Open Access Article
Open Access ArticleEmbedding DNA-based natural language in microbes for the benefit of future researchers†
Heqian
Zhang
 a,
Jiaquan
Huang
a,
Jiaquan
Huang
 a,
Xiaoyu
Wang
a,
Zhizeng
Gao
a,
Xiaoyu
Wang
a,
Zhizeng
Gao
 b,
Song
Meng
cde,
Hang
Li
b,
Song
Meng
cde,
Hang
Li
 f,
Shanshan
Zhou
g,
Shang
Wang
h,
Shan
Wang
i,
Xunyou
Yan
j,
Xinwei
Yang
k,
Xiaoluo
Huang
*l and
Zhiwei
Qin
f,
Shanshan
Zhou
g,
Shang
Wang
h,
Shan
Wang
i,
Xunyou
Yan
j,
Xinwei
Yang
k,
Xiaoluo
Huang
*l and
Zhiwei
Qin
 *a
*a
aCenter for Biological Science and Technology, Advanced Institute of Natural Sciences, Beijing Normal University, Zhuhai, Guangdong 519087, China. E-mail: z.qin@bnu.edu.cn
bSchool of Marine Sciences, Sun Yat-sen University, Southern Laboratory of Ocean Science and Engineering, Zhuhai, Guangdong 519000, China
cState Key Laboratory of Drug Research & Natural Products Research Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, 201203, China
dUniversity of Chinese Academy of Sciences, Beijing, 100049, China
eZhongshan Institute for Drug Discovery, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Zhongshan, Guangdong 528400, China
fSchool of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou, Guangdong 510006, China
gState Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, 100101, China
hCollege of Plant Science, Jilin University, Changchun, Jilin 130062, China
iState Key Laboratory of Microbial Technology, Shandong University, Qingdao, Shandong 266237, China
jCollege of Life Science, Langfang Normal University, Langfang, Hebei 065000, China
kSchool of Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, Henan 450001, China
lShenzhen Key Laboratory of Synthetic Genomics, Guangdong Provincial Key Laboratory of Synthetic Genomics, Key Laboratory of Quantitative Synthetic Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, Guangdong 518055, China. E-mail: huangxl@siat.ac.cn
First published on 14th October 2024
Abstract
Microorganisms are valuable resources as antibiotic producers, biocontrol agents, and symbiotic agents in various ecosystems and organisms. Over the past decades, there has been a notable increase in the identification and generation of both wild-type and genetically modified microbial strains from research laboratories worldwide. However, a substantial portion of the information represented in these strains remains scattered across the scientific literature. To facilitate the work of future researchers, in this perspective article, we advocate the adoption of the DNA-based natural language (DBNL) algorithm standard and then demonstrate it using a Streptomyces species as a proof of concept. This standard enables the sophisticated genome sequencing and subsequent extraction of valuable information encoded within a particular microbial species. In addition, it allows the access of such information for the continued research and applications even if a currently cultivated microbe cannot be cultured in the future. Embracing the DBNL algorithm standard promises to enhance the efficiency and effectiveness of microbial research, paving the way for innovative solutions and discoveries in diverse fields.
Introduction
In the 1670s, the pioneering Dutch scientist Antonie van Leeuwenhoek ushered in a new era in microbiology by observing the first microorganism through his ingeniously designed microscope. Over the centuries, knowledge derived from microbiology has evolved to play a pivotal role in diverse fields, such as medicine, agriculture, industry, and environmental science. As evidence of the remarkable progress in molecular biology and genetics, it nowadays has become increasingly common for research groups to readily engage in the genetic manipulation of specific microorganisms.The genetic landscape of microorganisms is characterized by its dynamism, shaped by myriad evolutionary forces, environmental exigencies, and stochastic occurrences. This complicated interplay dictates the preservation, loss, and acquisition of genetic traits within microbial populations over time, presenting formidable challenges to researchers. One significant hurdle arises from the inability to culture a substantial portion of microorganisms from diverse environments in laboratory settings.1,2 This prompts us to ask a pertinent question: can the microbes that are currently cultivable continue to be cultured in the distant future, for example, a century from now? In addition, since obtaining timely first-hand information on specific model organisms, whether wild-type or genetically modified, and unique microbial resources, is paramount, the following question then arises: where and how can such information be acquired?
Generally, the predominant research outcomes in the scientific community have relied heavily on extensive academic publications. However, the dissemination of such publications typically entails a relatively long time period. In light of recent advancements in digital technologies, a thought experiment has emerged: could the most crucial characteristics of any cultivated microbe be encoded and stored as DNA sequences within their live cells, with the ability to decode this information into a natural language readable by humans? This question takes on particular significance considering that such timely acquisition can now be achieved through straightforward sequencing and analysis – a technology that is no longer laborious and costly, but generally affordable and accessible.3
DNA digital data storage is the process of encoding and decoding binary data to synthesize strands of DNA.4 The synthesized DNA stores the aforementioned information in its host cell. Compared to canonical methods, DNA storage offers numerous advantages, including high storage density, extended storage time, minimal footprint, and low maintenance costs. Moreover, advancements in gene recombination technology enable the encryption of information within DNA, embedding it into biological cells.5 Because biological cells have the ability to replicate, knowledge stored in organisms can be passed down from one generation to another. In addition, information contained within biological cells cannot be accessed by computer hackers, who often operate through electronic-based systems, making this information potentially highly secret. These valuable properties make it a very promising medium for specialized data storage applications, such as scientific knowledge. However, the lack of a clear pipeline or guidelines for realistic data storage precludes its future deployment.
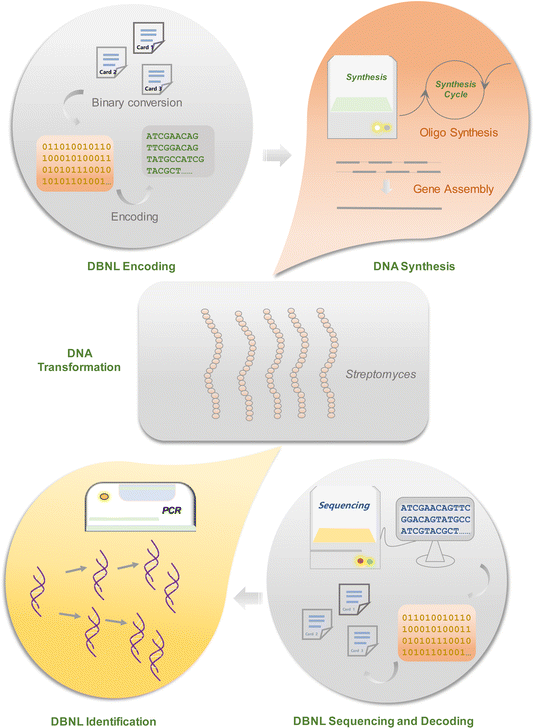
Here we introduce the “DNA-based natural language” (DBNL), which is embedded in microbial cells on the basis of DNA digital data storage. An illustrated diagram has been shown in Fig. 1. Our primary objective is to ensure the preservation of the key information of these bacteria when they are intentionally or unintentionally introduced into the environment. Simultaneously, we strive to provide valuable first-hand information for future researchers that is easily accessible through straightforward sequencing and analysis. The integration of DBNL into microbial systems addresses the urgent need for long-term data preservation, particularly as certain microbial strains become unculturable over time. It not only enhances data storage capabilities but also ensures the integrity and accessibility of essential biological information. By employing error-correction techniques and redundancy, DBNL serves as a promising solution for the challenges faced in microbial research, facilitating better data retrieval and analysis in future studies. We anticipated the embedded information will serve as a valuable resource, allowing researchers to learn the discoveries of their predecessors, particularly from a metagenomic perspective. This dimension holds substantial reference value for future researchers.
Results and discussion
Design of the DBNL criteria
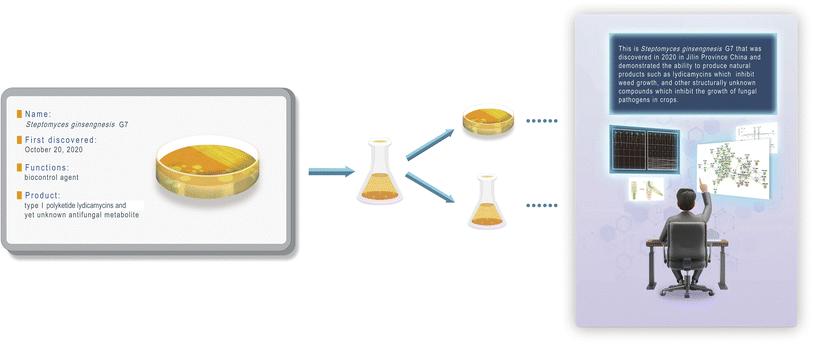
The DBNL criteria typically comprise crucial information of immense value to researchers across various disciplines. Given the diversity of research fields involved, it is essential to establish a common framework that allows for the uniform encoding and decoding of information across various applications. To address the inherent variability in the types of data that researchers seek to store, we propose a standardized encoding–decoding algorithm that prioritizes key attributes such as correctness, efficiency, robustness, scalability, generality, and security (see following sections). This algorithm will facilitate the seamless integration of DBNL into different research disciplines, ensuring compatibility and interoperability among diverse microbial systems. We also highlight the importance of creating a collaborative platform where researchers can share best practices and insights on data encoding and storage methodologies. By fostering a community dedicated to the standardization of DNA information storage, we can promote the effective use of DBNL and enhance its applicability in various fields, from natural product drug discovery to environmental microbiology.To further illustrate this perspective, we present a case study using the recently discovered rhizosphere species Streptomyces ginsengnesis G7 (see following sections). This strain is notable for its biocontrol potential, as it produces natural products that serve as both herbicides and antibiotics. Therefore, in the context of this strain, we aim to summarize pertinent information, focusing on its secondary metabolite production, antifungal activities, and herbicidal utilities.
Encoding DBNL standard into DNA sequences
To incorporate information into microorganisms, the DBNL standard must first be translated into DNA sequences using appropriate codec methods (Fig. 2). While this conversion is relatively straightforward, with A, T, C, and G represented by [00], [01], [11], and [10], it can result in a high number of single nucleotide homopolymers or regional variations in GC content. These issues could lead to significant failures in downstream processes such as DNA synthesis, amplification, and sequencing.6,7 A recent codec platform developed by Huang et al. offers a potential solution through an algorithm called “Wukong”, which facilitates flexible conversion between binary and DNA sequences.8 This algorithm begins by constructing a comprehensive codec rule library based on permutations and combinations of mappings between four binary bits and two nucleotides. Two distinct codec rules are then selected to encode the 2n-th and (2n − 1)-th coding units, establishing an initial direct conversion between binary bits and nucleotides. Since two nucleotides can encode four bits of information, this algorithm enables the integration of two to four binary strings into a single DNA sequence, allowing precise control over GC content and the occurrence of homopolymers. Additionally, this platform includes various user-friendly algorithms, such as DNA Fountain, which effectively manages biological constraints by processing segmented binary data to generate a new sequence that meets specified criteria for GC content and homopolymer length.Error correction is another critical aspect when encoding DBNL for microorganisms. Current methods include inserting error correction codes and introducing data redundancy into the encoded DNA sequences. Classic error correction codes, such as the Hamming code9 and the Reed–Solomon code,10 can address single nucleotide errors to some extent, but they struggle with significant fragment replacement or loss. For bacteria that replicate over extended periods, relying solely on an error correction code may not suffice. Therefore, in order to ensure high-fidelity data storage and retrieval in microorganisms, encoded DNA should be designed with additional redundancy. To this end, two practical approaches exist for this purpose: one is the classical computational “XOR” conversion, which can generate varying degrees of redundancy.8 Although no standard exists for the optimal level of redundancy to ensure stable data reproduction and high-precision decoding, a one-third redundancy can mitigate significant sequence loss. Another method is to create multiple copies of the data within a single cell; these DNA copies can repair each other in the event of specific faults.
Synthesizing DNA sequence stored with DBNL standard
Current DNA data storage technology encodes and stores computer data in the form of synthetic oligopools or genes.11–15 Synthetic oligopools are crafted through a high-throughput Chip-DNA synthesis process, often offering a cost-effective solution for storing large volumes of data.16 However, the conversion of thousands to millions of oligos from an oligopool into a specific microorganism proves both time-consuming and costly. To address this challenge, it's prudent to utilize longer synthetic DNA for embedding data into bacteria. Presently, commercial firms offer the production of synthetic DNA ranging from 1 kB to 10 kB at reasonable prices. This approach presents a viable method for effectively embedding data into microorganisms, offering a balance between cost efficiency and data storage capacity. In this work, the DBNL was encoded into a sequence consisting of 1800 base pairs.Transforming of DBNL-DNA into microorganisms
While a standardized approach for introducing DNA into microbes has yet to be established, emerging genome editing technology such as CRISPR/Cas has been proved to be a powerful tool for engineering a wide range of organisms.17,18 Additionally, the classic Red recombination method continues to hold value in various microbial engineering applications. A crucial consideration in the integration of DBNL-DNA is its placement within the microbial genome. As a foreign fragment, there is a risk of loss over extended cultivation periods. To mitigate this risk, integrating DBNL-DNA at multiple chromosomal loci is recommended. Although there is no universal solution, leveraging available engineering expertise with these organisms can guide the selection of appropriate genetic loci for integration. Furthermore, to enhance the stability and retention of DBNL-DNA, we suggest incorporating multiple copies into the target microorganisms. This strategy can increase the resilience of the integrated DNA and improve its persistence throughout cultivation cycles.Identification and sequencing DBNL-DNA from microorganism
Currently, genome sequencing technology offers the capability to decode an organism's genome at a remarkably low cost. This presents an excellent opportunity for identifying DBNL-DNA. However, the assembly of next-generation sequencing data from a microbe's genome can be time-consuming and prone to errors, particularly for species with high GC content. To overcome these challenges, it's advisable to sequence DBNL-DNA from PCR amplicons. Incorporating accessible primer sites into DBNL-DNA and keeping each fragment to a reasonable length can facilitate this process. For practical experimentation, a length of 3–10 kB is typically sufficient. Both Sanger sequencing and other emerging techniques can be employed to interpret the PCR amplicon of the DBNL-DNA sequence. This approach offers a reliable means of confirming the identity and integrity of the DBNL-encoded DNA, ensuring its accuracy for subsequent applications.Decoding DBNL information from microorganism
Once DBNL-DNA sequences are obtained from microorganisms, decoding the data can be facilitated by using the same program that encoded it. The above Wukong algorithm illustrates this capability, where data information can be decoded utilizing a specific data encoding algorithm along with the stored parameters. To streamline this process, it's recommended to adopt a standardized parameter associated with DBNL-DNA encoding. This standardization ensures that all DBNL-DNA information across different organisms can be interpreted with precision, minimizing the risk of errors stemming from incorrect parameter information. By employing a uniform parameter, the decoding process becomes more reliable and efficient. It's important to note that users may choose to apply additional compression or encryption processes to the DBNL information. Consequently, decoding such DNBL information necessitates the utilization of corresponding compression or encryption algorithms. This flexibility accommodates diverse user preferences while maintaining the integrity and security of the encoded data.Experimental validation of DNBL embedding in Streptomyces
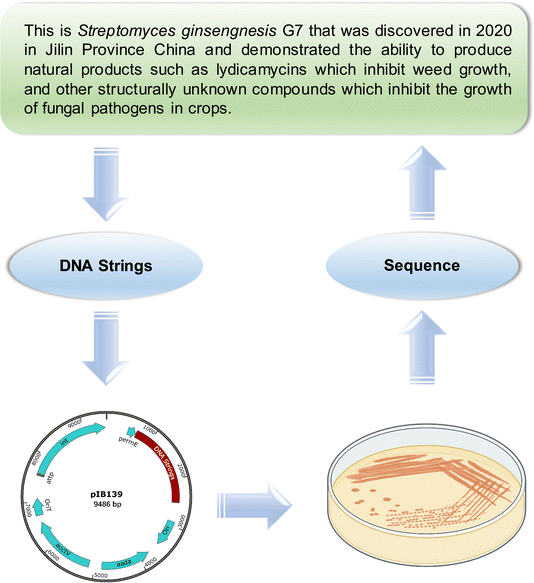
As a proof of concept, we wanted to imbue Streptomyces, a renowned producer of special metabolites, with a flexible context. Streptomyces species, which are common among actinomycetes, account for more than half of natural product antibiotics.19 The natural products they produced also have multifaceted applications, serving as herbicides and immunosuppressants. In this work, our focus was on S. ginsengnesis G7, a notable strain for its capacity to generate polyketide lydicamycins as potential herbicides.20–22 In addition, this strain produces other antifungal compounds whose chemical structures remain currently unidentified.Our project began with the compilation of a comprehensive summary outlining the primary functions of this biocontrol strain (Fig. 3). Using the Wukong encoder, we translated these insights into a DNA sequence (see ESI† for details). This sequence was synthesized and then integrated into the G7 chromosome using advanced protocols. Notably, this synthesized DNA sequence can be decoded into readable natural language through genome sequencing and decoding, ensuring its accessibility and comprehensibility.
To address potential concerns regarding the stability of foreign DNA, particularly in scenarios lacking selective pressure or neofunctionalization, we conducted genetic experiments wherein synthetic foreign DNA was integrated into the bacterial chromosome at the certain recombination site. For this purpose, we employed a bacterial φC31 integrase vector, pIB139, containing the attP attachment site, which corresponds to the attB site in the bacterial genome. Four of the recombined mutants were confirmed by PCR testing and subsequently underwent sequencing. We then sub-cultured these strains, harvested their spores and mycelia, and performed a repeat sequencing. Notably, no mutational changes were observed. These findings strongly suggest that our integration strategy ensures the stability of foreign DNA within the bacterial genome.
Conclusions
The utilization of digital DNA information storage has emerged as a promising technique with vast potential across various application scenarios. Recent successes have been observed in data archiving, information security, and synthetic biology. This perspective article introduces a proof of concept involving the embedding of DBNL in microbes, using a Streptomyces species as an illustrative example to demonstrate its accessibility. Such applications hold significant value for future researchers who may encounter a strain without any accompanying information, whether through re-exploration from the environment (in the case that the strain of interest is released to the environment) or through other means. This concept is also reminiscent of watermarking techniques for microbial strain protection. For instance, DBNL can serve as an effective “watermark” to safeguard valuable strains against theft. Additionally, the DBNL can encapsulate encrypted information, providing a robust layer of privacy for strain protection. Attempting to steal the strain would be challenging, as accessing all the “secrets” embedded within the DBNL would not be straightforward, thereby preventing the loss or theft of the strain.Although it may be impractical to categorize the entirety of the boundless array of microorganisms, the significance of pioneering work becomes evident when considering the dedication of more than 2200 laboratories spanning across 108 countries worldwide working on such efforts.23 Additionally, while the probability of mutations during DNA replication is indeed non-negligible, we believe these mutations typically result in point changes or duplications rather than the complete loss of exogenous DNA fragments. As such, this issue can be effectively managed by incorporating additional copies of the DNA sequences encoding the information. Moreover, we anticipate that these endeavours could be coupled with currently well-developed large language models, which would further contribute to error correction and increase the tolerance of bacterial DNA mutation and precision of encoded information.
While the current limitations in the amount of information that can be stored in DNA present significant challenges, they also drive innovation in this field. By condensing bespoke information and embracing new methodologies, such as automated synthesis platforms, researchers can not only reduce costs but also improve the efficiency of DNA storage solutions. Future efforts must focus on standardizing encoding techniques to facilitate interoperability across various applications and research disciplines. Such advancements will be crucial in overcoming the existing limitations and unlocking the full potential of DNA data storage in diverse scientific fields.
Finally, in light of the potential implications of employing DBNL standards, we recognize the importance of adhering to biosecurity and ethical guidelines. The integration of DBNL-encoded organisms into research and application contexts necessitates a thorough understanding of existing regulations surrounding genetically modified organisms (GMOs). It is essential that the development and use of these organisms comply with stringent biosecurity protocols to mitigate any unintended ecological or environmental consequences. Prior to deploying DBNL-encoded organisms, rigorous approval processes must be undertaken, which include comprehensive assessments conducted by relevant regulatory bodies. These processes will ensure that potential risks are adequately evaluated and managed. Moreover, we advocate for the establishment of a robust risk assessment framework that examines possible environmental impacts, particularly those related to biodiversity and ecosystem health. Addressing these considerations is crucial to fostering public trust and acceptance of GMO technologies. In addition, ethical principles should guide the development and application of DBNL in microbial systems, promoting responsible innovation and ensuring that research practices are aligned with societal values. By integrating bioethical considerations into the research and implementation phases, we can better prevent unintended consequences and enhance the sustainability of biotechnological advancements.
Taken together, these research undertakings, focused on cultivated microorganisms, underscore the imperative nature of such initiatives and should not be underestimated. Therefore, we call upon the community to commit to and actively engage in this field.
Data availability
The microbial materials are available upon request from the authors. For the Wukong algorithm, please see ref. 8.Author contributions
Heqian Zhang: conceptualization, methodology, writing – review & editing. Jiaquan Huang: conceptualization, methodology, writing – review & editing. Xiaoyu Wang: conceptualization, methodology, writing – review & editing. Zhizeng Gao: conceptualization, methodology, writing – review & editing. Song Meng: conceptualization, methodology, writing – review & editing. Hang Li: conceptualization, methodology, writing – review & editing. Shanshan Zhou: conceptualization, methodology, writing – review & editing. Shang Wang: conceptualization, methodology, writing – review & editing. Shan Wang: conceptualization, methodology, writing – review & editing. Xunyou Yan: conceptualization, methodology, writing – review & editing. Xinwei Yang: conceptualization, methodology, writing – review & editing. Xiaoluo Huang: conceptualization, methodology, writing – review & editing. Zhiwei Qin: conceptualization, methodology, project administration, writing – review & editing, funding acquisition.Conflicts of interest
The authors declare no conflict of interests.Acknowledgements
This work was supported by National Key Research and Development Program of China (2021YFF1201700), the National Natural Science Foundation of China (32170079), the Natural Science Foundation of Guangdong (2021A1515012026 and 2024A1515012593), Guangdong Talent Scheme (2021QN020100), Shenzhen Science and Technology Program (RCYX20221008092950122).References
- A. Oren and G. M. Garrity, ISME J., 2018, 12, 309–311 CrossRef.
- A. E. Murray, J. Freudenstein, S. Gribaldo, R. Hatzenpichler, P. Hugenholtz, P. Kämpfer, K. T. Konstantinidis, C. E. Lane, R. T. Papke and D. H. Parks, Nat. Microbiol., 2020, 5, 987–994 CrossRef CAS.
- J. M. Heather and B. Chain, Genomics, 2016, 107, 1–8 CrossRef CAS.
- L. Ceze, J. Nivala and K. Strauss, Nat. Rev. Genet., 2019, 20, 456–466 CrossRef CAS PubMed.
- S. Khan, M. W. Ullah, R. Siddique, G. Nabi, S. Manan, M. Yousaf and H. Hou, Int. J. Genomics, 2016, 2016, 2405954 Search PubMed.
- S. E. Van der Verren, N. Van Gerven, W. Jonckheere, R. Hambley, P. Singh, J. Kilgour, M. Jordan, E. J. Wallace, L. Jayasinghe and H. Remaut, Nat. Biotechnol., 2020, 38, 1415–1420 CrossRef CAS PubMed.
- T. P. Niedringhaus, D. Milanova, M. B. Kerby, M. P. Snyder and A. E. Barron, Anal. Chem., 2011, 83, 4327–4341 CrossRef CAS PubMed.
- X. Huang, J. Cui, W. Qiang, J. Ye, Y. Wang, X. Xie, Y. Li and J. Dai, iMeta, 2024, 3, e168 CrossRef PubMed.
- S. Zhang, B. Huang, X. Song, T. Zhang, H. Wang and Y. Liu, 3 Biotech, 2019, 9, 342 CrossRef.
- R. N. Grass, R. Heckel, M. Puddu, D. Paunescu and W. J. Stark, Angew. Chem., Int. Ed., 2015, 54, 2552–2555 CrossRef CAS.
- M. Lu, Y. Wang, W. Qiang, J. Cui, Y. Wang, X. Huang and J. Dai, Sci. China Life Sci., 2023, 66, 1447–1450 CrossRef CAS.
- Z. Ping, S. Chen, G. Zhou, X. Huang, S. J. Zhu, H. Zhang, H. H. Lee, Z. Lan, J. Cui, T. Chen, W. Zhang, H. Yang, X. Xu, G. M. Church and Y. Shen, Nat. Comput. Sci., 2022, 2, 234–242 CrossRef PubMed.
- Y. Erlich and D. Zielinski, Science, 2017, 355, 950–954 CrossRef CAS PubMed.
- N. Goldman, P. Bertone, S. Chen, C. Dessimoz, E. M. LeProust, B. Sipos and E. Birney, Nature, 2013, 494, 77–80 CrossRef CAS PubMed.
- G. M. Church, Y. Gao and S. Kosuri, Science, 2012, 337, 1628 CrossRef CAS.
- R. A. Hughes and A. D. Ellington, Cold Spring Harbor Perspect. Biol., 2017, 9, a023812 CrossRef.
- A. V. Anzalone, L. W. Koblan and D. R. Liu, Nat. Biotechnol., 2020, 38, 824–844 CrossRef CAS PubMed.
- M. Pacesa, O. Pelea and M. Jinek, Cell, 2024, 187, 1076–1100 CrossRef CAS.
- G. A. Quinn, A. M. Banat, A. M. Abdelhameed and I. M. Banat, J. Med. Microbiol., 2020, 69, 1040–1048 CrossRef CAS PubMed.
- X. Zhan, X. Li, Y. Zeng, S. Jiang, C. Pan, S. Pan, J. Huang, H. Zhang and Z. Qin, New J. Chem., 2024, 48, 525–528 RSC.
- H. Zhang, X. Li, S. Pan, J. Huang and Z. Qin, New J. Chem., 2023, 47, 12093–12100 RSC.
- J. Huang, X. Li, X. Zhan, S. Pan, C. Pan, J. Li, S. Fan, L. Zhang, K. Du, Z. Du, J. Zhang, H. Huang, J. Li, H. Zhang and Z. Qin, Plant Physiol., 2024, 194, 2709–2723 CrossRef CAS PubMed.
- T. F. O'Brien and J. Stelling, Biomedica, 2014, 34, 9–15 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00251b |
| This journal is © The Royal Society of Chemistry 2024 |