Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Toward safer and more sustainable by design biocatalytic amide-bond coupling†
Elisabeth
Söderberg
 ab,
Kerstin
von Borries
ab,
Kerstin
von Borries
 c,
Ulf
Norinder
c,
Ulf
Norinder
 d,
Mark
Petchey
d,
Mark
Petchey
 e,
Ganapathy
Ranjani
e,
Ganapathy
Ranjani
 ab,
Swapnil
Chavan
ab,
Swapnil
Chavan
 f,
Hanna
Holmquist
f,
Hanna
Holmquist
 g,
Magnus
Johansson
g,
Magnus
Johansson
 h,
Ian
Cotgreave
f,
Martin A.
Hayes
h,
Ian
Cotgreave
f,
Martin A.
Hayes
 e,
Peter
Fantke
e,
Peter
Fantke
 i and
Per-Olof
Syrén
i and
Per-Olof
Syrén
 *ab
*ab
aSchool of Chemistry, Biotechnology and Health, Science for Life Laboratory, KTH Royal Institute of Technology, Tomtebodavägen 23 171 65, Solna, Sweden. E-mail: per-olof.syren@biotech.kth.se
bSchool of Engineering Sciences in Chemistry, Biotechnology and Health, Department of Fibre and Polymer Technology, KTH Royal Institute of Technology, 114 28 Stockholm, Sweden
cQuantitative Sustainability Assessment, Department of Environmental and Resource Engineering, Technical University of Denmark, Bygningstorvet 115, 2800 Kgs., Lyngby, Denmark
dDepartment of Computer and Systems Sciences, Stockholm University, 164 25 Kista, Sweden
eDiscovery Sciences, BioPharmaR&D, AstraZeneca, 431 83 Mölndal, Sweden
fThe Research Institute of Sweden RISE, Chemical and pharmaceutical toxicology, 151 36 Södertälje, Sweden
gIVL Swedish Environmental Research Institute, Aschebergsgatan 44, 411 33 Göteborg, Sweden
hEarly CVRM Medicinal Chemistry, BioPharmaR&D, AstraZeneca, 431 83 Mölndal, Sweden
iSubstitute ApS, Graaspurvevej 55 2400 Copenhagen, Denmark
First published on 7th September 2024
Abstract
Amide bond synthesis is ranked as the second most important challenge in key green chemistry research areas identified by the ACS Green Chemistry Institute. While developing more sustainable amide bond forming reactions has been in focus, significantly less attention has been given to human toxicity and environmental aspects of the underlying amine and acid substrates and their corresponding coupled products, a potentially important contribution to the overall sustainability of the amide-bond-forming reactions. Here, we explore biocatalytic amide bond formation from a safer-and-more-sustainable-by-design perspective in which commercially available amines and acids as well as their corresponding amide products were evaluated in silico based on potential human toxicity and environmental fate and exposure. This in silico filtering resulted in a panel of 188 amine and 54 acid building blocks that could be classified as safe, referred to herein as “safechems”. To enable couplings of safechems, we generated a panel of robust and promiscuous ancestral ATP-dependent amide bond synthetases (ABS) using McbA from Marinactinospora thermotolerans SCSIO 00652 as a template. Ancestral ABS enzymes exhibited complementary specificities in the coupling of a representative safechem subset of 17 amines and 16 acids while showing an increased thermostability of up to 20 °C compared to the extant biocatalyst. Finally, the pool of safechems and their corresponding amides were evaluated by USEtox (the UNEP-SETAC toxicity model), analysing not only the intrinsic properties of the compounds but evaluating their complete impact pathway including fate, exposure and effects. The amides were in general predicted as more toxic compared to the starting acids and amines through non-additive effects, emphasising that focusing on the toxicity of the building blocks alone is not sufficient to strive towards low human and ecotoxicity impact. Pursuing a safer and more sustainable by design perspective in the implementation of safechems did not prevent us from generating an array of novel products with potentially potent applications as exemplified here by enzymatic synthesis of substructures that are part of drug candidates for e.g. cancer treatment.
Introduction
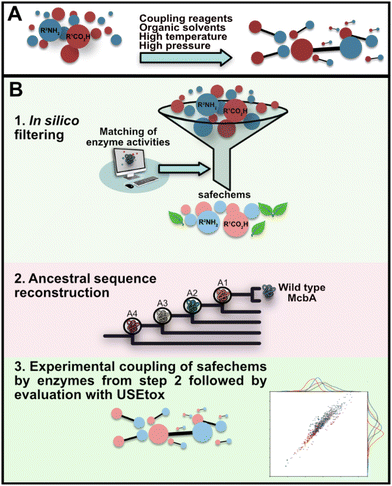
The generation of platform chemicals, agrochemicals, building blocks, and pharmaceuticals of uttermost importance for our everyday life can suffer from hazardous reaction conditions, toxicity of intermediates and products, and poor stoichiometries that do not fully align with the 12 principles of green chemistry.1–3 For an average chemical reaction, only a third of the starting raw material is converted into the desired product,1 and it is not uncommon that 25–100 fold more waste than the product is generated. In 2022, the European Commission launched the ‘Safe and Sustainable by Design’ (SSbD) framework to establish a unified set of criteria for chemicals and materials to fulfil the European Union's Chemical Strategy for Sustainability (CSS). The SSbD framework focuses on safeguarding environmental and human health and covers the entire lifecycle of the chemical or material, including production, application, and disposal.4 Yet, at present toxicity and environmental impacts of substrates used and those of the corresponding end- and by-products are rarely considered in green chemistry,1 especially in synthetic biology. We wondered whether a safer-by-design biocatalytic concept for amide bond synthesis could be applied, while at the same time allowing us to navigate within a diverse product chemical space (Fig. 1).The forefront of green chemistry consists of incorporating biocatalysis under mild reaction conditions in chemical manufacturing.5,6 With biocatalytic reactions, the use of organic solvents, coupling agents, and metals can be diminished, or even abolished.7,8 A prevalent example is amide bond synthesis: an important chemical transformation9,10 as amide bonds are common motifs in chemical products, polymers, and primary and secondary metabolites. Chemical amide bond synthesis relies on harsh reaction conditions and toxic reagents.11–14 Stoichiometric amounts of coupling reagents are often necessary which generates a considerable amount of waste.9,11 The reactions are often performed in organic solvents, and in some cases protecting groups are needed, requiring multiple steps to prevent reactive functional groups from undergoing changes during the amide bond formation process. Accordingly, amide bond synthesis is ranked as the second most important challenge in key green chemistry research areas identified by the ACS Green Chemistry Institute.12 Focus has been on developing more environmentally benign catalysts, and whereas biocatalytic amide-bond formation has received attention,11,15,16 toxicity, environmental fate and exposure of the building blocks and the products are rarely considered.
Herein, by applying hazard assessment with in silico screening and application of USEtox (the UNEP-SETAC toxicity model) in concert with enzyme engineering we explore a safer-by-design concept on a biocatalytic system. We aim for safer and more sustainable by design enzymatic amide-bond couplings in which large quantities of coupling reagents and volumes of organic solvents relative to the amide yield could be circumvented by biocatalytic transformation in aqueous media and in which toxicological effects of substrates and products are considered and optimised already at the design stage of process development (Fig. 1).
Results
Aromatic amines and acids were selected as model systems, as amides with aromatic substituents are frequently found in chemical building blocks in materials science and the chemical industry.17 We reasoned that initial filtering of all possible starting reagents from a hazard perspective would generate a reduced possible pool of safer starting materials, whilst still allowing for exploration of diverse chemical space. Lists of commercially available aromatic amines and carboxylic acids including SMILES strings were downloaded from commercial online catalogues. Starting from circa 105 unique acid and amine structures, we performed in silico hazard assessment, covering human toxicity (endocrine disruption, carcinogenic, mutagenic, and reproductive toxicity, max score being 26)18–21 and environmental fate and exposure (persistence, biodegradation, and bioconcentrations in fish, max score being 3).22,23 We've named the scores related to human toxicity “toxicity scores”, and the scores related to environmental fate and exposure “environmental scores”. When we discuss both the scores or the sum of them, they are referred to as “in silico hazard scores”. The analysis using established models which include predictions of potential interactions with various receptors resulted in predicted in silico hazard scores ranging from 0 to 29, a lower score implies a lesser cause of concern. Initially, we filtered the lists to only keep the aromatic amines and acids that had a in silico hazard score equal to or lower than six, resulting in an intermediary set of 3365 acids and 2482 amines (see supplementary dataset 1 available in raw data repository on Zenodo), corresponding to roughly half of all the aromatic compounds in the lists. The next step consisted of comparing the structures of amines and acids with reactivities of known enzymes from database searches to identify suitable biocatalysts.24Analysis of ATP-dependent amide bond synthetases identifies McbA as a suitable starting biocatalyst
Lipases and esterases are well-established enzymatic systems for acyl transfers but have a prohibitively low activity towards amides. Furthermore, their active site topologies and polarities are sometimes not compatible with the target molecule. For these reasons, we turned our attention towards amide-bond synthesising enzymes which are receiving increasing attention in green chemistry applications (Fig. 2A).25–29 Most of the amide-bond synthesising enzymes are ATP-dependent, where ATP is utilised to activate the carboxylic acid (or ester) substrate, forming either an acyl-adenylate or an acyl-phosphonate intermediate. ATP-dependent amide bond synthetases (ABS), a subgroup of the ANL-family (comprising acyl/aryl-CoA ligases, non-ribosomal peptide synthase adenylation domains, and luciferases), constitute an interesting group of acyl-adenylate forming enzymes. The enzymes belonging to these families consist of two domains, a larger N-terminal domain and a smaller C-terminal domain, and the active site is located between these two. After the entry of the acyl donor and the adenylation step, the two domains rotate relative to each other, creating a tunnel for the incoming nucleophile.30,31 For ANL enzymes, the incoming nucleophile can be e.g., coenzyme A or a phosphopantetheine, creating a thioester bond with the acyl substrate. Sequentially, the thioester intermediate is transferred to a condensation domain or an external enzyme to transform it into the final amide product. For ABS enzymes, the incoming nucleophile is an amine, leading directly to the formation of amide products without the thioester intermediate step, or the need for an external condensation enzyme (Fig. 2B). The ability of the ABS enzymes to catalyse the formation of acyl-adenylate intermediates and amide bonds within the same active site is of interest, as super-stoichiometric quantities of amine, substrate channelling and auxiliary proteins can be avoided. To search for suitable ABS templates, a literature study was performed together with a bioinformatics approach in which a phylogenetic tree of characterised ABS enzymes was constructed, with the responding product spectrum shown at the leaves of the tree (Fig. 2A). Ideally, a biocatalyst suitable for diverse coupling chemistries herein should display promiscuity in accepting various acids and amines at near stochiometric ratios. From the analysis shown in Fig. 2A, it is evident that the ABS enzyme McbA fulfils these criteria. McbA was discovered as the responsible amide bond synthesising enzyme in the biosynthesis of marinacarbolines in Marinactinospora thermotolerans SCSIO 00652, a strain found in marine sediment from the northern China Sea.32–34 McbA synthesises the amide bond between 1-acetyl-3-carboxy-β-carboline and β-phenethylamine, or tryptamine (Fig. 2A, panel A1). Extensive experimental coupling with different acids and amines has been done with this enzyme.25,33,35 In coupling reactions with 1-acetyl-3-carboxy-β-carboline, McbA has shown broad acceptance towards primary amines, aliphatic amines of varying carbon chain lengths, derivatives of tryptamine, β-phenethylamine, and benzylamines.33,35 Aniline, amino acids, activated anilines, and other aromatic amine nucleophiles had lower coupling rates to 1-acetyl-9H-β-carboline-3-carboxylic acid (Fig. 2A, panel A2–A4).33,35 Carboxylic acid substrates such as 1-acetyl-3-carboxy-β-carboline derivatives, indole carboxylic acids, and benzoic acid were successfully coupled to β-phenethylamine by McbA (Fig. 2A, panel A5).25 Altogether, the broad substrate tolerance of McbA towards both aromatic acids and amines and the need for only 1.5 equivalents of amine over acid make this enzyme a solid base in exploring a safe-by-design concept herein.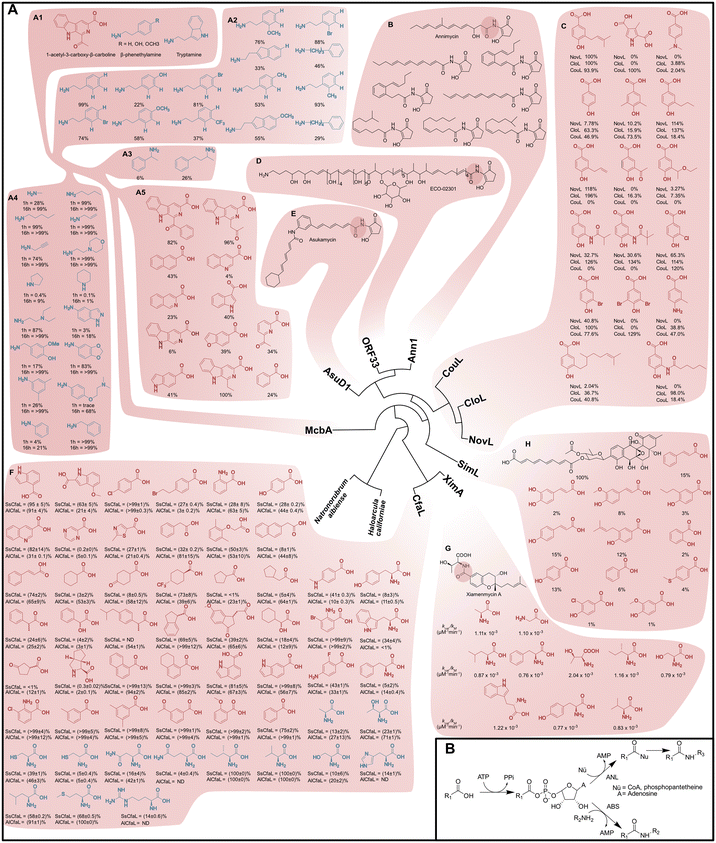 | ||
| Fig. 2 Phylogenetic tree of ABS and ABS-reminiscent enzymes and their substrate scope (amines in blue, acids in red, all measured by HPLC and from literature data). (A) (A1) Genuine substrates of McbA.47 (A2) Yields after 1 h at 37 °C; 0.2 mM 1-acetyl-3-carboxy-β-carboline, 1 mM ATP, 0.3 mM amine, 1 μM McbA.47 (A3) Yields after 24 h at 37 °C; 0.4 mM 1-acetyl-3-carboxy-β-carboline, 0.6 mM amine, 2 mM ATP, 1 mg mL−1 McbA.19 (A4) Yields after 1 h and 16 h at 37 °C; 0.4 mM 1-acetyl-3-carboxy-β-carboline, 2 mM ATP, 0.6 mM amine, 1 mg mL−1 McbA.20 (A5) Yields after 24 h at 37 °C; 0.4 mM acid, 0.6 mM β-phenethylamine, 2 mM ATP, 1 mg mL−1 McbA.19 (B) Ann1 enzyme's genuine product, annimycin,42 and amide products by Ann1 from Streptomyces asterosporus DSM 41452ΔMet.50 (C) Relative coupling yields of acids to aminocoumarin with enzymes NovL, CouL, and CloL after 30 min at 30 °C; 1 mM aminocoumarin, 1 mM acid, 5 mM ATP, 5 mM MnCl2, 0.5–2 μg enzyme.51,52 (D) Enzyme ORF33, amide bond synthetase of the antifungal agent ECO-02301.44,45 (E) The enzyme AsuD1 and its antimicrobial agent product asukamycin.43 (F) Screening results of carboxylic acids and amino acids accepted by CfaL enzymes from Streptomyces scabies and Azospirillum lipoferum. Yields after 20 h at 30 °C; 2 mM acid or 1 mM of 3-methylbenzoic acid, 5 mM L-Ile or 2 mM amino acid, 25 μM or 5 μM enzyme, 10 mM MgCl2.31 (G) Amide bond synthetase XimA and its anti-fibrotic drug candidate xiamenmycin A. The mutant XimA F201A was able to accept more amino acids, and kinetic parameters were measured at 30 °C in reactions with 5 mM MgCl2, 82.5 μg enzyme, different concentrations of ATP, amino acids, and xiamenmycin B.53,54 (H) The enzyme SimL's natural product the bacterial gyrase inhibitor Simocyclinone D8, and carboxylic acid substrate acceptance measured from reactions at 30 °C with 5 mM ATP, 5 mM MnCl2, 2 μg enzyme, 200 μM 3-amino-4,7-dihydroxy-8-methylcoumarin, 1 mM acid.55 (B) Reaction mechanisms of ANL (upper path) and ABS enzymes (lower path) are shown in the box. Catalysis occurs between the N-terminal and C-terminal domains in two partial reactions, first the adenylation of the carboxylate group to activate the substrate, and second a nucleophilic attack.35,36 | ||
In silico generation of safechems for enzymatic couplings
Our procedure to generate safechem building blocks that can be considered safer and amenable for enzymatic transformation by McbA is outlined in Fig. 3. Briefly, the intermediate pool of 2482 amines and 3365 acids resulting from the first step (Fig. 3, top, left) was filtered based on their morgan2 fingerprint structure similarity score to each known accepted substrate of McbA, herein called reference compounds (Fig. 2A, panels A1–A5).36 The compounds with top five and bottom five similarity scores for each reference compound were selected for further consideration, to extend possible substrate scope beyond substrates currently known to be efficiently converted by McbA. Any duplicates were removed, resulting in a panel of 54 acids and 188 amines. A second filtering step was subsequently performed to only keep the acids and amines that together formed in silico amides with a predicted toxicity score equal to or lower than nine and an environmental score equal to or lower than two. We heightened the threshold level for amides, as they scored as more harmful compared to acids and amines (further supported by the USEtox assessment, see below), and we wanted to have a substantial set of amines and acids to work with. The resulting panel of safechems (Fig. 3) consisted of 30 acids and 62 amines, out of which a subset of 16 of the acids and 17 of the amines were arbitrarily selected for further evaluation in experimental enzymatic couplings by McbA and engineered variants thereof.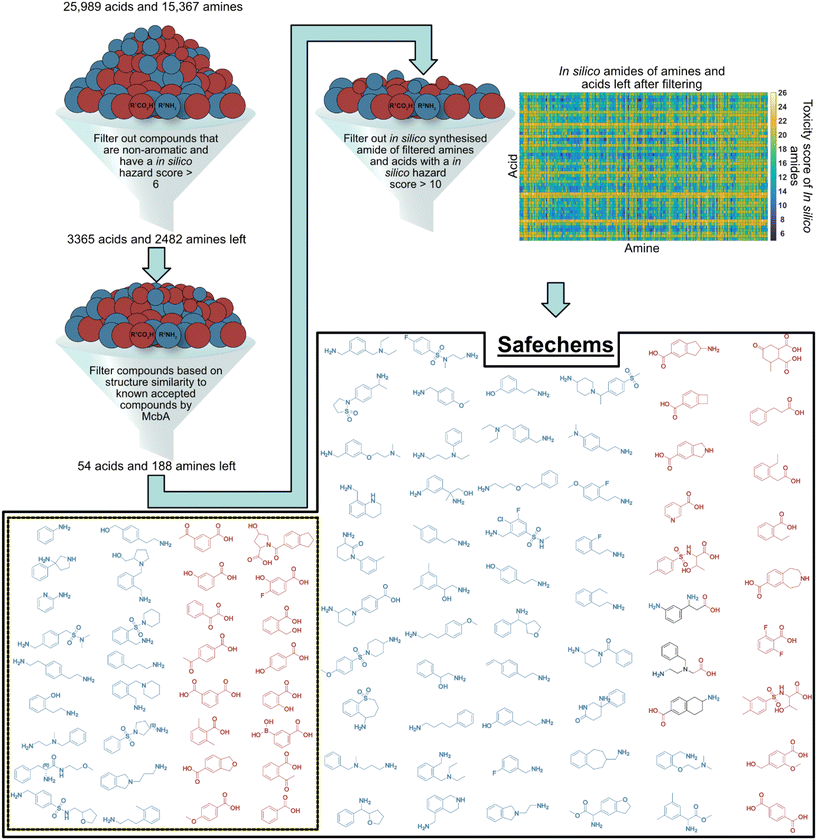 | ||
| Fig. 3 Generation of safechems amenable for couplings. The starting material was two lists with 25994 unique acids and 15374 unique amines. Each compound was given an in silico hazard score (0–29) based on published models,20 and filtered on aromaticy and in silico hazard score, which resulted in aromatic candidate substrates with an in silico hazard score equal to or lower than six. After the first filtration, the compounds were filtered based on the morgan2 fingerprint structure similarity score (0–1) to known accepted substrates (reference compounds shown in Fig. 2) by McbA. The compounds with the top five and bottom five similarity scores for each reference compound were selected to allow for expanded chemical diversity. Any duplicates were removed, and in silico amides were assembled from these remaining acids and amines. In silico amides with a toxicity score equal to or lower than nine and an environmental score equal to or lower than two were kept, resulting in a safechem library (box) composed of 30 acids and 62 amines. In the dashed square are the subset 16 safechem acids and 17 safechem amines that were arbitrarily selected to be used in the experimental coupling. | ||
Enzymatic coupling of safechems by ancestral McbAs
We reasoned that ancestral ATP-dependent amide bond synthetases (ABSs) using McbA as a template would be more robust and promiscuous than their modern counterparts, thus allowing for a wider range of couplings. Ancestral Sequence Reconstruction (ASR) is a bioinformatic technique employed to study potential molecular evolution and to generate sequences of putative ancestral proteins and enzymes.37,38 Utilising existing sequence data, a multiple sequence alignment is made from which a phylogenetic tree is constructed; allowing for computation of the ancestral sequence at the tree's nodes.38,39 It is commonly observed that ancestral enzymes exhibit improved robustness compared to their modern counterparts.40–43 Increased robustness is believed to stem from the importance of protein stability in accommodating mutations during evolution.44,45 Here, the McbA protein sequence from Marinactinospora thermotolerans SCSIO 00652 (GenBank: AGL76720.1) was used as a query when searching for related sequences in the non-redundant database using the NCBI protein blast tool. The final phylogenetic tree consisted of the query sequence, 27 sequences from bacteria, and two archaeal sequences which were used to root the tree. Four nodes in the phylogenetic tree were used to compute the ancestral sequences (A1–A4) by maximal likelihood. All ancestors (sequence identity given in Table S3, ESI†) were expressible and had an increased thermostability of up to circa 20 °C (Fig. S1, 2 and Tables S4, 5, ESI†). A full phylogenetic tree and amino acid alignments between McbA and the ancestors are given in the ESI (Fig. S3–5†). Enzymatic couplings of the subset safechem library were performed on a 20 μL scale in phosphate buffer (5% v/v DMSO) without shaking or ATP recycling in 384-well plates for 20 hours at 37 °C (Fig. 4, see Materials and methods). To characterise the performance of the enzymes, each amine and acid combination in the subset was tested for coupling, independent of whether the corresponding amide product was considered safe by our in silico hazard score. Accordingly, we performed additional toxicity assessments of these enzymatically coupled products. As discussed in detail below (see also Fig. 7 and Table S1†), this analysis verified that the predicted in silico hazard score of amides that were synthesised experimentally from the safechems shown in Fig. 4 did not deviate significantly from the in silico hazard score using the whole safechem library (shown in the box in thick lines in Fig. 3). All amide products were verified by mass spectrometry showing the expected mass (see Table S13†). Ancestors A3 and A4 exhibited no activity and were consequently excluded during the subsequent coupling process. Ancestor A1 displayed a notable reduction in activity, except for increased efficacy in coupling 2-naphthoic acid (1a) and interestingly the poor nucleophile aniline (11b). The experimental coupling results indicate that the activity of A1 is limited to acids 1a, 6a, 10a, and a few other acid and amine combinations (Fig. 4). McbA had in general higher conversion compared to both ancestors A1 and A2 at the coupling temperature used. However, a noteworthy shift in phenotypic characteristics appears to have transpired in ancestor A2 concerning amine specificity (Fig. 4). 70 expected amide masses were detected by UPLC-MS, and 38 of the amides had substantial diode array detector (DAD) peaks. 32 of the DAD detected amides have not been reported before according to a SciFinder search.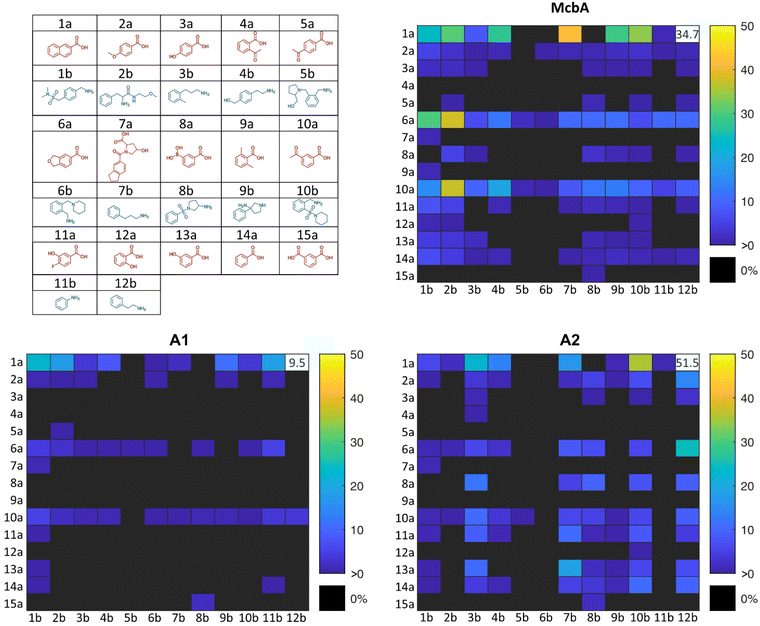 | ||
| Fig. 4 Accessible chemical space in the coupling of a subset of safechems (shown inside the dashed box of Fig. 3) by wild-type McbA and ancestors A1 and A2. Only the safechems that generated substantial DAD absorbance in two replicates are shown in this figure. Safechems not shown thus respond to reactions for which product generation could not be verified due to e.g. overlap of acid and potential amide peaks, weak UV signal, or no conversion displayed by any of the enzyme variants, respectively. Ancestors A3 and A4 were not active, hence not included. The conversions of the reactions were measured as the amide DAD peak area percentage of the total amide and acid DAD peak area. Values in the white boxes respond to the control reactions of 2-naphthoic acid (1a) and phenethylamine (12b) (see methods), and conversions shown for the control reactions respond to averages of several different measurements (see supplementary dataset 2 available in raw data repository on Zenodo. Link under Data availability). | ||
Preparative biocatalytic amide synthesis
As a proof of concept, three of the coupling reactions were scaled up to a preparative scale. As amide synthesis by McbA on a mg scale has been done previously,25,35 we first evaluated whether the new McbA variant A2 also is amenable to upscaling. Synthesis of 3-hydroxy-N-(3-phenylpropyl)benzamide by A2 was carried out in a 10 mL reaction with phosphate buffer (4% v/v DMSO), 8 mM 13a, 12 mM 7b, 16 mM ATP, 4 mU μL−1 inorganic pyrophosphatase, 4 mM MgCl2, and 0.7 mg mL−1 of A2. Inorganic pyrophosphatase was included in the reaction as inorganic pyrophosphate has been shown to inhibit McbA.35 The reaction proceeded for 60 h at 37 °C and 170 rpm, after which approximately 3.4 mM of the acid was left in the reaction as determined by HPLC-MS (Fig. 5, for the calibration curve of the acid, see Fig. S21 in the ESI†). The expected mass of the amide (256 m/z) was present in the HPLC-MS chromatogram, and the peak of the mass eluted at the same retention time as the amide standard (Fig. 5). The reaction mixture was prepared for crude NMR by extraction with ethyl acetate, rotary evaporation, and dissolving in DMSO-d6. The reaction sample as well as NMR control samples of 13a and 7b were analysed with 13C-NMR and 1H-NMR. In the 13C and 1H spectra of the crude sample from the biocatalytic reaction, there is evidence for amide bond formation (Fig. S8–10†). For full NMR and HPLC-MS chromatograms of the upscaled synthesis of 3-hydroxy-N-(3-phenylpropyl)benzamide and the amide standard, see Fig. S6–10 and S22–23.†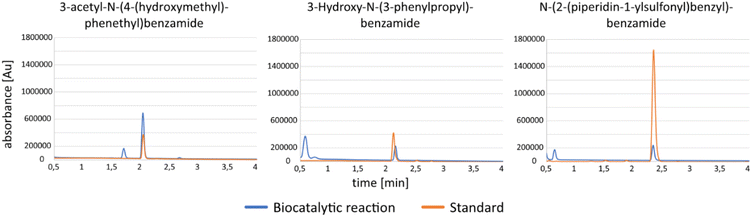 | ||
| Fig. 5 HPLC-MS of samples from preparative biocatalytic amide synthesis and amide standards. The expected masses of the amides (from left to right 298, 256, and 359m/z) were confirmed. | ||
With these findings, in the same reaction set up as the previous scale-up with A2, the synthesis of 3-acetyl-N-(4-(hydroxymethyl)phenethyl)benzamide (acid 10a and amine 4b) was performed with McbA as a catalyst and N-(2-(piperidin-1-ylsulfonyl)benzyl)benzamide (acid 14a and amine 10b) with A2 as a catalyst. The reactions were monitored by thin-layer chromatography and HPLC-MS. When there were roughly 0.7 mM 10a and 2.9 mM 14a left in their respective reactions (Fig. 5, for calibration curves of the acids, see Fig. S24 and S27†) and the masses of the amides were confirmed (298 and 359 m/z, see Fig. S25, 26 and S28, 29† for complete HPLC-MS chromatograms of samples from the biocatalytic reactions and amide standards), the amides were extracted from the reaction mixtures with DCM, and the organic layers were washed with a saturated NaHCO3 solution followed by 10% HCl acid solution, brine solution, drying over MgSO4, and concentrated under vacuum. Amide formation was confirmed by 13C-NMR and 1H-NMR (Fig. 6, Fig. S13–15 and S18–20†). The conversions were 91.2 and 54.0% and isolated yields were 58 and 42% for 3-acetyl-N-(4-(hydroxymethyl)phenethyl)benzamide and N-(2-(piperidin-1-ylsulfonyl)benzyl)benzamide.
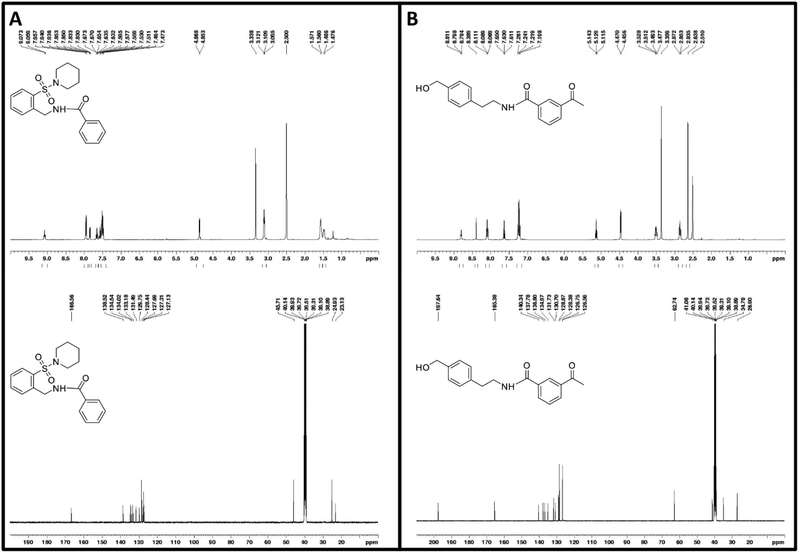 | ||
| Fig. 6 1H- and 13C-NMR spectrograms of purified products from the biocatalytic synthesis of (A) N-(2-(piperidin-1-ylsulfonyl)benzyl)benzamide and (B) 3-acetyl-N-(4-(hydroxymethyl)phenethyl)benzamide. | ||
Evaluation of safechems by USEtox
To support the in silico hazard scores and for complete impact pathway assessment, for each of the safechem building blocks (30 acids, 62 amines, and 255 amides) human toxicity and ecotoxicity impact potential expressed as characterisation factors (CFs) were retrieved using USEtox 2.12.46–48 The required input parameters were predicted using in silico methods, namely OPERA49 for basic chemical and fate-related properties, ECOSAR50 for ecotoxicity effects and CTV51 for human toxicity effects. Median CFs were derived by performing a Monte Carlo analysis with 500 draws for each chemical to account for uncertainty in the underlying prediction models. The results shown in Fig. 7 demonstrate that amides have a higher potential for toxic impacts as compared to their respective substrates, particularly for human toxicity impacts. However, we note, as in the initial screening, that the human toxicity and ecotoxicity impact of the amides is not simply the sum of their amine and acid moieties (Fig. S33†).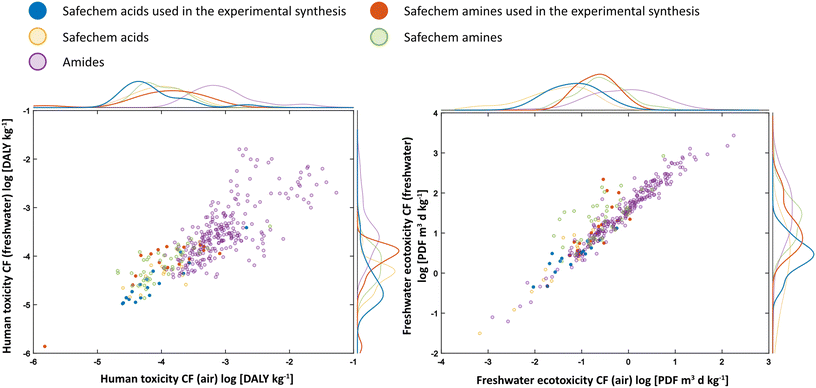 | ||
| Fig. 7 Density scatter plot of USEtox characterisation factors for toxicity potential impacts on humans and freshwater ecosystem of the safechem panel and their amide products. On the x-axis is the human/ecotoxicity CF for emission to air, and on the y-axis is the human/ecotoxicity CF for emission to freshwater. The amides were in general predicted as more harmful compared to the safechem amines and acids. The amides in this figure had an in silico hazard score equal to or lower than ten, and surpassed the filtering in Fig. 3. | ||
In addition, we compared the characterisation results of the safechems with a sample of 408 aromatic amines and 448 aromatic acids of the candidate building blocks that were previously filtered out based on their high in silico hazard scores. The results presented in Fig. 8 show that the filtering process caused a significant shift towards chemicals with lower toxicity impact potential, with median shifts of 0.88 and 0.83 log[PDF m3 d kg−1] towards lower ecotoxicity impact potential and median shifts of 0.44 and 0.39 log[DALY kg−1] towards lower human toxicity impact potential for acids and amines, respectively. While this demonstrates the usefulness of the screening approach based on intrinsic hazard and environmental parameters on persistence and bioaccumulation, it is important to note that some compounds with notable toxicity impact potentials were not detected during the initial screening. Specifically, 16 supposed safechems surpass the sample median of filtered-out candidates for human toxicity impacts, while seven do so for ecotoxicity impacts (Fig. 8). The latter set with a higher ecotoxicity CF displays higher ecotoxicity (as shown by lower ecotoxicity hazard concentrations) than other candidates, a blind spot in the in silico hazard scores that could be addressed by integrating ecotoxicity-specific flags for relevant species in future screenings. However, the 16 chemicals with higher human toxicity CF lack a direct trend linked to cancer or non-cancer effect concentrations. This implies a nuanced interaction with fate and exposure processes contributing to elevated CF for these chemicals compared to the sampled candidates. The sensitivity to the emission compartment seems more important for human toxicity compared to ecotoxicity, possibly due to more intricate fate processes (Fig. 7). This demonstrates the importance of quantitatively assessing the impact pathway even after thoroughly screening for critical hazards and influential fate properties to ensure the detection of all chemicals with substantial impact potential. For more information about input parameters and standard deviations, see Tables S6 and S7 in the ESI.†
 | ||
| Fig. 8 Density scatter plot of USEtox characterisation factors for toxicity potential impacts on humans and freshwater ecosystem of safechems and subset of the aromatic amines and acids that were filtered out (Fig. 3). On the x-axis is the human/ecotoxicity CF for emission to air, and on the y-axis is the human/ecotoxicity CF for emission to freshwater. Safechems which surpassed the sample median of filtered-out candidates for human toxicity and ecotoxicity impacts are annotated. | ||
Discussion
Chemical synthesis of amide bonds suffers from the use of large volumes of organic solvents and quantities of coupling reagents. Accordingly, significant attention has been given to catalyst and process development for enzyme- and chemical-catalysed amide bond synthesis. For instance, Freiberg and co-authors used dipyridyl dithiocarbonate as a coupling reagent in a 1-pot reaction under different neat aqueous reaction conditions with high isolated yields, with an almost one-to-one ratio of the coupling reagent and the carboxylic acid.52 Other examples include the development of the water-removable organic coupling reagent ynamide53 and the air and water-stable zirconium oxo cluster.54 The current state of the art in biocatalysis and green chemistry involves the incorporation of life cycle assessment (LCA) to evaluate environmental aspects of catalysis.55–57 In contrast, the hazards of substrates and products are less considered in green chemistry and biocatalysis. The European Commission initiative of SSbD is a step forward to create a common global foundation towards sustainable chemical manufacturing, including considerations of both chemical hazards and exposure, as well as other sustainability parameters. In our work, we have focused on identifying low-hazard building blocks for amide bond synthesis, but also counterposed the results against the complete impact pathway, as hazard-describing effects alone do not characterise toxic impact, which requires combining hazard with environmental fate and exposure. We ruled out amines and acids, as well as their potential amide products, based on their predicted hazard to human health and the environment. The predictions were made by models that have been developed from high-throughput datasets from receptors linked to molecular initiating events in combination with conformal prediction to quantify uncertainties, enabling prediction of potential interactions between chemicals and the receptors.18–23 This enabled us to identify safer building blocks that we name safechems and have lower in silico hazard scores. To complement the evaluation, we analysed the safechem panel and a subset of filtered-out aromatic amines, acids and their corresponding amides by another in silico based assessment, this time with USEtox.58 USEtox allows for the evaluation of a complete impact pathway covering fate, exposure, and effect on both humans and (freshwater) ecosystems. It quantifies impact potential relative to the chemical mass emitted, which would make it possible to also consider differences in yield or production efficiency in future analyses. The amides’ CFs were higher in both human toxicity and ecotoxicity compared to their corresponding amines and acids. This trend was not directly related to the additive toxicity impact potential of each amide's constituent acid and amine moieties nor an increase in toxicity at higher molecular weight (see Fig. S32 and S33†). This highlights the multifaceted nature of predicting the toxicity impact potential but also the potential for underestimating impacts if assessments solely rely on the profiles of the building blocks. Evaluating the toxicity of the building blocks and that of their potential product offers a more accurate representation of their potential impact. With the filtering process (Fig. 3), we were able to filter out compounds deemed to be associated with a higher impact by USEtox (Fig. 7). However, USEtox identified 23 chemicals with elevated impact potential that were not detected during the in silico hazard score evaluation process (Fig. 8). In our work towards safety and sustainability, we first used in silico filtering to reduce the risk of using or producing hazardous compounds towards humans and the environment, and secondly, evaluated the potential for harmful effects as a consequence of accumulation or distribution of the compounds in the environment and human populations.Even though biocatalysis has the benefits of operating under mild aqueous reaction conditions under ambient pressures and temperature conditions, it doesn't necessarily perform better from a life cycle perspective compared to chemical synthesis. Some of the main environmental impacts of biocatalysis are the energy and media consumption of the fermentation and downstream processes.59–62 It's therefore important to work with stable reusable enzymes. Directed evolution is one of the more acknowledged techniques to engineer enzymes. In an iterative process, evolutionary pressure is exerted on the gene of interest to create a library which is screened in search of improved variants.63 While this technique has successfully generated several interesting enzyme variants, ASR is a less laboratory-intensive alternative, an appealing feature as the development of a biocatalytic process can be a time-consuming process.62 ASR is a useful tool for generating robust ancestral variants of homologous proteins,40–42 hence we used this technique to create stable McbA ancestors. When we searched for homologous amino acid sequences of McbA, there were a few hits with high sequence similarities. This might be explained by the fact that McbA was discovered in a bacterium found in ocean sediment, which also highlights the potential of finding more interesting ABS enzymes in unexplored environments. The low sequence similarity between the sequences used to make the phylogenetic tree resulted in low sequence similarity between the wild-type McbA and the ancestors. Still, two of the four ancestors were active, and we achieved an increased thermostability by 18–31 °C, with the latter value corresponding to the second apparent Tm. The lack of activity or issues with the expression of older ancestors has been seen in previous studies.40,64,65 A plausible reason why ancestors A3 and A4 were inactive is that sequences of more ancient nodes more frequently contain incorrectly inferred sites.45,66,67
The exploratory synthesis with a subset of safechems was performed in an aqueous buffer with 5% DMSO with the ancestors and McbA. A1 exhibited very low activity, limited to a few acids. Interestingly, A1 did show a high capacity in coupling 2-naphthoic acid (1a) and the poor nucleophile aniline (12b). A2 seems to have undergone a phenotype switch, by having a broader acceptance towards amines instead of acids. McbA was able to convert amines and acids into their respective amide products with a higher conversion compared to A2. Nevertheless, A2's altered phenotype complements McbA, thus enabling us to explore a broader array of the safechems. Furthermore, A2's enhanced thermostability holds potential value from a sustainability perspective, as it can offer a longer half-life and facilitates biocatalysis at elevated temperatures, thereby boosting the reaction rate and the solubility of the substrates, thus decreasing the amount of water required.62,68,69 In this work, the synthesis was performed on a 20 μL scale, without shaking or any ATP recycling system. Cofactors for in vitro biocatalysis are economically expensive and environmentally burdening. However, a previous experiment successfully applied an ATP-recycling system in amide bond synthesis by McbA,35 and research has presented new ATP-recycling systems with inexpensive phosphate sources.70,71 The conversions of our reactions will most likely improve when performed at the catalytic temperature optimum of each enzyme variant and by implementing an ATP-recycling system and shaking.
To assess the suitability of A2 for preparative synthesis, we prepared a 10 mL reaction for the synthesis of 3-hydroxy-N-(3-phenylpropyl)benzamide, a commercially available amide that serves as a substructure in the phosphodiesterase inhibitor and cardiotonic agent 5-[1-(3,4-dimethoxybenzoyl)-3,4-dihydro-2H-quinolin-6-yl]-6-methyl-3,6-dihydro-1,3,4-thiadiazin-2-one.72,73 HPLC-MS analysis of the reaction confirmed that the product had the expected mass of the desired amide, and crude NMR spectrograms indicated amide bond formation. Encouraged by these findings, two additional preparative syntheses were performed with A2 and McbA, targeting one amide each. The products from the reactions were purified and amide bond formation was verified for both reactions by HPLC-MS, 1H-NMR and 13C-NMR analyses. These results demonstrate the applicability of the enzymes beyond microliter-scale synthesis.
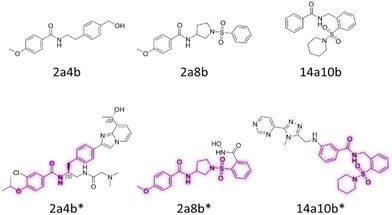
Importantly, the elimination of toxic building blocks does not necessarily restrict the identification of new compounds. Out of the 38 amides with substantial DAD peaks, 32 of them have not been recorded before at SciFinder. One of the detected documented amides, N-[2-[4-(hydroxymethyl)phenyl]ethyl]-4-methoxybenzamide (2a4b) is a substructure to a drug candidate for treatment of cellular proliferative diseases (2a4b*).74,75 Other examples of the detected documented amides are 4-methoxy-N-(1-(phenylsulfonyl)pyrrolidin-3-yl)benzamide (2a8b), an intermediate of the metalloprotease inhibitor (2a8b*),76 and N-(2-(piperidin-1-ylsulfonyl)benzyl)benzamide (14a10b), which is part of a cancer drug candidate associated with G-protein-coupled receptor kinase 2 expression (14a10b*) (Fig. 9).77
Conclusion
An important aspect of making chemical manufacturing less impactful when it comes to ecotoxicity and human toxicity effects is enabling the selection of safer substrates and aiming for safer products. By incorporating predictive models for human toxicity and environmental fate and exposure of compounds, alongside structural fingerprint similarity comparisons to known enzyme substrates, safer building blocks that are compatible with enzymatic transformations can be identified. In this study, biocatalytic amide bond formation from a safer and more sustainable by design perspective was explored. Our study shows:• The potential of combining in silico filtering with the application of USEtox to assess human toxicity and environmental fate and exposure of amines and acids and their amide products. Starting from 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 374 and 25
374 and 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 994 commercially available amines and acids, a safer chemical space was generated based on potential human toxicity and environmental fate and exposure of the substrates and related products. USEtox displayed the potential of this approach to significantly shift safechem building blocks towards chemicals with lower toxicity impact potential but also highlighted chemicals with notable toxicity impact potential that were not detected during the in silico filtering.
994 commercially available amines and acids, a safer chemical space was generated based on potential human toxicity and environmental fate and exposure of the substrates and related products. USEtox displayed the potential of this approach to significantly shift safechem building blocks towards chemicals with lower toxicity impact potential but also highlighted chemicals with notable toxicity impact potential that were not detected during the in silico filtering.
• The suitability of implementing ancestral homologs to McbA with a thermostability increase of 18–31 °C in the coupling of diverse building blocks; represented by 16 safechem acids and 17 safechem amines.
• The potential of discovery of novel products. 38 amides out of 272 possible were detected with substantial DAD peaks, out of which 32 represent uncharacterised structures.
We believe our study illuminates the potential to discover new compounds within a safe chemical space towards a safer and more sustainable by design framework using biocatalysis. Future chemical manufacturing with less dependency on toxic substances without compromising chemical diversity could thus be reached.
Materials and methods
Details of protein expression and purification, thermostability assessment, UPLC-MS, and characterisation of amide standards can be found in the ESI.†Ancestral reconstruction of McbA
To construct a phylogenetic tree to McbA, NCBI protein blast tool was used to search for related amino acid sequences in the non-redundant database, using GenBank accession no. AGL76720.1 as a query. The closest 250 hits were selected, and any redundant sequences or species, or modified amino acid sequences were removed. The remaining 27 sequences from bacteria, two outgroup sequences from two different archaea species (WP_161991522.1 Natronorubrum aibiense and WP007190185.1 Haloarcula californiae), and the query, were aligned in MAFFT v7.49 with the L-INS-i algorithm.78,79 Gap-rich regions in the alignment were removed using trimAl 1.2rev57,80 at which around 15% of all positions were removed. Using IQtree's model finder with 1000 bootstrap replicates, the Le-Gascuel model81 with invariable site plus discrete Gamma mode (gamma distribution 4)82 was suggested to be the most suitable model of evolution for the alignment. With PAML v4,83 the ancestral sequences of four different nodes were computed with maximum likelihood statistics.In silico screening – list of compounds with structures predicted to be safer
To create a low-toxic enzymatic amide bond synthesis pipeline, the aim was to select safer building blocks that together form safer in silico amides. The starting point was two lists consisting of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 993 acids and 21
993 acids and 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 243 amines. The acids and amines in each list were characterised in terms of potential interactions with endocrine disruption (23 in silico models),18 CMR toxicity (3 in silico models),19–21 persistence, biodegradation, and bioconcentrations in fish22,23 were characterised with models built using the same methodology as reported in a study.18 To classify these scores, we call the scores related to human toxicity “toxicity scores”, the scores related to environmental fate and exposure “environmental scores”, and generally when talking about both types of scores or the sum of them they are referred to as “in silico hazard scores”. All models are classification models and use the conformal prediction framework with Random Forest as a base classifier. They all operate at an 80% confidence level, i.e. accepting 20% prediction errors and confirmed by 10-fold cross-validation. The amines and acids were given a score depending on the sum of each unwanted predicted interaction from the 29 models. Dice-similarity scores (0–1), based on RDKit generated Morgan2 fingerprints (radius set to 2 and the fingerprint length to 1024),36 compared the structural similarities to already known accepted substrates of McbA (Fig. 2A, panel A1–A5). The amines and acids were filtered by only keeping the aromatic compounds that showed an in silico score equal to or lower than 6. Then, for each reference compound, the top five and bottom five dice similarity score amines and acids were saved, to ensure the candidate safechems would have a broad structural diversity to enable characterisation of the enzymes. Any duplicates in the compiled lists of top and bottom five dice similarity scoring amines and acids were removed. The remaining amines and acids were used to generate in silico amides, and in silico scores were computed for each in silico amide with the same models as used for the acids and amines. Amines and acids that formed in silico amides with a toxicity score equal to or lower than nine and an environmental score equal to or lower than two were decided to be tested against the wild-type and ancestral enzymes.
243 amines. The acids and amines in each list were characterised in terms of potential interactions with endocrine disruption (23 in silico models),18 CMR toxicity (3 in silico models),19–21 persistence, biodegradation, and bioconcentrations in fish22,23 were characterised with models built using the same methodology as reported in a study.18 To classify these scores, we call the scores related to human toxicity “toxicity scores”, the scores related to environmental fate and exposure “environmental scores”, and generally when talking about both types of scores or the sum of them they are referred to as “in silico hazard scores”. All models are classification models and use the conformal prediction framework with Random Forest as a base classifier. They all operate at an 80% confidence level, i.e. accepting 20% prediction errors and confirmed by 10-fold cross-validation. The amines and acids were given a score depending on the sum of each unwanted predicted interaction from the 29 models. Dice-similarity scores (0–1), based on RDKit generated Morgan2 fingerprints (radius set to 2 and the fingerprint length to 1024),36 compared the structural similarities to already known accepted substrates of McbA (Fig. 2A, panel A1–A5). The amines and acids were filtered by only keeping the aromatic compounds that showed an in silico score equal to or lower than 6. Then, for each reference compound, the top five and bottom five dice similarity score amines and acids were saved, to ensure the candidate safechems would have a broad structural diversity to enable characterisation of the enzymes. Any duplicates in the compiled lists of top and bottom five dice similarity scoring amines and acids were removed. The remaining amines and acids were used to generate in silico amides, and in silico scores were computed for each in silico amide with the same models as used for the acids and amines. Amines and acids that formed in silico amides with a toxicity score equal to or lower than nine and an environmental score equal to or lower than two were decided to be tested against the wild-type and ancestral enzymes.
Experimental coupling of safechems by McbA and ancestors A1–A4 in 384 well-plates
The reactions were prepared in 384-well polypropylene v-shaped plates from Greiner. In the plates, two replicates of each enzyme were screened against one safechem amine and 16 safechem acids. Exceptions were made for amines which we did not have enough of to test against all acids and enzymes. Besides the acids and amines from the filtering, phenethylamine and 2-naphthoic acid were included in the assay as control reactions. For each acid and amine combination, one no-enzyme control was prepared. The condition for each reaction was 20 μL of 1 mg ml−1 protein (10 mg ml−1 stock), 1 mM acid (60 mM stock in DMSO), 1.5 mM amine (60 mM stock in DMSO or 50 mM potassium phosphate pH 7.5), supplemented with 5 mM ATP together with 50 mM potassium phosphate pH 7.5 (5% v/v DMSO). For the no-enzyme controls, an equal amount of storage buffer was added instead of protein. Opentrons OT-2 Robot distributed the reaction buffer to each well in the reaction plate, while Mosquito High Volume robot (SPTlabtech) aliquoted amines, acids, protein, DMSO, and storage buffer from their respective mother plate to the reaction plate. Once the plates were prepared, the plates were sealed with aluminium film and incubated at 37 °C with no shaking for 20 hours.Preparative biocatalytic amide synthesis
Preparative synthesis of 3-hydroxy-N-(3-phenylpropyl)benzamide (3-hydroxybenzoic acid (13a) and 3-phenyl-1-propyamine (7b)), N-(2-(piperidin-1-ylsulfonyl)benzyl)benzamide (benzoic acid (14a) and [2-(piperidine-1-sulfonyl)phenyl]methanamine hydrochloride (10b)) by A2 and 3-acetyl-N-(4-(hydroxymethyl)phenethyl)benzamide (3-acetylbenzoic acid (10a) and [4-(2-aminoethyl)phenyl]methanol (4b)) by McbA were set up in 10 mL reactions in 50 mM phosphate buffer, DMSO (4% v/v), with 8 mM acid, 12 mM amine, 16 mM ATP, 4 mU μL−1 inorganic pyrophosphatase from Saccharomyces cerevisiae (Sigma-Aldrich), 4 mM MgCl2, and 0.7–1 mg mL−1 of enzyme. The reactions proceeded for up to 60 h at 37 °C and 170 rpm. Confirmation of the presence of the amide by expected m/z, and the amount of acid left in the reaction, was assessed by TLC and 305 nm UV peak area and calibration curves of acid standards using an Agilent 1260 Infinity II HPLC system equipped with an Agilent InfinityLab LC/MSD. Samples for HPLC-MS were quenched with 1.5 volumes of acetonitrile, and 3 μL was injected into an ACE C8 column (50 × 3 mm, 3 μm). The methods used lasted 3 minutes, a flow rate of 1 mL min−1 at 40 °C with an acetonitrile gradient from 10% to 97%, and 0.1% trifluoroacetic acid/H2O for the 3-hydroxy-N-(3-phenylpropyl)benzamide and 3-acetyl-N-(4-(hydroxymethyl)phenethyl)benzamide reactions, while the acetonitrile gradient for N-(2-(piperidin-1-ylsulfonyl)benzyl)benzamide reaction was 30% to 80%.The reaction mixture for the synthesis of 3-hydroxy-N-(3-phenylpropyl)benzamide was prepared for crude 1H-and 13C-NMR analysis by extraction with 30 mL of ethyl acetate three times. Residual water was removed with sodium sulphate, and rotary evaporation was employed to remove the ethyl acetate before dissolving the sample in DMSO-d6. NMR spectra of the reaction and acid and amine standards were recorded with a Bruker Avance AM 400 instrument at room temperature. The proton frequency was 400.13 MHz and the carbon frequency was 100.61 MHz. The chemical shifts were reported as δ values (ppm) of the residual DMSO-d6 (2.50 and 39.52 ppm) taken as a reference. The 3-phenyl-1-propylamine and 3-hydroxybenzoic acid standards were prepared by dissolving 45 mg of the compounds in 800 μL of DMSO-d6.
Reaction mixtures for the synthesis of N-(2-(piperidin-1-ylsulfonyl)benzyl)benzamide and 3-acetyl-N-(4-(hydroxymethyl)phenethyl)benzamide were extracted with DCM, and the organic layers were washed with a saturated NaHCO3 solution followed by 10% HCl acid solution, brine solution, dried over MgSO4, and concentrated under vacuum. The samples were dissolved in DMSO-d6 and recorded by 1H-and 13C-NMR as described above.
3-Acetyl-N-(4-(hydroxymethyl)phenethyl)benzamide: Rf = 0.42 (MeOH/CHCl3 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90)), yield: 13.7 mg (63.1%), nature: colourless solid. 1H NMR, 400 MHz, DMSO-d6: 8.80 (t, J = 5.6 Hz, 1H), 8.4 (s, 1H), 8.09 (t, J = 7.9 Hz, 2H), 7.63 (t, J = 7.3 Hz, 1H), 7.23 (dd, J = 16, 8.0 Hz, 4H), 5.12 (t, J = 5.7 Hz, 1H), 4.46 (d, J = 5.7 Hz, 2H), 3.50 (dd, J = 16, 6.6 Hz, 2H), 2.85 (t, J = 7.2 Hz, 2H), 2.64 (s, 3H). 13C NMR, 100 MHz, DMSO-d6: 197.6, 165.4, 140.3, 137.8, 136.8, 135.0, 131.7, 130.7, 128.9, 128.4, 126.8, 126.6, 62.7, 41.1, 34.8, 26.9. DEPT-135, 100 MHz, DMSO-d6: 131.5, 130.4, 128.6, 128.1, 126.5, 126.3, 62.5, 40.8, 34.5, 26.7. IR: 3397, 3300, 3063, 2881, 2917, 1675, 1629, 1599, 1539, 1469, 1424, 1176, 1092, 1075 cm−1.
90)), yield: 13.7 mg (63.1%), nature: colourless solid. 1H NMR, 400 MHz, DMSO-d6: 8.80 (t, J = 5.6 Hz, 1H), 8.4 (s, 1H), 8.09 (t, J = 7.9 Hz, 2H), 7.63 (t, J = 7.3 Hz, 1H), 7.23 (dd, J = 16, 8.0 Hz, 4H), 5.12 (t, J = 5.7 Hz, 1H), 4.46 (d, J = 5.7 Hz, 2H), 3.50 (dd, J = 16, 6.6 Hz, 2H), 2.85 (t, J = 7.2 Hz, 2H), 2.64 (s, 3H). 13C NMR, 100 MHz, DMSO-d6: 197.6, 165.4, 140.3, 137.8, 136.8, 135.0, 131.7, 130.7, 128.9, 128.4, 126.8, 126.6, 62.7, 41.1, 34.8, 26.9. DEPT-135, 100 MHz, DMSO-d6: 131.5, 130.4, 128.6, 128.1, 126.5, 126.3, 62.5, 40.8, 34.5, 26.7. IR: 3397, 3300, 3063, 2881, 2917, 1675, 1629, 1599, 1539, 1469, 1424, 1176, 1092, 1075 cm−1.
N-(2-(Piperidin-1-ylsulfonyl)benzyl)benzamide: Rf = 0.44 (MeOH/CHCl3 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90)), yield: 12.1 mg (78.2%), nature: colourless solid. 1H NMR, 400 MHz, DMSO-d6: 1H NMR, 400 MHz, DMSO-d6: 9.07 (t, J = 6.3 Hz, 1H), 7.95 (d, J = 7.1 Hz, 1H), 7.94 (d, J = 8.7 Hz, 1H), 7.84 (dd, J = 8.3, 1.4 Hz, 1H), 7.67–7.63 (m, 1H), 7.58 (t, J = 7.2 Hz, 1H), 7.53–7.47 (m, 4H), 4.86 (d, J = 6.0 Hz, 2H), 3.11 (t, J = 5.3 Hz, 4H), 1.57 (br, 4H), 1.47 (br, 2H). 13C NMR, 100 MHz, DMSO-d6: 166.6, 138.5, 134.5, 134.0, 133.2, 131.5, 129.8, 128.4, 127.7, 127.3, 127.1, 45.7, 39.6, 24.9, 23.1. DEPT-135, 100 MHz, DMSO-d6: 132.9, 131.2, 129.6, 128.2, 127.4, 127.1, 126.9, 45.5, 39.6, 24.7, 22.9. IR: 3327, 3069, 2925, 2852, 1638, 1542, 1489, 1337, 1311, 1162, 1052 cm−1.
90)), yield: 12.1 mg (78.2%), nature: colourless solid. 1H NMR, 400 MHz, DMSO-d6: 1H NMR, 400 MHz, DMSO-d6: 9.07 (t, J = 6.3 Hz, 1H), 7.95 (d, J = 7.1 Hz, 1H), 7.94 (d, J = 8.7 Hz, 1H), 7.84 (dd, J = 8.3, 1.4 Hz, 1H), 7.67–7.63 (m, 1H), 7.58 (t, J = 7.2 Hz, 1H), 7.53–7.47 (m, 4H), 4.86 (d, J = 6.0 Hz, 2H), 3.11 (t, J = 5.3 Hz, 4H), 1.57 (br, 4H), 1.47 (br, 2H). 13C NMR, 100 MHz, DMSO-d6: 166.6, 138.5, 134.5, 134.0, 133.2, 131.5, 129.8, 128.4, 127.7, 127.3, 127.1, 45.7, 39.6, 24.9, 23.1. DEPT-135, 100 MHz, DMSO-d6: 132.9, 131.2, 129.6, 128.2, 127.4, 127.1, 126.9, 45.5, 39.6, 24.7, 22.9. IR: 3327, 3069, 2925, 2852, 1638, 1542, 1489, 1337, 1311, 1162, 1052 cm−1.
USEtox – characterisation factors aggregating fate, exposure and human and ecotoxicological effects
To evaluate whether the applied screening based on intrinsic chemical properties led to a potential reduction in toxic impact, the selected safechems were assessed with USEtox 2.12, considering the entire impact pathway from emissions via fate and exposure to effects on humans and ecosystems. USEtox is the scientific consensus model for characterising human toxicity and ecotoxicity impacts, developed under the auspices of the Life Cycle Initiative hosted at UN Environment.58,84 Toxicity impact potentials in USEtox are expressed as characterisation factors (CF) per unit mass emitted into different emission compartments or in different product applications. We obtained the required chemical input parameters using in silico prediction methods, namely OPEn structure–activity/property Relationship App (OPERA)49 for basic chemical and fate-related properties, Ecological Structure Activity Relationships Program (ECOSAR)50 for ecotoxicity effects and Conditional Toxicity Value (CTV)51 for human toxicity effects. We performed a Monte Carlo analysis to account for the uncertainty in the underlying prediction models by varying each predicted value as a log10normal distribution with the mean defined as the predicted value and the standard deviation derived from the model's reported log10RMSE (see Table S6† for details). As the reliability of each prediction model can vary across chemicals depending on its applicability domain (AD), the standard deviation was scaled by uncertainty factors depending on the applicability of the model to a given chemical structure (see Table S7† for details). Where human toxicity effect data could not be predicted for certain chemicals, the missing values were conservatively imputed from the 95th percentile of the given effect parameter data across the safechems with available data within each chemical class (acids, amines, amides) and varied with the threefold standard deviation to consider the increase in uncertainty. Characterisation results were derived by taking the median CF that were computed from drawing 500 random samples from the input parameter distributions for each chemical. We compared the results obtained for the safechems with a sample of 848 candidate chemicals (448 acids and 440 amines) that had previously been filtered out based on high in silico scores. The candidate sample was created with a close-to-equal representation across in silico scores ranging from 6 to 29. The USEtox input parameter data and characterisation results were obtained for the sampled candidates following the same approach as for the safechems. We focused on the impacts of emissions on continental freshwater and rural air, as these were considered most applicable for a potential industrial synthesis setting. The mass emitted was considered equal across substrates and products, respectively, and characterisation results were therefore compared directly at the level of CF.Author contributions
ES performed experimental work, contributed to in silico screening, designed enzymes and experiments. KB designed and performed USEtox analyses under the supervision of HH and PF. UN performed and designed the in silico screening parts with contributions from SC and IC. MP synthesised the amide standard, planned experiments and contributed to experimental design. GR synthesised amide standards, purified amides from the upscaled biocatalytic reactions and did the characterisation. MH, MJ, HH, PF and POS supervised and designed research. All authors have edited, read and approved the manuscript.Data availability
The data supporting this article have been included as part of the ESI. ESI† includes supplementary data and NMR spectra and additional methods description.A raw data depository containing full in silico datasets used, nanoDSF raw data and HPLC raw data is available from Zenodo: https://zenodo.org/records/12704378?token=eyJhbGciOiJIUzUxMiJ9.eyJpZCI6ImFmZTJhZWQzLTgxZWMtNDA3Ni04NjU5LWQzYmVhMzhjMDMyNyIsImRhdGEiOnt9LCJyYW5kb20iOiJjMjFjY2MyZjY4YTI0ZjQxNmVkMDNlY2U1NWUwYWIxNyJ9.8lGn9M9PI8O_cR0Lft8tbtQHFmMH7vVoTQxOkMcACmMIvtqmF7qDn4djfoPxc65GuxLzkp-onrFNaOtHpHFDvw.
The raw data depository includes:
Supplementary dataset 1: in silico screening data of aromatic amines and acids; human- and ecotoxicity scores, and dice structure similarity scores to reference compounds of McbA.
Supplementary dataset 2: UPLC-MS data of experimental coupling reactions of Safechems with McbA, A1, and A2.
Supplementary dataset 3: USEtox data of safechems.
Supplementary dataset 4: Nano-DSF data of McbA and ancestors A1–A4.
.zip archives of HPLC raw data.
Conflicts of interest
MJ, MP, and MH are employees of AstraZeneca, Sweden. The rest of the authors do not declare any conflict of interest.Acknowledgements
We greatly acknowledge funding from The Swedish Foundation for Strategic Environmental Research MISTRA, program SafeChem 2018/11. Computations were enabled by resources provided by the National Academic Infrastructure for Supercomputing in Sweden (NAISS), partially funded by the Swedish Research Council (VR) through grant agreement no. 2022-06725. We greatly acknowledge the PDC Centre for High-Performance Computing at the Royal Institute of Technology and SNIC and NAISS (projects NAISS 2023/5-395, NAISS 2023/5-232, naiss2024-5-346). This work was also supported by the Swedish Foundation of Strategic Research (FFF20-0027). This work was financially supported by the PARC project (Grant No. 101057014) funded under the European Union's Horizon Europe Research and Innovation program.References
- J. B. Zimmerman, P. T. Anastas, H. C. Erythropel and W. Leitner, Designing for a green chemistry future, Science, 2020, 367(6476), 397–400 CrossRef CAS PubMed.
- P. T. Anastas, Green chemistry : theory and practice, Oxford Univ. Press, Oxford, Oxford, 1998 Search PubMed.
- P. T. Anastas and J. B. Zimmerman, Peer Reviewed: Design Through the 12 Principles of Green Engineering, Environ. Sci. Technol., 2003, 37(5), 94A–101A CrossRef PubMed.
- C. Caldeira, R. Farcal, C. Moretti, L. Mancini, H. Rauscher and J. Riego Sintes, et al., Safe and sustainable by design chemicals and materials: review of safety and sustainability dimensions, aspects, methods, indicators, and tools, Publications Office of the European Union, Luxembourg, 2022 Search PubMed.
- R. Kumar, M. J. Karmilowicz, D. Burke, M. P. Burns, L. A. Clark and C. G. Connor, et al., Biocatalytic reductive amination from discovery to commercial manufacturing applied to abrocitinib JAK1 inhibitor, Nat. Catal., 2021, 4(9), 775–782 CrossRef CAS.
- J. I. Ramsden, S. C. Cosgrove and N. Turner, Is it time for biocatalysis in fragment-based drug discovery?, Chem. Sci., 2020, 11(41), 11104–11112 RSC.
- R. A. Sheldon and J. M. Woodley, Role of Biocatalysis in Sustainable Chemistry, Chem. Rev., 2018, 118(2), 801–838 CrossRef CAS.
- R. A. Sheldon, The E factor at 30: a passion for pollution prevention, Green Chem., 2023, 25(5), 1704–1728 RSC.
- P. Schneider and G. Schneider, Privileged Structures Revisited, Angew. Chem., Int. Ed., 2017, 56(27), 7971–7974 CrossRef CAS PubMed.
- E. Vitaku, D. T. Smith and J. T. Njardarson, Analysis of the Structural Diversity, Substitution Patterns, and Frequency of Nitrogen Heterocycles among U.S, FDA Approved Pharmaceuticals, J. Med. Chem., 2014, 57(24), 10257–10274 CrossRef CAS PubMed.
- M. T. Sabatini, B. LeeT, H. F. Sneddon and T. D. Sheppard, A green chemistry perspective on catalytic amide bond formation, Nat. Catal., 2019, 2(1), 10–17 CrossRef CAS.
- M. C. Bryan, P. J. Dunn, D. Entwistle, F. Gallou, S. G. Koenig and J. D. Hayler, et al., Key Green Chemistry research areas from a pharmaceutical manufacturers’ perspective revisited, Green Chem., 2018, 20(22), 5082–5103 RSC.
- E. Valeur and M. Bradley, Amide bond formation: beyond the myth of coupling reagents, Chem. Soc. Rev., 2009, 38(2), 606–631 RSC.
- B. S. Jursic and Z. Zdravkovski, A Simple Preparation of Amides from Acids and Amines by Heating of Their Mixture, Synth. Commun., 1993, 23(19), 2761–2770 CrossRef CAS.
- A. Goswami and S. G. V. Lanen, Enzymatic strategies and biocatalysts for amide bond formation: tricks of the trade outside of the ribosome, Mol. BioSyst., 2015, 11(2), 338–353 RSC.
- J. Pitzer and K. Steiner, Amides in Nature and Biocatalysis, J. Biotechnol., 2016, 235, 32–46 CrossRef CAS PubMed.
- R. L. Myers, The 100 Most Important Chemical Compounds: A Reference Guide, Bloomsbury Publishing USA, 2007, p. 352 Search PubMed.
- M. Sapounidou, U. Norinder and P. L. Andersson, Predicting Endocrine Disruption Using Conformal Prediction – A Prioritization Strategy to Identify Hazardous Chemicals with Confidence, Chem. Res. Toxicol., 2023, 36(1), 53–65 Search PubMed.
- N. Fjodorova, M. Vračko, M. Novič, A. Roncaglioni and E. Benfenati, New public QSAR model for carcinogenicity, Chem. Cent. J., 2010, 4(1), S3 Search PubMed.
- K. Hansen, S. Mika, T. Schroeter, A. Sutter, A. ter Laak and T. Steger-Hartmann, et al., Benchmark Data Set for in Silico Prediction of Ames Mutagenicity, J. Chem. Inf. Model., 2009, 49(9), 2077–2081 CrossRef CAS.
- C. Jiang, H. Yang, P. Di, W. Li, Y. Tang and G. Liu, In silico prediction of chemical reproductive toxicity using machine learning, J. Appl. Toxicol., 2019, 39(6), 844–854 CrossRef CAS.
- J. Tunkel, P. H. Howard, R. S. Boethling, W. Stiteler and H. Loonen, Predicting ready biodegradability in the Japanese ministry of international trade and industry test, Environ. Toxicol. Chem., 2000, 19(10), 2478–2485 CrossRef CAS.
- S. Dimitrov, N. Dimitrova, T. Parkerton, M. Comber, M. Bonnell and O. Mekenyan, Base-line model for identifying the bioaccumulation potential of chemicals, SAR QSAR Environ. Res., 2005, 16(6), 531–554 CrossRef CAS PubMed.
- W. Finnigan, L. J. Hepworth, S. L. Flitsch and N. J. Turner, RetroBioCat as a computer-aided synthesis planning tool for biocatalytic reactions and cascades, Nat. Catal., 2021, 4(2), 98–104 CrossRef CAS PubMed.
- M. R. Petchey, A. Cuetos, B. Rowlinson, S. Dannevald, A. Frese and P. W. Sutton, et al., The Broad Aryl Acid Specificity of the Amide Bond Synthetase McbA Suggests Potential for the Biocatalytic Synthesis of Amides, Angew. Chem., 2018, 130(36), 11758–11762 CrossRef.
- M. Winn, M. Rowlinson, F. Wang, L. Bering, D. Francis and C. Levy, et al., Discovery, characterization and engineering of ligases for amide synthesis, Nature, 2021, 593(7859), 391–398 CrossRef CAS.
- M. Lubberink, W. Finnigan and S. L. Flitsch, Biocatalytic amide bond formation, Green Chem., 2023, 25(8), 2958–2970 RSC.
- M. Winn, S. M. Richardson, D. J. Campopiano and J. Micklefield, Harnessing and engineering amide bond forming ligases for the synthesis of amides, Curr. Opin. Chem. Biol., 2020, 55, 77–85 CrossRef CAS PubMed.
- M. R. Petchey and G. Grogan, Enzyme–Catalysed Synthesis of Secondary and Tertiary Amides, Adv. Synth. Catal., 2019, 361(17), 3895–3914 CrossRef CAS.
- M. A. Marahiel, T. Stachelhaus and H. D. Mootz, Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis, Chem. Rev., 1997, 97(7), 2651–2674 CrossRef CAS PubMed.
- A. M. Gulick, Conformational Dynamics in the Acyl-CoA Synthetases, Adenylation Domains of Non-ribosomal Peptide Synthetases, and Firefly Luciferase, ACS Chem. Biol., 2009, 4(10), 811–827 CrossRef CAS PubMed.
- Q. Chen, C. Ji, Y. Song, H. Huang, J. Ma and X. Tian, et al., Discovery of McbB, an enzyme catalyzing the β-carboline skeleton construction in the marinacarboline biosynthetic pathway, Angew. Chem., Int. Ed., 2013, 52(38), 9980–9984 CrossRef CAS PubMed.
- C. Ji, Q. Chen, Q. Li, H. Huang, Y. Song and J. Ma, et al., Chemoenzymatic synthesis of β-carboline derivatives using McbA, a new ATP-dependent amide synthetase, Tetrahedron Lett., 2014, 55(35), 4901–4904 CrossRef CAS.
- X. P. Tian, S. K. Tang, J. D. Dong, Y. Q. Zhang, L. H. Xu and S. Zhang, et al., Marinactinospora thermotolerans gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China Sea, Int. J. Syst. Evol. Microbiol., 2009, 59(5), 948–952 CrossRef CAS PubMed.
- M. R. Petchey, B. Rowlinson, R. C. Lloyd, I. J. S. Fairlamb and G. Grogan, Biocatalytic Synthesis of Moclobemide Using the Amide Bond Synthetase McbA Coupled with an ATP Recycling System, ACS Catal., 2020, 10(8), 4659–4663 CrossRef CAS PubMed.
- D. Rogers and M. Hahn, Extended-Connectivity Fingerprints, J. Chem. Inf. Model., 2010, 50(5), 742–754 CrossRef CAS PubMed.
- D. L. Trudeau, M. Kaltenbach and D. S. Tawfik, On the Potential Origins of the High Stability of Reconstructed Ancestral Proteins, Mol. Biol. Evol., 2016, 33(10), 2633–2641 CrossRef CAS PubMed.
- J. W. Thornton, Resurrecting ancient genes: experimental analysis of extinct molecules, Nat. Rev. Genet., 2004, 5(5), 366–375 CrossRef CAS.
- R. Merkl and R. Sterner, Reconstruction of ancestral enzymes, Perspect. Sci., 2016, 9, 17–23 CrossRef.
- D. A. Hueting, S. R. Vanga and P. O. Syrén, Thermoadaptation in an Ancestral Diterpene Cyclase by Altered Loop Stability, J. Phys. Chem. B, 2022, 126(21), 3809–3821 CrossRef CAS PubMed.
- N. M. Hendrikse, G. Charpentier, E. Nordling and P. Syrén, Ancestral diterpene cyclases show increased thermostability and substrate acceptance, FEBS J., 2018, 285(24), 4660–4673 CrossRef CAS PubMed.
- G. Gamiz-Arco, L. I. Gutierrez-Rus, V. A. Risso, B. Ibarra-Molero, Y. Hoshino and D. Petrović, et al., Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase, Nat. Commun., 2021, 12(1), 380 CrossRef CAS PubMed.
- T. Devamani, A. M. Rauwerdink, M. Lunzer, B. J. Jones, J. L. Mooney and M. A. O. Tan, et al., Catalytic Promiscuity of Ancestral Esterases and Hydroxynitrile Lyases, J. Am. Chem. Soc., 2016, 138(3), 1046–1056 CrossRef CAS PubMed.
- J. Zheng, N. Guo and A. Wagner, Selection enhances protein evolvability by increasing mutational robustness and foldability, Science, 2020, 370(6521), eabb5962 CrossRef CAS PubMed.
- G. N. Eick, J. T. Bridgham, D. P. Anderson, M. J. Harms and J. W. Thornton, Robustness of Reconstructed Ancestral Protein Functions to Statistical Uncertainty, Mol. Biol. Evol., 2017, 34(2), 247–261 CAS.
- A. D. Henderson, M. Z. Hauschild, D. van de Meent, M. A. J. Huijbregts, H. F. Larsen and M. Margni, et al., USEtox fate and ecotoxicity factors for comparative assessment of toxic emissions in life cycle analysis: sensitivity to key chemical properties, Int. J. Life Cycle Assess., 2011, 16(8), 701–709 CrossRef CAS.
- R. K. Rosenbaum, M. A. J. Huijbregts, A. D. Henderson, M. Margni, T. E. McKone and D. van de Meent, et al., USEtox human exposure and toxicity factors for comparative assessment of toxic emissions in life cycle analysis: sensitivity to key chemical properties, Int. J. Life Cycle Assess., 2011, 16(8), 710–727 CrossRef CAS.
- R. K. Rosenbaum, T. M. Bachmann, L. S. Gold, M. A. J. Huijbregts, O. Jolliet and R. Juraske, et al., USEtox—the UNEP-SETAC toxicity model: recommended characterisation factors for human toxicity and freshwater ecotoxicity in life cycle impact assessment, Int. J. Life Cycle Assess., 2008, 13(7), 532–546 CrossRef CAS.
- K. Mansouri, C. M. Grulke, R. S. Judson and A. J. Williams, OPERA models for predicting physicochemical properties and environmental fate endpoints, J. Cheminf., 2018, 10(1), 10 Search PubMed.
- R. T. Wright, K. Fay, A. Kennedy, K. Mayo-Bean, K. Moran-Bruce and W. Meylanet al., METHODOLOGY DOCUMENT for the ECOlogical Structure-Activity Relationship Model (ECOSAR) Class Program. The Office of Pollution Prevention and Toxics, U.S. Environmental Protection Agency, 2022, available from: https://www.epa.gov/system/files/documents/2022-03/methodology-document-v.2.2.pdf Search PubMed.
- J. A. Wignall, E. Muratov, A. Sedykh, K. Z. Guyton, A. Tropsha and I. Rusyn, et al., Conditional Toxicity Value (CTV) Predictor: An In Silico Approach for Generating Quantitative Risk Estimates for Chemicals, Environ. Health Perspect., 2018, 126(5), 057008 CrossRef PubMed.
- K. M. Freiberg, R. D. Kavthe, R. M. Thomas, D. M. Fialho, P. Dee and M. Scurria, et al., Direct formation of amide/peptide bonds from carboxylic acids: no traditional coupling reagents, 1-pot, and green, Chem. Sci., 2023, 14(13), 3462–3469 RSC.
- T. Liu, X. Zhang, Z. Peng and J. Zhao, Water-removable ynamide coupling reagent for racemization-free syntheses of peptides, amides, and esters, Green Chem., 2021, 23(24), 9916–9921 RSC.
- Y. Zhang, F. d. Azambuja and T. N. Parac-Vogt, Zirconium oxo clusters as discrete molecular catalysts for the direct amide bond formation, Catal. Sci. Technol., 2022, 12(10), 3190–3201 RSC.
- M. A. F. Delgove, A. B. Laurent, J. M. Woodley, S. M. A. De Wildeman, K. V. Bernaerts and Y. van der Meer, A Prospective Life Cycle Assessment (LCA) of Monomer Synthesis: Comparison of Biocatalytic and Oxidative Chemistry, ChemSusChem, 2019, 12(7), 1349–1360 CrossRef CAS PubMed.
- R. Rosa, R. Spinelli, P. Neri, M. Pini, S. Barbi and M. Montorsi, et al., Life Cycle Assessment of Chemical vs Enzymatic-Assisted Extraction of Proteins from Black Soldier Fly Prepupae for the Preparation of Biomaterials for Potential Agricultural Use, ACS Sustainable Chem. Eng., 2020, 8(39), 14752–14764 CrossRef CAS.
- M. Becker, A. Ziemińska-Stolarska, D. Markowska, S. Lütz and K. Rosenthal, Comparative Life Cycle Assessment of Chemical and Biocatalytic 2′3′-Cyclic GMP-AMP Synthesis, ChemSusChem, 2023, 16(5), e202201629 CrossRef CAS PubMed.
- P. Fantke, W. A. Chiu, L. Aylward, R. Judson, L. Huang and S. Jang, et al., Exposure and toxicity characterization of chemical emissions and chemicals in products: global recommendations and implementation in USEtox, Int. J. Life Cycle Assess., 2021, 26(5), 899–915 CrossRef CAS PubMed.
- P. H. Nielsen, K. M. Oxenbøll and H. Wenzel, Cradle-to-gate environmental assessment of enzyme products produced industrially in denmark by novozymes A/S, Int. J. Life Cycle Assess., 2007, 12(6), 432–438 CrossRef CAS.
- S. Kim, C. Jiménez-González and B. E. Dale, Enzymes for pharmaceutical applications—a cradle-to-gate life cycle assessment, Int. J. Life Cycle Assess., 2009, 14(5), 392–400 CrossRef CAS.
- J. Kenthorai Raman, V. Foo Wang Ting and R. Pogaku, Life cycle assessment of biodiesel production using alkali, soluble and immobilized enzyme catalyst processes, Biomass Bioenergy, 2011, 35(10), 4221–4229 CrossRef.
- F. Gallou, H. Gröger and B. H. Lipshutz, Status check: biocatalysis; its use with and without chemocatalysis. How does the fine chemicals industry view this area?, Green Chem., 2023, 25(16), 6092–6107 RSC.
- F. H. Arnold, Directed Evolution: Bringing New Chemistry to Life, Angew. Chem., Int. Ed., 2018, 57(16), 4143–4148 CrossRef CAS PubMed.
- L. Lei, L. Zhao, Y. Hou, C. Yue, P. Liu and Y. Zheng, et al., An Inferred Ancestral CotA Laccase with Improved Expression and Kinetic Efficiency, Int. J. Mol. Sci., 2023, 24(13), 10901 CrossRef CAS PubMed.
- P. Babkova, E. Sebestova, J. Brezovsky, R. Chaloupkova and J. Damborsky, Ancestral Haloalkane Dehalogenases Show Robustness and Unique Substrate Specificity, ChemBioChem, 2017, 18(14), 1448–1456 CrossRef CAS PubMed.
- R. N. Randall, C. E. Radford, K. A. Roof, D. K. Natarajan and E. A. Gaucher, An experimental phylogeny to benchmark ancestral sequence reconstruction, Nat. Commun., 2016, 7(1), 12847 CrossRef PubMed.
- T. Matsumoto, H. Akashi and Z. Yang, Evaluation of Ancestral Sequence Reconstruction Methods to Infer Nonstationary Patterns of Nucleotide Substitution, Genetics, 2015, 200(3), 873–890 CrossRef PubMed.
- J. A. Iannuzzelli, J. P. Bacik, E. J. Moore, Z. Shen, E. M. Irving and D. A. Vargas, et al., Tuning Enzyme Thermostability via Computationally Guided Covalent Stapling and Structural Basis of Enhanced Stabilization, Biochemistry, 2022, 61(11), 1041–1054 CrossRef CAS PubMed.
- R. J. Fox, S. C. Davis, E. C. Mundorff, L. M. Newman, V. Gavrilovic and S. K. Ma, et al., Improving catalytic function by ProSAR-driven enzyme evolution, Nat. Biotechnol., 2007, 25(3), 338–344 CrossRef CAS PubMed.
- M. Tavanti, J. Hosford, R. C. Lloyd and M. J. B. Brown, ATP regeneration by a single polyphosphate kinase powers multigram-scale aldehyde synthesis in vitro, Green Chem., 2021, 23(2), 828–837 RSC.
- G. A. Strohmeier, A. Schwarz, J. N. Andexer and M. Winkler, Co-factor demand and regeneration in the enzymatic one-step reduction of carboxylates to aldehydes in cell-free systems, J. Biotechnol., 2020, 307, 202–207 CrossRef CAS PubMed.
- J. White, J. A. Lee, N. Shah and C. H. Orchard, Differential effects of the optical isomers of EMD 53998 on contraction and cytoplasmic Ca2+in isolated ferret cardiac muscle, Circ. Res., 1993, 73(1), 61–70 CrossRef CAS PubMed.
- R. J. Solaro, G. Gambassi, D. M. Warshaw, M. R. Keller, H. A. Spurgeon and N. Beier, et al., Stereoselective actions of thiadiazinones on canine cardiac myocytes and myofilaments, Circ. Res., 1993, 73(6), 981–990 CrossRef CAS PubMed.
- X. Qian, W. L. Ashcraft, W. Jianchao, Y. Bing, H. Jiang, G. Bergnes and M. P. Bradley, et al., Preparation of 3-chloro-4-isopropoxybenzamide and 3-cyano-4-isopropoxybenzamide derivatives as inhibitors of mitotic kinesins. US2007149516, 2007 Search PubMed.
- X. Qian, A. McDonald, H. J. Zhou, N. D. Adams, C. A. Parrish and K. J. Duffy, et al., Discovery of the First Potent and Selective Inhibitor of Centromere-Associated Protein E: GSK923295, ACS Med. Chem. Lett., 2010, 1(1), 30–34 CrossRef CAS PubMed.
- C. Villamil, G. Decrescenzo, M. Brent, R. Shashidhar, L. Bedell and T. Barta, et al., Sulfonyl aryl or heteroaryl hydroxamic acid compounds, 7th edn, US2003191317A1, 2003 Search PubMed.
- J. B. Hurov, A. Lantermann, H. Xu, E. L. P. Chekler, G. Piizzi and K. M. Cuoto, et al., GRK2 Inhibitors and uses thereof. US20230009608A1, 2023 Search PubMed.
- K. Katoh, K. Misawa, K. i. Kuma and T. Miyata, MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform, Nucleic Acids Res., 2002, 30(14), 3059–3066 CrossRef CAS PubMed.
- K. Katoh and D. M. Standley, MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability, Mol. Biol. Evol., 2013, 30(4), 772–780 CrossRef CAS PubMed.
- S. Capella-Gutiérrez, J. M. Silla-Martínez and T. Gabaldón, trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses, Bioinformatics, 2009, 25(15), 1972–1973 CrossRef PubMed.
- S. Q. Le and O. Gascuel, An Improved General Amino Acid Replacement Matrix, Mol. Biol. Evol., 2008, 25(7), 1307–1320 CrossRef CAS PubMed.
- X. Gu, Y. X. Fu and W. H. Li, Maximum likelihood estimation of the heterogeneity of substitution rate among nucleotide sites, Mol. Biol. Evol., 1995, 12(4), 546–557 CAS.
- Z. Yang, PAML 4: phylogenetic analysis by maximum likelihood, Mol. Biol. Evol., 2007, 24(8), 1586–1591 CrossRef CAS PubMed.
- M. Owsianiak, M. Z. Hauschild, L. Posthuma, E. Saouter, M. G. Vijver and T. Backhaus, et al., Ecotoxicity characterization of chemicals: Global recommendations and implementation in USEtox, Chemosphere, 2023, 310, 136807–136807 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc03665d |
| This journal is © The Royal Society of Chemistry 2024 |