Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Digichem: computational chemistry for everyone†
Oliver S.
Lee
 ab,
Malte C.
Gather
ab,
Malte C.
Gather
 *bc and
Eli
Zysman-Colman
*bc and
Eli
Zysman-Colman
 *a
*a
aOrganic Semiconductor Centre, EaStCHEM School of Chemistry, University of St Andrews, St Andrews, KY16 9ST, UK. E-mail: eli.zysman-colman@st-andrews.ac.uk
bOrganic Semiconductor Centre, SUPA School of Physics and Astronomy, University of St Andrews, St Andrews, KY16 9SS, UK
cHumboldt Centre for Nano- and Biophotonics, Department of Chemistry, University of Cologne, Greinstr. 4-6, 50939 Köln, Germany. E-mail: malte.gather@uni-koeln.de
First published on 16th August 2024
Abstract
We describe a new tool for the efficient management of computational chemistry. Digichem is a program that automates and simplifies nearly the entire computational pipeline, including large-scale batch submission of calculations, analysis and results parsing, the generation of 3D density plots and 2D graphs of calculation data, storage and retrieval of calculation results to a database, and automated handling of multi-step jobs. The program is designed to reduce the tedium and likelihood of human error for researchers of all skill-levels but is particularly targeted towards novice users who otherwise may find the barrier to entry to computational chemistry unnecessarily high. To date, this program has been used to successfully run and analyse over 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individual calculations, evidencing its usefulness and utility. The Digichem program is presently released under a free-to-use license, and components of the Digichem system are additionally available under an open-source license.
000 individual calculations, evidencing its usefulness and utility. The Digichem program is presently released under a free-to-use license, and components of the Digichem system are additionally available under an open-source license.
Introduction
Since its inception in the 1920s, computational chemistry has had a revolutionary impact on the way chemists perform research.1 Once limited to describing the bonding interactions of the hydrogen molecule,2 modern computational chemistry can accurately predict a multitude of molecular properties for systems containing hundreds or thousands of atoms,3–6 including molecular orbitals and their energies, molecular vibrations, electronic excited states, transition states, nuclear magnetic resonance (NMR) shielding and coupling constants, and reaction pathways. Much of this explosion in scale was driven by the seminal works of Hohenberg, Kohn, and Sham,7,8 building upon the works of many others,9–12 in developing the fast but accurate methodology that is known as density functional theory (DFT). Today, a plethora of computational models are available to the theoretician. This not only includes various flavours of DFT, including the original local-density approximation functionals, the nowadays ubiquitous hybrid functionals such as B3LYP13–16 and PBE0,17–19 and the modern double-hybrid functionals of Truhlar,20 Grimme21 and others, but also post Hartree-Fock (HF) methods, such as Møller–Plesset perturbation theory22,23 and coupled cluster theory.24–27 In the latter case, the popularity of these post-HF wavefunction based methods has been greatly accelerated by time-saving approximations, such as the resolution of the identity (RI) approximation,28–34 and the related chain of sphere exchange algorithm (COSX),35,36 permitting their application to larger systems than has been possible previously. These developments, amongst others, have led to computational chemistry being routinely used to understand natural phenomena that are otherwise inscrutable, and to predict the properties of molecules, either as a screening tool to help guide synthetic efforts, or to provide additional evidence for synthetic pathways or molecular properties.Over its lifetime, the accuracy, speed, and breadth of problems that can be addressed by computational chemistry has been steadily expanded. However, comparatively little has been done to improve the usability, accessibility, or productivity of computational chemistry. Most computational tasks are still too demanding in terms of memory, hard-drive space, and central-processing unit (CPU) cores to be performed on a personal computer and completed in a reasonable amount of time. Instead, most computational chemistry is relegated to distributed, remote super-computing clusters. These clusters invariably operate without a graphical user interface, forcing the user to interact solely via the command line. For experienced computing users, this setup enables faster and more efficient workflows because of its inherent support for scripting, but for many non-expert users the opposite is true, and the lack of a familiar interface can make even basic computing tasks, such as opening a file, arduous. Further, solving a computational chemistry problem rarely involves executing only a single program. Instead, it typically consists of several programs operating in sequence, which together make-up a pipeline, each program taking the output from the last as input to the next. The core of this pipeline is the computational chemistry program, or engine, which solves the computational chemistry problem itself, but other steps are equally important in order to correctly set up the computing environment and analyse the results (Fig. 1). These steps may include, but are not limited to: (1) drawing of the molecules/systems of interest; (2) choice of computational parameters (level of theory, CPU/memory usage, molecular properties of interest, etc.); (3) uploading of the molecule files to the cluster where the calculations are performed; (4) interfacing to the cluster queuing system; (5) running the computational chemistry engine; (6) regular monitoring to determine if the calculation has completed; (7) analysis of output files to determine if the calculation succeeded correctly (for example, checking for convergence of the wavefunction); (8) downloading of the completed calculation results; (9) extraction of results from output files; (10) optional post-processing of results, such as the generation of orbital density plots; (11) collation and further processing/analysis.
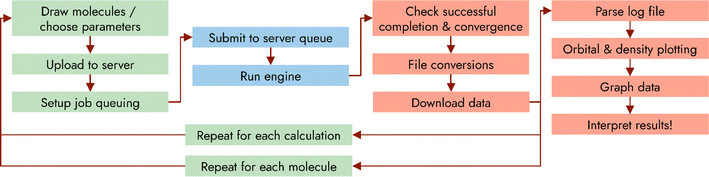 | ||
| Fig. 1 Flow-diagram of an example of the quantum chemistry pipeline. Green tasks relate to setup, blue to executing the computational chemistry engine, and red to analysis. | ||
This pipeline is complex and difficult to learn because it is made up of many different individual programs, and the user must learn how to use all of these programs correctly before any results can be obtained. Complicating matters is that most of the setup is performed using text files, with little-to-no validation, and error reporting varies greatly in quality between the different programs. Further, it is normally necessary to perform the pipeline multiple times for each molecule because the properties of interest to the chemist (e.g., HOMO–LUMO gap) are dependent on the correct molecular geometry being obtained first, and the geometry optimisation calculation which provides this must normally be run separately. The process must then be repeated again for each molecule in the study, and so large computational studies can quickly swell to thousands of individual calculations.
Together, these problems place an enormous learning burden on the would-be computational chemist and make the field challenging to break into. Even for more experienced users, the daily process of performing computations can be tedious, time consuming and error prone, because so much of the process needs to be curated manually. Many computational chemists will know the anguish of discovering a mistake in an input file only after the calculation has returned with useless data, wasting days of CPU time and demanding the entire process be started again. Even when performed correctly, this process is inefficient. Many parts of the pipeline, most noticeably between separate calculation steps, require manual intervention by the chemist to set up the next stage. This results in the overall execution time being limited not by the speed of the computer, but by how attentive the user is in checking the calculation progress. For computational screens involving many molecules, the inefficiencies are multiplied because only one molecule can be prepared by the user at a time. For very large computational projects, this essentially mandates that the scientist writes their own code to help automate the process, but this is not a skill that is accessible to all, nor one that is necessarily linked to scientific proficiency.
Existing solutions
Many researchers use computational chemistry as a tool to support their experimental research, rather than working full time in the field itself. In these studies, the number of computations is typically lower, and so the impact of the inefficiencies of the computational chemistry pipeline is lessened. In these cases, many scientists will choose to accept the status quo. For researchers who are unable to overcome the learning barrier themselves, they may instead be able to outsource computational tasks to collaborators, thus relieving themselves personally of the burden. For scientists looking to conduct more complex studies however, or who do not yet have a well-established network of collaborators to rely upon, this situation is not tenable. In these cases, it is common for the researcher to develop programs or scripts to assist them in managing the pipeline. In the simplest case, a group may share a set of standardised input templates, which each researcher then modifies to suit their needs. In this way, the need to write the whole input file from scratch is removed. More advanced programs may automatically merge parts of the input file together, for example to combine a standardised section specifying the method of the calculation with a customizable section containing the geometry for each molecule. For large-scale computational screens, this sort of automation is near-essential because of the number of files that need to be prepared. Sometimes, these in-house developed codes will mature to the point where they will be shared with other researchers, but in many cases they will only be available to members of the same research group or close collaborators. This results in a large duplication of effort because multiple researchers are attempting to solve the same problem. The generalizability of the scripts can also be an issue, and they may need to be substantially modified to address a new problem. This demands a significant investment of time at the start of each new project. While human error should be reduced because of the automation they offer, care needs to be taken in modifying these programs, because any errors that are introduced can propagate to many calculations. Finally, the quality of these programs depends on how much time the researcher can dedicate to maintaining them, and this is not always in abundance.Alternatively, or in addition, researchers can use an existing code or product. As examples, CalcUS,37 WebMO,38 ChemCompute39 and MolCalc40 are all web-based, calculation submission platforms. They provide tools to draw and/or upload molecular structures, choose calculation options, and submit a calculation to a number of backend computational chemistry programs. They also offer common analysis options, such as the parsing of molecular orbital energies, the plotting of molecular orbitals and the visualization of vibrational modes, although the exact feature set differs between each program. They all interface to a queuing system, either coming bundled with an internal queuing manager (CalcUS) or interfacing to external programs, such as SLURM41 (WebMO). CalcUS and WebMO are targeted towards academic research, while ChemCompute and MolCalc are designed for undergraduate teaching. CalcUS and MolCalc are free and open-source software, while ChemCompute is offered as a free-to-use service only, and WebMO is commercial, although it does offer a free basic version with a reduced feature set. Winmostar42 is a similar calculation submission platform, but is designed to be run on the user's desktop rather than in a web-browser, and is also commercial. Maestro,43 Spartan,44 TMoleX45 and GaussView46 are graphical user-interfaces (GUIs) to the computational engines Jaguar,47 Q-Chem,48 Turbomole,49 and Gaussian,50 respectively, and each is specific to its own engine. Maestro is typically distributed as part of the commercial Schrodinger platform, along with Jaguar, which, according to the authors,43 is targeted primarily towards researchers in drug discovery. Spartan is sold as a standalone product, with Q-Chem bundled inside of it, while GaussView is sold as an addon for Gaussian, and TMoleX is included in the purchase of Turbomole. All four offer common analysis features, such as parsing of molecular orbital energies, vibrational frequencies, and excited states, along with more advanced functionality such as the plotting of molecular orbital densities. They differ somewhat in their support for submitting calculations. Both GaussView and TMoleX can either write input files to the hard drive, or call the underlying engine directly, if installed on the same machine. TMoleX additionally supports calling remote installations of Turbomole. Spartan and Maestro, on the other hand, are designed to perform the calculation internally, without calling an external engine, although both can additionally interface to remote installations. Molden,51 Gabedit52 and Avogadro53 are more general tools and interface to a number of backend programs. They offer the usual analysis functions such as molecular orbital plotting, as well as input file creation tools, but do not manage the running of the calculation itself. There are also a number of libraries and support tools available that the intrepid computational chemist can incorporate into their own scripts and programs. Open Babel54,55 is a tool for the interconversion of chemistry file formats, while cclib56,57 provides parsing of calculation output. Both are available as a python application-programming interface (API) and a command-line tool. RDKit,58 CDK59–62 and ASE63 are three extensive libraries for performing computational chemistry, with RDKit and CDK focusing on analysis and post-processing, and ASE63,64 on performing atomistic simulations. They are mainly accessed via an API, but RDKit and ASE also offer a number of standalone programs. Finally, AQME65 is a collection of workflows designed to automate various repetitive computational chemistry tasks.
Despite the plethora of programs and libraries that are available, a complete single solution is not readily available. Many of the codes that are described above offer excellent support for either analysis or submission management, but none offers sufficient support for both for it to be used as a standalone tool. The user must still navigate multiple different programs in order to manage the complete pipeline. In particular, none of the tools that we have explored offer comprehensive support for large-scale computational screens, where the number of individual computations may reach the thousands, and in these cases the scientist is still expected to develop their own scripts. Many of the programs that offer a more complete set of tools may also be too expensive for a subset of researchers or exist only as programming libraries and cannot be used without the scientist having coding experience. To address these problems, we have developed Digichem, a complete computational management tool suitable for computational chemists of all skill-levels, but particularly directed at novice users, that we will now describe in more detail.
Program scope
Computational chemistry is a broad term. Included in this definition is the study of both molecular and periodic (i.e., crystalline) systems, using techniques such as quantum chemistry (QC), molecular mechanics (MM), molecular dynamics (MD), mixed QC/MM methods,66 Monte Carlo67 simulations and, increasingly, machine learning (ML). While some concepts are common to multiple different branches of computational chemistry, the way in which each type of calculation is performed, and the results obtained, can differ substantially. In Digichem, we have chosen to focus on the application of quantum chemistry to molecular systems. This includes common wavefunction methods (HF, MP, CC) as well as DFT, using atomic-orbital type basis sets. We do not explicitly include or exclude certain methods (e.g., different functionals), but rather interface with the functionality that is already presented in each computational engine. Currently, the program supports interfacing with the Gaussian,50,68 Turbomole,69 and ORCA70 programs. All common molecular calculations are supported, including single-point energies, geometry optimisations (including of excited states), vibrational frequencies, excited states (via both linear-response type methods71,72 and ΔSCF73 for the first triplet state), and nuclear magnetic resonance chemical shifts and coupling constants. The exact feature set supported varies from program to program, and naturally depends on the functionality of the underlying computational engine. The program does not currently support calculations involving multiple structures, most noticeably transition state (saddle point) type calculations, but we look forward to incorporating these in the future.General design and program flow
Digichem is a python program that runs directly on the computational server. It is designed to entirely replace (or abstract) the manual process with a single interface, reducing the steps the user must undertake to the minimum possible (Fig. 2). | ||
| Fig. 2 Flow-diagram of the Digichem pipeline. Green tasks relate to setup, blue to executing the computational engine, and red to analysis. | ||
Each calculation conceptually begins with the scientist choosing a number of molecules and some properties to study, and this is true for both Digichem and the manual process. However, unlike the manual process, Digichem does not limit the user to one type of input file (traditionally this would be the file-type expected by the computational engine) so the user can use whatever molecular sketching software they prefer. The computational parameters, which normally are included with the molecular geometry in the same file, are also stored separately in a program-agnostic format, or they can be later chosen from an internal library (see below). The user then uploads their chosen molecules to the server and runs the digichem command. All the chosen molecules are submitted simultaneously with one command, and the computation for each molecule is run in parallel. For each molecule, Digichem will create an appropriate folder structure (one folder per molecule, one sub-folder per calculation) and create the necessary input files for both the calculation program and resource manager, based on the calculation options the user chose earlier. Digichem will then run the calculation program, and simultaneously log the program's resource usage (CPU load, memory, file-space, etc.), which can be useful information for benchmarking or troubleshooting. Once complete, Digichem parses the output file to extract the calculation results and to determine whether the calculation completed without error, which is based on a number of criteria (see below). Assuming the calculation was successfully completed, Digichem will then perform any post-processing tasks (e.g., converting Gaussian checkpoint files (.chk) to their formatted variant (.fchk)), render any density plots, draw any 2D graphs, and write the numerical results to comma-separated value (CSV) files. The raw output files from the calculation are also preserved, allowing the user to perform their own custom post-processing if they wish. All the processed data are then compiled into a single PDF report for the user to read later. Finally, if any additional calculation types were requested, Digichem will take the output geometry and repeat the entire process for the next calculation type, and so-on until all requested calculations have been performed.
Digichem is split into distinct sub-modules to handle each part of the calculation pipeline. All the sub-modules are accessed from a single command on the command-line, which is digichem, followed by a keyword to identify the sub-module. Each of the sub-modules can be controlled through a command-line interface, which is quicker for power-users, while the most important sub-modules also support a graphical interactive interface powered by the Urwid library,74 which can be accessed by specifying the ‘-I’ (interactive) option after the sub-module name.
The interactive interface can also be launched directly by specifying the ‘digichem interactive’ command (Fig. 3), from which all major facets of the program can be accessed. For novice users, this is the only command they are required to learn. The graphical interface formats text and other non-alphanumeric characters, such as the Unicode box drawing characters,75 to appear like graphical widgets such as buttons, checkboxes, text-entry fields, and scrollable windows. These interactive elements are more familiar to users of traditional GUIs than the opaque command-line. Because the library powering this interface only uses the functionality already available in the text console,74 this interface does not require a full graphical software stack (e.g., X-server) on the remote cluster. All access to Digichem is provided via the secure-shell (SSH) protocol, which is already the default way users interact with their calculation servers, and it is therefore equally accessible from client machines running Windows, MacOS, or Linux. Digichem does not require any firewall ports to be opened, except for SSH port 22, which is already required to provide access to the cluster, in contrast to web-server designs.37,38,40 The currently supported sub-modules are: digichem submit, for calculation submission; digichem convert, for managing input data types, digichem result, for parsing and analysing completed calculation results; digichem report, for generating portable document format (PDF) reports of completed results; digichem image, for generating graphical data from completed calculations; and digichem database, for managing historical calculation data.
Installation
Although written primarily in the python programming language, Digichem is distributed as a self-contained binary produced by the PyInstaller program.76 This ‘frozen’ package contains all of the dependencies required for Digichem to run, including the Python interpreter itself. This type of distribution has the advantage that the end user doesn't need to install any other software or Python packages, greatly streamlining the installation process. The only software that Digichem depends upon that is not provided are the computational engines themselves. The Digichem packages are currently compiled on three different versions of the CentOS operating system (7.9, 8.5, and Stream-8), which in our experience are by far the most common operating systems used on computational clusters at present. Other Linux distributions are not explicitly tested but should also be supported so long as they are no older than CentOS-7.9, which was last released in 2020 and has now reached end-of-life. We will continue to update our list of target operating systems as usage patterns change.For the end-user, the installation process consists of only four steps:
(1) Download the most recent version of Digichem from our Github repository.77 The program can either be downloaded directly onto the computational server (using wget), or downloaded first to the user's personal computer and then uploaded to the server using the secure copy protocol (SCP) or file-transfer protocol (FTP).
(2) Extract the archive using the ‘tar’ command.
(3) Run the included install script to install the necessary symbolic links.
(4) Run the included setup script to discover the available server queues and computational engines.
The installation procedure is further explained in a detailed, step-by-step fashion within our user documentation.78 Depending on the end-user's needs, Digichem can either be installed personally (i.e., only for the current user), which does not require server admin privileges or super-user access, or to a shared location in which a single Digichem installation can serve multiple users on the same server. We believe we have simplified the installation procedure as much as possible considering the lack of a graphical user interface on the computational servers. For users who have had any prior experience working with the secure-shell protocol (SSH, the standard method users interact with computational servers), the steps should be familiar and easy to follow. However, we also acknowledge for complete novices, who may have never used a command line before, even these steps may be overwhelming. We intend to address this shortcoming in a future version by providing a fully graphical, automated installation procedure, should there prove to be sufficient demand.
Calculation submission
Input files
In most computational engines, the molecular geometry and the calculation options (the method) are combined into a single input file, but in Digichem we made the conscious decision to separate them. This makes it easy to reuse a computational methodology across multiple molecules, as is required in a large-scale computational screen, and to repeat a calculation in the future with a different structure. Both the method and coordinate files are written in the non-binary YAML format,79 which can be edited using any text editor and is more intuitive than many of the calculation engine native formats. In addition to the molecular geometry, the coordinate file also specifies the overall charge and spin multiplicity of the system, something which is missing from many popular cheminformatics formats (XYZ, for example), and this is preserved across Digichem calculations. Both files are calculation engine independent, which means that any geometry or method file is compatible with any of the calculation engines that Digichem supports, so long as the chosen calculation options are supported by the underlying calculating engine (Fig. S5†). To improve interoperability, we have also made efforts to standardize calculation options across different engines, for example supporting both PBE0 and the Gaussian specific PBE1PBE as names for this popular hybrid functional.17,19 Digichem does not currently offer a 3D molecule builder, because this is not possible within the confines of a terminal interface, but instead supports all of the common cheminformatics file formats. These are automatically converted to the internal Digichem format, using either the external obabel program54 or the internal Digichem parser, permitting the user to use whichever 3D sketching software they prefer. A full description of the supported file formats in available in Table S4.† Files can also be manually interconverted with the digichem convert sub-module.The digichem submit command
Submission in Digichem is achieved using the digichem submit sub-module. Any number of coordinate and/or method files can be specified at once. Each coordinate file will spawn a calculation to be run in parallel with the other coordinate files, while each method will be queued up to be performed in series using the output geometry from the previous calculation step (Fig. 4). Once submitted, all the selected calculations will be started automatically, and no further interaction by the user is required. Digichem currently supports the Gaussian,50,68 Orca70 and Turbomole49 programs, and can handle submitting to different programs one after another. This process can be repeated near-infinitely and is limited only by the resources available on the underlying cluster. In this way, computational screens of any size can be handled in an identical manner. Parallel calculation submission and resource management is handled by external queuing software, and Digichem currently supports both SLURM41 and PBS.80 Options to control shared resources, such as the amount of memory, number of parallel CPUs, and maximum job execution time, can all be configured as part of the method file. Digichem has options to ensure that the resources requested by the calculation, such as the amount of memory, do not exceed those requested from the scheduler.Submission with the interactive interface
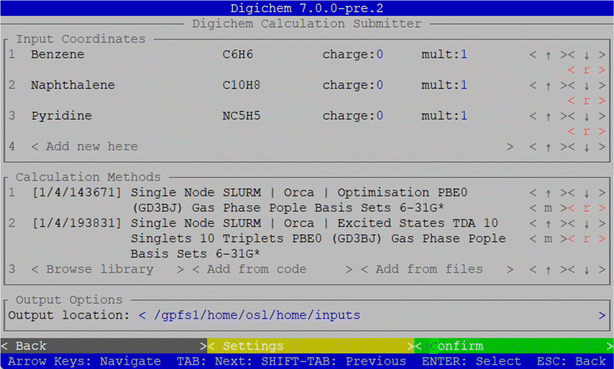
The digichem submit sub-module greatly simplifies the submission process for intermediate and advanced users, but for novice users the requirement to use the command-line and to write method files by hand is not ideal. For this scenario, the interactive submission interface is more suitable, which can be accessed using the digichem submit -I command (Fig. 5), or from the main menu.From this screen, the user can choose all the parameters of the calculation without using the command-line or generating the files manually. The starting geometries can be selected from a file-picker type interface (Fig. S6a†), similar to how files are selected in any commonly used GUI application, and the charge and multiplicity can be adjusted for each using text fields. The empirical formula of each molecule is also displayed to permit the user to check that the correct structure has been loaded. The calculation methods, meanwhile, can be selected from one of the approximately 240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pre-built calculations stored in Digichem's internal library, which cover the calculations of optimised geometries, vibrational frequencies, single-point energies and gradients, excited states, and NMR properties (Fig. S6c and d†). Multiple calculation methods can be chosen, and each will be performed in series, and the interface provides buttons to re-order the methods to ensure they are performed in the correct sequence. Each calculation method is associated with a unique numerical identifier (ID) consisting of three parts separated by a forward-slash, which in order correspond to: (1) the SLURM/PBS queue, (2) the calculation engine, and (3) the calculation itself. For example, the optimisation calculation shown in Fig. 5 has the unique code 1/4/143674. These calculation codes, if known in advance, can be used to quickly select a calculation from the internal library without navigating the interactive hierarchy, and can be specified on the command line to further speed up the submission process for advanced users. The library provides options for the most popular DFT functionals, including PBE0
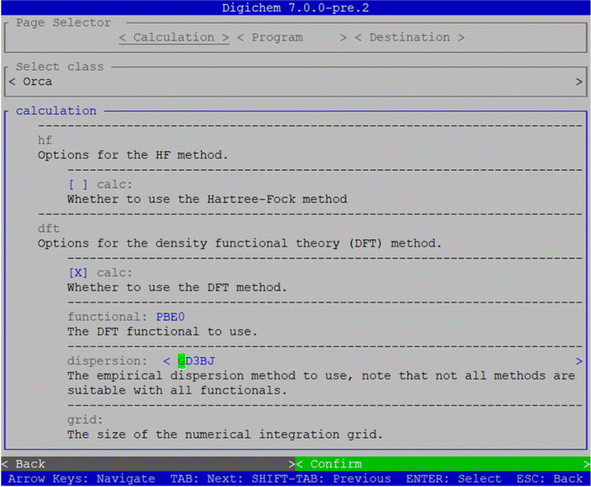
000 pre-built calculations stored in Digichem's internal library, which cover the calculations of optimised geometries, vibrational frequencies, single-point energies and gradients, excited states, and NMR properties (Fig. S6c and d†). Multiple calculation methods can be chosen, and each will be performed in series, and the interface provides buttons to re-order the methods to ensure they are performed in the correct sequence. Each calculation method is associated with a unique numerical identifier (ID) consisting of three parts separated by a forward-slash, which in order correspond to: (1) the SLURM/PBS queue, (2) the calculation engine, and (3) the calculation itself. For example, the optimisation calculation shown in Fig. 5 has the unique code 1/4/143674. These calculation codes, if known in advance, can be used to quickly select a calculation from the internal library without navigating the interactive hierarchy, and can be specified on the command line to further speed up the submission process for advanced users. The library provides options for the most popular DFT functionals, including PBE0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17,19 and B3LYP,13–16 as well as more advanced methods such as Møller–Plesset and coupled-cluster theory, a variety of common solvents and the most common basis sets in the Pople,81 Karlsruhe,82 and correlation-consistent families.83 We expect that, in most cases, the Digichem library will provide all the calculation options that a novice computational chemist would require, but the interactive interface also provides options to load a method file in case the user wishes to write a method from scratch or re-use a method from a previous calculation. Finally, Digichem provides an interface to interactively modify the calculation options from any loaded method (Fig. 6). This interface groups related options together and provides help messages and validation, where appropriate, to help novice users find and understand each calculation option. In this way, a user could select an existing method from the library and modify it to suit their needs; for example, increasing the number of requested CPUs so that the calculation will run more quickly. Once the calculation has been set up to the user's specifications, all queued molecules will be submitted to the queuing manager simultaneously once the green ‘confirm’ button is pressed (Fig. 5).
17,19 and B3LYP,13–16 as well as more advanced methods such as Møller–Plesset and coupled-cluster theory, a variety of common solvents and the most common basis sets in the Pople,81 Karlsruhe,82 and correlation-consistent families.83 We expect that, in most cases, the Digichem library will provide all the calculation options that a novice computational chemist would require, but the interactive interface also provides options to load a method file in case the user wishes to write a method from scratch or re-use a method from a previous calculation. Finally, Digichem provides an interface to interactively modify the calculation options from any loaded method (Fig. 6). This interface groups related options together and provides help messages and validation, where appropriate, to help novice users find and understand each calculation option. In this way, a user could select an existing method from the library and modify it to suit their needs; for example, increasing the number of requested CPUs so that the calculation will run more quickly. Once the calculation has been set up to the user's specifications, all queued molecules will be submitted to the queuing manager simultaneously once the green ‘confirm’ button is pressed (Fig. 5).
File storage and monitoring
Folder structure
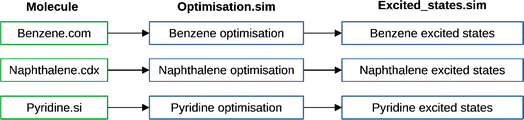
For each calculation submitted, Digichem automatically partitions the calculation data into a hierarchy of files and folders (Fig. 7). At the top of the hierarchy, each molecule is self-contained to its own directory. Within this, each individual calculation is stored in a separate sub-directory, which is named after the calculation in question. Digichem ensures that no two calculations can be performed in the same sub-directory, even if the same calculation is submitted multiple times. In this case, a number is appended to the directory name to distinguish it, and this logic is race-condition free. Within each calculation sub-directory, a third tier of directories store the calculation data. These directories are as follows: ‘Flags’, which conveys information about the current status of the calculation (see below); ‘Input’, which stores input files both for the specific calculation engine and Digichem; ‘Logs’, which stores log messages from Digichem and monitors CPU and memory usage (see below); ‘Output’, which stores raw output from the calculation engine, including the log file and any checkpoint binary files; ‘Results’, which stores formatted calculation data in text format, including CSV; and ‘Report’, which contains the PDF report and any associated image data. | ||
| Fig. 7 Example of the Digichem directory hierarchy for three molecules (benzene, naphthalene and pyridine) and two calculations (a geometry optimisation and a TDA-DFT excited-states calculation). | ||
The Results and Report folders will naturally only be created after the calculation has completed, as they only contain completed calculation data. A separate ‘scratch’ directory is also created and managed for each calculation, which is used by many computational engines for input/output (IO) intensive reading and writing. This scratch directory is normally stored outside of the rest of the calculation directory hierarchy to take advantage of faster and/or larger physical file storage media. Each scratch directory is also ensured to be unique for each calculation and is automatically removed following the completion of the corresponding calculation. This hierarchy of folders is managed entirely by the program, and greatly simplifies data management and storage.
Calculation monitoring
Calculation monitoring is traditionally achieved by: (1) checking the status of the job using the relevant queuing manager, for example with the squeue command; and (2) checking the final lines of the calculation engine log file, for example using the less or tail commands. Digichem provides an alternative that does not depend on external tools, and is more convenient for novice users, by means of the Flags sub-directory. Within this folder are a number of text files with distinctive names. These files are all empty, but the name of each file conveys information about the status of the calculation, and new files are created, and old ones are deleted, as certain milestones in the calculation are hit. At the beginning of the calculation, the PENDING flag will be the only file present, which indicates the calculation is currently in the queue, and awaiting resources. Once the calculation reaches the top of the queue and execution begins, the PENDING flag will be deleted and replaced with the STARTED flag. Once the calculation is complete, this in turn will be replaced with the SUCCESS flag, and the POST phase will begin, and so on. In this manner, it is possible to monitor the entire process of the calculation from the file-explorer, without using any external tools. The currently supported file flags, and their meanings, are detailed in Table 1.| Flag name | Description |
|---|---|
| PENDING | The calculation has been submitted but has not yet begun. Most commonly, this occurs because the calculation is waiting in the queue for server resources |
| STARTED | The calculation has begun. This flag persists even after the calculation has completed |
| RUNNING | The calculation is currently ongoing. This flag is removed once the calculation has completed |
| SUCCESS | The calculation has completed successfully |
| CONVERGED | The optimisation converged successfully; only relevant to geometry optimisations |
| NOT_CONVERGED | The optimisation did not converge successfully; only relevant to geometry optimisations |
| CLEANUP | The main calculation has finished, and Digichem is currently cleaning up |
| ERROR | The calculation has stopped because an error occurred |
| POST | The main calculation has finished, and Digichem is currently performing post-analysis. This largely involves writing the PDF report and any associated image files |
| DONE | All work on the calculation folder is complete, and Digichem will make no further changes. The calculation folder can be safely moved, downloaded or deleted |
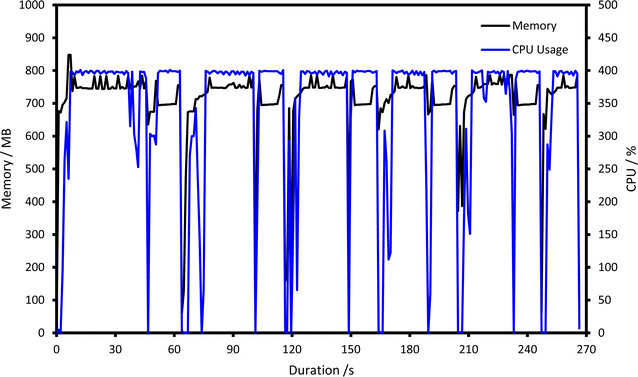
The CPU usage, memory usage and read/write speed of the calculation over time is of importance to both users of computational chemistry and designers of new software, as it helps to identify ‘bottlenecks’ (i.e., the slow part) of a calculation, and to diagnose out-of-memory failures. Digichem provides calculation profiling for all of its supported calculation engines, and records information such as CPU load, memory consumption, and read/write speed. This information is continuously logged to a CSV file as the calculation progresses, so it can be inspected even before the calculation has completed, and the profiling frequency can be manually adjusted to favour either higher resolution (more profiling steps) or lower file size (fewer profiling steps). The CPU and memory statistics for an example optimisation calculation are shown graphically in Fig. 8, which clearly details the short, large decreases in CPU usage surrounding a new energy calculation step starting.
Calculation analysis
Check for calculation completion
After each calculation has completed Digichem then automatically performs parsing of the calculation log file. In the first instance, Digichem determines whether the calculation has completed successfully, which can normally be detected by the presence of a trigger line at the end of the calculation (for example, “****ORCA TERMINATED NORMALLY****” for Orca, or “Normal termination of Gaussian” for Gaussian), as well as by checking if the exit code of the calculation engine is 0. The correct convergence of the density or wavefunction is also checked according to the criteria set by each computational program, and for a geometry optimisation the optimisation must have converged to a minimum for the calculation to be considered successful. The success/failure status of the program is used to set the SUCCESS or ERROR file flags, as appropriate, and is also used to determine whether to continue to the next calculation in the queue. If the calculation is not successful, no further calculations in the chain will be submitted. This prevents errors being accidently propagated to subsequent calculations and wasting CPU time.The calculaton of vibrational frequencies is a popular method to check whether a geometry optimisation has fully converged. If negative (i.e., imaginary) frequencies are present, then this typically suggests the geometry has not converged to a local minimum. In the current version of Digichem, negative frequencies are treated as a warning and are presented in red in the generated PDF report to draw attention, but they do not stop further program execution. This is because: (1) imaginary frequencies do not always indicate a geometry optimisation has failed to converge, and in certain types of calculations (transition-state geometry optimisations, for example) they are expected; and (2) the user may decide that the error introduced by the incorrect geometry is negligible (particularly if the negative frequency is very small) and should be ignored, rather than resubmitting. Of course, the user is free to change the molecular geometry and resubmit the calculation if they choose.
Log file parsing
If the calculation was successful, Digichem then proceeds to perform a more complete analysis of the output. The calculation log file is parsed using the cclib56 library, which is able to extract various numerical results, such as orbital energies and vibrational frequencies, as well as vital calculation metadata, including the DFT functional/calculation method, basis set and solvent. Additional data, which is not present explicitly in the calculation output, is also calculated by Digichem. This includes simulated absorption spectra, using Gaussian-broadened electronic excited states or vibrational frequencies, simulated NMR spectra, fluorescence decay rates, the dissymmetry factor of transition dipole moments,84 and spin–orbit coupling, the latter of which is calculated using a modified version of the PySOC program.85 The processed data is saved to a number of text files in the Results directory of the calculation. The most important of these results, such as the calculation metadata, HOMO/LUMO energies and geometry information are available in the summary results file (Fig. S9†). This file is written in a plain-text format which allows it to be easily read using simple text editors, including those available on the command line (nano, vi, emacs, etc.), and thus provides an immediate overview of the completed calculation data. For further processing with other programs or tools, the individual calculation results are saved to a number of tabular CSV files, which can be opened using common spreadsheet management programs or graphing software. This gives the user the opportunity to perform their own analysis and graphing; however, Digichem also generates graphs of the most important results automatically (described below). Together, these text-based result files can greatly facilitate an author's ability to conform to FAIR data practices because the files are easily read and are program-independent. Additionally, all the results processed from the completed calculation are stored in a Digichem-specific YAML file. This file can be read by the ‘result’, ‘report’ and ‘database’ sub-modules to load the full-set of calculation results, without having to re-parse the calculation log file. Finally, the output geometry of the calculation is stored in the Digichem-specific .si format and the program-independent .xyz format, allowing for easy reuse in future calculations or deposition in ESI.† All of the automatically generated result files, and their contents, are detailed in Table 2.| File | Type | Contents |
|---|---|---|
| Absorptions | .csv | Simulated UV-vis absorption spectrum using Gaussian-broadened excited states, plotted on an energy (eV) scale |
| Atoms | .csv | Atoms of the studied molecule and their output geometry |
| Beta | .csv | Orbital energies and symmetries of any beta orbitals |
| CC | .csv | Coupled cluster (CC) energies, including at each optimisation step if relevant |
| ES | .csv | Electronic excited state (ES) energies, multiplicities and other data |
| IR | .csv | Simulated IR absorption spectrum using Gaussian-broadened vibrational frequencies |
| MP | .csv | Møller–Plesset (MP) energies, including at each optimisation step if relevant |
| NMR | .csv | Nuclear magnetic resonance (NMR) data. This file contains calculated NMR shielding, and a matrix of coupling constants between each of the non-magnetically equivalent atoms of the molecule. Additional CSV files are also created of simulated NMR spectra, with and without simulated decoupling, using Gaussian-broadened peaks |
| Orbitals | .csv | Orbital energies and symmetries. If the calculation uses unrestricted orbitals (or otherwise contains both alpha and beta orbitals), this file will contain only the alpha orbitals |
| SCF | .csv | Self-consistent field (SCF) energies, which normally correspond to calculations at the Hartree–Fock or DFT level, including at each optimisation step if relevant |
| SOC | .csv | Spin–orbit coupling (SOC) matrix between each calculated excited singlet and triplet state |
| Summary | .csv | Single-row overview of the calculation metadata and principal results |
| Summary | .txt | The same information as above, but presented in a human-readable text format |
| UV-vis | .csv | Simulated UV-vis absorption spectrum using Gaussian-broadened excited states, plotted on a wavelength (nm) scale |
| Vibrations | .csv | Calculated vibrational frequencies |
| Geometry | .si | Output geometry in a Digichem-native format, including charge and multiplicity |
| Geometry | .xyz | Output geometry in a program-independent format, not including charge or multiplicity |
| Result | .sir | Complete processed output from the calculation, in a lossless Digichem-native format, can be used for further processing |
The digichem result command
In addition to the automated parsing that is performed by Digichem at the end of each calculation, it is also possible to parse any calculation log file on demand. This is achieved with the ‘digichem result’ sub-module, which is capable of writing any of the result files that are normally generated automatically. This allows the user, for example, to analyse a calculation result that was not submitted by Digichem, and still obtain the same data. Meanwhile, the desired output format can be selected by specifying the relevant option after the command, for example CSV can be selected with the ‘-c’ option, or the text summary format with ‘-s’.Often, the scientist is only interested in a subset of the results from the calculation output. Digichem supports the extraction of targeted data through filters, which can reference any part of the nested hierarchy of data that Digichem stores internally. For example, the filter ‘-f orbitals’ will return all orbital information associated with the calculation, while ‘-f orbitals:HOMO’ will only return information related to the HOMO, and ‘-f orbitals:HOMO:energy’ will only return the energy of the HOMO, and so on. In addition, Digichem supports not only parsing results from a single calculation result file, but also many simultaneously by specifying multiple output files after the ‘digichem result’ command. In this way, the results from entire studies can be collated into a single spreadsheet file, and specific results can be extracted using the commands described above, giving the user the flexibility to acquire any result they require with ease.
Data storage
Result databases
Processing large datasets is one of the more daunting and tedious tasks faced by the computational chemist. As their size grows, datasets, tables, and CSV files become increasingly cumbersome to navigate and process, and a true database quickly becomes advantageous. Databases (through querying) allow huge amounts of data to be searched quickly and can store results more efficiently (taking less file space) than the raw calculation output files. This means that past calculation data can be stored for long periods of time. Digichem maintains an internal database of calculation data that is stored locally on the computational server. By default, this database is accessible only by the user who submitted the calculations, but shared databases (on the same server) can also be established to permit collaboration between members of the same group. Once each calculation is complete, the results are automatically stored in each of the configured databases. These databases can be later queried to retrieve data matching certain criteria using the ‘database’ submodule (see below). Digichem also provides tools to insert data, using ‘digichem database insert’, delete data, using ‘digichem database delete’ and copy data, using the ‘digichem database slice’ command, which allows for convenient and efficient sharing of datasets between researchers. Digichem supports both the JSON-based TinyDB86 backend for human-readable data storage, and the binary-based Mongita87 backend for more efficient storage. Both backend programs operate using file-locks to manage concurrent access, and so can be safely used from multiple processes simultaneously.In addition to the typical calculation metadata, Digichem also assigns a unique ID to each calculation result. These IDs are calculated from the checksum of the corresponding log file, meaning they are deterministic, and are used to prevent an identical calculation from being inserted into the same database twice. In addition, each calculation has a ‘history’ attribute, which can optionally contain the ID of the calculation that occurred before it and thus generated the geometry for the following calculation. For example, the ‘history’ attribute of an excited-states calculation might contain the ID of the geometry optimisation that preceded it. In this way, the researcher can easily determine the chain of calculations that lead to each result.
Database queries
The ‘search’, ‘delete’ and ‘slice’ commands all support a simple query language to select which calculation results to interrogate. Each query starts by identifying an attribute to search against, for example the HOMO energy, followed by a comparison operator, the most common of which are ‘<’ (less than), ‘≤’ (less than or equal to), ‘ = ’ (equal to), ‘ = = ’ (exactly equal to, which is case sensitive for text), ‘>’ (greater than) or ‘≥’ (greater than or equal to). Digichem also supports more advanced queries to allow comparison of a single item in a list, and for molecular substructure searching. Each query is then completed with a value to search against. As a full example, the command ‘digichem database main search orbitals:values:any:label==HOMO:energy:value <−0.5’ would retrieve all calculation results from the main Digichem database with a HOMO energy of less than −0.5 eV. Multiple queries can be specified simultaneously and combine in a logical AND fashion, which permits querying for results that fall between a given range. The current version of the program does not yet offer a graphical interface to the database searching operations, mostly because of the design challenges in writing an interface for a complex query language. We intend to incorporate such a feature in a future release.Calculation reports
For experienced practitioners, the tables of data generated by the ‘result’ and ‘database’ commands are an efficient way to access the results of a calculation or study. However, for novice users, these tables may be daunting and confusing because the data they contain is presented without context. If the user does not understand the headings of the table, then the data within it are useless. Likewise, if the user is unaware of the correct name of the datum they require, they cannot use the filter tools to extract it because they do not know what to query. Even for the experienced users, there are some data that must be visualized to be understood. A classic example for computational chemistry would be an orbital density plot, which shows the electron density distribution of an orbital throughout the molecule, and these types of results must be shown graphically. Even for data that are purely numerical, the user must typically format or produce graphs of the results before they can be understood or shared with a collaborator. This work is time-consuming, yet can be automated.Automated PDF reports
To achieve this, Digichem provides the ‘report’ sub-module. Following from the successful completion of a calculation, Digichem will automatically generate a single PDF document that presents all the parsable results of that calculation. This report is presented in a style designed to mimic that of a scientific journal article (Fig. 9), as we envisioned that this format should be familiar to scientists and thus easy to navigate. A selection of example calculation reports is included in the ESI.†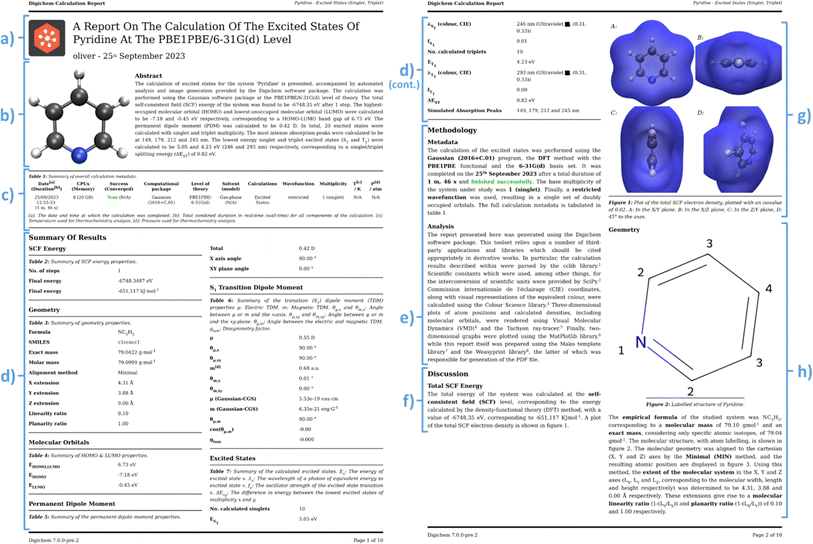 | ||
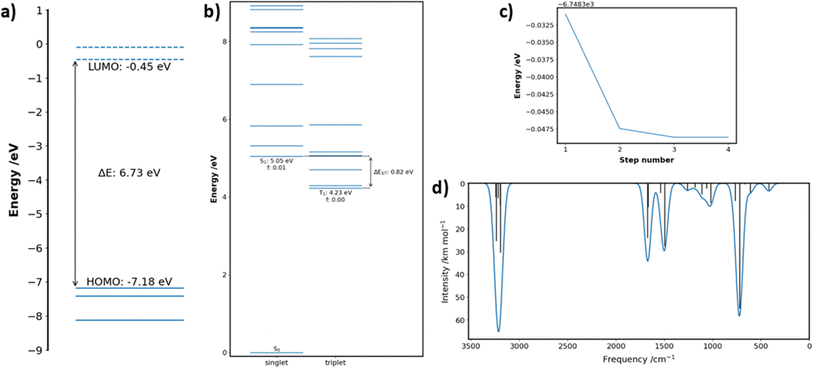
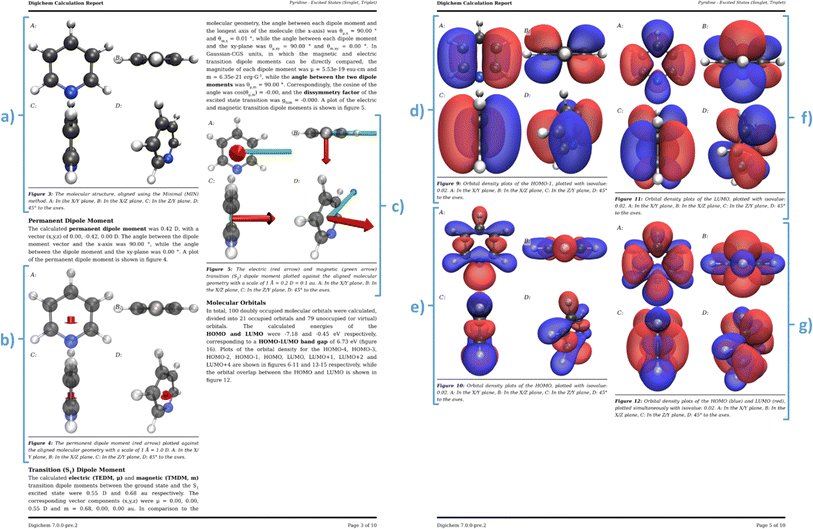
| Fig. 9 Excerpts from an example calculation report generated by Digichem. The excited states of pyridine at the PBE0/6-31G* level of theory using the Tamm Dancoff approximation were calculated. (a) Header section, (b) abstract summary and 3D rendered image of the molecule, (c) calculation metadata, (d) result summary tables, (e) methodology section, (f) start of the result discussion section, (g) 3D rendered of the total electron density at the DFT level, (h) start of the geometry discussion section, including 2D drawing of the molecule. The full PDF report is available in the ESI.† | ||
The report begins with a header section that contains the name of the molecule and the type of calculation that was performed, including which properties were calculated and at what level of theory. This is exemplified in Fig. 9a, which summarizes the calculation of the excited states of pyridine, calculated at the DFT level using the PBE0 functional (named PBE1PBE in Gaussian terminology) and the 6-31G(d) basis set. This section is followed by the ‘abstract’ (Fig. 9b), which contains a brief textual summary of the most important results of the calculation. This again contains the molecule name and level of theory, but also included are important numerical results, such as the HOMO–LUMO gap, and the energies of the lowest energy singlet and triplet excited states. In place of the traditional graphical abstract, the Digichem report includes a 3D rendered image of the geometry of the molecule. Next is a table of the metadata of the calculation (Fig. 9c), which includes the calculation engine used (in this case, Gaussian 16) and the execution time. Next is the summary section (Fig. 9d), in which headline results from the calculation are displayed in a tabular format. The data presented here are identical to those shown in the summary text result file generated by the ‘result’ sub-module, but here additional captions and more descriptive headings provide context to the tables. A methodology section follows (Fig. 9e), which contains a textual description of the same metadata shown in Fig. 9c, followed by a brief description of the analysis performed by Digichem itself. This latter section also includes references to the various libraries used to help generate the report, such as Weasyprint,88 Mako,89 and cclib.56 The final section shown is the discussion, which occupies the bulk of the report. Visible here is the discussion of the total system energy (Fig. 9f), with an accompanying 3D plot of the total electron density of the system at the PBE0 level (Fig. 9g), and a discussion of the molecular geometry (Fig. 9h) including a 2D drawing of the molecule produced by the RDKit library.58
Individual sub-section discussions cover all the results from the calculation that Digichem is able to parse, which includes total system energies (at the SCF/DFT, MP and/or CC levels), molecular geometry, orbitals, permanent and/or transition dipole moments (PDM/TDM), the latter of which includes the calculated dissymmetry factor between the electronic and magnetic TDM,84 if both are present, electronic excited states, and vibrational frequencies. Each discussion section is populated with important numerical results and provides general context for the results, to help the user to better understand the data that they have computed. We note that we do not use artificial intelligence or machine learning models to write any part of the report or interpret the results, they are compiled entirely from predetermined templates.
2D graphs
Where relevant, these discussion are aided by graphical representations of the data, which are drawn using the Matplotlib library.90 This includes a graph of the total system energy plotted against each optimisation step gap (Fig. 10c), which is useful for diagnosing convergence problems, a graph of the HOMO–LUMO and close-lying orbitals gap (Fig. 10a), and a graph of the electronic excited-state energies (Fig. 10b). Digichem is also capable of simulating UV-Vis absorption/emission, IR absorption and NMR spectra, by applying a Gaussian line-broadening function to the calculated electronic excited states, vibrational frequencies (Fig. 10c) and/or NMR shielding parameters and coupling constants.3D renders
In addition to these graphs, the Digichem calculation report sub-module can create three-dimensional renders of the molecular geometry (Fig. 11a), which can be optionally augmented with a plot of the permanent dipole moment (Fig. 11b) and/or a selected transition dipole moment (Fig. 11c). In the case of the TDM, both the electric and magnetic components are plotted, if available, as the relative orientation of both is an important parameter for the design of circularly-polarized light (CPL) emitters,84 amongst other applications. Digichem can also generate renders of electron densities, and currently supports orbital densities, total SCF density, spin-density (for open-shell systems), natural-transition orbital densities, and excited-state difference densities. The choice of which orbitals are plotted can be configured by the user, and by default includes the densities of the HOMO (Fig. 11e), the LUMO (Fig. 11f), the HOMO–LUMO pair together (Fig. 11g), and any orbitals that have a significant contribution to an electronic excited-state transition. The ‘significance’ of each orbital is based on the probability of an electronic transition involving the orbital; by default, orbitals involved in transitions with >20% probability are deemed to be significant and are included, although this value can be adjusted by the user. The orientation of the molecule in each render is determined by one of three rotation algorithms (described further in the ESI†) and each scene is rendered from four different perspectives to allow the researcher to view the molecule from multiple angles, these are: (1) along the z-axis; (2) along the y-axis; (3) along the x-axis; and (4) at 45° to the three axes. This ensures that at least one of the rendered angles shows the molecule clearly in the majority of cases. Of course, the user can still choose to render the molecule manually from a custom angle if necessary, using either their normal molecular viewing software or the Digichem tools. Digichem currently supports two rendering engines for generating these images: VMD,91,92 which is well-established software in the field of computational chemistry, but is only free for academic use; and Blender,93 using the Beautiful Atoms plugin,94 which by contrast is less well established but is open-source. All the examples showcased here are rendered using VMD, but the content is the same regardless of the engine used; only the visual style of the render is different.The digichem report and image commands
In the same manner as the ‘result’ sub-module, the report program has both an automated and manual implementation, and reports can be generated at any time using the ‘digichem report’ command. As for the ‘result’ program, this sub-module can also be used to parse calculations that were not submitted with Digichem, and so it is not necessary to re-submit old, completed calculations only to generate a new report. Regardless of whether it is generated automatically, or through the ‘digichem report’ command, each report contains not only the final PDF file, but also individual image files for every graph and 3D render used within it. These images are available in both a portable, low-quality JPEG format, and a publication-ready PNG format, and can be readily included in the user's own works. Digichem also offers the ‘digichem image’ command to generate only single images for situations in which the entire report is not needed or the user requires an image that is not included by default. For example, the command ‘digichem image output.log --image HOMO−10’ can be used to render the HOMO−10, which would typically be absent from the automatically generated report, while the command ‘digichem image output.log --list’ can be used to list all the available images that Digichem can render from a given calculation.Together, the data contained in each calculation report should be sufficient for the needs of computational chemistry users of all experience levels, and in many cases, they remove the requirement for the scientist to use external tools to process or format the calculation output. The automatic generation of publication-ready 3D renders saves considerable time for the chemist, both because this task is tedious, and because the renders themselves take time to compute. Lastly, the portable nature of the PDF means it is ideally suited for the sharing of results with collaborators and colleagues, and because all the results of the calculation are included within it (at least to the extent parse-able by Digichem), it is rare that the raw calculation output needs to be distributed.
Performance
A full benchmarking study of the time required to render each 3D image, parse a completed calculation log file, and fully generate a PDF report is presented in the ESI.† In summary, the time required to parse each log file depends mainly on the size of the file and the speed of the underlying file system, but even large log files (29 MB, 420![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individual lines) are parsed in less than 30 seconds. Each 3D image takes on average 10–30 seconds to render, with the rendering time decreasing on hardware with faster file system read/write speeds. The time required to write each PDF file is only a few seconds, with most of the time taken to generate each report being spent on rendering the constituent images. The total time taken by Digichem to perform all post-processing (log file parsing, image rendering, graph and result file writing, PDF generation) is generally less than 10 min (Table S3†), which is significantly less time than what would be required to manually undertake these tasks.
000 individual lines) are parsed in less than 30 seconds. Each 3D image takes on average 10–30 seconds to render, with the rendering time decreasing on hardware with faster file system read/write speeds. The time required to write each PDF file is only a few seconds, with most of the time taken to generate each report being spent on rendering the constituent images. The total time taken by Digichem to perform all post-processing (log file parsing, image rendering, graph and result file writing, PDF generation) is generally less than 10 min (Table S3†), which is significantly less time than what would be required to manually undertake these tasks.
Program status and future work
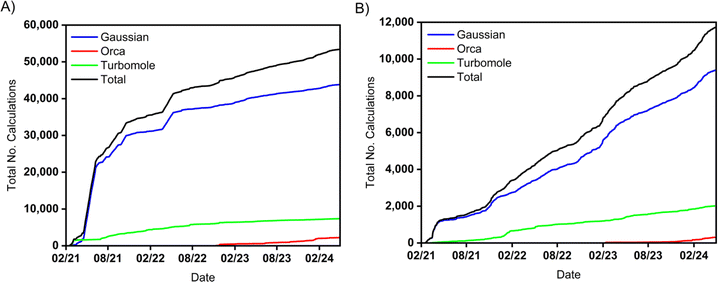
Digichem has been in development for over three years, during which time it has been extensively tested by the Zysman-Colman research group. Since mid-February 2021 (when we first began recording usage information) to the end of April 2024, Digichem has been successfully used to submit 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 406 calculations (Fig. 12a), an average of 46 calculations per day. By the time of writing of this article, Digichem has entirely replaced the manually-operated calculation pipeline for nearly all of the group's research. The program currently supports three computational engines, which are Gaussian,50 Turbomole,49 and Orca,70 with the latter only being introduced in November of 2022 (Fig. 12, red line). Due to its more recent inclusion, some of the more advanced analysis features are not yet supported for Orca, including natural transition orbitals and difference density plots, and these are features we intend to incorporate soon. We are actively considering the support of new computational engines, particularly those that offer complementary types of calculations that are not supported by the current roster of backend programs. In particular, the Python native computational package PySCF95 appears to be an ideal candidate, considering it shares the same programming language as Digichem, it has an open-source license, and it supports a wide-range of double-hybrid DFT functionals, which have recently shown great promise for the prediction of challenging molecular properties in the field of thermally activated delayed fluorescence.96 Meanwhile, we are continuing to expand the range of metadata that Digichem can parse, and in a forthcoming version we will add support for recognising the different excited states methodologies (e.g., TDA-DFT vs. TD-DFT), as well as pertinent performance data, such as the number of CPUs and the amount of memory that was allocated to each calculation. We are also looking to expand upon the program's support for in-series calculation queuing, as Digichem is not currently able to automatically submit multiple calculations from one completed calculation in a branching fashion. Finally, we intend to develop an additional web-based interface to the program to further increase the approachability of computational chemistry, which would be particularly appreciated by the novice user.
406 calculations (Fig. 12a), an average of 46 calculations per day. By the time of writing of this article, Digichem has entirely replaced the manually-operated calculation pipeline for nearly all of the group's research. The program currently supports three computational engines, which are Gaussian,50 Turbomole,49 and Orca,70 with the latter only being introduced in November of 2022 (Fig. 12, red line). Due to its more recent inclusion, some of the more advanced analysis features are not yet supported for Orca, including natural transition orbitals and difference density plots, and these are features we intend to incorporate soon. We are actively considering the support of new computational engines, particularly those that offer complementary types of calculations that are not supported by the current roster of backend programs. In particular, the Python native computational package PySCF95 appears to be an ideal candidate, considering it shares the same programming language as Digichem, it has an open-source license, and it supports a wide-range of double-hybrid DFT functionals, which have recently shown great promise for the prediction of challenging molecular properties in the field of thermally activated delayed fluorescence.96 Meanwhile, we are continuing to expand the range of metadata that Digichem can parse, and in a forthcoming version we will add support for recognising the different excited states methodologies (e.g., TDA-DFT vs. TD-DFT), as well as pertinent performance data, such as the number of CPUs and the amount of memory that was allocated to each calculation. We are also looking to expand upon the program's support for in-series calculation queuing, as Digichem is not currently able to automatically submit multiple calculations from one completed calculation in a branching fashion. Finally, we intend to develop an additional web-based interface to the program to further increase the approachability of computational chemistry, which would be particularly appreciated by the novice user.
Availability and licensing
We have demonstrated through our continuing usage that Digichem is ready for use in active research environments. To facilitate this, the Digichem project has been split into two main components. Core components of the program (Digichem-core) have been released as a python library under the permissive, open-source BSD-3-clause license. This library is freely available for any purpose and can be incorporated into computational workflows by the user. Currently, the library contains functionality pertaining to results parsing, image generation (both 2D graphs and 3D density plots), file interconversion, simulated spectroscopy (vibrational frequencies, nuclear magnetic resonances etc.), and other miscellaneous functions. Meanwhile, the full program is available in a closed-source format (i.e., compiled binary only) that contains bundled dependencies (produced using PyInstaller76). At present, the interactive interface, PDF report generation, database management, and calculation submission features are limited to the complete program only and are not available in the open-source library. This program is released under a timed license that is free to use for any purpose, but automatically expires after a set duration (currently set to 3 months from release). New releases (compiled automatically every night) are automatically upgraded with a new license with a new expiry. We have chosen this licensing model for two reasons:• To ensure users remain up-to-date with recent releases. As the software license expires every 3 months, this ensures the user updates the program at least this frequently. This is important, particularly during this rapid development phase, to ensure that crucial bug fixes (as well as new features) are distributed to end-users. We have chosen 3 months as a compromise between convenience (to not force constant updates) and recentness.
• All software requires updates, maintenance, and development to remain useful and relevant. In a fully open-source project, this requires a critical mass of committed developers to sustain, many (if not all) of whom are not directly paid for their time. This can be difficult to achieve, especially in academia, as evidenced by the examples of software that do not see updates past their initial publication, and/or are no longer available.97–99 We are committed to the continued development of Digichem, and acknowledge that this cannot be done for free, forever. By adopting this licensing scheme, we are able to explore funding strategies for future development, either through commercialisation, sponsorship, or other means.
Digichem-core is available from ref. 100, while the full Digichem program is available from ref. 77.
There is no functionality to automatically update the software in the current version, but we believe that the installation procedure is extremely simple (see above) and does not require administrator or super-user privileges. This means a ‘normal’ user can download and update their copy of Digichem easily and on their own. We are working to develop an automated update procedure that we intend to release in a future version of the program.
We also acknowledge that some research projects may last longer than the three-month license window, and researchers may want to continue using the same version of Digichem to ensure the consistency of the results obtained. To address this, Digichem has adopted semantic versioning. In this scheme, each version ‘number’ is split into three parts (e.g., 7.1.0), and each indicates what has changed compared to the previous version. An increase to the first number (7) indicates a backwards-compatibility breaking change, the second number (1) indicates a new feature (or features) that are fully backwards-compatible, and the last number (0) indicates a bug fix or other changes that do not introduce new functionality. This means that all versions of Digichem 7.x.x, for example, will produce the same results (although later versions may have extra functionality). We have additionally implemented an automated testing suite that compares the data parsed with each version to a set of expected results, and builds will only proceed if the results match. When a new major version of Digichem is released (version 8.x.x, for example), we will continue to update the license of the previous versions (7.x.x and below), so a researcher is able to use the same major version number throughout their study.
Conclusions
We have developed a program designed to automate and simplify the computational chemistry pipeline. We have included tools that reduce the tedium, duration, and likelihood of errors in performing calculation submission, management, and analysis for studies of all sizes, but we have particularly focused on performing large-scale computational screens where these issues are normally exasperated. The program is designed to be used by computational chemists of all skill levels and experience, but we expect it to be of particular value to novice users, who would normally find the process of learning the intricacies of the pipeline the most daunting. We have extensively tested this program over a period of more than three years, and the future direction of the project has been outlined. Through the continued development of Digichem, we continue to strive towards making computational chemistry accessible for all.Data availability
The code for Digichem-core can be found at https://github.com/Digichem-Project/digichem-core, and for the Digichem program at https://github.com/Digichem-Project/build-boy. The version of the code employed for this study is version 7.0.0-pre.2.Conflicts of interest
The authors O. S. L. and E. Z.-C. are currently exploring avenues to commercialize parts of the software described in this article.Acknowledgements
The authors would like to acknowledge Mr Vlad Tataranu for his advice, and Dr Ettore Crovini, Dr Tomas Matulaitis, and Dr Campbell Mackenzie for their assistance in testing early beta versions of the program. This research was financially supported by the European Research Council under the European Union's Horizon 2020 Framework Programme (FP/2014-2020)/ERC grant agreement no. 640012 (ABLASE). We thank the Quantum-Materials Centre for Doctoral Training at the University of St Andrews for support.References
- Y. Cao, J. Romero, J. P. Olson, M. Degroote, P. D. Johnson, M. Kieferová, I. D. Kivlichan, T. Menke, B. Peropadre, N. P. D. Sawaya, S. Sim, L. Veis and A. Aspuru-Guzik, Chem. Rev., 2019, 119, 10856–10915 CrossRef CAS PubMed.
- W. Heitler and F. London, Z. Phys., 1927, 44, 455–472 CrossRef CAS.
- J. Liu and X. He, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2023, 13, e1650 CAS.
- J. M. Soler, E. Artacho, J. D. Gale, A. García, J. Junquera, P. Ordejón and D. Sánchez-Portal, J. Phys.: Condens. Matter, 2002, 14, 2745–2779 CrossRef CAS.
- S. Grimme and P. R. Schreiner, Angew. Chem., Int. Ed., 2018, 57, 4170–4176 CrossRef CAS PubMed.
- J. Wilhelm, D. Golze, L. Talirz, J. Hutter and C. A. Pignedoli, J. Phys. Chem. Lett., 2018, 9, 306–312 CrossRef CAS PubMed.
- W. Kohn and L. J. Sham, Phys. Rev., 1965, 140, A1133–A1138 CrossRef.
- P. Hohenberg and W. Kohn, Phys. Rev., 1964, 136, B864–B871 CrossRef.
- L. H. Thomas, Math. Proc. Cambridge Philos. Soc., 1927, 23, 542–548 CrossRef CAS.
- E. Fermi, Rend. Accad. Naz. Lincei, 1927, 6, 602–607 CAS.
- P. A. M. Dirac, Math. Proc. Cambridge Philos. Soc., 1930, 26, 376–385 CrossRef CAS.
- J. C. Slater, Phys. Rev., 1951, 81, 385–390 CrossRef CAS.
- S. H. Vosko, L. Wilk and M. Nusair, Can. J. Phys., 1980, 58, 1200–1211 CrossRef CAS.
- P. J. Stephens, F. J. Devlin, C. F. Chabalowski and M. J. Frisch, J. Phys. Chem., 1994, 98, 11623–11627 CrossRef CAS.
- A. D. Becke, J. Chem. Phys., 1993, 98, 1372–1377 CrossRef CAS.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS PubMed.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1997, 78, 1396 CrossRef CAS.
- C. Adamo and V. Barone, J. Chem. Phys., 1999, 110, 6158–6170 CrossRef CAS.
- Y. Zhao, B. J. Lynch and D. G. Truhlar, J. Phys. Chem. A, 2004, 108, 4786–4791 CrossRef CAS.
- S. Grimme, J. Chem. Phys., 2006, 124, 034108 CrossRef.
- D. Cremer, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2011, 1, 509–530 CAS.
- C. Møller and M. S. Plesset, Phys. Rev., 1934, 46, 618–622 CrossRef.
- F. Coester, Nucl. Phys., 1958, 7, 421–424 CrossRef.
- F. Coester and H. Kümmel, Nucl. Phys., 1960, 17, 477–485 CrossRef CAS.
- J. Čížek and J. Paldus, Int. J. Quantum Chem., 1971, 5, 359–379 CrossRef.
- J. Čížek, J. Chem. Phys., 1966, 45, 4256–4266 CrossRef.
- A. M. Burow, M. Sierka and F. Mohamed, J. Chem. Phys., 2009, 131, 214101 CrossRef PubMed.
- C. Hättig, J. Chem. Phys., 2003, 118, 7751–7761 CrossRef.
- C. Hättig and A. Köhn, J. Chem. Phys., 2002, 117, 6939–6951 CrossRef.
- C. Hättig and F. Weigend, J. Chem. Phys., 2000, 113, 5154–5161 CrossRef.
- C. Hättig, A. Hellweg and A. Köhn, Phys. Chem. Chem. Phys., 2006, 8, 1159–1169 RSC.
- M. Sierka, A. Hogekamp and R. Ahlrichs, J. Chem. Phys., 2003, 118, 9136–9148 CrossRef CAS.
- F. Weigend, A. Köhn and C. Hättig, J. Chem. Phys., 2002, 116, 3175–3183 CrossRef CAS.
- R. Izsák and F. Neese, J. Chem. Phys., 2011, 135, 144105 CrossRef.
- R. Izsák, A. Hansen and F. Neese, Mol. Phys., 2012, 110, 2413–2417 CrossRef.
- R. Robidas and C. Y. Legault, J. Chem. Inf. Model., 2022, 62, 1147–1153 CrossRef CAS.
- W. F. Polik and J. R. Schmidt, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2022, 12, e1554 CAS.
- M. J. Perri and S. H. Weber, J. Chem. Educ., 2014, 91, 2206–2208 CrossRef CAS.
- J. H. Jensen and J. C. Kromann, J. Chem. Educ., 2013, 90, 1093–1095 CrossRef CAS.
- A. B. Yoo, M. A. Jette and M. Grondona, in Job Scheduling Strategies for Parallel Processing, ed. D. Feitelson, L. Rudolph and U. Schwiegelshohn, Springer Berlin Heidelberg, Berlin, Heidelberg, 2003, vol. 2862, pp. 44–60 Search PubMed.
- Winmostar V11 X-Ability Co. Ltd, Tokyo, Japan, 2023 Search PubMed.
- Schrödinger Release 2023-3, Maestro Schrödinger, LLC, New York, NY, 2023 Search PubMed.
- Spartan 20 Wavefunction, Inc., Irvine, 2023 Search PubMed.
- C. Steffen, K. Thomas, U. Huniar, A. Hellweg, O. Rubner and A. Schroer, J. Comput. Chem., 2010, 31, 2967–2970 CrossRef CAS.
- R. Dennington, T. A. Keith and J. M. Millam, GaussView, Version 6, Semichem Inc., Shawnee Mission, KS, 2016 Search PubMed.
- A. D. Bochevarov, E. Harder, T. F. Hughes, J. R. Greenwood, D. A. Braden, D. M. Philipp, D. Rinaldo, M. D. Halls, J. Zhang and R. A. Friesner, Int. J. Quantum Chem., 2013, 2110–2142 CrossRef CAS.
- Y. Shao, Z. Gan, E. Epifanovsky, A. T. B. Gilbert, M. Wormit, J. Kussmann, A. W. Lange, A. Behn, J. Deng, X. Feng, D. Ghosh, M. Goldey, P. R. Horn, L. D. Jacobson, I. Kaliman, R. Z. Khaliullin, T. Kuś, A. Landau, J. Liu, E. I. Proynov, Y. M. Rhee, R. M. Richard, M. A. Rohrdanz, R. P. Steele, E. J. Sundstrom, H. L. Woodcock, P. M. Zimmerman, D. Zuev, B. Albrecht, E. Alguire, B. Austin, G. J. O. Beran, Y. A. Bernard, E. Berquist, K. Brandhorst, K. B. Bravaya, S. T. Brown, D. Casanova, C.-M. Chang, Y. Chen, S. H. Chien, K. D. Closser, D. L. Crittenden, M. Diedenhofen, R. A. DiStasio, H. Do, A. D. Dutoi, R. G. Edgar, S. Fatehi, L. Fusti-Molnar, A. Ghysels, A. Golubeva-Zadorozhnaya, J. Gomes, M. W. D. Hanson-Heine, P. H. P. Harbach, A. W. Hauser, E. G. Hohenstein, Z. C. Holden, T.-C. Jagau, H. Ji, B. Kaduk, K. Khistyaev, J. Kim, J. Kim, R. A. King, P. Klunzinger, D. Kosenkov, T. Kowalczyk, C. M. Krauter, K. U. Lao, A. D. Laurent, K. V. Lawler, S. V. Levchenko, C. Y. Lin, F. Liu, E. Livshits, R. C. Lochan, A. Luenser, P. Manohar, S. F. Manzer, S.-P. Mao, N. Mardirossian, A. V. Marenich, S. A. Maurer, N. J. Mayhall, E. Neuscamman, C. M. Oana, R. Olivares-Amaya, D. P. O'Neill, J. A. Parkhill, T. M. Perrine, R. Peverati, A. Prociuk, D. R. Rehn, E. Rosta, N. J. Russ, S. M. Sharada, S. Sharma, D. W. Small, A. Sodt, T. Stein, D. Stück, Y.-C. Su, A. J. W. Thom, T. Tsuchimochi, V. Vanovschi, L. Vogt, O. Vydrov, T. Wang, M. A. Watson, J. Wenzel, A. White, C. F. Williams, J. Yang, S. Yeganeh, S. R. Yost, Z.-Q. You, I. Y. Zhang, X. Zhang, Y. Zhao, B. R. Brooks, G. K. L. Chan, D. M. Chipman, C. J. Cramer, W. A. Goddard, M. S. Gordon, W. J. Hehre, A. Klamt, H. F. Schaefer, M. W. Schmidt, C. D. Sherrill, D. G. Truhlar, A. Warshel, X. Xu, A. Aspuru-Guzik, R. Baer, A. T. Bell, N. A. Besley, J.-D. Chai, A. Dreuw, B. D. Dunietz, T. R. Furlani, S. R. Gwaltney, C.-P. Hsu, Y. Jung, J. Kong, D. S. Lambrecht, W. Liang, C. Ochsenfeld, V. A. Rassolov, L. V. Slipchenko, J. E. Subotnik, T. Van Voorhis, J. M. Herbert, A. I. Krylov, P. M. W. Gill and M. Head-Gordon, Mol. Phys., 2015, 113, 184–215 CrossRef CAS.
- D. Oelkrug, H. J. Egelhaaf and J. Haiber, Thin Solid Films, 1996, 284–285, 267–270 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16, Revision C.01, 2016 Search PubMed.
- G. Schaftenaar and J. H. Noordik, J. Comput.-Aided Mol. Des., 2000, 14, 123–134 CrossRef CAS.
- A.-R. Allouche, J. Comput. Chem., 2011, 32, 174–182 CrossRef CAS.
- M. D. Hanwell, D. E. Curtis, D. C. Lonie, T. Vandermeersch, E. Zurek and G. R. Hutchison, J. Cheminf., 2012, 4, 17 CAS.
- N. M. O'Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch and G. R. Hutchison, J. Cheminf., 2011, 3, 33 Search PubMed.
- N. M. O'Boyle, C. Morley and G. R. Hutchison, Chem. Cent. J., 2008, 2, 5 CrossRef PubMed.
- N. M. O'Boyle, A. L. Tenderholt and K. M. Langner, J. Comput. Chem., 2008, 29, 839–845 CrossRef PubMed.
- The author O. S. L. has contributed code to the open-source cclib project.
- RDKit: Open-source cheminformatics, https://www.rdkit.org Search PubMed.
- C. Steinbeck, C. Hoppe, S. Kuhn, M. Floris, R. Guha and E. Willighagen, Curr. Pharm. Des., 2006, 12, 2111–2120 CrossRef CAS PubMed.
- C. Steinbeck, Y. Han, S. Kuhn, O. Horlacher, E. Luttmann and E. Willighagen, J. Chem. Inf. Comput. Sci., 2003, 43, 493–500 CrossRef CAS PubMed.
- E. L. Willighagen, J. W. Mayfield, J. Alvarsson, A. Berg, L. Carlsson, N. Jeliazkova, S. Kuhn, T. Pluskal, M. Rojas-Chertó, O. Spjuth, G. Torrance, C. T. Evelo, R. Guha and C. Steinbeck, J. Cheminf., 2017, 9, 33 Search PubMed.
- J. W. May and C. Steinbeck, J. Cheminf., 2014, 6, 3 Search PubMed.
- A. Hjorth Larsen, J. Jørgen Mortensen, J. Blomqvist, I. E. Castelli, R. Christensen, M. Dułak, J. Friis, M. N. Groves, B. Hammer, C. Hargus, E. D. Hermes, P. C. Jennings, P. Bjerre Jensen, J. Kermode, J. R. Kitchin, E. Leonhard Kolsbjerg, J. Kubal, K. Kaasbjerg, S. Lysgaard, J. Bergmann Maronsson, T. Maxson, T. Olsen, L. Pastewka, A. Peterson, C. Rostgaard, J. Schiøtz, O. Schütt, M. Strange, K. S. Thygesen, T. Vegge, L. Vilhelmsen, M. Walter, Z. Zeng and K. W. Jacobsen, J. Phys.: Condens. Matter, 2017, 29, 273002 CrossRef PubMed.
- The author O. S. L. has contributed code to the open-source ASE project.
- J. V. Alegre-Requena, S. Sowndarya S. V., R. Pérez-Soto, T. M. Alturaifi and R. S. Paton, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2023, e1663 CAS.
- L. W. Chung, W. M. C. Sameera, R. Ramozzi, A. J. Page, M. Hatanaka, G. P. Petrova, T. V. Harris, X. Li, Z. Ke, F. Liu, H.-B. Li, L. Ding and K. Morokuma, Chem. Rev., 2015, 115, 5678–5796 CrossRef CAS PubMed.
- E. Paquet and H. L. Viktor, BioMed Res. Int., 2015, 2015, 1–18 CrossRef.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision D.01, 2013 Search PubMed.
- R. Ahlrichs, M. Bär, M. Häser, H. Horn and C. Kölmel, Chem. Phys. Lett., 1989, 162, 165–169 CrossRef CAS.
- F. Neese, F. Wennmohs, U. Becker and C. Riplinger, J. Chem. Phys., 2020, 152, 224108 CrossRef CAS PubMed.
- E. K. U. Gross and W. Kohn, in Advances in Quantum Chemistry, Elsevier, 1990, vol. 21, pp. 255–291 Search PubMed.
- S. Hirata and M. Head-Gordon, Chem. Phys. Lett., 1999, 314, 291–299 CrossRef CAS.
- A. Hellman, B. Razaznejad and B. I. Lundqvist, J. Chem. Phys., 2004, 120, 4593–4602 CrossRef CAS PubMed.
- I. Ward and Contributors, Urwid, https://urwid.org/index.html Search PubMed.
- Unicode Consortium, Unicode Stand. Version 15-1, 2023, pp. , pp. 250–257 Search PubMed.
- Collaborators, PyInstaller, https://github.com/pyinstaller/pyinstaller Search PubMed.
- O. S. Lee and E. Zysman-Colman, Build-boy, https://github.com/Digichem-Project/build-boy Search PubMed.
- Getting started — Digichem 6.1.0 documentation, https://doc.digi-chem.co.uk/introduction/index.html#how-to-install (accessed 17 July 2024) Search PubMed.
- K. Simonov and Contributors, PyYAML (version 6.0), https://pyyaml.org/ Search PubMed.
- OpenPBS Altair Engineering Inc., 2023 Search PubMed.
- R. Ditchfield, W. J. Hehre and J. A. Pople, J. Chem. Phys., 1971, 54, 724–728 CrossRef CAS.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297 RSC.
- T. H. Dunning, J. Chem. Phys., 1989, 90, 1007–1023 CrossRef CAS.
- H. Kubo, T. Hirose, T. Nakashima, T. Kawai, J. Hasegawa and K. Matsuda, J. Phys. Chem. Lett., 2021, 12, 686–695 CrossRef CAS PubMed.
- X. Gao, S. Bai, D. Fazzi, T. Niehaus, M. Barbatti and W. Thiel, J. Chem. Theory Comput., 2017, 13, 515–524 CrossRef CAS PubMed.
- M. Siemens, msiemens/tinydb, https://github.com/msiemens/tinydb Search PubMed.
- S. Rogowski, scottrogowski/mongita, https://github.com/scottrogowski/mongita Search PubMed.
- K. Community, Weasyprint: The Awesome Document Factory, https://weasyprint.org Search PubMed.
- M. Bayer, Mako Templates for Python, https://www.makotemplates.org Search PubMed.
- J. D. Hunter, Comput. Sci. Eng., 2007, 9, 90–95 Search PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graph., 1996, 14, 33–38 CrossRef CAS PubMed.
- J. Stone, PhD thesis, Computer Science Department, University of Missouri-Rolla, 1998.
- Blender Online Community, Blender – a 3D modelling and rendering package Blender Foundation, Blender Institute, Amsterdam, 2023 Search PubMed.
- X. Wang, T. Tian and U. Aschauer, Beautiful Atoms: A python library for the manipulation and visualization of atomistic structure using blender (Version 2.1.0), https://github.com/beautiful-atoms/beautiful-atoms Search PubMed.
- Q. Sun, X. Zhang, S. Banerjee, P. Bao, M. Barbry, N. S. Blunt, N. A. Bogdanov, G. H. Booth, J. Chen, Z.-H. Cui, J. J. Eriksen, Y. Gao, S. Guo, J. Hermann, M. R. Hermes, K. Koh, P. Koval, S. Lehtola, Z. Li, J. Liu, N. Mardirossian, J. D. McClain, M. Motta, B. Mussard, H. Q. Pham, A. Pulkin, W. Purwanto, P. J. Robinson, E. Ronca, E. R. Sayfutyarova, M. Scheurer, H. F. Schurkus, J. E. T. Smith, C. Sun, S.-N. Sun, S. Upadhyay, L. K. Wagner, X. Wang, A. White, J. D. Whitfield, M. J. Williamson, S. Wouters, J. Yang, J. M. Yu, T. Zhu, T. C. Berkelbach, S. Sharma, A. Yu. Sokolov and G. K.-L. Chan, J. Chem. Phys., 2020, 153, 024109 CrossRef CAS PubMed.
- M. Kondo, Chem. Phys. Lett., 2022, 804, 139895 CrossRef CAS.
- S. G. Chiodo and M. Leopoldini, Comput. Phys. Commun., 2014, 185, 676–683 CrossRef CAS.
- V. Sojo, A. Peraza, F. Ruette, M. Sánchez, A. E. Acosta and J. Comput, Methods Sci. Eng., 2012, 12, 397–406 Search PubMed.
- H. Borkent, J. Van Rooij, O. Stueker, I. Brunberg and G. Fels, J. Chem. Educ., 2003, 80, 582 CrossRef CAS.
- O. S. Lee and E. Zysman-Colman, Digichem-core, https://github.com/Digichem-Project/Digichem-core Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00147h |
| This journal is © The Royal Society of Chemistry 2024 |