with MVDA and mechanistic based MPC.70
However, building non-linear mathematical models to be used in NMPC using classical DoE approaches is impractical. This is because classical DoE does not capture the profiles of the factors, but rather explores its corners. As such, it does not take in consideration process dynamics as it uses end of process measurements disregarding the process evolution over time. The intensified Design of Experiments (iDoE) framework was designed to address this issue and enable consideration of process dynamics.71,72 Moreover, hybrid semi-parametric models can be developed to account for nonlinear behaviour of bioprocesses54 by combining existing process knowledge to the process control dynamics. The use of hybrid semi-parametric models for bioprocess control has been discussed by over a decade.73 In the study, von Stosch provides a general model agnostic framework that can be implemented to incorporate process knowledge and control parameters.
Some of the issues of NMPC relate to its reliance on accurate process models with predictive power and its inefficient computational time. However, the aforementioned framework addresses this issue, as an accurate model can be developed with varying levels of a priori process knowledge by integrating empirical process control dynamics. Nevertheless, one of the major limitations of MPC comes from the difficulty in tuning its parameters, such as the control horizon, which dictates the number of future control moves to be optimised, the prediction horizon, which is the number of future time steps predicted, and the weighting matrices, which balance the importance of different control objectives.74 However, what favours NMPC applications in ICBs is that its use of nonlinear online optimisation algorithm that can take up to several minutes, is acceptable for mammalian bioprocesses due to the lengthy bioprocessing time (typically in weeks).46,67 Another pertinent concern is the lack of explicit economic considerations in classical NMPC algorithms, which often present conflicting objective functions to the ones used in Economic MPC (EMPC). As such, traditionally, economic optimisation has been performed by higher-level systems which determine the appropriate set-points for the NMPC controller.74
For this goal, Real Time Optimisation (RTO) can be used to provide optimal references which aim to maximise profitability.74 RTO systems have been widely adopted in other industries, as evidenced by many successful implementations over recent decades.75 These systems typically use first-principles models updated in real-time with process data measurements followed by a re-run of the optimisation process. These optimisation algorithms are run less often than in MPC as they might run once every hour. In cases where the optimisation interval significantly exceeds the closed-loop process dynamics, steady-state models are often used for optimisation.75 This dual-step approach to RTO is both straightforward and widely used, but can impede the achievement of the optimum of the actual plant. The main issue arises from the lack of integration between the model update and the optimisation phase, particularly when there is a discrepancy between the plant and the model75 often referred to as plant-model mismatch which leads to convergence towards the wrong optimum.
There are various approaches to address this issue. One approach consists of using recalibration algorithms to re-estimate the parameters of the model to better match the plant outputs, thereby making the real time model adaptive and converge towards an optimal operation. However, this method is not effective if the model used has structural mismatches, i.e. incorrect assumptions and simplifications which can come from neglecting potentially dominant transport phenomena or reactions that have not been accounted for. To address structural mismatches, discrepancy models and trust regions can be implemented to converge to the true plant optimal conditions.75 On one hand, discrepancy models are data driven models that learn the discrepancy between the plant's real data and the model's predictions. By incorporating this error model into the optimisation process, they can continually refine the model based on new plant data, leading to more accurate predictions and better optimisation results.75 Trust regions, on the other hand, have been discussed in a pivotal study from Del Rio Chanona et al.75 These can be incorporated to manage how far the optimisation algorithm should ‘trust’ the model at each step. The idea is to restrict the optimisation steps to regions where the model was more likely to be accurate, thus avoiding significant errors due to model inaccuracies.76
Another direction explored in the literature to address the lack of first-principles models with the right structure as defined in Section 2.3 in early stages of the QbD process is the use of ML. Intelligent control systems (ICS) such as Artificial Neural Networks (ANN) and their applications to the biomanufacturing industry have been discussed extensively in the literature.67,77 Some of the success in the adoption of ICLs comes from modeling CPPs as a sequence of time series measurements. Recurrent Neural Networks (RNN) are an ANN architecture that has excelled at modeling sequences. Its application in control systems has seen better performance than PID controllers since, given enough data, they can model more complex dynamics and thus track set points more efficiently. As such, ICS have often been adopted in model predictive control for nonlinear system identification.78 This is especially true for architectures such as Long Short-Term Memory (LSTMs) which can better handle long-term dependencies in sequences. One of the most established nonparametric models used in control is the Nonlinear AutoRegressive network with eXogenous inputs (NARX), which can be used in systems identification to model the plants dynamics. NARX have been used to represent numerous discrete-time nonlinear models used in NMPC such as Hammerstein and Wiener models, Volterra models, and polynomial autoregressive moving average model with exogenus inputs (polynomial ARMAX).74 These have been applied by Sargantanis and Karim et al.79 to control Dissolved Oxygen (DO) in a bioprocess as early as 1999.
A more modern ICS method which sees the application of RNN in conjunction with Reinforcement Learning (RL) was studied by Petsagkourakis et al.80 The study applies Policy Gradient methods from RL to update a control policy batch-to-batch, using a recurrent neural network (RNN). The proposed RL strategy is compared to the traditional NMPC, showing advantages in handling uncertainties and computational efficiency. However, the main issue with RL which is true for ICS in general, is that they are data hungry. This issue was addressed by Petsagkourakis via transfer learning to train the model offline first, which sped up the online learning phase, thus requiring significantly less new data to tune the RL model. Additionally, apprenticeship learning, and inverse RL can be used to provide an initial example to the RL agent, thereby obtaining similar results. Another major issue with the application of RL in control systems is that it performs poorly at handling constrains. This is why RL has seen little use in industrial biomanufacturing processes where optimal operating points often lie within hard constraints. In a study by Pan et al.81 a novel algorithm that combines trust regions, GPs, and acquisition functions to solve the constrained RL problem efficiently and reliably has been discussed. It could be argued that, despite the computational drawbacks introduced by incorporating RTO in RL, its application in biomanufacturing, where bioprocesses can span weeks, remains acceptable.
Altogether, ICS applications remain an active area of research. NMPC can be considered the best candidate to unlock automation in ICBs and implement QbC.
3 Digital twins construction and use cases
DT development in biomanufacturing is an iterative process, where each iteration is driven by a business objective and can span the entire drug product life-cycle, from process development to continued process verification (CVP). It is important to highlight that DTs are a digital solution, therefore, to develop this successfully, embracing IT operating models is crucial. Thus, a version-controlled single source of truth (SST) of the DT for the entire biomanufacturing process should be maintained throughout its life cycle. Additionally, concepts from IT such as Continuous Integration and Continuous Deployment (CI/CD) can be useful in integrating disparate models owned by various teams into the SST.3.1 Digital twin construction
As mentioned in Section 2.3, digital twin development in the bioprocess industry begins with identifying an adequate first principal model. This section will focus on mechanistic models for cell-based UPS; however, the underlying modelling principles discussed are also applicable to cell-free bioprocesses where kinetic parameters and model structures can be adjusted in the same way.Fig. 4a illustrates the three main stages of digital twin development. The first step consists of developing initial models, usually steady-state or dynamic models,82 that can be derived from macroscopic mass balances.83 These models may use empirical relations to describe parameters such as growth rate or product formation rate and can be fit using adequate experimental designs as discussed and represent a static model of the process.
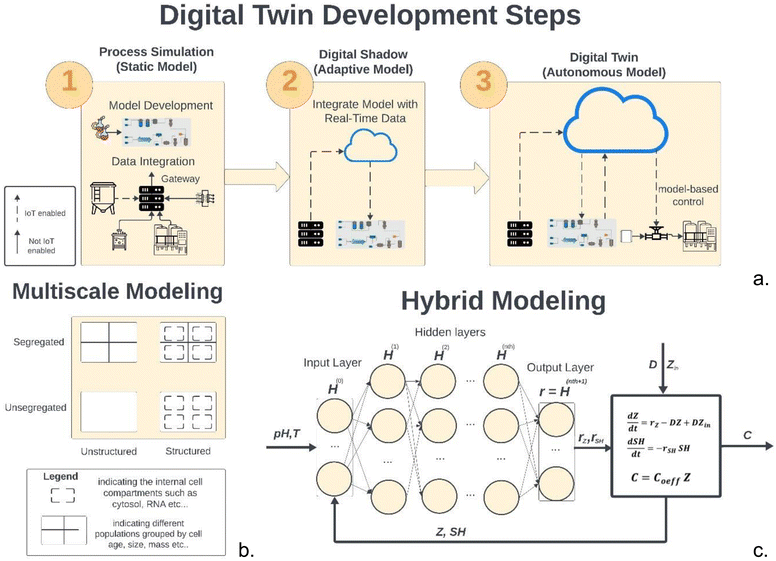 | ||
| Fig. 4 (a) Illustration of the steps required to develop a digital twin. The first step involves integration of data from various pieces of equipment and developing a model that can simulate the process. The second step involves connecting the data streams integrated to a gateway to connect to the model and adjusting the model to be re-calibrated in real time. The final step involves connecting the process control systems to the adaptive models to implement model-based control strategies. (b) Representation of hybrid modelling using a DNN and a first-principal model adapted from ref. 48. (c) Various categories of mathematical models used to describe cell cultures. In general, a model which accounts for the biotic phase referred to as a structured model, whilst a model which considered population differences referred to as segregated. | ||
The second step usually consists of developing a digital shadow of the process.82 This step can begin as soon as real-time data from similar processes or scaled-down versions of the bioprocess become available. To connect real-time data to the static models, OPC UA and other IIoT communication protocols can be used as mentioned in the Section 1.1 and in the review by Chen et al.84 Consequently, a digital shadow can be developed by making the mechanistic models adaptive to the process data and recalibrate its parameters in real time. Least squares is a widely used method for parameter estimation. However, it is not suitable for real-time parameter estimation as it requires the entire data set to be available before the estimation can be performed. In contrast, real-time parameter estimation requires the use of recursive algorithms that update the parameter estimates as new data become available. This can be achieved by applying state estimation algorithms such as Recursive Least Squares (RLS) or Kalman Filters, more specifically, Extended Kalman Filter (EKF) and Unscented Kalman Filters (UKF). According to Wan et al.,85 UKF can deal better with higher order nonlinearities whilst being as computationally efficient as EFK. For those interested, this reference provides a comprehensive discussion on the application and advantages of UKF.85 The third step consists of using the real-time adaptive digital shadow to control the operation. This can be achieved using popular PCS software such as Emmerson Delta-V that can control UPS86 and provide support for OPC UA and model-based control methods. As mentioned earlier, the evidence shows that MPC is the most appropriate strategy for real-time control. For this step, an adaptive MPC is often required to close the loop and use the digital twin to perform feedback and feed forward control. In the study by Markana et al.,87 EFK was combined with an Economic Model Predictive Control for a fed-batch reactor. In another work by Nimmegeers et al.,88 an algorithm which combines MPC with moving-horizon estimation was combined with UKF to obtain comparable results.
Overall, the use of state estimation algorithms for monitoring and process control in real time can demonstrate satisfactory performance, however, these methods are often computationally expensive to run in real time. Additionally, RLS and Kalman Filter recalibrate the model to provide good estimates of the local state of the system. However, historical patterns in the data might not be captured due to the recalibration window not being large enough. As mentioned in Section 2.4, ML models can capture these patterns very effectively. More specifically, ANNs are currently the most popular ML technique in bioprocess engineering.77 One of the most common applications of ML in DT development is illustrated in Fig. 4a. This involves combining macroscopic material balance equations of extracellular species with ML modeling method, predominantly shallow Feed Forward Neural Networks (FFNNs). These FFNNs are often configured with a single hidden layer often using the hyperbolic tangent as an activation function.89 This technique is often referred to as hybrid semi-parametric modelling (HSM) or grey box modelling and has been discussed extensively in the literature, most notably the work of von Stosch et al.90–94 on use case and refinement of the technique. This technique gained popularity since prediction accuracy of hybrid models has been always proven superior to the mechanistic models.89 HSM can significantly speed up process development especially at the early stages where data for the new bioprocess is scarce and the model structure is not well-understood. In scenarios where the latter is a limiting factor, a parallel model structure, where the data-driven models are trained using the same inputs used for the mechanistic model, can be used to rectify its predictions.94 However, as more knowledge about the process becomes available and the mechanistic model structure is improved, the serial structure should be preferred as it can perform better than the parallel.94 Trade-offs in the model structure selection have been explored in two reviews on the technique.90,94 Other notable improvements have been highlighted93 where bagging and stacking methods have been discussed to improve model validation and performance. In bagging, the data are re-partitioned several times, one model is developed on every partition, and then, the models are aggregated. The stacking is an ensemble method in which the contributions of each neural network to the final prediction are weighted according to their performance on the input domain.93 Deep Neural Networks based on convolution neural networks (CNN), LSTM and physics-informed neural networks (PINNs) have also been reported in the literature with promising results.83
However, even though significant model improvements can be obtained through this methodology, in later stages of the process development, explaining the relationship in the data from first principles becomes increasingly important as it can lead to better extrapolation. Opportunities for structural improvements of the bioprocess macroscopic models are given by an increase in the variability rather than the volume of the data. Through technologies such as ‘omics' and system-biology, which have gained popularity around the early 2000s95 a large amount of data regarding intracellular activity can be obtained. These data have been organised into several online databases with information regarding interactions and metabolic pathways.96 This can provide a good infrastructure to build multiscale mechanistic models that are generally regarded in the literature as a good candidate for DT.97,98Fig. 4c illustrates how mechanistic bioprocess models can be classified. This includes unstructured, structured, segregated, and unsegregated.55,99 Structured models take into account the intracellular activities in a fermentation process including metabolic fluxes, cellular division, and substrate uptake mechanisms. Segregated models, on the other hand, describe cell cultures as heterogeneous populations and attempts to group cells with identical properties such as age, size, or mass, together. Segregated models can be developed from population balance models (PBM). On the other hand, there are two main approaches to developing structured models. Using kinetic models which consider intracellular compartments such as cytosol, mitochondria, or nucleus,95 these are often referred to as Genome Scale Models (GEM). GEMs can be derived from assuming well-mixed compartments95 and all cells are equal and change as one with time.99 However, the balance is performed over the concentrations of intracellular species (metabolites, proteins, RNA/DNA, and other chemical constituents), which are involved in a very complex network of physio-chemical transformations.95
Alternatively, Flux Balance Analysis (FBA), avoids the definition of kinetic rates.95 This is a popular technique from system-biology that defines the function of optimizing based on a cellular objective.98 Examples include maximization of growth rate, ATP production, minimization of ATP consumption, or minimization of NADH production.98 In the FDA, intracellular metabolic fluxes (fluxome) are estimated based on some experimental data,96 such as the consumption/production rates of extracellular compounds.96 Isotopomer experiments, in which labelled substrates are administered to the cells and the fate of the label is further analysed by nuclear magnetic resonance (NMR) or mass spectrometry (MS),96 can also provide useful information on intracellular flux distributions that can be included in the model.96 However, FBA is less rigorous as it does not have knowledge of the kinetics95 and thus it often leads to underestimating the rates of certain sets of intracellular reactions.98 To derive a digital twin using a structured segregated model, various approaches could be attempted under the umbrella term of multiscale modelling. The first approach could be to integrate an intracellular model into the macroscopic mass balance.95
However, given the very large number of species and reactions involved in intracellular processes,98 it is critical to reduce Genome Scale Models (GEMs) to the reactor operating conditions.95 Alternatively, Population Balance Models (PBM) could be integrated with macroscopic balance equations by modifying the reaction rates to include the influence of traits (such as age, mass, size of cell populations). The most promising technique has been the former since, as empirical evidence shows, segregated model capabilities can be simulated from accurate intracellular structured models. A pivotal study100 in which a structured dynamic model describing whole animal cell metabolism was developed. The model was capable of simulating cell population dynamics (segregated model characteristics), the concentrations of extracellular and intracellular viral components, and the heterologous product titres. The dynamic simulation of the model in batch and fed-batch mode gave good agreement between model predictions and experimental data. The model considers the concentration of about 40 key metabolites, including fatty acids, amino acids, sugars, and intermediates of metabolic pathways like glycolysis and the tricarboxylic acid cycle (TCA).
However, the aforementioned structured segregated model is use case-specific, and developing a multi-product digital twin out of such a technique would be considerably inefficient. In the work of Moser et al.,97 an interesting case for a digital twin development framework via a Generalised Structured Modular Model (GSM) which can be applied to various cell lines has been proposed. This was achieved by dividing the cell into six compartments that enabled capture of two key physicochemical phenomena involved in culture processes at the cellular level, transmembrane transport, which governs nutrient uptake and product secretion, and the metabolic reactions in the intracellular environment.98 In the study, Moser represents the substrates by grouping it into carbon (SC), nitrogen (SN) and amino acid substrates (SAA). These substrates are used in the model for biomass and product formation, as well as for maintenance metabolism. Furthermore, several interconnected submodels are embedded in the developed GSM model, which makes use of a sigmoid function as a simple flag mechanism to enable the decoupling of submodels.97 Thus, potential effects of factors like pH or temperature on rates or yield coefficients can be integrated into the model structure and will be discriminated against by the sigmoid function through parametrisation when those effects are negligible.
An obvious point to highlight is that with the increased complexity, the model structure warrants adjustments to be used in RTO. Overall, DTs can provide high-fidelity simulations and more accurately predict operational changes and extreme scenarios which are impractical to measure from experimentation, thereby solving many problems related to imbalanced data sets in ML. As such, DTs can often be used to simulate a large amount of data to train agents or build surrogate models. This technique enables the integration of Computational Fluid Dynamics (CFD) models and thus the entire abiotic (cell environment) conditions of the bioprocess. In a study conducted by Del Rio Chanona et al.,101 a CFD model was used to generate data points for different scenarios of photobioreactor design and operation, which were then used to train a convolutional neural network (CNN) as a surrogate model. The CNN was able to learn the complex fluid dynamics and kinetic mechanisms of the CFD model and predict the system performance under new design conditions. Simulating training data for a surrogate model could also be used for system identification and state space modelling.102
In addition, simplifications of the full DT by different submodels can be used in MPC. Unlike in intracellular kinetic models which have no dynamic predictive power,103 dynamic FBA has been incorporated into a nonlinear MPC application by coupling the intracellular metabolism with the reactor kinetics.98 Population balance models have also been used in MPC control.104 In the case of training an agent, this can be achieved through deep reinforcement learning by simulating stochastic data using the digital twin.80 These capabilities allow for various use cases, which will be discussed in the next section.
3.2 Digital twins use cases
Fig. 5 shows some of the use cases of DT in bioprocess development and operation. As discussed earlier, there are many examples of the use of digital twins in the literature in model-based54 and model-assisted55 Design of Experiments and for digital process design in line with QbDD expectations.52In operation, digital twins can provide a lot of value when integrated within the existing industrial automation stack.105 By integrating MES data to the DT, these can assist with all the aspects of manufacturing operations, from predicting maintenance to Out of Spec batches. Moreover, higher level metrics such as Overall Equipment Effectiveness (OEE) can be leveraged for advanced automation. Additionally, using ERP data, material shortages can be predicted and accounted for in operation. ERP data can also help provide financial context to what-if-analysis studies, which can inform operational changes and provide overall support with drug product life-cycle management in CVP. The adaptive capabilities discussed earlier will allow the DT to improve over time and more accurately represent the various aspects of the physical systems.
Nevertheless, as mentioned earlier, DTs can be used to train AI models that can control operations autonomously. A crucial example of the use of RL in APC was showcased by Yokogawa demonstrating the successful autonomous operation of a distillation column for 35 days.106 Yokogawa Electric Corporation and ENEOS Materials Corporation have tested an AI algorithm called Factorial Kernel Dynamic Policy Programming (FKDPP) to control a distillation column at an ENEOS Materials chemical plant. The AI algorithm has demonstrated a high level of performance while controlling the distillation column for almost an entire year.106 The AI controlled the distillation column directly, stabilising quality, achieving high yield, and saving energy.106
The AI solution could control distillation operations that were beyond the capabilities of existing control methods (PID control/APC) and had necessitated manual control of valves based on the judgements of experienced plant personnel.106 The AI maintained stable control of the liquid levels and maximised the use of waste heat, even in winter and summer weather, with external temperatures changes by about 40 °C, by eliminating the production of off-spec products, the autonomous control AI reduced fuel, labour, and other costs, and made efficient use of raw materials.106 While producing good quality products that met shipping standards, the AI autonomous control reduced steam consumption and CO2 emissions by 40% compared to conventional manual control.
4 Digital transformation in bioprocessing 4.0
Digital transformation in biomanufacturing refers to the process of going from semi-automated or paper-based manufacturing to autonomous, digital manufacturing. This process revolves around the adoption of an IIoT architecture that makes all the events within the business accessible to the right people in real-time. However, digital transformation is not a project and should not be treated as such, neither in its engineering execution nor in its business evaluation. Therefore, for a successful digital transformation plan, it is crucial to achieve technological advancements with a short time to value, ensuring a quick ROI to prove the business value of the initiative early on. This can be achieved by working in an agile manner and utilising early and frequent stakeholder feedback to guide technological development. This iterative and gradual approach was adopted by companies such as Novartis, when developing a centralised view for their clinical trials, namely Nerve Live107 and Pfizer, in an attempt to make manufacturing more productive using less resources. This allows management to comprehend the value of each phase in the digital transformation strategy as it demonstrates tangible business results throughout its evolution. This methodology aligns with IT project management philosophies and lean manufacturing, emphasising continuous improvement and short, iterative innovation cycles which is a major shift from the current waterfall approach which many engineering projects adopt in biomanufacturing. In the literature and industry there are numerous case studies of digital transformations, three primary ways to execute on such a plan have been observed:4.1 Partnering with integrators for customisable solutions
Companies are increasingly collaborating with systems integrators to develop customisable digital solutions. An example of this approach is Tulip which has embraced the Industry 4.0 paradigm by focusing on operator-centric solutions and collaborating with the ISPE to align their offerings with the Pharma 4.0 vision. For this solution, the integrator provides the necessary infrastructure, allowing for tailored customisation based on specific needs of operators and managers. Körber Pharma are also providing diverse Industry 4.0 solutions to integrate with existing disparate systems. With the Werum PAS-X MES Software Suite, that integrates with the existing infrastructure, VR/AR tools for remote equipment troubleshooting (such as Xpert View) and personnel training (with Line Optimiser). Additionally, using their platform teams can deploy digital twins on top of the MES giving them its full digital context. Similarly, Merck's Bio4C ProcessPad™ which can collect data from multiple sources including process data from batches, ERP, MES, LIMS, Historians, process equipment, and manual sources into a single, validated data source whilst keeping their records compliant with 21 CFR part 11. Bio4C™ Suite is based on their “4C strategy” (control, connect, collect, and collaborate), designed to provide aids in gradual digital transformation via different functional capabilities. This model is becoming the preferred method for digital transformation in the pharmaceutical industry as it requires low to no level of digital maturity to begin with. Another noteworthy partnership between GlaxoSmithKline (GSK), Atos and Siemens lead to the development of a digital twin of the mRNA vaccine manufacturing process.108 This was equipped with PAT sensors and in silico models to run optimisations online and simulate process changes.4.2 Purchasing off-the-shelf solutions
Many companies opt for ready-made digital solutions available in the market. Notable examples of providers in this category include Siemens, Sartorius, Emerson, and Cytiva. For example, Cytiva's GoSilico® software can be used to calibrate digital twins of downstream chromatography columns. These off-the-shelf solutions offer a range of standardised digital tools that can be directly implemented into existing systems in a plug and play fashion. However, this solution often requires some existing internal competence with Industry 4.0 technologies and a clear vision on how these tools integrate with the existing infrastructure. Additionally, they might not be customisable enough to deal with specific edge-cases at the plant.4.3 Developing in-house solutions
Some companies choose to build their digital solutions internally. Notable examples include Novartis as mentioned earlier with Nerve Live and also data42, a data lake initiative that aims to enhance the understanding of diseases and patient insights using clinical trial data. Similarly, GSK has developed its technology stack, indicating a trend towards internal development for more tailored and company-specific digital solutions. However, these solutions often require a deep understanding of digital tools which is not applicable for most biomanufacturing companies.The digital integration of the data across the enterprise will potentially provide opportunities for big data analytics, as ANN can be used to analyse these various data sources and find patterns in a higher dimensional space that we are capable of analysing. This could lead to the transformative productivity gains experienced in the previous decades.
5 Conclusion and future perspective
The transition from an Integrated Continuous Biomanufacturing (ICB) platform to a smart factory represents a significant, yet manageable, shift. While moving from batch to continuous processes posed considerable challenges, the key technological hurdles for smart factory integration have already been addressed in other industries. Solutions such as OPC UA for interoperability and connectivity, cloud computing for data storage in data lakes provided by cloud service providers, and the established AI algorithms for big data analysis have laid a strong foundation. Additionally, this evolution is supported by numerous roadmaps and frameworks and a wide range of business partnerships.However, since its inception, Industry 4.0 has become a bit of a buzzword and is hard to justify an expectation of the orders of magnitude in productivity gains promised by any industrial revolution. This could be attributed to the poor execution of the strategies. Although manufacturing processes are interconnected, perhaps the lack of a central hub and FAIR data standardisation across the enterprise, might prevent companies from experiencing the true gains in productivity. Perhaps, there is a disconnect between strategy and technology. The future factory aims to enhance productivity in industries where agility, product personalisation, and diversity are the desired business outcomes, such as in biomanufacturing, rather than focusing solely on high production volumes. This could also be attributed to the fact that a smart factory makes operation exponentially more agile over time and with the current slowing down of drug approval rates, the effects of increasing availability of high-quality historical data still need to be experienced. Ultimately, it is relatively early to make any definitive statements about the impact of Industry 4.0 since it was launched in less than 15 years ago.
Another clear observation is that the transition from the third to the fourth industrial revolution seems more evolutionary than revolutionary. The paradigms of quality by digital design (QbDD) and quality by control (QbC) which have been identified in the literature and explored in this manuscript, are essentially advanced iterations of quality by design (QbD). QbDD accelerates process development using digital tools, it is the result of a natural expansion of the PAT toolbox. On the other hand, QbC refers to autonomous operation within a pre-defined design space which could also be considered an evolution of continuous automation. Compared to the shift from quality by testing (QbT) to QbD, these are not revolutions but natural progressions in utilizing PAT for developing and operating validated processes more automatically. However, the use of AI algorithms might necessitate regulatory alignment and adjustments in process validation to ensure quality risk mitigation, this will also arguably bring revolutionary modes of operation which are still unclear.
What is clear is that biomanufacturing's future lies in multi-product integrated intensified continuous platforms that are modular, flexible, and capable of delivering a broad range of biologics. Bioprocessing 4.0 should encompass the shift from in vivo or in vitro to in silico process development and control, with plant-wide modeling and simulation at its core. Companies will develop unique digital twins encapsulating all process knowledge, updated as more insights are gained. Experimental teams will perform only information rich experiments suggested by the model, and the digital twin will be used for autonomous operation within validated ranges. Engineers will focus on troubleshooting and advanced analysis, supported by AR and VR technologies integrated with data from MES and centralised data hubs. Handovers will become less risky with access to contextualised data from the cloud and training new personnel will be increasingly less costly as workflows and SOPs will be digitised.
I4.0 technologies have achieved the appropriate level of technical maturity to be integrated in today's biomanufacturing operations, as evidenced by the abundant availability of commercially available I4.0 solutions. This represents a paradigm shift towards more agile, proactive, and reactive biomanufacturing operations, driven by advanced digital technologies and data integration, fundamentally transforming the industry's approach to process development and control.
Abbreviations
| AI | Artificial intelligence |
| ALVEN | Algebraic learning via elastic net |
| ANOVA | Analysis of variance |
| APC | Advanced process control |
| AR | Augmented reality |
| CAPA | Corrective and preventive action |
| CDMO | Contract development and manufacturing organization |
| CPP | Critical process parameter |
| CQA | Critical quality attribute |
| CVP | Continued process verification |
| DCS | Distributed control system |
| DoE | Design of experiments |
| DT | Digital twin |
| ERP | Enterprise resource planning |
| FBA | Flux balance analysis |
| FDA | Food and drug administration |
| FMEA | Failure mode and effects analysis |
| FKDPP | Factorial kernel dynamic policy programming |
| GEM | Genome scale model |
| GMP | Good manufacturing practice |
| ICB | Integrated continuous biomanufacturing |
| ICH | International council for harmonisation of technical requirements for pharmaceuticals for human use |
| IIoT | Industrial internet of things |
| ISPE | International society for pharmaceutical engineering |
| IT | Information technology |
| KPI | Key performance indicator |
| MES | Manufacturing execution system |
| ML | Machine learning |
| MIMO | Multi-input multi-output |
| MPC | Model predictive control |
| MQTT | Message queuing telemetry transport |
| MVDA | Multivariate data analysis |
| NMPC | Nonlinear model predictive control |
| OEE | Overall equipment effectiveness |
| OPC UA | Open platform communication unified architecture |
| OT | Operations technology |
| PAR | Process acceptance range |
| PAT | Process analytical technology |
| PCS | Process control system |
| PQS | Pharmaceutical quality system |
| PID | Proportional-integral-derivative |
| PLS | Projection to latent structures |
| PP | Process parameter |
| PPQ | Process performance qualification |
| QbC | Quality by control |
| QbD | Quality by design |
| QbDD | Quality by digital design |
| QbT | Quality by testing |
| QTPP | Quality target product profile |
| R&D | Research and development |
| RL | Reinforcement learning |
| RLS | Recursive least squares |
| ROI | Return on investment |
| RSM | Response surface methodology |
| RTRT | Real-time release testing |
| SOP | Standard operating procedure |
| SOTA | State of the art |
| SCADA | Supervisory control and data acquisition |
| SST | Single source of truth |
| UPS | Upstream processing |
| UKF | Unscented Kalman filters |
| VR | Virtual reality |
Data availability
No new data exists to share.Author contributions
Conceptualization and writing original draft, K. I. and P. Z. M. Writing – review & editing, all authors. Supervision, J. C., Z. K. and P. Z. M.Conflicts of interest
The authors declare no competing interests.Acknowledgements
P. Z. M. acknowledges support from University College London Research Opportunity Scholarship (UCL-ROS)/H. Walter Stern Scholarship for funding K. I's PhD studentship. J. L. C., Z. K. and P. Z. M. also acknowledge support from the Wellcome Leap R3 Program.References
- M. Demesmaeker, D. Kopec and A. M. Arsénio, Bioprocessing 4.0 – Where Are We with Smart Manufacturing in 2020?, Technical Report, 2020, https://www.pharmoutsourcing.com Search PubMed.
- M. Hermann, T. Pentek and B. Otto, Design Principles for Industrie 4.0 Scenarios, 2016 49th Hawaii International Conference on System Sciences (HICSS), 2016, pp. 3928–3937, DOI:10.1109/HICSS.2016.488.
- N. Sarah Arden, A. C. Fisher, K. Tyner, L. X. Yu, S. L. Lee and M. Kopcha, Industry 4.0 for pharmaceutical manufacturing: Preparing for the smart factories of the future, Int. J. Pharm., 2021, 602, 120554 CrossRef PubMed.
- M. Demesmaeker, D. Kopec and A. M. Arsénio, Biologics CDMO Trends and Opportunities in China Building Future-Proof Supply Chains with Graph Technology Decentralization: A Direct Approach to Clinical Trials, Technical Report, 2020, https://www.AjiBio-Pharma.com Search PubMed.
- A. Kumar, I. A. Udugama, C. L. Gargalo and K. V. Gernaey, Why is batch processing still dominating the biologics landscape? Towards an integrated continuous bioprocessing alternative, Processes, 2020, 8(12), 1–19, DOI:10.3390/pr8121641.
- G. Subramanian, Process Control, Intensification, and Digitalisation in Continuous Biomanufacturing, 2021 Search PubMed.
- G. Kumar, M. Koch, A. Arsenio and J. Wagner, Bioprocessing 4.0: Simplifying the Journey, BioProcess International, 2021, https://www.bioprocessintl.com/sponsored-content/simplifying-the-bioprocessing-4-0-journey Search PubMed.
- Step. Committee for Medicinal Products for Human Use, ICH Guideline Q13 on Continuous Manufacturing of Drug Substances and Drug Products, Technical Report, 2023, https://www.ema.europa.eu/contact Search PubMed.
- M. K. Maruthamuthu, S. R. Rudge, A. M. Ardekani, M. R. Ladisch and M. S. Verma, Process Analytical Technologies and Data Analytics for the Manufacture of Monoclonal Antibodies, Trends Biotechnol., 2020, 38(10), 1169–1186 CrossRef CAS PubMed.
- A. Ashoori, B. Moshiri, A. Khaki-Sedigh and M. R. Bakhtiari, Optimal control of a nonlinear fed-batch fermentation process using model predictive approach, J. Process Control, 2009, 19(7), 1162–1173, DOI:10.1016/j.jprocont.2009.03.006.
- K. Pandey, M. Pandey, V. Kumar, U. Aggarwal and B. Singhal, Bioprocessing 4.0 in biomanufacturing: paving the way for sustainable bioeconomy, Syst. Microbiol. Biomanuf., 2024, 4, 407–424 CrossRef CAS.
- C. L. Gargalo, I. Udugama, K. Pontius, P. C. Lopez, R. F. Nielsen, A. Hasanzadeh, S. Soheil Mansouri, C. Bayer, H. Junicke and K. V. Gernaey, Towards smart biomanufacturing: a perspective on recent developments in industrial measurement and monitoring technologies for bio-based production processes, J. Ind. Microbiol. Biotechnol., 2020, 47(11), 947–964, DOI:10.1007/s10295-020-02308-1.
- I. A. Udugama, M. Öner, P. C. Lopez, C. Beenfeldt, C. Bayer, J. K. Huusom, K. V. Gernaey and G. Sin, Towards Digitalization in Bio-Manufacturing Operations: A Survey on Application of Big Data and Digital Twin Concepts in Denmark, Front. Chem. Eng., 2021, 3, 2673–2718 Search PubMed.
- R. Vatankhah Barenji, Y. Akdag, B. Yet and L. Oner, Cyber-physical-based PAT (CPbPAT) framework for Pharma 4.0, Int. J. Pharm., 2019, 567, 0378–5173 Search PubMed.
- A. Ouranidis, T. Vavilis, E. Mandala, C. Davidopoulou, E. Stamoula, C. K. Markopoulou, A. Karagianni and K. Kachrimanis, mRNA Therapeutic Modalities Design, Formulation and Manufacturing under Pharma 4.0 Principles, Biomedicines, 2022, 10(1), 50 CrossRef CAS PubMed.
- G. A. Van Den Driessche, D. Bailey, E. O. Anderson, M. A. Tarselli and L. Blackwell, Improving protein therapeutic development through cloud-based data integration, SLAS Technol., 2023, 28(5), 293–301, DOI:10.1016/j.slast.2023.07.002.
- C. Alarcon and C. Shene, Fermentation 4.0, a case study on computer vision, soft sensor, connectivity, and control applied to the fermentation of a thraustochytrid, Comput. Ind., 2021, 128, 103431, DOI:10.1016/j.compind.2021.103431.
- D. P. Wasalathanthri, R. Shah, J. Ding, A. Leone and Z. J. Li, Process analytics 4.0: A paradigm shift in rapid analytics for biologics development, Biotechnol. Prog., 2021, 37(4), e3177 CAS.
- T. Savage, N. Basha, J. McDonough, O. K. Matar and E. A. del Rio Chanona, Machine Learning-Assisted Discovery of Novel Reactor Designs, arXiv, 2023, preprint, arXiv:2308.08841, http://arxiv.org/abs/2308.08841 Search PubMed.
- D. Lutkemeyer, I. Poggendorf, T. Scherer, J. Zhang, A. Knoll and J. Lehmann, First steps in robot automation of sampling and sample management during cultivation of mammalian cells in pilot scale, Biotechnol. Prog., 2000, 16(5), 822–828, DOI:10.1021/bp0001009.
- W. Dedden, K. Sauermann, and C. Wölbeling, Working Groups Plug & Produce, Technical Report, 2021 Search PubMed.
- J. Clovis Kabugo, S. Liisa Jämsä-Jounela, R. Schiemann and C. Binder, Industry 4.0 based process data analytics platform: A waste-to-energy plant case study, Int. J. Electr. Power Energy Syst., 2020, 115, 105508, DOI:10.1016/j.ijepes.2019.105508.
- M. N. Karim and S. L. Rivera, Comparison of feed-forward and recurrent neural networks for bioprocess state estimation, Comput. Chem. Eng., 1992, 16, S369–S377, DOI:10.1016/S0098-1354(09)80044-6.
- I. Carla Reinhardt, Dr J. C. Oliveira and D. T. Ring, Current Perspectives on the Development of Industry 4.0 in the Pharmaceutical Sector, J. Ind. Inf. Integr., 2020, 18, 100131 Search PubMed.
- J. David Contreras, J. Isidro Garcia and J. D. D. Pastrana, Developing of industry 4.0 applications, Int. J. Online Eng., 2017, 13(10), 30–47, DOI:10.3991/ijoe.v13i10.7331.
- R. Iqbal, F. Doctor, B. More, S. Mahmud and U. Yousuf, Big Data analytics and Computational Intelligence for Cyber–Physical Systems: Recent trends and state of the art applications, Future Generat. Comput. Syst., 2020, 105, 766–778, DOI:10.1016/j.future.2017.10.021.
- ICH, Q10 Pharmaceutical Quality System, Technical Report, 2009, http://www.fda.gov/cder/guidance/index.htmhttp://www.fda.gov/cber/guidelines.htm Search PubMed.
- D. P. Wasalathanthri, M. S. Rehmann, Y. Song, Y. Gu, L. Mi, C. Shao, L. Chemmalil, J. Lee, S. Ghose, M. C. Borys, J. Ding and Z. J. Li, Technology outlook for real-time quality attribute and process parameter monitoring in biopharmaceutical development—A review, Biotechnol. Bioeng., 2020, 117(10), 3182–3198 CrossRef CAS PubMed.
- M. Muldbak, C. Gargalo, U. Krühne, I. Udugama and K. V. Gernaey, Digital Twin of a pilot-scale bio-production setup, Comput.-Aided Chem. Eng., 2022, 49, 1417–1422, DOI:10.1016/B978-0-323-85159-6.50236-0.
- BioPhorum, A Best Practice Guide to using the BioPhorum Digital Plant Maturity Model and Assessment Tool Connect Collaborate Accelerate, 2019 Search PubMed.
- I. Imanol Arzac, M. Vallerio, C. Perez-Galvan and F. J. Navarro-Brull. Industrial Data Science for Batch Manufacturing Processes, Technical Report, 2022 Search PubMed.
- Nature, Scientists applaud plans for UK-style advanced research agency in Japan, Nature, 2023, https://www.nature.com/articles/d41586-023-02688-1 Search PubMed.
- Körber Pharma, Revolutionize Your Pharma Training: Reduce Errors and Enhance Learning Efficiency with Augmented Reality, 2024, https://www.koerber-pharma.com/en/blog/revolutionize-your-pharma-training-reduce-errors-and-enhance-learning-efficiency-with-augmented-reality Search PubMed.
- K. A. Esmonde-White, M. Cuellar, C. Uerpmann, B. Lenain and I. R. Lewis, Raman spectroscopy as a process analytical technology for pharmaceutical manufacturing and bioprocessing, Anal. Bioanal. Chem., 2017, 409(3), 637–649, DOI:10.1007/s00216-016-9824-1.
- I. A. Udugama, S. Badr, K. Hirono, B. X. Scholz, Y. Hayashi, M. Kino-oka and H. Sugiyama, The role of process systems engineering in applying quality by design (QbD) in mesenchymal stem cell production, Comput. Chem. Eng., 2023, 172, 108144, DOI:10.1016/j.compchemeng.2023.108144.
- S. Sachio, C. Kontoravdi and M. M. Papathanasiou, A model-based approach towards accelerated process development: A case study on chromatography, Chem. Eng. Res. Des., 2023, 197, 800–820, DOI:10.1016/j.cherd.2023.08.016.
- S. Wold, K. Esbensen and P. Geladi, Principal Component Analysis, Chemom. Intell. Lab. Syst., 1987, 2, 37–52, DOI:10.1016/0169-7439(87)80084-9.
- P. Nomikos and J. F. MacGregor, Monitoring batch processes using multiway principal component analysis, AIChE J., 1994, 40(8), 1361–1375, DOI:10.1002/aic.690400809.
- P. Nomikos and J. F. Macgregor, American Society for Quality Multivariate SPC Charts for Monitoring Batch Processes, Technical Report, 1995 Search PubMed.
- Sartorius, Umetrics® Suite of data analytics software, 2024, website, https://www.sartorius.com/en/products/process-analytical-technology/data-analytics-software Search PubMed.
- US FDA, Pharmaceutical CGMPS for the 21 st Century—A Risk-Based Approach Final Report, Technical Report, 2004 Search PubMed.
- ICH, International Conference on Harmonisation of Technical RequireMents for Registration of Pharmaceuticals for Human Use Pharmaceutical Development Q8(R2), Technical Report, 2009 Search PubMed.
- A. Schmidt, H. Helgers, F. Lukas Vetter, S. Zobel-Roos, A. Hengelbrock and J. Strube, Process Automation and Control Strategy by Quality-by-Design in Total Continuous mRNA Manufacturing Platforms, Processes, 2022, 10(9), 1783, DOI:10.3390/pr10091783.
- FDA, Guidance for Industry PAT – A Framework for Innovative Pharmaceutical Development, manufacturing, and Quality Assurance, Technical Report, 2004, http://www.fda.gov/cvm/guidance/published.html Search PubMed.
- ICH, Q10 Pharmaceutical Quality System, Technical Report, 2009, http://www.fda.gov/cder/guidance/index.htmhttp://www.fda.gov/cber/guidelines.htm Search PubMed.
- W. Sommeregger, B. Sissolak, K. Kandra, M. von Stosch, M. Mayer and G. Striedner, Quality by control: Towards model predictive control of mammalian cell culture bioprocesses, Biotechnol. J., 2017, 12(7), 1600546 CrossRef PubMed.
- M. von Stosch, R. M. C. Portela and C. Varsakelis, A roadmap to AI-driven in silico process development: bioprocessing 4.0 in practice, Curr. Opin. Chem. Eng., 2021, 33, 100692 CrossRef.
- J. Pinto, J. R. C. Ramos, R. S. Costa and R. Oliveira, Hybrid Deep Modeling of a GS115 (Mut+) Pichia pastoris Culture with State–Space Reduction, Fermentation, 2023, 9(7), 643, DOI:10.3390/fermentation9070643.
- S. Daniel, Z. Kis, C. Kontoravdi and N. Shah, A blueprint for quality by digital design to support rapid RNA vaccine process development, manufacturing & supply, Vaccine Insights, 2022, 1(4), 219–233, DOI:10.18609/vac.2022.33.
- I. Walsh, M. Myint, T. Nguyen-Khuong, Y. S. Ho, S. K. Ng and M. Lakshmanan, Harnessing the potential of machine learning for advancing “Quality by Design” in biomanufacturing, mAbs, 2022, 14(1), 2013593, DOI:10.1080/19420862.2021.2013593.
- D. van de Berg, Z. Kis, C. Fredrik Behmer, K. Samnuan, A. K. Blakney, C. Kontoravdi, R. Shattock and N. Shah, Quality by design modelling to support rapid RNA vaccine production against emerging infectious diseases, npj Vaccines, 2021, 6(1), 65, DOI:10.1038/s41541-021-00322-7.
- Z. Kis, K. Tak, D. Ibrahim, S. Daniel, D. van de Berg, M. M. Papathanasiou, B. Chachuat, C. Kontoravdi and N. Shah, Quality by design and techno-economic modelling of RNA vaccine production for pandemic-response, Comput.-Aided Chem. Eng., 2022, 49, 2167–2172, DOI:10.1016/B978-0-323-85159-6.50361-4.
- J. Möller, K. B. Kuchemüller, T. Steinmetz, K. S. Koopmann and R. Pörtner, Model-assisted Design of Experiments as a concept for knowledge-based bioprocess development, Bioprocess Biosyst. Eng., 2019, 42(5), 867–882, DOI:10.1007/s00449-019-02089-7.
- B. Bayer, R. D. Diaz, M. Melcher, G. Striedner and M. Duerkop, Digital twin application for model-based doe to rapidly identify ideal process conditions for space-time yield optimization, Processes, 2021, 9(7), 1109 CrossRef.
- C. Herwig, R. Pörtner, and J. Möller, Digital Twins Applications to the Design and Optimization of Bioprocesses, Technical Report, 2021, http://www.springer.com/series/10 Search PubMed.
- C. Hutter, M. von Stosch, M. N. Cruz Bournazou and A. Butté, Knowledge transfer across cell lines using Hybrid Gaussian Process models with entity embedding vectors, arXiv, 2020, 118, 4389–4401, DOI:10.1002/bit.27907 , http://arxiv.org/abs/2011.13863.
- H. Narayanan, M. F. Luna, M. von Stosch, M. Nicolas Cruz Bournazou, G. Polotti, M. Morbidelli, A. Butté and M. Sokolov, Bioprocessing in the Digital Age: The Role of Process Models, Biotechnol. J., 2020, 15(1), 1900172 CrossRef CAS PubMed.
- A. R. Ferreira, J. M. L. Dias, M. Von Stosch, J. Clemente, A. E. Cunha and R. Oliveira, Fast development of Pichia pastoris GS115 Mut+ cultures employing batch-to-batch control and hybrid semiparametric modeling, Bioprocess Biosyst. Eng., 2014, 37(4), 629–639, DOI:10.1007/s00449-013-1029-9.
- S. S. Rosa, D. Nunes, L. Antunes, D. M. F. Prazeres, M. P. C. Marques and A. M. Azevedo, Maximizing mRNA vaccine production with Bayesian optimization, Biotechnol. Bioeng., 2022, 119(11), 3127–3139, DOI:10.1002/bit.28216.
- G. Franceschini and S. Macchietto, Model-based design of experiments for parameter precision: State of the art, Chem. Eng. Sci., 2008, 63(19), 4846–4872, DOI:10.1016/j.ces.2007.11.034.
- P. Kroll, A. Hofer, S. Ulonska, J. Kager and C. Herwig, Model-Based Methods in the Biopharmaceutical Process Lifecycle, Pharm. Res., 2017, 34(12), 2596–2613 CrossRef CAS PubMed.
- F. Galvanin, S. Macchietto and F. Bezzo, Model-based design of parallel experiments, Ind. Eng. Chem. Res., 2007, 46(3), 871–882, DOI:10.1021/ie0611406.
- A. Bensoussan, R. Braatz, J. Chen, A. Chiuso, Y. Dong, G. Feng, B. Gopaluni, Z.-P. Jiang, Y. Jin, J. H. Lee, Q. Liu, L. Qiu, J. Rawlings, V. Zavala, J. Wang, C. Yang, F. You, H. Zhao and Q. Zhu, IFAC Workshop Series: Workshop Series on Control Systems and Data Science Towards Industry 4.0, 2021 Search PubMed.
- S. Mascia, P. L. Heider, H. Zhang, R. Lakerveld, B. Benyahia, P. I. Barton, R. D. Braatz, C. L. Cooney, J. M. B. Evans, T. F. Jamison, K. F. Jensen, A. S. Myerson and B. L. Trout, End-to-end continuous manufacturing of pharmaceuticals: Integrated synthesis, purification, and final dosage formation, Angew. Chem., Int. Ed., 2013, 52(47), 12359–12363, DOI:10.1002/anie.201305429.
- W. Sun and R. D. Braatz, ALVEN: Algebraic learning via elastic net for static and dynamic nonlinear model identification, Comput. Chem. Eng., 2020, 143, 107103 CrossRef CAS.
- S. Ganesh, Q. Su, Z. Nagy and G. Reklaitis, Advancing smart manufacturing in the pharmaceutical industry. in Smart Manufacturing: Applications and Case Studies, Elsevier, 2020, pp. 21–57, DOI:10.1016/B978-0-12-820028-5.00002-3.
- A. S. Rathore, S. Mishra, S. Nikita and P. Priyanka, Bioprocess control: Current progress and future perspectives, Life, 2021, 11(6), 557 CrossRef PubMed.
- K. A. Esmonde-White, M. Cuellar and I. R. Lewis, The role of Raman spectroscopy in biopharmaceuticals from development to manufacturing, Anal. Bioanal. Chem., 2022, 414, 969–991 CrossRef CAS PubMed.
- M. Pappenreiter, S. Döbele, G. Striedner, A. Jungbauer and B. Sissolak, Model predictive control for steady-state performance in integrated continuous bioprocesses, Bioprocess Biosyst. Eng., 2022, 45(9), 1499–1513, DOI:10.1007/s00449-022-02759-z.
- A. Armstrong, K. Horry, T. Cui, M. Hulley, R. Turner, S. S. Farid, S. Goldrick and D. G. Bracewell, Advanced control strategies for bioprocess chromatography: Challenges and opportunities for intensified processes and next generation products, J. Chromatogr. A, 2021, 1639, 461914, DOI:10.1016/j.chroma.2021.461914.
- M. von Stosch and M. J. Willis, Intensified design of experiments for upstream bioreactors, Eng. Life Sci., 2017, 17(11), 1173–1184, DOI:10.1002/elsc.201600037.
- B. Bayer, M. Duerkop, G. Striedner and B. Sissolak, Model Transferability and Reduced Experimental Burden in Cell Culture Process Development Facilitated by Hybrid Modeling and Intensified Design of Experiments, Front. Bioeng. Biotechnol., 2021, 9, 2296–4185 Search PubMed.
- M. Von Stosch, R. Oliveira, J. Peres and S. F. De Azevedo, A general hybrid semiparametric process control framework, J. Process Control, 2012, 22(7), 1171–1181, DOI:10.1016/j.jprocont.2012.05.004.
- M. A. Henson, Nonlinear Model Predictive Control: Current Status and Future Directions, Technical Report, 1998 Search PubMed.
- E. A. Del Rio Chanona, J. E. Alves Graciano, E. Bradford and B. Chachuat, Modifier-adaptation schemes employing Gaussian processes and trust regions for real-time optimization, in IFAC – PapersOnLine, Elsevier B.V., 2019, vol. 52, pp. 52–57, DOI:10.1016/j.ifacol.2019.06.036.
- E. A. del Rio-Chanona, P. Petsagkourakis, E. Bradford, J. Eduardo Alves Graciano and B. Chachuat, Real-Time Optimization Meets Bayesian Optimization and Derivative-Free Optimization: A Tale of Modifier Adaptation, Comput. Chem. Eng., 2020, 147, 107249 CrossRef , http://arxiv.org/abs/2009.08819.
- M. Mowbray, T. Savage, C. Wu, Z. Song, B. A. Cho, E. A. Del Rio Chanona and D. Zhang, Machine learning for biochemical engineering: A review, Biochem. Eng. J., 2021, 172, 108054, DOI:10.1016/j.bej.2021.108054.
- W. M. Kouw, Information-Seeking Polynomial NARX Model-Predictive Control Through Expected Free Energy Minimization, IEEE Control Syst. Lett., 2023, 1, DOI:10.1109/LCSYS.2023.3347190 , URL https://ieeexplore.ieee.org/document/10373893/.
- L. G. Sargantanis and M. N. Karim, Variable structure NARX models: Application to dissolved-oxygen bioprocess, AIChE J., 1999, 45(9), 2034–2045, DOI:10.1002/aic.690450920.
- P. Petsagkourakis, I. O. Sandoval, E. Bradford, D. Zhang and E. A. del Rio-Chanona, Reinforcement learning for batch bioprocess optimization, Comput. Chem. Eng., 2020, 133, 106649 CrossRef CAS.
- E. Pan, P. Petsagkourakis, M. Mowbray, D. Zhang and A. del Rio-Chanona, Constrained Model-Free Reinforcement Learning for Process Optimization, Comput. Chem. Eng., 2020, 154, 10746 Search PubMed , http://arxiv.org/abs/2011.07925.
- H. Helgers, A. Hengelbrock, A. Schmidt and J. Strube, Digital twins for continuous mrna production, Processes, 2021, 9(11), 1967, DOI:10.3390/pr9111967.
- R. Agharafeie, J. R. C. Ramos, J. M. Mendes and R. Oliveira, From Shallow to Deep Bioprocess Hybrid Modeling: Advances and Future Perspectives, Fermentation, 2023, 9(10), 922 CrossRef CAS.
- Y. Chen, O. Yang, C. Sampat, P. Bhalode, R. Ramachandran and M. Ierapetritou, Digital twins in pharmaceutical and biopharmaceutical manufacturing: A literature review, Processes, 2020, 8(9), 1088 CrossRef.
- E. A. Wan and R. Van Der Merwe, The Unscented Kalman Filter for Nonlinear Estimation, Technical Report, 2000 Search PubMed.
- J. S. Alford, Bioprocess control: Advances and challenges, Comput. Chem. Eng., 2006, 30(10–12), 1464–1475, DOI:10.1016/j.compchemeng.2006.05.039.
- A. Markana, N. Padhiyar and K. Moudgalya, Multi-criterion control of a bioprocess in fed-batch reactor using EKF based economic model predictive control, Chem. Eng. Res. Des., 2018, 136, 282–294, DOI:10.1016/j.cherd.2018.05.032.
- P. Nimmegeers, D. Vercammen, S. Bhonsale, F. Logist and J. Van Impe, Metabolic reaction network-based model predictive control of bioprocesses, Appl. Sci., 2021, 11(20), 9532, DOI:10.3390/app11209532.
- J. Pinto, J. R. C. Ramos, R. S. Costa, S. Rossell, P. Dumas and R. Oliveira, Hybrid deep modeling of a CHO-K1 fed-batch process: combining first-principles with deep neural networks, Front. Bioeng. Biotechnol., 2023, 11, 1237963, DOI:10.3389/fbioe.2023.1237963.
- A. M. Schweidtmann, D. Zhang and M. von Stosch, A review and perspective on hybrid modeling methodologies, Digit. Chem. Eng., 2024, 10, 100136, DOI:10.1016/j.dche.2023.100136 , URL https://linkinghub.elsevier.com/retrieve/pii/S2772508123000546.
- M. Sokolov, M. von Stosch, H. Narayanan, F. Feidl and A. Butté, Hybrid modeling — a key enabler towards realizing digital twins in biopharma?, Curr. Opin. Chem. Eng., 2021, 34, 100715 CrossRef.
- M. von Stosch, S. Davy, K. Francois, V. Galvanauskas, J. M. Hamelink, A. Luebbert, M. Mayer, R. Oliveira, R. O'Kennedy, P. Rice and J. Glassey, Hybrid modeling for quality by design and PAT-benefits and challenges of applications in biopharmaceutical industry, Biotechnol. J., 2014, 9(6), 719–726, DOI:10.1002/biot.201300385.
- J. Pinto, C. Rodrigues de Azevedo, R. Oliveira and M. von Stosch, A bootstrap-aggregated hybrid semi-parametric modeling framework for bioprocess development, Bioprocess Biosyst. Eng., 2019, 42(11), 1853–1865, DOI:10.1007/s00449-019-02181-y.
- M. von Stosch, R. Oliveira, J. Peres and S. Feyo de Azevedo, Win-parametric modeling in process systems engineering: Past, present and future, Comput. Chem. Eng., 2014, 60, 86–101, DOI:10.1016/j.compchemeng.2013.08.008.
- J. Pinto, J. Antunes, J. Ramos, R. S. Costa and R. Oliveira, Modeling and optimization of bioreactor processes, in Current Developments in Biotechnology and Bioengineering: Advances in Bioprocess Engineering, Elsevier, 2022, pp. 89–115, DOI:10.1016/B978-0-323-91167-2.00016-2.
- A. P. Teixeira, N. Carinhas, J. M. L. Dias, P. Cruz, P. M. Alves, M. J. T. Carrondo and R. Oliveira, Hybrid semi-parametric mathematical systems: Bridging the gap between systems biology and process engineering, J. Biotechnol., 2007, 132(4), 418–425, DOI:10.1016/j.jbiotec.2007.08.020.
- A. Moser, C. Appl, S. Brüning and V. C. Hass, Mechanistic Mathematical Models as a Basis for Digital Twins, Adv. Biochem. Eng./Biotechnol., 2021, 176, 133–180, DOI:10.1007/10_2020_152.
- M. Monteiro, S. Fadda and C. Kontoravdi, Towards advanced bioprocess optimization: A multiscale modelling approach, Comput. Struct. Biotechnol. J., 2023, 21, 3639–3655, DOI:10.1016/j.csbj.2023.07.003.
- E. Anane, Model-based Strategies for Scale-down Studies in Fed-batch Cultivation of Escherichia coli Expressing Proinsulin, Technical Report, 2019 Search PubMed.
- J. D. Jang, C. S. Sanderson, L. C. L. Chan, J. P. Barford and S. Reid, Structured modeling of recombinant protein production in batch and fed-batch culture of baculovirus-infected insect cells, Cytotechnology, 2000, 34(1–2), 71–82, DOI:10.1023/a:1008178029138.
- E. Antonio del Rio-Chanona, J. L. Wagner, H. Ali, F. Fiorelli, D. Zhang and K. Hellgardt, Deep learning-based surrogate modeling and optimization for microalgal biofuel production and photobioreactor design, AIChE J., 2019, 65(3), 915–923, DOI:10.1002/aic.16473.
- E. Bradford, A. M. Schweidtmann, D. Zhang, K. Jing and E. Antonio del Rio Chanona, Dynamic modeling and optimization of sustainable algal production with uncertainty using multivariate Gaussian processes, Comput. Chem. Eng., 2018, 118, 143–158, DOI:10.1016/j.compchemeng.2018.07.015.
- S. Badr and H. Sugiyama, A PSE perspective for the efficient production of monoclonal antibodies: integration of process, cell, and product design aspects, Curr. Opin. Chem. Eng., 2020, 27, 121–128 CrossRef.
- M. Karimi Alavijeh, I. Baker, Y. Y. Lee and S. L. Gras, Digitally enabled approaches for the scale up of mammalian cell bioreactors, Digit. Chem. Eng., 2022, 4, 100040 CrossRef.
- I. Errandonea, S. Beltrán and S. Arrizabalaga, Digital Twin for maintenance: A literature review, Comput. Ind., 2020, 123, 103316 CrossRef.
- Yokogawa, Reinforcement-learning AI marks significant advances in Industrial Autonomy Artificial intelligence algorithm based on Factorial Kernel Dynamic Policy Programming (FKDPP) joins PID and Advanced Process Control (APC) to deliver new levels of autonomous operations, Technical Report, 2022 Search PubMed.
- L. A. Finelli and V. Narasimhan, Leading a Digital Transformation in the Pharmaceutical Industry: Reimagining the Way We Work in Global Drug Development, Clin. Pharmacol. Ther., 2020, 108(4), 756–761 CrossRef PubMed.
- A. G. Siemens, Press Release: Realizing the Digital Transformation in the Process Industries, Technical Report, 2021 Search PubMed.
| This journal is © The Royal Society of Chemistry 2024 |