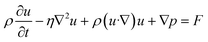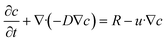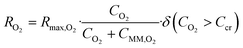Open Access Article
Open Access ArticleSliceChip: a benchtop fluidic platform for organotypic culture and serial assessment of human and rodent pancreatic slices†
Charles G.
Alver
 ac,
Silvia
Álvarez-Cubela
b,
Isabella
Altilio
b,
Emily
Hutchison
a,
Emma
Warrner
ac,
Silvia
Álvarez-Cubela
b,
Isabella
Altilio
b,
Emily
Hutchison
a,
Emma
Warrner
 a,
Mariana E.
Viso
a,
Mariana E.
Viso
 a,
Giana
Vitale
a,
David
Oliver
a,
Ricardo L.
Pastori
*b,
Juan
Dominguez-Bendala
*b and
Ashutosh
Agarwal
a,
Giana
Vitale
a,
David
Oliver
a,
Ricardo L.
Pastori
*b,
Juan
Dominguez-Bendala
*b and
Ashutosh
Agarwal
 *abd
*abd
aDepartment of Biomedical Engineering, University of Miami, Coral Gables, FL, USA. E-mail: A.agarwal2@miami.edu; Tel: +1 305 243 8925
bDiabetes Research Institute, University of Miami Miller School of Medicine, Miami, FL, USA. E-mail: RPastori@med.miami.edu; JDominguez2@med.miami.edu; Tel: +1 305 243 5349 Tel: +1 305 243 4092
cMedical Scientist Training Program, University of Miami Miller School of Medicine, Miami, FL, USA
dDesai Sethi Urology Institute, University of Miami Miller School of Medicine, Miami, FL, USA
First published on 8th January 2024
Abstract
Enzymatically isolated pancreatic islets are the most commonly used ex vivo testbeds for diabetes research. Recently, precision-cut living slices of human pancreas are emerging as an exciting alternative because they maintain the complex architecture of the endocrine and exocrine tissues, and do not suffer from the mechanical and chemical stress of enzymatic isolation. We report a fluidic pancreatic SliceChip platform with dynamic environmental controls that generates a warm, oxygenated, and bubble-free fluidic pathway across singular immobilized slices with continuous deliver of fresh media and the ability to perform repeat serial perfusion assessments. A degasser ensures the system remains bubble-free while systemic pressurization with compressed oxygen ensures slice medium remains adequately oxygenated. Computational modeling of perfusion and oxygen dynamics within SliceChip guide the system's physiomimetic culture conditions. Maintenance of the physiological glucose dependent insulin secretion profile across repeat perfusion assessments of individual pancreatic slices kept under physiological oxygen levels demonstrated the culture capacity of our platform. Fluorescent images acquired every 4 hours of transgenic murine pancreatic slices were reliably stable and recoverable over a 5 day period due to the inclusion of a 3D-printed bioinert metallic anchor that maintained slice position within the SliceChip. Our slice on a chip platform has the potential to expand the useability of human pancreatic slices for diabetes pathogenesis and the development of new therapeutic approaches, while also enabling organotypic culture and assessment of other tissue slices such as brain and patient tumors.
Introduction
For over 40 years, researchers have used enzymatically isolated human pancreatic islets for studying diabetes pathogenesis and the development of new therapeutic approaches.1,2 However, these models capture only a small part of the complex architecture of the pancreas and the whole-organ (exocrine and endocrine) changes that occur in diabetes.3,4 More recently, living slices of human pancreatic tissues have been used to study the interplay of exocrine and endocrine tissues in their conserved natural environment.5,6 Recent improvements to slice culture conditions have even expanded the use of slices to study real-time islet regeneration.7Though human pancreatic slices are excellent anatomical and functional models of the pancreas, long-term slice cultures can be difficult to manage, and serial assessments can be particularly challenging.6 Pancreatic slices require a highly oxygenated environment due to their elevated oxygen consumption rate. Further, cell metabolites can quickly build up around the slices, impacting cellular feedback loops.8 More importantly, digestive enzymes produced by the slices rapidly deteriorate the cellular structure of the slices in static cultures.9 Hence, long-term slice cultures require frequent medium changes to prevent cellular degradation.7 For functional assessments of slices, glucose stimulated insulin secretion (GSIS) studies typically utilize multiple slices in a non-sterile open well, limiting the ability to repeatedly analyze the response of single slices.10 Cultures with multiple free-floating slices are also difficult to repeatedly image without compromising sterility and reliability because slices tend to move within culture during medium changes and imaging. As a result, capturing high-time resolution images (every ∼4–6 hours for several days) requires extensive manual intervention to transfer slices between incubators and imaging microscopes with significant risk of contamination during transfer and movement artifacts during serial imaging. As such, microfluidic organ on chip platforms, like those developed by our lab and others, can engineer near-physiological oxygen and perfusion environments for maintenance of slice viability, and the study of pancreas physiology by hormone secretion and serial fluorescence imaging.11,12
Organs on chips (OoC) have traditionally been constructed as low volume microfluidic platforms from polydimethylsiloxane (PDMS), an optically transparent, gas-permeable and low-cost silicone polymer.11 PDMS based devices cannot typically accommodate large clearances required for manual seeding, culture, and recovery of primary ex vivo tissue and also suffer from drug absorption within PDMS and from in-line bubble formation due to difficulties with the control of dissolved gases over long periods of operation.13,14 The natural gas permeability of PDMS also makes it difficult to control the level of oxygen, as dissolved oxygen will tend to equilibrate with its surroundings.15 We describe a novel pancreatic slice on a chip (SliceChip) that features optimized design of convective fluid flows, use of bio-inert and non-absorbent materials, reversible assembly for manual loading and unloading of pancreatic slices, and straightforward integration with commercial imaging systems. We also report the design of a fluidic control platform that allows efficient switching between media reservoirs, exquisite control over dissolved oxygen, and bubble-free and leak-free operation, which is critical for long term microfluidic culture. Through the combination of SliceChip and fluidic control platform, we show a benchtop device capable of long-term culture and serial functional assessment with concurrent real-time imaging of primary human and murine pancreatic slices.
Materials & methods
Computational fluid dynamics analyses
Finite element analysis (FEA) was performed using COMSOL Multiphysics 5.3a (COMSOL Inc., Sweden). A 3D model of the fluid channel with an embedded approximation of a pancreatic slice was exported from SolidWorks as a STEP file and imported into COMSOL. The laminar flow and transport of dilute species physics modules were used to compute the fluid dynamics of the microfluidic chip, glucose transport in the system, and oxygen consumption in the pancreatic slice. The fluid in the channel was defined as an incompressible fluid with flow and solute diffusion governed respectively by the Navier–Stokes equation:| ∇·u = 0 |
where c is the species concentration, D is the diffusion coefficient, R is the reaction rate, and u is the velocity field.
The volumetric flow rate used across our fluid dynamics models was selected by a time-dependent study of glucose transport in the chip with the flow rate set by a parametric sweep (20–80 μl min−1). This model was used to calculate the effective glucose washout time at different flow rates and identify the ideal flow rate. Fluid velocity and pressure profiles were generated and plotted across the channel within a steady state model. Shear stress (τ) across the pancreatic slice was computed using Newtons law:
| τ = γ·η |
The steady state equilibrium oxygen distribution and consumption in the pancreatic slice was modeled using an ovoid approximation of a pancreatic slice that assumes a solid mass of tissue with uniform maximal oxygen consumption rate (OCR).16 The multi-physics module was used to feed the flow profile generated in the laminar flow module into the transport of dilute species module. Oxygen diffusivity within each part of the model was represented by the diffusive permeability αD, the product of effective diffusivity D, and the Bunsen solubility coefficient α, for the respective model part. Using diffusive permeability allowed our model to directly solve for oxygen concentration in terms of oxygen partial pressure and eliminated the need for a partition condition between the fluid and tissue phase of the model.7 Oxygen consumption was modeled as a consumption reaction within the pancreatic slice as defined by the Michaelis–Menten consumption rate:
Design and fabrication of microfluidic chip
The microfluidic chip was designed using SolidWorks 2020 (Dassault Systèmes, France). Chips were machined from clear 1/8′′ thick polymethyl methacrylate (PMMA) labs (McMaster-Carr, Elmhurst, IL) on an MDX-540 CNC milling machine (Roland, Japan). Unique top and bottom pieces were separately milled using a rotary clamp to mill features onto both sides of each piece. A 30 W CO2 laser engraver (Epilog Laser, Golden, CO) was used to cut the chips to their final shape. Silicone gaskets made in-house from a two-part silicone epoxy (Duraseal 1533, Cotronics Corp.) were attached to the lower face of the top chip using a dual-sided adhesive film (PS-1340, Polymer Science). The completed microfluidic chip can be assembled and sealed using a commercially available microfluidic chip clamp (Micronit Microfluidics, The Netherlands).Design and fabrication of slice anchors
The biocompatible slice anchors were designed using SolidWorks 2020 (Dassault Systèmes, France) and sent to a commercial 3D printing manufacturer (Protolabs, Maple Plain, MN) where they were printed by selective laser sintering using a biocompatible stainless steel 316 L powder.18System assembly
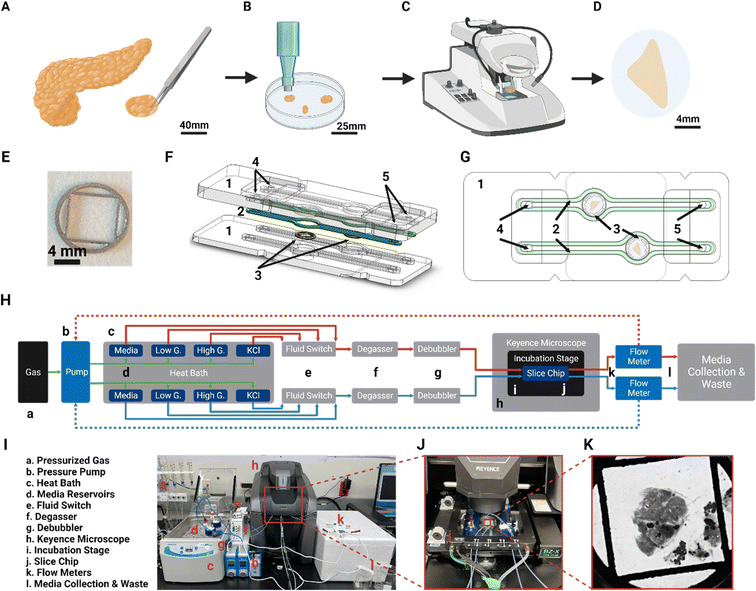
A complete stepwise protocol, including tubing lengths, manufacturer names, and part numbers, for the system assembly is available in Protocol S1.† A brief description is included here. Prior to assembly, each portion of the system was sterilized and gathered in a culture hood. Fluidic tubing, bubble traps, analytic selector valves, degassing chambers, flow units, and the microfluidic chip were sterilized by passing, in order, 70% ethanol, sterile distilled water, and clean air through them. Media bottles, GL-45 pressure caps, Micronit clamp and ferrules, pneumatic tubing and connections were autoclaved on a 60 minute gravity cycle. After rinsing and drying or autoclaving, all parts were UV-irradiated for 30 minutes before assembly.Within a culture hood, media bottles (filled with prewarmed base media and GSIS stimulants), sealed with GL-45 pressure caps (Fluigent, France, Cat# RESCAP-PCK) and pneumatically connected in series, were fluidically connected in parallel to the analytic selector valve. Outlet tubing from the valve passed into the following in series: a degassing chamber, bubble trap (Diba Omnifit, Cat# UX-21940-39), Micronit clamp, and flow units (Fluigent, France, Cat# FLU-S-D). Once the fluidic pathway was assembled, the bottom of the chip was placed within the Micronit clamp to load a single pancreatic slice into each channel. A fine brush of 2/0 size (Electron Microscopy Sciences, Hatfield, PA, Cat# 66100-20) was used to move a single slice into the 8 mm diameter chip well. A slice anchor was placed on top of the slice with the slice centered in the inner square opening. Once the slice was secured, the top chip was placed, and the clamp was closed.
The entirety of the sealed fluidic system was moved from the culture hood to a counter containing a pre-heated 37 °C bead bath, Keyence BZ-X810 microscope (Keyence, Itasca, IL), a Flow EZ pressure pump (Fluigent, France, Cat# LU-FEZ-0345) pressurized by a composite 95% O2 5% CO2 gas cannister (Airgas Inc., Radnor, PA), and a preheated 37 °C Tokai Hit stage-top incubator (Tokai Hit, Japan, Cat# STFX). The Micronit clamp was placed inside of the stage-top incubator, and this complex was placed into the Keyence microscope. Media and GSIS solutions were connected to the pressure pump and placed within the bead bath.
Medium degassing
Medium pressurized and equilibrated with 95% O2 and 5% CO2 composite gas was degassed to prevent super saturation using a two-chamber DEGASi Plus Micro Degasser (Biotech, Sweden). Degassing chambers utilize gas-permeable tubing housed inside of a vacuum chamber to remove dissolved gases from the fluid flowing through the tubing. Residual levels of dissolved gases are proportional to the flow rate of the fluid and inversely proportional to the internal chamber volume. A standard 100 μl internal volume chamber and an in-house modified 65 μl chamber were used to generate two levels of residual oxygen in separate flow channels with equal flow rates.Oxygen measurement
Oxygen measurements within the organ chip were conducted using an optical oxygen sensor placed into the top of a modified organ chip. An oxygen spot sensor attached to a luer lock was placed into a hole on the upper face of the top chip, allowing it to directly access the fluidic channel. The partial pressure of oxygen was measured each minute over the course of 16 h via a multichannel oxygen meter (PreSens, Germany) connected to the spot sensors by polymer optical fibers. Measurements were taken at 37 °C.Human pancreas slicing & conditioning
Human pancreatic tissue slices were obtained as pancreas biopsies from Prodo Labs (Aliso Viejo, CA) or tissue slices as part of the University of Florida and nPOD's human pancreatic tissue slice optimization initiative. De-identified demographic data for each of the donors is provided in Table S2.† Human pancreatic slices were generated following previously established protocols and cut to the standard slice thickness of 120 μm.6,18 Sections of human pancreas, approximately 0.5 cm3 blocks, were imbedded in an agarose solution inside of a 35 mm Petri dish at 37 °C. A 7 mm round biopsy punch was then used to remove a cylindrical section of tissue and agarose. Using a vibratome, 120 μm-thick slices were taken off of the cylinders and placed into a slice washing media comprised of DPBS (Sigma Aldrich, St. Louis, MO, Cat# D8537), 1× B27-minus Insulin (Invitrogen, Carlsbad, CA, Cat# A1895602), 1× penicillin–streptomycin–amphotericin B solution (Sigma Aldrich, St. Louis, MO, Cat# A5955), 100 μg ml−1 trypsin inhibitor from Glycine max (Sigma Aldrich, St. Louis, MO, Cat# T6522), 10 μg ml−1 aprotinin (Sigma Aldrich, St. Louis, MO, Cat# A6106), 10 μg ml−1 chymostatin (solubilized initially in DMSO; Sigma Aldrich, St. Louis, MO, Cat# 11004638001), and 5.5 mM D-glucose (Sigma Aldrich, St. Louis, MO, Cat# G8644).Prior to transfer into the microfluidic chip, slices were cultured for 24 h in an uncoated AirHive Dish (Biorep, Miami, FL) using the culture medium previously described.7 A complete experimental timeline for human slices is shown in Fig. 4A. Human slice culture medium contained basal BrainPhys neuronal medium without Phenol Red (Stemcell Technologies, Vancouver, BC, Cat# 05791) supplemented with 2% B27 supplement minus-insulin (Invitrogen, Carlsbad, CA, Cat# A1895601), 1% penicillin–streptomycin–amphotericin B solution (Sigma Aldrich, St. Louis, MO, Cat# A5955), 1% Glutamax supplement (Invitrogen, Carlsbad, CA, Cat# 35050061), L-glutamic acid 3.7 μg ml−1 (Sigma Aldrich, St. Louis, MO, Cat# 49449), 5.5 mM final D-glucose concentration (Sigma Aldrich, St. Louis, MO, Cat# G8644), 100 μg ml−1 trypsin inhibitor from Glycine max (Sigma Aldrich, St. Louis, MO, Cat# T6522), 10 μg ml−1 aprotinin (Sigma Aldrich, St. Louis, MO, Cat# A6106), 10 μg ml−1 chymostatin (solubilized initially in DMSO; Sigma Aldrich, St. Louis, MO, Cat# 11004638001), and 1% HEPES buffer (Invitrogen, Carlsbad, CA, Cat# 15630080). The same culture medium was used within the microfluidic chip for the duration of the slice culture.
Transgenic mouse models
Animal experiments were conducted under the supervision and oversight of the University of Miami Institutional Animal Care and Use Committee (IACUC) and Division of Veterinary Resources (DVR) at the University of Miami. In order to create the INS2-Cre/mTmG reporter, we crossed B6.Cg-Tg (Ins2-Cre) 25Mgn/J (INS2-Cre; Jackson Labs, Bar Harbor, ME, Cat# 003573) with B6.Gt (ROSA) 26Sortm4 (ACTB-tdTomato,-EGFP) Luo/J (mTmG; Jackson Labs, Bar Harbor, ME, Cat# 007676). In the resulting mouse, all insulin-producing cells express EGFP, while all non-insulin-producing cells express fluorescent tdTomato. INS2-Cre/mTmG mice were utilized for pancreatic tissue slicing. All mice were housed in specific pathogen-free (SPF) conditions at the DVR's animal care facility. They were maintained on a 12 h light/dark cycle with ad libitum access to standard irradiated chow and filtered drinking water.Mouse pancreas slicing & conditioning
Murine slices were generated and processed in accordance with a previously published method.6 Prior to transfer into the microfluidic chip, murine slices were cultured for between 24–96 hours in an uncoated AirHive Dish.7 Murine slice medium consists of a custom-made Waymouth's MB 752/1 medium (with L-glutamine; without D-glucose; Biological Industries, Cromwell, CT, Cat# 06-1110-01-1 A) supplemented with 11 mM D-glucose (Sigma Aldrich, St. Louis, MO, Cat# G8644), 1% heat-inactivated FBS (Invitrogen, Carlsbad, CA, Cat# 16140063), 100 μg ml−1 trypsin inhibitor from Glycine max (Sigma Aldrich, St. Louis, MO, Cat# T6522), 10 μg ml−1 aprotinin (Sigma Aldrich, St. Louis, MO, Cat# A6106), 10 μg ml−1 chymostatin (solubilized initially in DMSO; Sigma Aldrich, St. Louis, MO, Cat# 11004638001), and 1× penicillin–streptomycin–amphotericin B solution (Sigma Aldrich, St. Louis, MO, Cat# A5955). All experiments with murine slices were completed under normal oxygen levels.Dynamic glucose stimulated insulin secretion studies
Dynamic glucose stimulated insulin secretion (GSIS) experiments were performed on individual slices within the SliceChip. The base perfusion solution was a Krebs buffer with 125 mM NaCl, 5.9 mM KCl, 1.28 mM CaCl2, 1.2 mM MgCl2, 25 mM HEPES, and 0.1% bovine serum albumin, at pH 7.4 and held at 37 °C. Separate buffered solutions each containing 3 mM glucose (G), 16.7 mM G, 10 μM carbachol (CCH) (Sigma Aldrich, St. Louis, MO, Cat# 212385-M), or 30 mM potassium chloride (KCl) were connected in parallel with the base culture media to the system's pressure pump and an analytic selector valve (IDEX Health & Science, Carlsbad, CA, Cat# 7060). The selector valve allowed for the manual selection of perfusates without having to break the sterility of the system. Slices were initially perfused for 60 minutes at 80 μl min−1 with 3 mM G to equilibrate before GSIS studies. Following the equilibration step, slices were perfused in 30 minute intervals with the following sequence: 3 mM G, 16.7 mM G, 3 mM G, CCH, 3 mM G; on the final day of experiments, KCl was added as a final interval. An experimental timeline of the GSIS perfusions is shown in Fig. 4B. Effluent samples were manually collected over three-minute intervals from the outlet tubing in a 96 well plate held at 4 °C. Following GSIS studies, the selector valve was set back to the base medium, except on the final day of experiments, when the slices were removed from the system for live/dead staining and insulin immunostaining. Perfusates were stored at −80 °C until analyzed using commercially available ultrasensitive Human-Insulin ELISA kits (Mercodia Inc., Winston Salem, NC, Cat# 10-1132-01) and colorimetric amylase assays (Abcam, United Kingdom, Cat# ab102523). As a control, static slice cultures kept in the standard culture dish were stimulated with the aforementioned solutions as a static incubation following the same timeline laid out above, and the entirety of the incubation solution was kept for analysis consistent with the perfusate analysis.Viability assessment
Human pancreatic slices were incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 fluorescein diacetate (FDA) (Invitrogen, Carlsbad, CA, Cat# F1303) in PBS (Sigma Aldrich, St. Louis, MO, USA Cat# P3813) to indicate live cells and 1
100 fluorescein diacetate (FDA) (Invitrogen, Carlsbad, CA, Cat# F1303) in PBS (Sigma Aldrich, St. Louis, MO, USA Cat# P3813) to indicate live cells and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 propidium iodide (PI) (Invitrogen, Carlsbad, CA, Cat# P1304MP) in PBS (Sigma Aldrich, St. Louis, MO, USA Cat# P3813) and counterstained with a 1
10 propidium iodide (PI) (Invitrogen, Carlsbad, CA, Cat# P1304MP) in PBS (Sigma Aldrich, St. Louis, MO, USA Cat# P3813) and counterstained with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 Hoechst solution (Invitrogen, Carlsbad, CA, Cat# H1339) in HEPES buffer (Invitrogen, Carlsbad, CA, Cat# 15630080). Slices are incubated in Hoechst solution for 15 min at room temperature. They were then washed with 1× PBS for 1 min and transferred into a 1
500 Hoechst solution (Invitrogen, Carlsbad, CA, Cat# H1339) in HEPES buffer (Invitrogen, Carlsbad, CA, Cat# 15630080). Slices are incubated in Hoechst solution for 15 min at room temperature. They were then washed with 1× PBS for 1 min and transferred into a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 FDA/PI solution to incubate for 3 min. Slices were then washed with 1× PBS for 10 min and moved to a 35 mm dish with a number-zero confocal grade crystal (MatTek Corporation, Ashland, MA, Cat# P35G-0-10-C) bottom for imaging.
1 FDA/PI solution to incubate for 3 min. Slices were then washed with 1× PBS for 10 min and moved to a 35 mm dish with a number-zero confocal grade crystal (MatTek Corporation, Ashland, MA, Cat# P35G-0-10-C) bottom for imaging.
Immunostaining and imaging
Tissue slices were washed 2× for 5 min in 1× phosphate buffered saline (PBS), pH 7.4 (Sigma Aldrich, St. Louis, MO, USA Cat# P3813), and fixed in 4% paraformaldehyde solution overnight. Slices were then washed 10× for 5 minutes in 1× phosphate buffered saline (PBS), pH 7.4 (Sigma Aldrich, St. Louis, MO, USA Cat# P3813). Permeabilization was done for 30 minutes using 0.3% Triton (Sigma Aldrich, St. Louis, MO, USA Cat# T9284-500 ml) in 1× PBS, pH 7.4 (Sigma Aldrich, St. Louis, MO, USA Cat# P3813). After this, slices were placed in a blocking buffer containing dH2O, 5% normal donkey serum (Jackson Labs, Bar Harbor, ME, Cat# H-400), 0.1% Triton, and 1× power block (Biogenex, San Ramon, CA, Cat# HK0855K). Primary antibodies for insulin (Ready to Use Polycolonal Guinea Pig anti-insulin, DAKO, Cat# IR002), amylase (Polyclonal Rabbit anti-amylase, Sigma Aldrich, Cat# A8273), and glucagon (Monoclonal Mouse anti-glucagon, R&D Systems, Cat# MAB1249) was dissolved in the blocking buffer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, and 1
200, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 respectively) and incubated for 48 h at 4 °C. The primary antibody was then removed, and slices were washed 7× for 5 min with 1× PBS, pH 7.4, 0.1% Triton. Secondary antibody solutions (1
500 respectively) and incubated for 48 h at 4 °C. The primary antibody was then removed, and slices were washed 7× for 5 min with 1× PBS, pH 7.4, 0.1% Triton. Secondary antibody solutions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400) were made with Alexa Fluor 488, 594, and 647 donkey anti-primary antibody and 1
400) were made with Alexa Fluor 488, 594, and 647 donkey anti-primary antibody and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 4′, 6-diamidino-2-phenylindole (DAPI) (Thermo Fisher/Life Technologies, Waltham, MA, Cat# D1306) in blocking buffer. Samples were incubated with secondary antibodies for 90 minutes and then washed 7× for 5 min with 1× PBS, pH 7.4. To get the least amount of background fluorescence, an additional wash was performed overnight at 4 °C with 1× PBS, pH 7.4. For imaging, slices were mounted on crystal slides (Staining Kit, Arcturus, Cat# Kit0420) with ProLong mounting reagent (ProLong Gold antifade reagent, Invitrogen, Cat# P36934) and sealed with a cover slip (Micor cover glass, VWR, Cat# 48393). Imaging was acquired using a Keyence BZ-X810 fluorescent microscope. Z-stack depth ranged between 120–130 μm depending on the slice thickness and condition while the number of images in the Z-stack ranged between 15–25 with a step size ranging between 2.5–4 μm. Insulin staining was used to determine the approximate area of islets following culture in the SliceChip. The fluorescent area was calculated using the BZ-X810 analyzer hybrid cell count module. To prevent bias in viability studies, we utilized ImageJ/FIJI to perform quantitative analysis of cell populations, using total area calculations for fluorescent area of live/dead regions and taking the viability as the ratio of live area to total area (live area + dead area).
400 4′, 6-diamidino-2-phenylindole (DAPI) (Thermo Fisher/Life Technologies, Waltham, MA, Cat# D1306) in blocking buffer. Samples were incubated with secondary antibodies for 90 minutes and then washed 7× for 5 min with 1× PBS, pH 7.4. To get the least amount of background fluorescence, an additional wash was performed overnight at 4 °C with 1× PBS, pH 7.4. For imaging, slices were mounted on crystal slides (Staining Kit, Arcturus, Cat# Kit0420) with ProLong mounting reagent (ProLong Gold antifade reagent, Invitrogen, Cat# P36934) and sealed with a cover slip (Micor cover glass, VWR, Cat# 48393). Imaging was acquired using a Keyence BZ-X810 fluorescent microscope. Z-stack depth ranged between 120–130 μm depending on the slice thickness and condition while the number of images in the Z-stack ranged between 15–25 with a step size ranging between 2.5–4 μm. Insulin staining was used to determine the approximate area of islets following culture in the SliceChip. The fluorescent area was calculated using the BZ-X810 analyzer hybrid cell count module. To prevent bias in viability studies, we utilized ImageJ/FIJI to perform quantitative analysis of cell populations, using total area calculations for fluorescent area of live/dead regions and taking the viability as the ratio of live area to total area (live area + dead area).
Time-lapse imaging
Slices of INS2-Cre/mTmG mouse pancreas were generated and kept in the SliceChip for 5 days for time-lapse fluorescent imaging. During this time, murine slices were perfused with media at 80 μl min−1 and imaged every 4 or 12 hours within the Keyence BZ-X810 fluorescent microscope with a Z-stack height of 105–120 and step height of 5–15 across a 32–40-tile grid. Post-acquisition, a full focus stitch of the Z-stack and tiles was generated using the built-in z-stack and stitching functions of the Keyence BZ-X810.Statistical methods
Statistical analyses were performed in GraphPad Prism 9.5.1 software (GraphPad Software, San Diego, CA). A Student's t-test was used to directly compare GSIS AUC results (flow conditions n = 3 slices, static n = 2 slices). A paired t-test was used to compare intra-condition AUC between 3 mM glucose and 16.7 mM glucose stimulation (n = 3 slices). A confidence interval of 95% was considered significant with a designation of *P ≤ 0.05 and **P ≤ 0.01. Results at or slightly above the traditional levels of significance have their P values directly reported and nonsignificant values are shown as ns in graphs. An ordinary one-way ANOVA with Tukey's multiple comparisons test was used to assess statistical significance in the viability of slices in static or perfusion culture. Linear regression analysis was used to assess the trend of fluorescent area degradation in serial imaging and was constrained to an area of 1 at the onset of imaging.Results
SliceChip design
The design of the SliceChip aims to maintain a human or murine pancreatic slice in culture without limiting our ability to visualize slices, perform serial perfusions, or recover tissue samples after experimentation. Human pancreatic tissue slices are generated (Fig. 1) by precision slicing a piece of human donor pancreas, while maintaining the native architecture and distribution of endocrine and exocrine cells into 120 μm thick live sections.6 With a majority of pancreatic islets being smaller than 120 μm with an average diameter of 108.92 as recently reported, a slice thickness of 120 μm allows for an adequate number of intact islets within a slice and is the standard for pancreatic slices.9,19,20 Donor tissue that has been cleaned and sectioned into small chunks is imbedded into agarose (Fig. 1A) and a biopsy punch is used to remove agarose-tissue cylinders for slicing with a vibratome (Fig. 1B and C). Slices are removed from the vibratome and cultured in 35 mm dishes before placement into the SliceChip.Utilizing a similar design as previous organ on chips developed by our lab,12,21 SliceChip was designed in 3D CAD software to feature two 8 mm diameter, 1.5 mm deep culture wells with independent fluid pathways (Fig. 1F and G), allowing for concurrent analysis of two slices under different conditions. SliceChip is comprised of two interlocking optically clear PMMA pieces fabricated by subtractive manufacturing. Unlike PDMS-based chips, our resealable SliceChip can easily be opened after experiments to remove the slices, and then cleaned, sterilized, and reused. A silicone polymer gasket adhered to the top piece of the chip provides a hermetic seal around the fluid channels once the chip is clamped in the Micronit clamp. The central culture wells are slightly offset from each other to allow each well to be clearly visible once clamped. A set of holes in the top chip piece positionally correspond to holes in the clamp that can be used as fluidic inlets and outlets (Fig. 1F and G).
Anchor design
To ameliorate movement artifacts in slice images taken under flow conditions, we developed an anchor using 3D CAD software to secure the slice without the limitations to medium and solute diffusion caused by traditional fixation methods, such as secondarily imbedding slices in a hydrogel.22 The anchor is an 8 mm diameter disk of 0.5 mm thickness that features a central 25 mm2 square opening where the bulk of the tissue slice rests in culture (Fig. 1E). The anchor is 3D printed via selective laser sintering at a commercial 3D printer. To minimize pressure imparted onto the slice by the anchor, it is designed to apply force only to the agarose that remains on the outside of the tissue slices. An additional facet of protection is that the anchor features a differentially thick body to prevent the slice from moving beyond where it was initially placed. The outer ring of the anchor extends a full 0.5 mm while the inner mesh is 0.375 mm thick, providing a gap below the mesh that approximates the thickness of human or murine pancreatic slices. Because the slices are in contact with the anchors for an extended period of time, the anchors were printed out of stainless steel 316 L, which highly biocompatible and is often used in implantable medical devices.18Computation analysis of fluid dynamics
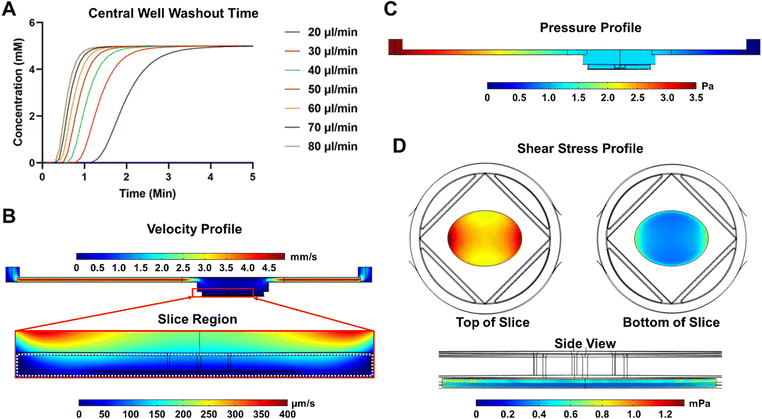
The fluid dynamic profile of the SliceChip was modeled in COMSOL multiphysics software. Because a major goal of this platform was to be able to perform GSIS experiments during the slice culture, a model was developed to compute the glucose washout time in the chip to facilitate choosing the volumetric flow rate to be used in the chip. The washout time was computed at various flow rates (20–80 μl min−1) using a multiphysics model that linked the flow of the liquid within the chip and the diffusion of glucose. The system began at 0 mM glucose concentration, and a step function was used to rapidly bring the inlet concentration to 5 mM; the average concentration above the slice was recorded and plotted over time (Fig. 2A). We aimed to have a total time from inlet to outlet of less than three minutes so that we could collect samples with a high degree of time resolution while still easily linking discrete collection samples to infusion times. 80 μl min−1 was chosen as the flow rate because the central well washout time was approximately 1.2 minutes. Factoring in the system's dead volume of 140 μl, 80 μl min−1 resulted in an inlet to outlet time of 2.95 minutes, where 70 μl min−1 or lower would have resulted in times over 3 minutes. 80 μl min−1 is also within the range of flow rates previously used for GSIS experiments.7,12Using the selected volumetric flow value of 80 μl min−1, velocity, pressure, and shear stress profiles were generated for the SliceChip (Fig. 2B–D). In the velocity profile (Fig. 2B) we see that, despite fast-moving fluid in the inlet and outlet channels, the fluid moving through the larger well and around the pancreatic slice is significantly slower and does not exceed 200 μm s−1. The system demonstrates a uniform pressure gradient across the chip (Fig. 2C). Fluid flow within the chip also generates shear stress around the slice (Fig. 2D). Shear forces are greatest on the edges of the top surface of the slice where the surface of the slice runs perpendicular to the flow direction. Most of the slice experiences a shear stress below 1 mPa and even at its maximum of 1.2 mPa, the shear stress is well below the levels that have been reported to cause damage to islets.23,24
Degasser incorporation and oxygen residuals
Bubble formation within the fluid pathway can be detrimental to extended culture in an organ chip, limiting or blocking fluid flow and drying out cells.14 A system such as ours, in which flow is generated by a pressure gradient, is especially prone to bubble formation owing to the parameters that control gaseous solubility in a fluid as described by Henry's Law.| C = kP |
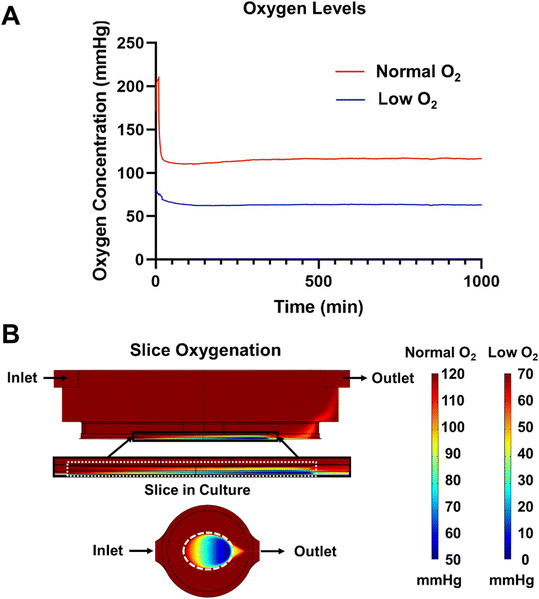
Initial experiments were ended before 24 hours due to excessive bubble formation that halted flow in the system. To prevent bubble formation, we prevented the oversaturation of the medium by providing a slight temperature gradient (37–30 °C) between the medium reservoirs and the culture well. While the increased solubility due to temperature did initially slow bubble formation, it was not enough to eliminate the bubbles. From here, we addressed the other component of Henry's Law, partial pressure. We used a degassing chamber that applies a vacuum across a gas permeable fluid tube to eliminate dissolved gas within the flowing medium. The efficiency of the degasser was directly related to the length of the degasser tubing and the flow rate of the medium per manufacturer reporting. While a counter-intuitive solution given our desire to increase the oxygen delivery to the pancreatic slices, degassing the medium resulted in bubble-free media over extended culture times. However, this did create a new problem—the residual oxygen level in degassed media would be lower than desirable for slice cultures.7 Using a SliceChip modified to include an oxygen sensor (Fig. S2†), we measured the residual pressure of oxygen to be 0 mmHg in the central well, unacceptable for culturing tissue. To address this problem, we supplemented the amount of oxygen initially dissolved in the media by pressurizing the medium with a composite gas made up of 95% O2 and 5% CO2 to maintain pH buffers. Repeat analysis of the residual oxygen level with the oxygen-supplemented and pressurized medium resulted in a partial pressure of 70 mmHg (Fig. 3A). To bring the residual in line with atmospheric levels, we modified the degasser tubing length, removing 35 mm. This resulted in a residual of 120 mmHg (Fig. 3A). By modulating the length of the degasser tubing and oxygen doping the medium with pressurized composite gas, we were able to effectively create a bubble-free system with variable oxygen levels.
Computational analysis of oxygen consumption
Adequate oxygenation of pancreatic slices has been shown to be important to the maintenance of slice cultures.7 We aimed to maintain the normal oxygen tension inside of a slice within the physiological range of 30–50 mmHg, preferably at or above the upper limit given the prior islet survival benefits shown for islets cultured in supraphysiological oxygen.16,26,27 As such, we created a 3D COMSOL model of steady state oxygen consumption within our SliceChip using the transport of dilute species module. The inflow of oxygen into the culture well was linked by the Multi-Physics model to the chip's fluid dynamics computed by the laminar flow module. A slice mimic at the bottom of the culture well was assigned a maximal oxygen consumption rate (OCR) of 0.03 mol m−3 s−1 consistent with prior oxygen consumption models and studies.28 The modeled slice was immobile with a small (10 μm) gap underneath it to mimic the height difference between the gap under the anchor and the thickness of the slice that allowed media to flow under the slice. Oxygenation in the slice was modeled with an input of low oxygen (70 mmHg) and normal oxygen (120 mmHg) as provided by using the standard degasser and modified degasser, respectively (Fig. 3B). An oxygen level below 0.1 mmHg was considered to be anoxic.7,28 Neither normal nor low oxygen systems produced regions below this threshold, with respective minimums of 52.1 mmHg and 2.1 mmHg. The model shows that the oxygen level in the slice forms a gradient with the concentration decreasing as we move from the inlet to outlet side of the slice, from outside to inside, and from top to bottom. The computational model shows the equilibrium gradient within the SliceChip and is consistent over the course of the culture. The overall residual oxygen within the steady state consumption model showed a similar operating range and levels (115–52 mmHg and 65–2 mmHg for normal and low oxygen systems, respectively) comparable to previously reported successful slice cultures.7System assembly
Traditionally, the environmental conditions of OoC cultures are controlled by an incubator; however, this limits the ability to continuously image cultures without disturbing their environment. Commonly used GSIS perfusion assemblies also require the removal of slices from an incubator, limiting the ability to perform serial assessments.6 To create a system by which we can continuously image, perfuse, and stimulate slices, we have adapted traditional OoC methods to create a platform that maintains the sterility, temperature, and O2 and CO2 levels of the SliceChip and perfusates within a Keyence BZ-X810 fluorescent microscope. To ensure sterility within the fluid pathway, all parts of the system are pre-sterilized and assembled in a sterile culture hood. A schematic and a real-world depiction of the platform components and fluid pathways are shown in Fig. 1H and I, respectively.The use of a Flow EZ pressure pump to pressurize medium reservoirs and drive fluid through the chip also requires the maintenance of a hermetic seal on each of the parallel fluidically independent SliceChip channels. A pressurized composite gas canister (95% O2 and 5% CO2) supplied the pressure for the pressure pumps and controlled the gas composition of the solutions used in the platform. Pressure caps fabricated by Fluigent sealed each of the media reservoirs and held the tubing that linked into the platform's fluidic pathway. At the start of each new SliceChip culture, we added all of the solutions, including those for GSIS experiments, to each of the fluidic pathways and stored them in a 37 °C bead bath with the jars for each pathway connected by a singular pneumatic pressure line with independent fluid outlets. Each of these outlet tubes met at a fluid switch that selected one of the input lines and directed it into the singular outlet. Fluid continued through a degasser and bubble trap that removed excess dissolved gas and large inline bubbles, respectively. Each fluid channel utilized an independent degasser such that the oxygenation level of each slice's medium could be altered, and different oxygenation conditions can be used concurrently for slices from the same individual. After the medium was free of bubbles, it moved into the SliceChip itself. The chip was clamped within the Micronit clamp, and that apparatus was kept inside of a stage top heater that maintained the system's temperature at 30 °C in accordance with previously reported slice culture protocols.7 The clamp-heater system was placed inside of a fluorescent microscope and held there for the duration of the experiment. The stage heater housing had spaces for the SliceChip's fluidic inlet and outlet tubing to enter and exit the heating area. While these openings would normally break the sterility of a culture held inside of a stage top heater, our platform did not break the fluid pathway that was hermetically sealed from the pressurized medium to collection outlet. The only part of our system exposed to the lab atmosphere was the outside of the tubing, which does not interface with the culture itself. On its way out of the SliceChip, medium from each fluid channel passed through a Flow Unit that monitored the volumetric flow rate. These flow meters fed the flow rate back to the pressure pumps to create a feedback loop that altered the pressure level so that flow was maintained at a steady 80 μl min−1. To assess the effects of oxygenation on slices and demonstrate the independence of the SliceChip's fluidic pathways, the system was set up so that one fluid lane passed through a modified degasser, providing a normal oxygen level, and the other passed through an unmodified degasser, providing a low oxygen level. The combination of a temperature- and gaseous environment-controlled, hermetically sealed fluid pathway with a variable fluid input housed inside of a fluorescent microscope created a system that could continuously image and perform GSIS experiments.
Repeat assessment of glucose stimulated insulin secretion (GSIS)
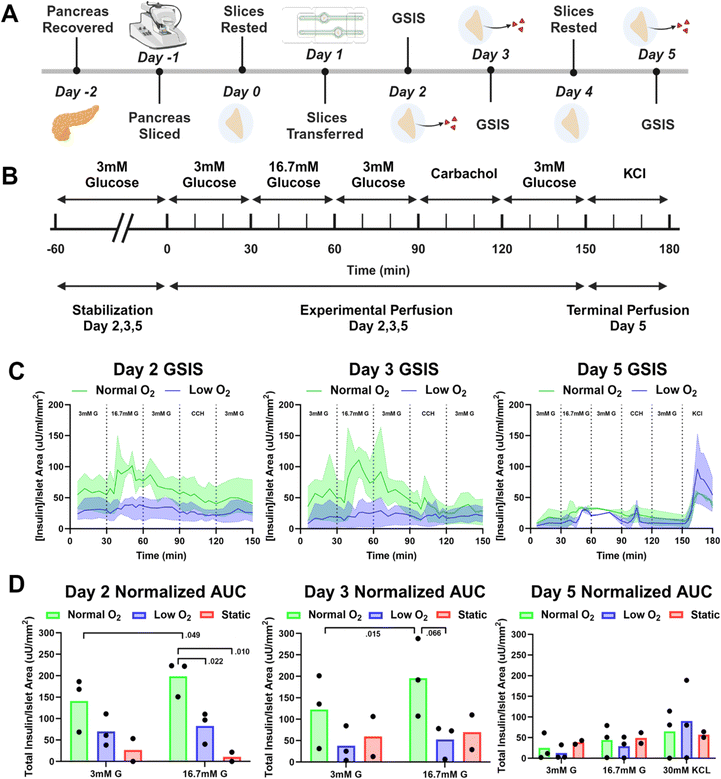
GSIS metrics are a standard method used to monitor pancreatic islets' time-dependent release of insulin in response to glucose stimulation. Traditionally these are endpoint experiments in which a set of multiple slices are moved from a sterile incubator to a non-sterile platform, perfused, and then immediately fixed and stained. Compared to this method of performing GSIS assessments of pancreatic slices, our sterile SliceChip provides a unique ability to perform repeat GSIS assessments of the same pancreatic slice over the course of multiple days. SliceChip's independent fluid pathways allowed us to concurrently run GSIS assessments with slices from single individuals held under different oxygen conditions to assess the effects of oxygenation on GSIS. Insulin secretion profiles, normalized to slice islet area (values shown in Fig. 6B), for slices cultured in normal and low oxygen conditions, are summarized in Fig. 4C. The mean and range of data are represented by the solid line and shaded region, respectively. On days 2 and 3, the slices kept under normal oxygen levels appeared to demonstrate a physiological insulin spike in response to glucose stimulation while those cultured with a lower oxygen level appeared to show a blunted response. The level of responsiveness between slices on days 2 and 3 appears similar. By contrast, day 5 showed an insulin spike in both normal and low oxygen conditions though with a decreased amplitude as compared to days 2 and 3. Assessment of the AUC during the period before, during, and directly after glucose stimulation was used to further investigate slice responsiveness under different oxygen conditions (Fig. 4D). An unpaired Student's t-test of the AUC of high glucose stimulation on days 2 and 3 demonstrated significance between the insulin secretion of normal and low oxygen conditions (P < 0.05 and P = 0.066 for days 2 and 3, respectively, n = 3 slices). While there appears to be a difference between the normal and low oxygen conditions for the high glucose interval on day 5, statistical analysis is unclear (P = 0.61, n = 3 slices). A separate analysis looked to compare the AUC of high glucose stimulation between static control slices and slices held under normal oxygen flow conditions, finding statistical significance on day 2 (P < 0.01, flow n = 3 slices, static n = 2 slices) and lacking significance on days 3 and 5 (P = 0.19, 0.877, respectively, flow n = 3 slices, static n = 2 slices). However, it should also be noted that the static culture had a lower n than the perfusion cultures, and this may impact statistical reliability. Because current GSIS methods are terminal for slices, the control slices had to undergo GSIS in a static incubation to allow for repeated assessment of the same slices function over multiple days and this method may not provide comparable GSIS results to those previously reported.7 A paired t-test of the AUC of 3 mM and 16.7 mM glucose stimulation within each oxygen condition, showed that slices kept under normal oxygen demonstrated a significant difference between insulin secretion under 3 mM and 16.7 mM glucose on days 2 and 3 (p = 0.049, p = 0.015, days 2 and 3 respectively, n = 3 slices), and no significance under low oxygen (p = 0.408, p = 0.4756, days 2 and 3 respectively, n = 3 slices). Day 5 analysis for both conditions was not significant (p = 0.240, p = 0.236, normal oxygen and low oxygen respectively, n = 3 slices). AUC stimulation indices (ratio of 16.7 mM G AUC to 3 mM G AUC) were calculated to evaluate the difference in insulin secretion between the low and high glucose stimulation phases. Each condition exhibited a stimulation index >1 (more insulin released during the high glucose phase than the low glucose phase). During days 2 and 3, slices in normal oxygen conditions had higher stimulation index than those in low oxygen. However, day 5 showed the opposite. Mean values and standard deviations for the stimulation indices are reported in Fig. S1.† Importantly, we have demonstrated that our SliceChip is capable of executing repeat GSIS perfusion assessments and recovering measurable analyte levels from the same singular pancreatic slice over multiple days.Viability
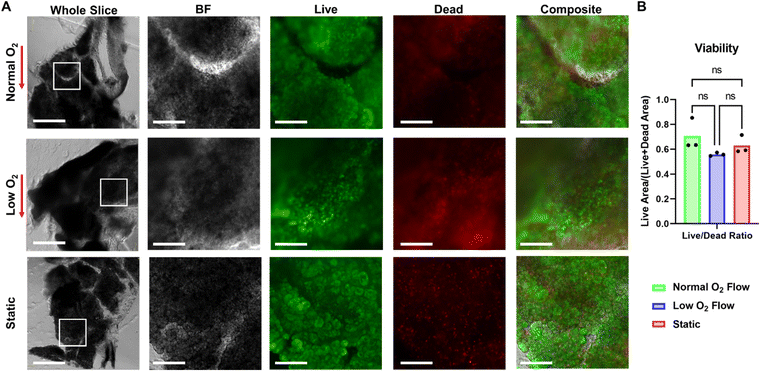
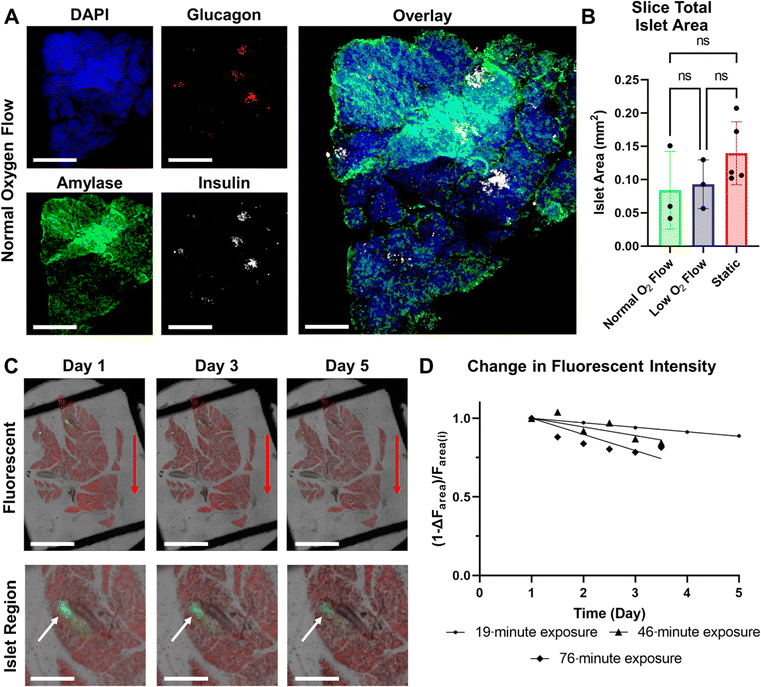
Following a 5 day incubation period that included three 3 hour perfusion or static GSIS experiments, the viability of slices kept in static culture was compared to those kept in SliceChips under normal and low oxygen conditions. The fluorescent area of live and dead cells was used to compute a ratio of live area to total cell area (live area and dead area) as a marker of tissue viability and is shown in Fig. 5B.6,7 ANOVA testing showed no significance between each of the culture conditions. Representative images of the post-culture live/dead stains for normal and low oxygen conditions as well as a static culture are presented in Fig. 5A. Though we report viability of pancreatic slices as a function of all of the cells within a slice, this is not an entirely representative metric of an individual slice's responsivity to glucose stimulation. Additional imaging shown in Fig. 6A demonstrates the maintenance of stainable insulin, glucagon, and amylase in slices following culture in the SliceChip as a visual marker for the endocrine and exocrine pancreatic tissue subtypes. Colocalization of insulin and glucagon producing cells within individual islets is evident on visual inspection. While some degree is expected due to the structure of the pancreatic islet, its prominence here can also be attributed to the fluorescent images being a full-focus consolidation of a 120 μm tall Z-stack of the 3D pancreatic slice. Continued insulin secretion in relation to high glucose throughout suggests islet tissue viability. Though exocrine function was assessed by carbachol stimulation as part of the GSIS experiment protocol, levels of secreted amylase were below the assay detection limit of 0.2 mU. However, the presence of amylase following immunostaining suggests maintenance of the exocrine tissues.Imaging
To assess our SliceChip platform's ability to take time-lapse fluorescent images of pancreatic slices, we generated pancreatic slices from a transgenic mouse model that allowed us to fluorescently visualize live insulin-producing cells via EGFP and non-insulin-producing cells with tdTomato. Composite stitches of full focus Z-stack images taken every 4 hours with a total exposure time of 19 minutes per imaging period were compiled, demonstrating the stability of fluorescent signal over the course of the 5 day culture. Composite brightfield/EGFP/tdTomato images from days 1, 3, and 5 are shown in Fig. 6C with a magnified region depicting an islet below the whole slice image. Over the full duration of the culture, we did note a decrease in fluorescent intensity, suggesting some expected photobleaching. However, a clear fluorescent image of the tissue was still recoverable. The degree of photobleaching was quantified as the loss in recoverable fluorescence by measuring the total recoverable fluorescent area at numerous time points and calculating the residual fluorescent area from the baseline, Fig. 6D. Additional slices were imaged with varying exposure lengths and imaging intervals (46 minutes of exposure every 12 hours and 76 minutes of exposure every 12 hours) to assess the impact of exposure time on photobleaching within the SliceChip. Linear regression analysis showed a loss of 2.9%, 5.5%, and 10.3% of the total fluorescent area each day in the 19, 46, and 76 minute exposures, respectively. A less than 3% daily loss was deemed acceptable given that this degree of photobleaching is correctable through post-acquisition processing.29Discussion
SliceChip is a novel benchtop culture system, capable of extended human pancreatic slice culture as well as repeated GSIS experiments and imaging analysis at the single slice level. To facilitate these experimental techniques outside of standard incubators, we created a platform that allows for the dynamic control of the temperature and gaseous environment supplied to slices held within a fluorescent microscope. An incubation stage that fits within a Keyence microscope provides thermal control of the SliceChip itself, while a bead bath warms media and other perfusates. Bubble formation during organ on chip operation is a major issue that has typically been solved by using open systems or gas permeable membranes, which perform well and maintain sterility within an incubator.30,31 However, our bench top device necessitated the development of a new method to maintain a sterile, bubble-free fluidic pathway. Initial bubble formation in prototype devices was caused by medium saturation and subsequent decreases in solubility due to temperature and pressure changes. We found that a degassing chamber effectively eliminated bubbling by decreasing the saturation of dissolved oxygen and other gaseous species in our perfusates. Traditionally, these chambers are used in cases where a low concentration of dissolved gases in a liquid has minimal impact on results, such as in chromatography or automated cell profiling.32 However, in the culture of live tissue samples, such as human pancreatic slices, it is imperative that the device can maintain physiologically appropriate levels of dissolved oxygen (30–50 mmHg within a pancreatic slice) as well as adequate levels of dissolved carbon dioxide to buffer the media. Using a pressurized canister of composite gas as the source of pressure for our pressure pump regulator, we doped the concentration of dissolved oxygen and carbon dioxide within the medium to maintain appropriate levels following degassing. Within the tissue slices, oxygenation levels accounting for slice oxygen consumption were maintained within the 30–50 mmHg range consistent with in vivo pancreatic oxygen tension.26 With this solution, extended cultures inside of the sealed organ chip remained bubble-free and received a physiological level of oxygen.7,26 Future studies should explore further modification of degasser length or gas composition to provide a higher degree of oxygenation control within systems.While computational modeling of the oxygen consumption demonstrated no observable regions of necrotic tissue in our normal and low oxygen conditions, we sought to address the impact of the differential oxygen levels on human pancreatic slices via their insulin responsiveness to glucose as a test of the system's ability to meaningfully control oxygenation. Repeat GSIS perfusion assessments of the same pancreatic slices held under normal and low oxygen conditions showed that a low oxygen environment appeared to blunt the release of insulin in response to high concentration of glucose in the early days of culture, whereas those cultured in normal oxygen conditions demonstrated a repeated physiologically relevant spike in response to high glucose throughout the full culture period. However, after extended culture, this differential response was less apparent, and the slices kept under low oxygen conditions appeared to regain the expected physiological response to glucose stimulation. On day 5, we observed signs of contamination in the input basal medium, which may have resulted in day 5 results to be an outlier. However, the solutions used for GSIS and the SliceChip did not show signs of contamination. As such, our ability to establish an effect of oxygenation on glucose responsiveness on day 5 remains limited in our study. More importantly, the continued responsiveness of slices kept under normal oxygen conditions demonstrates the ability of our system to perform repeat perfusion experiments and recover measurable levels of analytes from the same slice over multiple days. The observed difference in GSIS results in our SliceChip between normal and low oxygen conditions is understandable given the loss of function in islets held under low oxygen conditions that has been previously observed.7,12 Additionally, while the general viability and islet area was not significantly different between slices, it is not unexpected to still see a difference in their response to glucose stimulation. Prior studies have shown that the islets held under different oxygen tensions demonstrate a difference in glucose sensitivity before significant differences in islet viability and size.12,27 Another important consideration is that, in previously reported GSIS experiments, 3–4 slices were used simultaneously for analysis in terminal experiments.5,7,10,33 Having multiple slices per GSIS experiment masks the inherent variability of an individual slices and its islets, limiting the ability to analyze changes in individual slice function. Similarly, using a new set of pancreatic slices for each experiment limits the ability to study the effects of repeat GSIS stimulation on an individual pancreatic slice, a metric that has yet to be reported. In contrast, the repeated GSIS measurements taken with the SliceChip herein represent a unique repeated observation of the same individual slices over time. While we utilized our perfusion system solely for GSIS experiments, the use of a selector valve to change between sterile media and other solutions also allows for serial injection of pharmaceuticals and other therapeutics. As such, this platform has the potential to be used in the future to observe changes in single slice behaviors in response to drug treatments and other medical therapies.
Aside from the perfusion capabilities of our SliceChip platform, the maintenance of our SliceChip within a fluorescent microscope provides a host of live-cell imaging opportunities. Here we demonstrated the ability of our SliceChip to culture transgenic fluorescent murine slices over an extended period and to image them at a high magnification and with greater frequency than realistically practical if done manually. While a degree of photobleaching was apparent, changes to the imaging methodology to limit imaging time—including imaging over smaller regions, lower Z-stack height, lower exposure times, and decreased imaging frequency—are expected to effectively lessen the extent of this problem and could be optimized depending on the desired imaging result. Additionally, the inclusion of fluorescent live-cell markers or adenoviral markers, such as those used to assess calcium signaling or changes in gene expression, can expand the platform's imaging capabilities to assess intracellular calcium dynamics within pancreatic slices or potential cell type conversions.5–7,10,34 With the use of our slice anchor, slices kept within the SliceChip remained stable throughout the duration of experiments, effectively eliminating the movement artifacts often present across repeat whole slice imaging, and also decreasing the effort needed to align slice images taken on different days. The ability to take frequent, repeat, stable images without moving from an incubator to a microscope provides a sterile and reliable way to image changes to the architecture of tissue slices due to changes in cell populations while eliminating movement artifacts that may confound imaging assessments.
While we have directly described a system for the analysis of human pancreatic slices, the SliceChip system's functionalities—dynamic environmental controls, repeat serial perfusion, and stable live-cell fluorescent imaging—provide a unique opportunity for other kinds of tissue slices. Active research for ex vivo tissue samples is an ongoing effort with opportunities for new platforms to expand experimental capabilities.35,36 Current systems that focus on singular experimental aspects lack the combination of extended cultures, serial perfusions, easily recoverable tissues, and continuous imaging found in our SliceChip.35–37 Additionally, our use of a resealable PMMA-based organ chip eliminates many of the problems associated with traditional PDMS-based organ chips, such as drug retention and gaseous diffusion.11,13 In combination with its bubble-free design and environmental controls, we believe our system could easily be adapted to culture a variety of tissue slices and to expand their use for basic science and pharmaceutical research.
Conclusion
We developed a novel benchtop acrylic-based, bubble-free, pancreatic SliceChip platform that can maintain a sterile human pancreatic slice culture within a fluorescent microscope while running repeated serial perfusion experiments. Dynamic environmental controls allowed us to serially analyze insulin secretion in response to differential levels of glucose stimulation for singular slices. We observed the maintenance of a physiological response to high levels of glucose in slices kept under physiologically appropriate oxygen levels as well as the lack of a response under low oxygen levels. The design and use of a slice anchor within our platform in conjunction with the fluorescent microscope housing allowed us to take high frequency repeat fluorescent images with minimal photobleaching, a high degree of image stability, and minimal human effort. The unique capabilities and dynamic environment of the SliceChip platform provides an opportunity to study a variety of tissue slices beyond the pancreatic tissues studied here. This platform is a versatile tool for the study of ex vivo tissues.Conflicts of interest
The authors have no conflicts of interests to disclose.Acknowledgements
We acknowledge the individuals and families of those who donated their organs for their invaluable contribution to science. We would also like to thank Kaycie Alver for invaluable grammatical guidance. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Number F31DK132916 (CA), 1R01DK130846-01 (JD-B/RP/AA), U01DK120393 (JD-B/RP), and UH3DK122638 (AA).References
- J. S. Kaddis, B. J. Olack, J. Sowinski, J. Cravens, J. L. Contreras and J. C. Niland, JAMA, 2009, 301, 1580–1587 CrossRef CAS PubMed.
- N. J. Hart and A. C. Powers, Diabetologia, 2019, 62, 212–222 CrossRef CAS PubMed.
- T. P. Foster, B. Bruggeman, M. Campbell-Thompson, M. A. Atkinson, M. J. Haller and D. A. Schatz, Endocr. Pract., 2020, 26, 1505–1513 CrossRef PubMed.
- H. R. Prasanna Kumar, H. B. Gowdappa, T. Hosmani and T. Urs, Indian J. Endocrinol. Metab., 2018, 22, 121–125 CrossRef CAS PubMed.
- A. Marciniak, C. Selck, B. Friedrich and S. Speier, PLoS One, 2013, 8, e78706 CrossRef CAS PubMed.
- A. Marciniak, C. M. Cohrs, V. Tsata, J. A. Chouinard, C. Selck, J. Stertmann, S. Reichelt, T. Rose, F. Ehehalt, J. Weitz, M. Solimena, M. Slak Rupnik and S. Speier, Nat. Protoc., 2014, 9, 2809–2822 CrossRef CAS PubMed.
- M. M. F. Qadir, S. Alvarez-Cubela, J. Weitz, J. K. Panzer, D. Klein, Y. Moreno-Hernandez, S. Cechin, A. Tamayo, J. Almaca, H. Hiller, M. Beery, I. Kusmartseva, M. Atkinson, S. Speier, C. Ricordi, A. Pugliese, A. Caicedo, C. A. Fraker, R. L. Pastori and J. Dominguez-Bendala, Nat. Commun., 2020, 11, 3265 CrossRef CAS PubMed.
- C. A. Fraker, S. Alvarez, P. Papadopoulos, J. Giraldo, W. Gu, C. Ricordi, L. Inverardi and J. Dominguez-Bendala, Stem Cells, 2007, 25, 3155–3164 CrossRef CAS PubMed.
- T. Liang, S. Dolai, L. Xie, E. Winter, A. I. Orabi, N. Karimian, L. I. Cosen-Binker, Y. C. Huang, P. Thorn, M. S. Cattral and H. Y. Gaisano, J. Biol. Chem., 2017, 292, 5957–5969 CrossRef CAS PubMed.
- J. K. Panzer, H. Hiller, C. M. Cohrs, J. Almaca, S. J. Enos, M. Beery, S. Cechin, D. M. Drotar, J. R. Weitz, J. Santini, M. K. Huber, M. Muhammad Fahd Qadir, R. L. Pastori, J. Dominguez-Bendala, E. A. Phelps, M. A. Atkinson, A. Pugliese, A. Caicedo, I. Kusmartseva and S. Speier, JCI Insight, 2020, 5(8), e134525 CrossRef PubMed.
- D. E. Ingber, Nat. Rev. Genet., 2022, 23, 467–491 CrossRef CAS PubMed.
- S. N. Patel, M. Ishahak, D. Chaimov, A. Velraj, D. LaShoto, D. W. Hagan, P. Buchwald, E. A. Phelps, A. Agarwal and C. L. Stabler, Sci. Adv., 2021, 7(7), eaba5515 CrossRef CAS PubMed.
- B. J. van Meer, H. de Vries, K. S. A. Firth, J. van Weerd, L. G. J. Tertoolen, H. B. J. Karperien, P. Jonkheijm, C. Denning, A. P. IJzerman and C. L. Mummery, Biochem. Biophys. Res. Commun., 2017, 482, 323–328 CrossRef CAS PubMed.
- L. A. Low and D. A. Tagle, Exp. Biol. Med., 2017, 242, 1573–1578 CrossRef CAS PubMed.
- V. Palacio-Castaneda, N. Velthuijs, S. Le Gac and W. P. R. Verdurmen, Lab Chip, 2022, 22, 1068–1092 RSC.
- P. Buchwald, Theor. Biol. Med. Modell., 2009, 6, 5 CrossRef PubMed.
- C. A. Fraker, S. Cechin, S. Alvarez-Cubela, F. Echeverri, A. Bernal, R. Poo, C. Ricordi, L. Inverardi and J. Dominguez-Bendala, Cell Transplant., 2013, 22, 1723–1733 Search PubMed.
- K. Szwajka, J. Zielinska-Szwajka and T. Trzepiecinski, Materials, 2023, 16, 3460 CrossRef CAS PubMed.
- G. Da Silva Xavier, J. Clin. Med., 2018, 7(3), 54 CrossRef PubMed.
- P. Buchwald, X. Wang, A. Khan, A. Bernal, C. Fraker, L. Inverardi and C. Ricordi, Cell Transplant., 2009, 18, 1223–1235 Search PubMed.
- G. Lenguito, D. Chaimov, J. R. Weitz, R. Rodriguez-Diaz, S. A. Rawal, A. Tamayo-Garcia, A. Caicedo, C. L. Stabler, P. Buchwald and A. Agarwal, Lab Chip, 2017, 17, 772–781 RSC.
- M. W. Tibbitt and K. S. Anseth, Biotechnol. Bioeng., 2009, 103, 655–663 CrossRef CAS PubMed.
- A. Galli, M. Algerta, P. Marciani, C. Schulte, C. Lenardi, P. Milani, E. Maffioli, G. Tedeschi and C. Perego, Cells, 2020, 9(2), 413 CrossRef PubMed.
- M. Klak, P. Kowalska, T. Dobrzanski, G. Tymicki, P. Cywoniuk, M. Gomolka, K. Kosowska, T. Bryniarski, A. Berman, A. Dobrzyn, W. Sadowski, B. Gorecki and M. Wszola, Micromachines, 2021, 12(3), 304 CrossRef PubMed.
- R. Viadero, in Encyclopedia of Water, 2019, pp. 1–7, DOI:10.1002/9781119300762.wsts0096.
- H. Komatsu, C. Cook, C. H. Wang, L. Medrano, H. Lin, F. Kandeel, Y. C. Tai and Y. Mullen, PLoS One, 2017, 12, e0183780 CrossRef PubMed.
- H. Komatsu, F. Kandeel and Y. Mullen, Pancreas, 2018, 47, 533–543 CrossRef CAS PubMed.
- T. M. Suszynski, K. R. Mueller, A. C. Gruessner and K. K. Papas, Transplant. Proc., 2014, 46, 1960–1962 CrossRef CAS PubMed.
- N. B. Vicente, J. E. D. Zamboni, J. F. Adur, E. V. Paravani and V. H. Casco, J. Phys.: Conf. Ser., 2007, 90, 012068 CrossRef.
- M. Ishahak, J. Hill, Q. Amin, L. Wubker, A. Hernandez, A. Mitrofanova, A. Sloan, A. Fornoni and A. Agarwal, Front. Bioeng. Biotechnol., 2020, 8, 581163 CrossRef PubMed.
- D. McDuffie, C. G. Alver, B. Suthar, M. Helm, D. Oliver, R. A. Burgess, D. Barr, E. Thomas and A. Agarwal, Lab Chip, 2023, 23, 3106–3119 RSC.
- L. E. Borm, A. Mossi Albiach, C. C. A. Mannens, J. Janusauskas, C. Ozgun, D. Fernandez-Garcia, R. Hodge, F. Castillo, C. R. H. Hedin, E. J. Villablanca, P. Uhlen, E. S. Lein, S. Codeluppi and S. Linnarsson, Nat. Biotechnol., 2023, 41, 222–231 CAS.
- W. H. Li, Semin. Cell Dev. Biol., 2020, 103, 14–19 CrossRef CAS PubMed.
- M. Doke, S. Álvarez-Cubela, D. Klein, I. Altilio, J. Schulz, L. Gonçalves, J. Almaça, C. A. Fraker, A. Pugliese, C. Ricordi, M. M. F. Qadir, R. L. Pastori and J. Domínguez-Bendala, Cell Metab., 2023,(11), 1944–1960 CrossRef CAS PubMed.
- F. C. Garcia-Garcia, P. L. Candarlioglu, J. D. Porter, D. E. Davies, E. J. Swindle and H. Morgan, Organs-on-a-Chip, 2022, 4, 100020 CrossRef CAS.
- P. Herreros, S. Tapia-Gonzalez, L. Sanchez-Olivares, M. F. Laguna Heras and M. Holgado, Int. J. Mol. Sci., 2022, 23(5), 2549 CrossRef CAS PubMed.
- N. Rafiei, M. G. Moghadam, A. Au, R. Regeenes, S. Chidambaram, T. Liang, Y. Wang, C. M. Yip, H. Gaisano and J. V. Rocheleau, Biofabrication, 2022, 14, 041001 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00850a |
| This journal is © The Royal Society of Chemistry 2024 |