Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Leveraging GPT-4 to transform chemistry from paper to practice†
Wenyu
Zhang
 a,
Mason A.
Guy
a,
Jerrica
Yang
a,
Lucy
Hao
a,
Junliang
Liu
b,
Joel M.
Hawkins
c,
Jason
Mustakis
a,
Mason A.
Guy
a,
Jerrica
Yang
a,
Lucy
Hao
a,
Junliang
Liu
b,
Joel M.
Hawkins
c,
Jason
Mustakis
 c,
Sebastien
Monfette
c,
Sebastien
Monfette
 *c and
Jason E.
Hein
*c and
Jason E.
Hein
 *abde
*abde
aDepartment of Chemistry, The University of British Columbia, Vancouver, BC V6T 1Z1, Canada. E-mail: jhein@chem.ubc.ca
bTelescope Innovations Corp., Vancouver, BC, Canada
cPfizer Worldwide Chemical Research and Development, Pfizer Inc., Groton, Connecticut 06340, USA. E-mail: Sebastien.Monfette@pfizer.com
dDepartment of Chemistry, University of Bergen, Norway
eAcceleration Consortium, University of Toronto, Toronto, ON, Canada
First published on 3rd October 2024
Abstract
Large Language Models (LLMs) have revolutionized numerous industries as well as accelerated scientific research. However, their application in planning and conducting experimental science, has been limited. In this study, we introduce an adaptable prompt-set with GPT-4, converting literature experimental procedures into actionable experimental steps for a Mettler Toledo EasyMax automated laboratory reactor. Through prompt engineering, we developed a 2-step sequential prompt: the first prompt converts literature synthesis procedures into step-by-step instructions for reaction planning; the second prompt generates an XML script to communicate these instructions to the EasyMax reactor, automating experimental design and execution. We successfully automated the reproduction of three distinct literature-based synthetic procedures and validated the reactions by monitoring and characterizing the products. This approach bridges the gap between text-to-procedure transcription and automated execution, and streamlines literature procedure reproduction.
Introduction
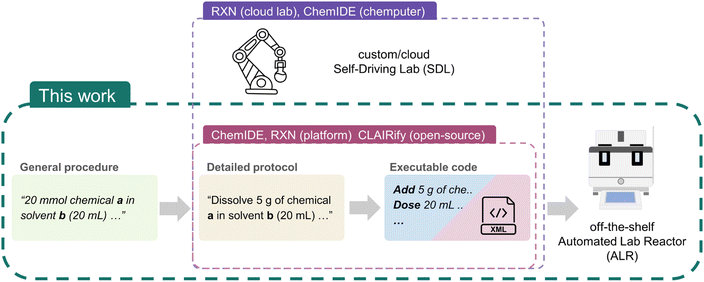
Self-Driving Labs (SDLs) integrate robotic automation with machine learning (ML) to explore chemical space. SDLs accelerate research and discovery across various disciplines, including organic synthesis,1–3 materials chemistry,4–6 photo-7,8 and electrochemistry.9–11 Automation liberates human researchers from time-consuming and repetitive tasks, while closed-loop optimization algorithms and/or computer vision further reduce the need for human oversight.7,12–14 However, the broader adoption of SDLs remains challenging because the required expertise in engineering and programming are beyond the scope of many chemistry laboratories. For example, programming an established automation system to execute a single, well-known chemical reaction can be a laborious exercise.14,15 Additionally, SDLs still require significant human involvement, skill, and time in the design of experiments and in mapping simple chemical actions to complex robot movements.16 The lack of standardization also poses a barrier in adaptability and transferability of SDLs development.Recognizant of this, the Cronin group introduced  ,17 a tool that transforms common procedures written in natural language to a domain-specific chemical descriptive language (χDL).18,19 This approach generalized the vast majority of chemistry related tasks, with the goal of facilitating standardization and transfer of chemical procedures for SDLs. This translation used a Natural Language Processing (NLP) algorithm, called SynthReader, to link text to their action entities and extract action details using pattern recognition. To support standardization, χDL is hardware-agnostic, and depends on SDLs to have a layer that expands the high-level χDL actions to basic hardware instructions or map these actions to programming functions. For example, χDL has enabled the successful transfer of synthetic protocols between different hardware platforms in different countries, and applied to the discovery of organic solid-state laser gain materials.20,21
,17 a tool that transforms common procedures written in natural language to a domain-specific chemical descriptive language (χDL).18,19 This approach generalized the vast majority of chemistry related tasks, with the goal of facilitating standardization and transfer of chemical procedures for SDLs. This translation used a Natural Language Processing (NLP) algorithm, called SynthReader, to link text to their action entities and extract action details using pattern recognition. To support standardization, χDL is hardware-agnostic, and depends on SDLs to have a layer that expands the high-level χDL actions to basic hardware instructions or map these actions to programming functions. For example, χDL has enabled the successful transfer of synthetic protocols between different hardware platforms in different countries, and applied to the discovery of organic solid-state laser gain materials.20,21
IBM's RXN for Chemistry (RXN) is a cloud-based SDL platform, supporting remote execution of a designed procedure on the automation lab.22 The platform also supports text-to-RXN procedure, a translation tool using the transformer model that promotes user-friendly experimental design and reaction planning.23 Despite the executable commands varying from ChemIDE/χDL in task names and arguments, both platforms support transcription of natural language to their executable languages, demonstrating that experimental design through natural language is more intuitive and enabling better transferability between SDLs.
Artificial Intelligence (AI) and Large Language Models (LLMs) have attracted significant public attention since the release of chat Generative Pre-trained Transformer (GPT) 3.5 from OpenAI.24 LLMs, trained on extensive datasets, provide comprehensive text responses to user requests upon various tasks. In chemistry particularly, LLMs have shown promise in reaction and molecular structure prediction owing to their expansive knowledge bases and complex pattern recognition capabilities.25–27 Embedding a chemistry-specific knowledge base into LLMs can noticeably improve the general performance in chemistry-specific queries.28–30 For example,  is a LLM chemistry agent used to predict organic molecule structures with desirable properties and assist with the planning and execution of their synthesis.29
is a LLM chemistry agent used to predict organic molecule structures with desirable properties and assist with the planning and execution of their synthesis.29
While LLMs can assist with interpreting experimental data and providing theoretical insights, they are traditionally incapable of conducting physical laboratory tasks, such as mixing reagents or operating lab equipment. This limitation can be overcome with their ability to assist in this text-to-procedure transcription with comprehensive understanding. For example, a comparable accuracy to transformer models can be achieved using GPT-3.5 with few-shot learning, by including several ground-truth instances in the prompt.31,32 Yoshikawa et al. proposed  , which generates χDL code from descriptive instructions using zero-shot GPT coupled with a verifier that iteratively prompts error messages in the chat.33 This LLM-based planning tool was later implemented to
, which generates χDL code from descriptive instructions using zero-shot GPT coupled with a verifier that iteratively prompts error messages in the chat.33 This LLM-based planning tool was later implemented to  , an AI voice assistant system on an electrochemistry-specialized SDL configuration, allowing interaction with the SDL in natural language.11 Additionally, Coscientist, a multi-LLM intelligent agent (GPT-4), used function calling with an embedded function pool. The system selects the most appropriate function according to the prompt to generate scripts in Emerald Cloud Lab (ECL) Symbolic Lab Language (SLL) and Opentrons Python application programming interface (API).34
, an AI voice assistant system on an electrochemistry-specialized SDL configuration, allowing interaction with the SDL in natural language.11 Additionally, Coscientist, a multi-LLM intelligent agent (GPT-4), used function calling with an embedded function pool. The system selects the most appropriate function according to the prompt to generate scripts in Emerald Cloud Lab (ECL) Symbolic Lab Language (SLL) and Opentrons Python application programming interface (API).34
Nonetheless, the implementation of LLMs in SDLs remains inaccessible for most chemists due to the lack of available and readily useable prompt methodologies for experimental design, and/or the labor-intensive development involved in using open-source LLM models. Platforms like  and
and  , while available for text-to-procedure translation, require specific SDL configurations to execute the generated code locally due to their hardware-agnostic nature. Moreover, some general procedures often lack exact mass or volume quantities, limiting the efficacy of current machine-readable transcription techniques or services. Indeed, this is also a laborious and time-consuming practice for human researchers.
, while available for text-to-procedure translation, require specific SDL configurations to execute the generated code locally due to their hardware-agnostic nature. Moreover, some general procedures often lack exact mass or volume quantities, limiting the efficacy of current machine-readable transcription techniques or services. Indeed, this is also a laborious and time-consuming practice for human researchers.
Recognizing these current limitations in text-to-procedure transcription and challenges in local SDLs execution, we aim to create an easy-to-use LLM prompt set for literature-to-procedure execution on an off-the-shelf automated laboratory reactor (ALR; Fig. 1). We selected the Mettler Toledo (MT) EasyMax because its software provides a user-friendly experimental design interface (iControl) and automatic design import from Extensible Markup Language (XML) (iC Data Center). We used ChatGPT-4 web-version to facilitate a no-code chemical protocol-to-procedure transcription, making it easy to be adopted by chemists with no coding expertise. We demonstrated the capability of this approach with three synthetic methods from the literature: (1) a detailed nucleophilic aromatic substitution reaction (SNAr) protocol; (2) a general hydrazine synthesis procedure and (3) an autonomous Curtius rearrangement monitoring protocol, showcasing different use case scenarios. Our two-prompt approach first transforms the literature protocol into a detailed stepwise procedure and subsequently to machine-executable XML scripts that communicated with MT EasyMax reactor. We further examined the generation robustness in length limits and accuracy as well as the code generation transferability in Python-based SDLs.
Methods
Selection of LLM
We chose GPT-4 over other generative AI platforms or other open-source models due to public accessibility, calculation accuracy, task-driven performance and token limit. First, OpenAI's ChatGPT family is one of the most accessible generative AI systems. The interaction with a GPT model through a web User Interface (UI) has no installation or hardware requirements (unlike options that require an API). GPT-4 also exhibits superior performance in chemistry-related tasks, including its enhanced accuracy in reaction prediction and structure elucidation.25 Furthermore, GPT-4's web browsing capability and integrated Python code interpreter35 enabled us to leverage its analytical capabilities for chemical phase lookup and advanced calculation tasks. Previous GPT models like GPT-3.5-turbo or out-of-the-box open-source models might result incorrect or missing output when there is a need of information lookup.Prompt engineering
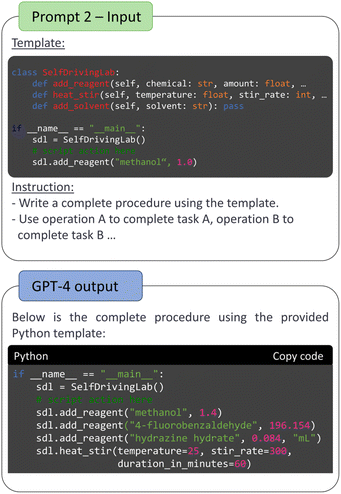
We developed a two-step sequential prompt to convert literature text into a machine-readable format using ChatGPT-4. The first prompt transformed general experimental procedures/protocols from the literature to detailed, structured, and step-by-step procedures. The second prompt transcribed the output from the first prompt to machine readable tailored XML scripts that can communicate with iControl. This sequential prompt approach also allows for the integration of additional prompts in either step for potential correction in molecular weight lookup and syntax error fixing.The first prompt in Fig. 2 (“prompt 1 – input” box) focused on breaking the translation task into three subtasks, called “instructions” in the prompt. This was inspired by how human researchers replicate a literature procedure, specifically contextualization, calculation and step organization as the “prelab” exercises. The first instruction requires the LLM to retrieve information to comprehensively understand the reaction before performing calculations. The second instruction specifically asks to find the reagent phase and calculate the actual addition mass or volume, significantly improving the mathematical accuracy of the chemical amounts in the stepwise procedure. Lastly, the third subtask is to rewrite the protocol to a step-by-step procedure with detailed chemical quantities, breaking down an unstructured procedure into modular and explicit steps. This prompt is customizable and can be adjusted to provide additional details, such as scale adjustments, chemical properties like liquid densities, or vendor information for reagents. An example of requesting a density search and formatting results in a markdown table can be found in ESI S3.2.†
The focus of the second prompt (Fig. 2 “prompt 2 – input” box) is to teach GPT-4 the EasyMax domain-specific XML structure. Although XML format in general can be familiar to LLMs, this iControl tailored structure and hierarchy are unseen to GPT-4. A minimal experimental design over iControl can be divided into two elements: an operation sequence  and a chemicals list
and a chemicals list  . The XML template includes the schema and contains necessary operations (heat, stir, add, dose, wait and end) organized in the
. The XML template includes the schema and contains necessary operations (heat, stir, add, dose, wait and end) organized in the  container. It also includes sample entries for solid and liquid chemicals in the
container. It also includes sample entries for solid and liquid chemicals in the  element, showcasing the iControl domain-specific XML's structure and hierarchy. The instructions in this prompt aim to map the operation names to the experimental operations, especially to differentiate solvent dosing from other reagent additions (solid or liquid) as the operation name may not be self-explanatory. The instruction also requests the generation of a 128 bit Universally Unique IDentifier (UUID) for every operation to replace the
element, showcasing the iControl domain-specific XML's structure and hierarchy. The instructions in this prompt aim to map the operation names to the experimental operations, especially to differentiate solvent dosing from other reagent additions (solid or liquid) as the operation name may not be self-explanatory. The instruction also requests the generation of a 128 bit Universally Unique IDentifier (UUID) for every operation to replace the  placeholder in the XML template. This step is important for iControl experiments, as it ensures each chemical or operation is distinctly identifiable and traceable during execution. For example, the add operation identifies the chemical to be added from the chemical list by referencing its
placeholder in the XML template. This step is important for iControl experiments, as it ensures each chemical or operation is distinctly identifiable and traceable during execution. For example, the add operation identifies the chemical to be added from the chemical list by referencing its  . Lastly, the instruction indicates that all chemicals used in the experiment should be defined in
. Lastly, the instruction indicates that all chemicals used in the experiment should be defined in  to ensure correct reference in reagent addition operations. Detailed prompts used can be found in ESI Table S2.†
to ensure correct reference in reagent addition operations. Detailed prompts used can be found in ESI Table S2.†
ALR configuration
Our experimental setup incorporated a Mettler Toledo EasyMax 102 synthesis workstation, equipped with an overhead stirrer and a SP-50 dosing unit for liquid handling. We opted for this commercialized ALR over custom or prototype SDL modules for its reliability, availability, and well-maintained user-friendly software families. We utilized the MT AutoChem software iControl 6.2 complemented by the iC Data Center 6.2 with Electronic Laboratory Notebook (ELN)-enabled capacity (detailed setup in ESI Section S4.2†) to allow experiment design and automatic XML import from a designated folder. Once the LLM-generated XML file was moved to the folder, it should appear to the iControl ELN session if there was no syntax error (ESI Fig. S13†). The experimental design interface in iControl can help visualize scripted operations, allowing easy fine-tuning. In case of any syntax error, the iC Data Center would create an error message file in the same folder that can be used to prompt the LLM for correction in the same dialogue (ESI Fig. S6†). This error correction approach is similar that of , enabling GPT to correct the syntax error by understanding the error message.33
, enabling GPT to correct the syntax error by understanding the error message.33
Execution and monitoring
The imported iControl experiment was then started without any modifications. Reagent addition operations were completed according to the pop-up window manually. Three reactions taken from literature were monitored using online high-performance liquid chromatography (HPLC) previously demonstrated with high reproducibility.3,36 The products were then characterized using nuclear magnetic resonance (NMR) spectroscopy. Detailed experiment and monitoring information are in ESI S1.†Results and discussion
Case study 1: SNAr aminolysis
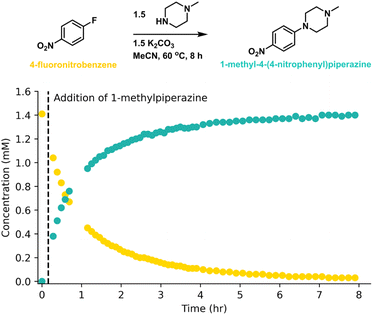
The transcription capabilities of GPT-4 was firstly evaluated with a SNAr reaction that was also done on a EasyMax 102 synthesis workstation.37 In this case, the literature protocol provided stir rate, temperature ramping detail and the mass and/or volume of the necessary reagent to carry out the reaction. We purposely let GPT-4 carry out the calculations using the standard prompt (ESI Table S2†), which matched the information from the literature. Following the output of the detailed stepwise procedure, each step is successfully mapped to iControl operations according to the given rules, see Fig. 3. The reaction was executed without modification and was monitored with online-HPLC (Fig. 4). We then confirmed the identity of 1-methyl-4-(4-nitrophenyl)piperazine via1H NMR spectroscopy (ESI Fig. S14†). The complete prompt and response generated by GPT-4 are in ESI Table S3.†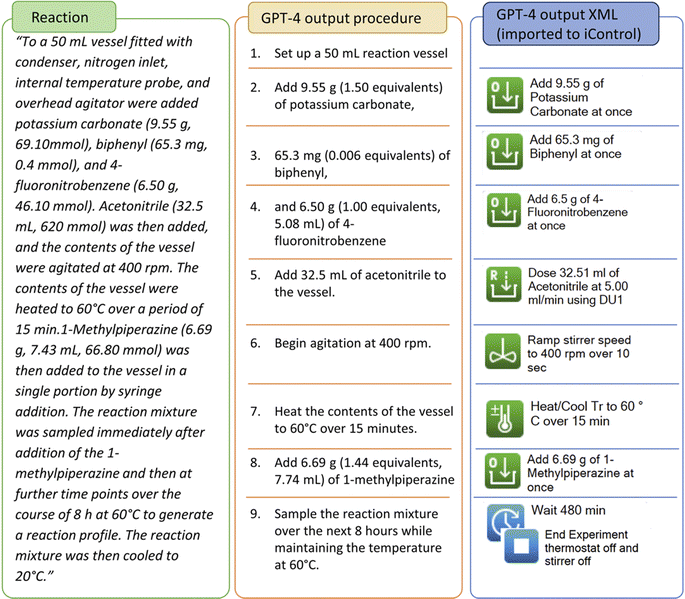 | ||
| Fig. 3 Literature procedure of SNAr aminolysis, stepwise procedure and iControl operations generated by GPT-4. | ||
Case study 2: hydrazone synthesis
Although the detailed experimental protocol with exact reagent amounts is sometimes required by the journals, which ease the preparation and calculation in reproducing the method, this is not invariably the case, especially in general procedure. Conventional tools, such as and
and  , are not capable of performing calculations of masses from mole or equivalence. The synthesis of hydrazone is a simple and straightforward reaction that we used to demonstrate the use case of performing calculation using GPT-4. In the general procedure of hydrazone synthesis,38 the amount of aldehyde is given in moles and equivalent to accommodate the various aldehyde species. Using 4-fluorobenzaldehyde as a test case, GPT-4 successfully outputted the correct molecular weight and calculated the correct mass of 4-fluorobenzaldehyde, see Fig. 5. Note that the identified step 3 involves both temperature and stirring information as these two steps are described in one sentence with no additional ramping instruction, like case study 1. Despite the identified single-step action, the output from second prompt successfully converted this step to
, are not capable of performing calculations of masses from mole or equivalence. The synthesis of hydrazone is a simple and straightforward reaction that we used to demonstrate the use case of performing calculation using GPT-4. In the general procedure of hydrazone synthesis,38 the amount of aldehyde is given in moles and equivalent to accommodate the various aldehyde species. Using 4-fluorobenzaldehyde as a test case, GPT-4 successfully outputted the correct molecular weight and calculated the correct mass of 4-fluorobenzaldehyde, see Fig. 5. Note that the identified step 3 involves both temperature and stirring information as these two steps are described in one sentence with no additional ramping instruction, like case study 1. Despite the identified single-step action, the output from second prompt successfully converted this step to  and
and  operations. The complete response generated by GPT-4 and the experiment design XML are in ESI Table S4.†
operations. The complete response generated by GPT-4 and the experiment design XML are in ESI Table S4.†
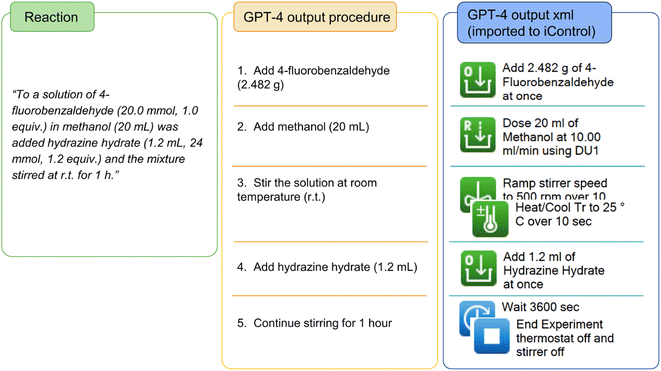 | ||
| Fig. 5 Detailed textual procedure of synthesis of hydrazone and stepwise procedure and iControl operations generated by GPT-4. | ||
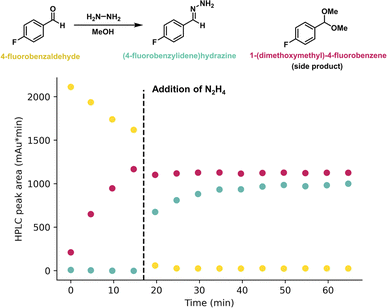
Following creation of the iControl protocol, the reaction was executed and monitored using online-HPLC and by plotting the peak areas of 4-fluorobenzaldehyde and (4-fluorobenzylidene)hydrazine over time (Fig. 6). The addition of the hydrazine hydrate was delayed to determine the pre-reaction composition of the benzaldehyde solution. A significant (4-fluorobenzylidene)hydrazine formation is presented at the first sampling point after the addition of hydrazine hydrate at 17 minutes. Formation of the intended product, (4-fluorobenzylidene)hydrazine, was also confirmed by 1H NMR spectroscopy. Acetal (1-dimethoxymethyl)-4-fluorobenzene is also present as an expected by-product (ESI Fig. S15†).
Case study 3: Curtius rearrangement
In this example of autonomous monitoring of Curtius rearrangement using NMR spectroscopy,39 the experimental protocol includes detailed documentation of monitoring techniques. Notably, the timing of reagent addition was determined by referencing the number of spectra collected. The complexity of the sampling technique may pose a barrier to proof-of-concept experimentations without autonomous monitoring tools. However, by prompting for calculation of actual reagent addition time using the sampling time interval, GPT-4 can comprehensively interpret the protocol and effectively generate a stepwise procedure as well as an iControl XML script with estimated reaction time (Fig. 7). The iControl design was executed, and the reaction was monitored with online-HPLC (Fig. 8, zoomed initial 7 hours data in ESI Fig. S2†). Note that the time course data had separate signals for 4-fluorobenzoyl azide formation that was not reflected in 19F NMR in the literature.39 The addition of hexafluoroisopropanol was delayed to 31 hours after an observed plateau of nitrene consumption. The carbamate formation after Curtius rearrangement was not observed with online-HPLC, but was characterized by 19F NMR spectroscopy with sampling before and after the addition of hexafluoroisopropanol (HFIPA) (ESI Fig. S16†). The complete prompt and response generated by GPT-4 are in ESI Table S5.†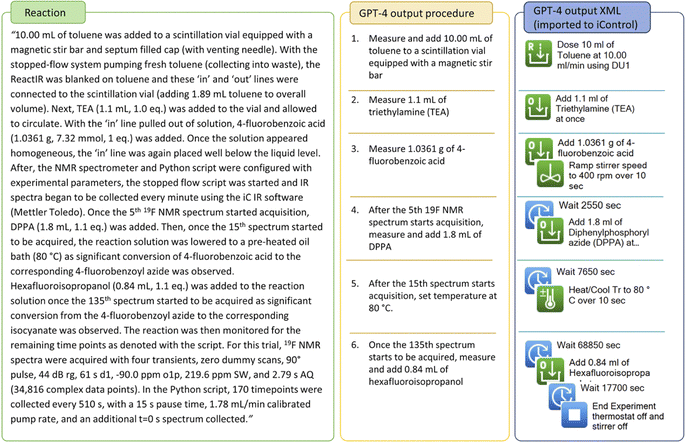 | ||
| Fig. 7 Literature procedure of Curtius rearrangement, stepwise procedure and iControl operations generated by GPT-4. | ||
 | ||
| Fig. 8 Online HPLC monitoring of 4-fluorophenyl isocyanate formation with DPPA addition at 42.5 min, and heat at 169.5 min. | ||
Robustness evaluation
The generation length limit were evaluated using the recently published metal-free C–N cross-coupling procedure.40 This method, including 9 reagent additions, 4 temperature changes and 3 reaction time settings, leading to a total of 18 iControl operations (including stir and end operations). Depending on the operation type, the token usage of one additional operation is 100 to 240.41 All solid reagents in this procedure were given in moles or equivalence, which may increase token usage during calculation steps in the first output. Assuming no additional prompt was used other than the base example, the resulting XML with the structured prompt would result in an estimated usage of 5140 tokens, which is beyond the ∼4000 limits in recent GPT models. This means that lengthy protocols need to be broken down into smaller portions, which could impact reliability of the translation. In 10 conversations during late April to June, 2024, we observed a higher occasion of multiple missing steps or unfinished scripting. For cases where operations or chemical elements are missing, the fix rate is promising when prompting for full procedure or full chemical list (ESI Table S11†). Due to the token limit, the maximum length of procedure is ideally within ∼120 words or 15 steps.The occurrence of syntax error is generally low because of the provided XML template, and they are often resolved effectively with the systematic error messages from iC Data Center. However, exceptions occur with missing element-ending tag (‘>’) (example in ESI Fig. S8†). In these instances, manual correction in text/code editor is more effective because this issue pertains more to text markup formatting than to scripting performance.
Using the same experimental protocol, the density lookup ability of GPT-4 shows 100% accuracy in 5 conversations that specifically requested density lookups. Although GPT-4 was demonstrated to have great understand of chemical information including molecular weight,27 the lookup results in prompt 1 cannot output the correct molecular weight for 5,6-dichloropyrazine-2,3-dicarbonitrile in all 10 conversations, resulting the incorrect mass output even though using the correct equation (ESI Table S12†). This error may stem from incorrect formula identification and a lack of chemistry knowledge, as GPT-4 did not activate web browsing feature, possibly related to recent cases of web searching reluctancy.42 However, this can be fixed by prompting “search the molecular weight” and/or providing the chemical formula to leverage general chemistry knowledge. Additionally, an external chemical database embedding can also enhance the lookup accuracy.
Aside from the molecular weight discrepancy, GPT-4 accurately converted chemical amounts from mole to mass in all 10 conversations (ESI Table S12†). Importantly, the code interpreter feature was not activated in any of those instances, highlighting that these calculations were performed based on the model's capabilities. Recent evaluations have demonstrated GPT-4's near human-level accuracy in more complex calculus problems, suggesting that its performance in basic chemical mass calculations should be reliably accurate.43,44 The generated iControl operations remains consistent across conversations, ensuring correct sequence of steps and parameters despite variations in unit and decimal places (ESI Table S13†).
Scalability and transferability
The EasyMax domain-specific XML schema adheres to the S88 format, a standardization of the control philosophy in hardware control. While hierarchy and operation parameters may vary between manufacturers, the fundamental philosophy remains consistent, aligning with the S88 baseline. Therefore, this prompt engineering approach should be transferable as long as the prompt can map experimental actions (e.g., heating, stirring, dosing) to operation names. Beyond the XML format, this approach should support scripting experimental procedures in any general-purpose programming languages. With more familiarity to the language and its function calling ability,45 the LLM can script series of functions to effectively plan and execute sequential tasks.As a proof of concept, we demonstrate the translation of hydrazone synthesis (case study 2) on Python-based SDLs.46,47 Methods such as heating, stirring and dosing were tailored to resemble the configuration of an ALR setup, with manual addition of reagents other than solvents. The second prompt was adapted using the Python method definitions with updated mapping instructions (Fig. 9). The resulting Python script accurately mapped functions for all the steps in hydrazine synthesis, showcasing the transferability of the prompt engineering approach across different instruments and programming languages (full conversation in ESI Table S7 and 8†). Beside direct execution with Python-based SDLs, the functions can also serve as a backend for scripting operations in domain-specific XML (ESI Table S9†). This permits the potential transcription of more complex procedures with fewer token usages, as Python function calls do not require hierarchical templates like XML. With more sophisticated SDLs, potentially designed for material synthesis, there is the prospect of fully autonomous pipelines bridging the gap between literature and product.
Limitations
Although this solution can streamline the calculation and scripting process for replicating a literature method, it relies on human intervention for prompt engineering, XML file saving, error correction and manual reagent addition during experiment. Some key parameters, such as stirring rate and ramping rate are sometimes not addressed in the literature and may require users' judgement to edit default values in XML template, or generated XML files before or after importing to iControl. The XML generation accuracy also depends on the operation mapping rules in prompt 2, where new rules need to be established for the addition of new workups or operations. Due to the hardware constraints, such as temperature range, stirring rate limit and vessel capacity, it's important yet challenging for LLMs to design experiments that strictly adhere to these rules. Additionally, the response may encounter an unexpected pause due to high server load or token limit, potentially requiring a “continue” prompt to resume the generation. It's worth noting that GPT performance can vary across different releases, and its behaviour may evolve over time.Lastly, although this approach is designed to reproduce literature methods, concerns around the safety and ethics of using LLMs remain, particularly when it comes to generating scripts for dangerous or unethical reactions. Most instrumental constraints (e.g., temperature) are managed by the safety protections of the ALR.  has proposed a warning system to flag dangerous reactions, enhancing lab safety.29 However, when utilizing ChatGPT with prompt engineering, preventing the generation or interpretation of hazardous scripts largely depends on the pre-training safeguards implemented by the generative AI provider (e.g., OpenAI).
has proposed a warning system to flag dangerous reactions, enhancing lab safety.29 However, when utilizing ChatGPT with prompt engineering, preventing the generation or interpretation of hazardous scripts largely depends on the pre-training safeguards implemented by the generative AI provider (e.g., OpenAI).
Conclusions
In this study, we have developed a prompt engineering solution to translate technical methodologies to stepwise procedures and machine-readable scripts in one dialogue. In the first prompt, GPT-4 demonstrates its proficiency in performing calculations, chemical phase lookup and generating steps with precise quantities in the procedure. With the XML template and mapping rules that associate solid, liquid dosing, heat and stir actions to the operation names in XML, GPT-4 can generate a complete XML script with reasonable accuracy and minimal human intervention. We demonstrated the execution of the XML script utilizing an EasyMax 102 workstation and confirmed the successful product formation using online HPLC and NMR. Overall, this approach streamlines the fundamental research necessity in reproducing literature methods with accessible hardware and services. While showcasing the possibility of using LLMs, there are still challenges in applying LLMs to more complex procedures or SDLs. To deal with less common or complicated reagents, Retrieval Augmented Generation (RAG)48 approach can be utilized, with an LLM such as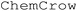 ,29 which can aid in increasing chemical accuracy and correctness. Finally, future work can also focus on developing a fully autonomous literature to product pipeline across various disciplines and SDLs configurations.
,29 which can aid in increasing chemical accuracy and correctness. Finally, future work can also focus on developing a fully autonomous literature to product pipeline across various disciplines and SDLs configurations.
Data availability
The prompts, generated XML files and Python codes can be found at: https://gitlab.com/heingroup/gpt-xml-translation.Author contributions
Conceptualization, J. E. H. and S. M.; prompt design, J. E. H., S. M., J. M. H. (before December 15th 2023) and W. Z.; prompt tuning, all authors; experimentation, M. A. G., J. Y. and J. L.; coding, W. Z. and L. H.; writing – original draft, W. Z.; writing – review & editing, all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge Rama El-khawaldeh, Paloma Prieto and Dr Joshua Derasp for their insightful guidance and conversation during the preparation of this manuscript. The authors acknowledge Mettler-Toledo AutoChem for their generous donation of process analytical equipment (EasyMax 102) and extend special thanks to Bridey Flynn for her invaluable software support. Financial support for this work was provided by Pfizer Global Research, The University of British Columbia, the Canada Foundation for Innovation (CFI-35883), the Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2021-03168, Discovery Accelerator Supplement), and the Canada First Research Excellence Fund (CFREF2022-00042).References
- M. Christensen, L. P. E. Yunker, F. Adedeji, F. Häse, L. M. Roch, T. Gensch, G. Dos Passos Gomes, T. Zepel, M. S. Sigman, A. Aspuru-Guzik and J. E. Hein, Data-science driven autonomous process optimization, Commun. Chem., 2021, 4, 112 CrossRef PubMed.
- M. Christensen, Y. Xu, E. E. Kwan, M. J. Di Maso, Y. Ji, M. Reibarkh, A. C. Sun, A. Liaw, P. S. Fier, S. Grosser and J. E. Hein, Dynamic sampling in autonomous process optimization, Chem. Sci., 2024, 15, 7160–7169 RSC.
- J. Liu, Y. Sato, F. Yang, A. J. Kukor and J. E. Hein, An Adaptive Auto-Synthesizer using Online PAT Feedback to Flexibly Perform a Multistep Reaction, Chem.: Methods, 2022, 2, e202200009 CAS.
- B. P. MacLeod, F. G. L. Parlane, A. K. Brown, J. E. Hein and C. P. Berlinguette, Flexible automation accelerates materials discovery, Nat. Mater., 2022, 21, 722–726 CrossRef CAS PubMed.
- C. C. Rupnow, B. P. MacLeod, M. Mokhtari, K. Ocean, K. E. Dettelbach, D. Lin, F. G. L. Parlane, H. N. Chiu, M. B. Rooney, C. E. B. Waizenegger, E. I. De Hoog, A. Soni and C. P. Berlinguette, A self-driving laboratory optimizes a scalable process for making functional coatings, Cell Rep. Phys. Sci., 2023, 4, 101411 CrossRef CAS.
- N. J. Szymanski, B. Rendy, Y. Fei, R. E. Kumar, T. He, D. Milsted, M. J. McDermott, M. Gallant, E. D. Cubuk, A. Merchant, H. Kim, A. Jain, C. J. Bartel, K. Persson, Y. Zeng and G. Ceder, An autonomous laboratory for the accelerated synthesis of novel materials, Nature, 2023, 624, 86–91 CrossRef CAS PubMed.
- A. Slattery, Z. Wen, P. Tenblad, J. Sanjosé-Orduna, D. Pintossi, T. Den Hartog and T. Noël, Automated self-optimization, intensification, and scale-up of photocatalysis in flow, Science, 2024, 383, eadj1817 CrossRef CAS PubMed.
- C. P. Haas, S. Biesenroth, S. Buckenmaier, T. Van De Goor and U. Tallarek, Automated generation of photochemical reaction data by transient flow experiments coupled with online HPLC analysis, React. Chem. Eng., 2020, 5, 912–920 RSC.
- K. Laws, M. Tze-Kiat Ng, A. Sharma, Y. Jiang, A. J. S. Hammer and L. Cronin, An Autonomous Electrochemical Discovery Robot that Utilises Probabilistic Algorithms: Probing the Redox Behaviour of Inorganic Materials, Chemelectrochem, 2024, 11, e202300532 CrossRef CAS.
- I. Oh, M. A. Pence, N. G. Lukhanin, O. Rodríguez, C. M. Schroeder and J. Rodríguez-López, The Electrolab: An open-source, modular platform for automated characterization of redox-active electrolytes, Device, 2023, 1, 100103 CrossRef.
- K. Darvish, M. Skreta, Y. Zhao, N. Yoshikawa, S. Som, M. Bogdanovic, Y. Cao, H. Hao, H. Xu, A. Aspuru-Guzik, A. Garg and F. Shkurti, ORGANA: A Robotic Assistant for Automated Chemistry Experimentation and Characterization, arXiv, 2024, preprint, arXiv:2401.06949, DOI:10.48550/arXiv.2401.06949.
- M. Abolhasani and E. Kumacheva, The rise of self-driving labs in chemical and materials sciences, Nat. Synth., 2023, 2, 483–492 CrossRef.
- R. El-khawaldeh, M. Guy, F. Bork, N. Taherimakhsousi, K. N. Jones, J. M. Hawkins, L. Han, R. P. Pritchard, B. A. Cole, S. Monfette and J. E. Hein, Keeping an “eye” on the experiment: computer vision for real-time monitoring and control, Chem. Sci., 2024, 15, 1271–1282 RSC.
- B. P. MacLeod, F. G. L. Parlane and C. P. Berlinguette, How to build an effective self-driving laboratory, MRS Bull., 2023, 48, 173–178 CrossRef.
- M. Christensen, L. P. E. Yunker, P. Shiri, T. Zepel, P. L. Prieto, S. Grunert, F. Bork and J. E. Hein, Automation isn't automatic, Chem. Sci., 2021, 12, 15473–15490 RSC.
- H. G. Martin, T. Radivojevic, J. Zucker, K. Bouchard, J. Sustarich, S. Peisert, D. Arnold, N. Hillson, G. Babnigg, J. M. Marti, C. J. Mungall, G. T. Beckham, L. Waldburger, J. Carothers, S. Sundaram, D. Agarwal, B. A. Simmons, T. Backman, D. Banerjee, D. Tanjore, L. Ramakrishnan and A. Singh, Perspectives for self-driving labs in synthetic biology, Curr. Opin. Biotechnol., 2023, 79, 102881 CrossRef CAS PubMed.
- S. H. M. Mehr, M. Craven, A. I. Leonov, G. Keenan and L. Cronin, A universal system for digitization and automatic execution of the chemical synthesis literature, Science, 2020, 370, 101–108 CrossRef CAS PubMed.
- S. Steiner, J. Wolf, S. Glatzel, A. Andreou, J. M. Granda, G. Keenan, T. Hinkley, G. Aragon-Camarasa, P. J. Kitson, D. Angelone and L. Cronin, Organic synthesis in a modular robotic system driven by a chemical programming language, Science, 2019, 363, eaav2211 CrossRef CAS PubMed.
- ChemIDE, https://croningroup.gitlab.io/chemputer/xdlapp/ Search PubMed.
- F. Strieth-Kalthoff, H. Hao, V. Rathore, J. Derasp, T. Gaudin, N. H. Angello, M. Seifrid, E. Trushina, M. Guy, J. Liu, X. Tang, M. Mamada, W. Wang, T. Tsagaantsooj, C. Lavigne, R. Pollice, T. C. Wu, K. Hotta, L. Bodo, S. Li, M. Haddadnia, A. Wołos, R. Roszak, C. T. Ser, C. Bozal-Ginesta, R. J. Hickman, J. Vestfrid, A. Aguilar-Granda, E. L. Klimareva, R. C. Sigerson, W. Hou, D. Gahler, S. Lach, A. Warzybok, O. Borodin, S. Rohrbach, B. Sanchez-Lengeling, C. Adachi, B. A. Grzybowski, L. Cronin, J. E. Hein, M. D. Burke and A. Aspuru-Guzik, Delocalized, asynchronous, closed-loop discovery of organic laser emitters, Science, 2024, 384, eadk9227 CrossRef CAS PubMed.
- R. Rauschen, M. Guy, J. E. Hein and L. Cronin, Universal chemical programming language for robotic synthesis repeatability, Nat. Synth., 2024, 3, 488–496 CrossRef.
- IBM RXN for Chemistry, https://rxn.res.ibm.com/ Search PubMed.
- A. C. Vaucher, F. Zipoli, J. Geluykens, V. H. Nair, P. Schwaller and T. Laino, Automated extraction of chemical synthesis actions from experimental procedures, Nat. Commun., 2020, 11, 3601 CrossRef PubMed.
- OpenAI, https://openai.com/ Search PubMed.
- T. Guo, K. Guo, B. Nan, Z. Liang, Z. Guo, N. V. Chawla, O. Wiest and X. Zhang, What can Large Language Models do in chemistry? A comprehensive benchmark on eight tasks, arXiv, 2023, preprint, arXiv:2305.18365, DOI:10.48550/arXiv.2305.18365.
- K. M. Jablonka, Q. Ai, A. Al-Feghali, S. Badhwar, J. D. Bocarsly, A. M. Bran, S. Bringuier, L. C. Brinson, K. Choudhary, D. Circi, S. Cox, W. A. De Jong, M. L. Evans, N. Gastellu, J. Genzling, M. V. Gil, A. K. Gupta, Z. Hong, A. Imran, S. Kruschwitz, A. Labarre, J. Lála, T. Liu, S. Ma, S. Majumdar, G. W. Merz, N. Moitessier, E. Moubarak, B. Mouriño, B. Pelkie, M. Pieler, M. C. Ramos, B. Ranković, S. G. Rodriques, J. N. Sanders, P. Schwaller, M. Schwarting, J. Shi, B. Smit, B. E. Smith, J. Van Herck, C. Völker, L. Ward, S. Warren, B. Weiser, S. Zhang, X. Zhang, G. A. Zia, A. Scourtas, K. J. Schmidt, I. Foster, A. D. White and B. Blaiszik, 14 examples of how LLMs can transform materials science and chemistry: a reflection on a large language model hackathon, Digit. Discov., 2023, 2, 1233–1250 RSC.
- K. Hatakeyama-Sato, N. Yamane, Y. Igarashi, Y. Nabae and T. Hayakawa, Prompt engineering of GPT-4 for chemical research: what can/cannot be done?, Sci. Technol. Adv. Mater., 2023, 3, 2260300 Search PubMed.
- D. Zhang, W. Liu, Q. Tan, J. Chen, H. Yan, Y. Yan, J. Li, W. Huang, X. Yue, W. Ouyang, D. Zhou, S. Zhang, M. Su, S. Zhong and Y. Li, A Chemical Large Language Model, arXiv, 2024, preprint, arXiv:2402.06852, DOI:10.48550/arXiv.2402.06852.
- A. M. Bran, S. Cox, O. Schilter, C. Baldassari, A. D. White and P. Schwaller, Augmenting large language models with chemistry tools, Nat. Mach. Intell., 2024, 6, 525–535 CrossRef PubMed.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, ChatGPT Chemistry Assistant for Text Mining and the Prediction of MOF Synthesis, J. Am. Chem. Soc., 2023, 145, 18048–18062 CrossRef CAS PubMed.
- Z. Zeng, Y.-C. Nie, N. Ding, Q.-J. Ding, W.-T. Ye, C. Yang, M. Sun, W. E, R. Zhu and Z. Liu, Transcription between human-readable synthetic descriptions and machine-executable instructions: an application of the latest pre-training technology, Chem. Sci., 2023, 14, 9360–9373 RSC.
- W. Zhang, Q. Wang, X. Kong, J. Xiong, S. Ni, D. Cao, B. Niu, M. Chen, Y. Li, R. Zhang, Y. Wang, L. Zhang, X. Li, Z. Xiong, Q. Shi, Z. Huang, Z. Fu and M. Zheng, Fine-tuning large language models for chemical text mining, Chem. Sci., 2024, 15, 10600–10611 RSC.
- N. Yoshikawa, M. Skreta, K. Darvish, S. Arellano-Rubach, Z. Ji, L. Bjørn Kristensen, A. Z. Li, Y. Zhao, H. Xu, A. Kuramshin, A. Aspuru-Guzik, F. Shkurti and A. Garg, Large language models for chemistry robotics, Auton. Robots, 2023, 47, 1057–1086 CrossRef.
- D. A. Boiko, R. MacKnight, B. Kline and G. Gomes, Autonomous chemical research with large language models, Nature, 2023, 624, 570–578 CrossRef CAS PubMed.
- Code Interpreter, https://platform.openai.com/docs/assistants/tools/code-interpreter Search PubMed.
- Y. Sato, J. Liu, A. J. Kukor, J. C. Culhane, J. L. Tucker, D. J. Kucera, B. M. Cochran and J. E. Hein, Real-Time Monitoring of Solid–Liquid Slurries: Optimized Synthesis of Tetrabenazine, J. Org. Chem., 2021, 86, 14069–14078 CrossRef CAS PubMed.
- I. W. Ashworth, L. Frodsham, P. Moore and T. O. Ronson, Evidence of Rate Limiting Proton Transfer in an S N Ar Aminolysis in Acetonitrile under Synthetically Relevant Conditions, J. Org. Chem., 2022, 87, 2111–2119 CrossRef CAS PubMed.
- J. Poh, D. N. Tran, C. Battilocchio, J. M. Hawkins and S. V. Ley, A Versatile Room-Temperature Route to Di- and Trisubstituted Allenes Using Flow-Generated Diazo Compounds, Angew. Chem., Int. Ed., 2015, 54, 7920–7923 CrossRef CAS PubMed.
- T. Maschmeyer, L. P. E. Yunker and J. E. Hein, Quantitative and convenient real-time reaction monitoring using stopped-flow benchtop NMR, React. Chem. Eng., 2022, 7, 1061–1072 RSC.
- P. S. Fier and S. Kim, Transition-Metal-Free C–N Cross-Coupling Enabled by a Multifunctional Reagent, J. Am. Chem. Soc., 2024, 146, 6476–6480 CrossRef CAS PubMed.
- Tokenizer, https://platform.openai.com/tokenizer Search PubMed.
- GPT4 not browsing the web or is very reluctant to do so, https://community.openai.com/t/gpt4-not-browsing-the-web-or-is-very-reluctant-to-do-so/688884 Search PubMed.
- S. Frieder, L. Pinchetti, A. Chevalier, R.-R. Griffiths, T. Salvatori, T. Lukasiewicz, P. C. Petersen and J. Berner, Mathematical Capabilities of ChatGPT, arXiv, 2023, preprint, arXiv:2301.13867, DOI:10.48550/arXiv.2301.13867.
- A. Gandolfi, GPT-4 in Education: Evaluating Aptness, Reliability, and Loss of Coherence in Solving Calculus Problems and Grading Submissions, Int. J. Artif. Intell. Educ., 2024 DOI:10.1007/s40593-024-00403-3.
- K. Lin, C. Agia, T. Migimatsu, M. Pavone and J. Bohg, Text2Motion: from natural language instructions to feasible plans, Auton. Robots, 2023, 47, 1345–1365 CrossRef.
- N. Depner, Master thesis, The Univeristy of British Columbia, 2024.
- Self-driving Solubility, https://gitlab.com/heingroup/self-driving-solubility Search PubMed.
- P. Lewis, E. Perez, A. Piktus, F. Petroni, V. Karpukhin, N. Goyal, H. Küttler, M. Lewis, W. Yih, T. Rocktäschel, S. Riedel and D. Kiela, Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks, arXiv, 2021, preprint, arXiv:2005.11401, DOI:10.48550/arXiv.2005.11401.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00248b |
| This journal is © The Royal Society of Chemistry 2024 |