Open Access Article
Open Access ArticleReversibly-bonded microfluidic devices for stable cell culture and rapid, gentle cell extraction†
Xiaohan
Feng
 a,
Zehaoyu
Wu
a,
Zehaoyu
Wu
 b,
Lily Kwan Wai
Cheng
b,
Yang
Xiang
c,
Ryohichi
Sugimura
c,
Xuyan
Lin
b,
Lily Kwan Wai
Cheng
b,
Yang
Xiang
c,
Ryohichi
Sugimura
c,
Xuyan
Lin
 *bd and
Angela Ruohao
Wu
*bd and
Angela Ruohao
Wu
 *abe
*abe
aDepartment of Chemical and Biological Engineering, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong SAR. E-mail: angelawu@ust.hk
bDivision of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong SAR. E-mail: xlinas@connect.ust.hk
cLi Ka Shing Faculty of Medicine, School of Biomedical Sciences, University of Hong Kong, Hong Kong SAR, China
dCenter for Engineering Material and Reliability, Guangzhou HKUST Fok Ying Tung Research Institute, Guangzhou, China
eState Key Laboratory of Molecular Neuroscience, The Hong Kong University of Science and Technology, Hong Kong SAR
First published on 21st June 2024
Abstract
Microfluidic chips have emerged as significant tools in cell culture due to their capacity for supporting cells to adopt more physiologically relevant morphologies in 3D compared with traditional cell culture in 2D. Currently, irreversible bonding methods, where chips cannot be detached from their substrates without destroying the structure, are commonly used in fabrication, making it challenging to conduct further analysis on cells that have been cultured on-chip. Although some reversible bonding techniques have been developed, they are either restricted to certain materials such as glass, or require complex processing procedures. Here, we demonstrate a simple and reversible polydimethylsiloxane (PDMS)–polystyrene (PS) bonding technique that allows devices to withstand extended operations while pressurized, and supports long-term stable cell cultures. More importantly, it allows rapid and gentle live cell extraction for downstream manipulation and characterization after long-term on-chip culturing, and even further subculturing. Our new approach could greatly facilitate microfluidic chip-based cell and tissue cultures, overcoming current analytical limitations and opening up new avenues for downstream uses of on-chip cultures, including 3D-engineered tissue structures for biomedical applications.
Introduction
Microfluidic devices have seen extensive utilization in the domain of cellular biology.1,2 For instance, 3D tumor models, cell-interaction models, and organ-on-chips have been successfully constructed using microfluidics for elucidating biological mechanisms and drug discovery.3–5 The conventional polydimethylsiloxane (PDMS) chip is usually irreversibly bonded to the substrate and therefore cannot be “opened” unless the device is destroyed.6 This makes such devices suitable for long-term cell culture, but poses difficulties for efficient extraction of a substantial number of viable cells from the device for downstream manipulation, even with the aid of trypsin. This obstacle therefore restricts the approaches to analyze the cells in microfluidic devices, confining them to imaging, bulk molecular analysis, or simple on-chip staining assays.Achieving reversible bonding between PDMS and the substrate is a possible solution to this limitation, and efforts have been made in this direction. While the tight irreversible bonding of devices can be readily achieved by plasma treatment and chemical modification such as 1% APTES treatment,7,8 developing stable, long-term methods for reversible bonding still remains challenging. Existing methods for reversible bonding work on a limited range of device–substrate material pairs, or require complex processing procedures. For instance, the simplest way to create reversible bonds is to increase the thickness of PDMS and bonding to glass without plasma treatment,9 but this method is less robust and suffers from low-pressure tolerance, with a high risk of leakage under pressurized or prolonged usage, which makes it non-ideal for long-term cell culture or scenarios that require pressurized pumping and control. More recently, a glass-based device manufacturing approach was proposed for achieving reversible bonding and gentle cell extraction.10 The reversible bonds in this approach are created by water dehydration between two glass slips with high cleanliness, therefore requiring neutral detergents and continued exertions of external force for bond formation. Also, this method is not compatible with PDMS-based devices, which are widely used for cell-based studies. The demand for a versatile and simple method of reversible bonding that is applicable to commonly used device-substrate materials thus still persists.
Another common approach to create reversible bonds is to provide a sacrificial layer within devices, which can be designed with multiple materials, such as poly(methyl methacrylate) (PMMA), polycarbonate (PC), polystyrene (PS), and polyethylene terephthalate (PET).11–13 For example, Thompson et al. used adhesive tape for creating reversible bonds within complex devices,12 but the low manufacturing throughput and the yet-determined biocompatibility limit the applicability of this method. Similarly, a silicone-based soft skin adhesive could be mixed with PDMS to create a sacrificial layer between the PDMS device and its substrate for long-term cell culture,13 but the low-adhesion property of this adhesive may influence cell attachment on devices, thereby affecting cellular biomechanics or functions on-chip. Other strategies, such as wax bonding,14 changing substrates,15 clamping,16,17 reducing curing agents,18 vacuum bonding,19 and sandwich bonding,20 all contribute to the development of reversible bonding devices for scientific research and applications. However, the scaled production of these methods may be limited due to their intricate manufacturing process or the requirement of implementing external assistance. Moreover, this poses increased difficulty in implementing the aforementioned approaches to create complex designs for cellular-related applications, let alone that no efficient live cell extraction has been reported using these methods.
In this study, we present a new approach for fabricating reversibly-bonded microfluidic devices (referred to hereafter as “reversible devices” for convenience; similarly, hereafter “irreversible devices” refers to conventional PDMS-glass devices whose substrates cannot be manually separated from the cover slabs), which is achieved by treating thermoplastics (PS focused) with a low-concentration (≤0.5%) (3-aminopropyl)triethoxysilane (APTES) solution. The low-concentration APTES solution introduces a small proportion of amine groups for covalent bonding, and it roughens the PS surface thereby enhancing van der Waals forces, overall resulting in a stable and reversible bonding between PS and PDMS (Fig. 1). The procedure requires no other equipment besides a benchtop plasma cleaner and can be performed outside the cleanroom, making it accessible to most laboratories that manufacture PDMS devices and compatible with most PDMS-based device fabrication workflows. We demonstrate the biocompatibility of this type of reversibly bonded device by performing long-term and stable cell culture on-chip with high cell viability, as well as showing tumor spheroid formation on-chip as an additional application. Moreover, the biocompatibility of this surface treatment is illustrated through vascular cell culture and vascular network formation experiments on-chip. Next, cells were retrieved from the chip by rapid hand-peeling of the PDMS slab and the high efficiency of live cell extraction was verified by cell counting, cell recovery experiments, and flow cytometry. Finally, we used a device design with multiple compartments to co-culture different cell types, showing the capability of the reversibly bonded device to accommodate 2D and 3D cell culture, which further extends the versatility of this bonding method. Overall, this reversible bonding method offers a robust and scalable chip fabrication process with high biocompatibility, and allows for gentle cell extraction from microfluidic devices. The viable cells extracted from the environment as engineered within reversible devices can further contribute to the understanding of cellular behaviors and the mechanisms behind.
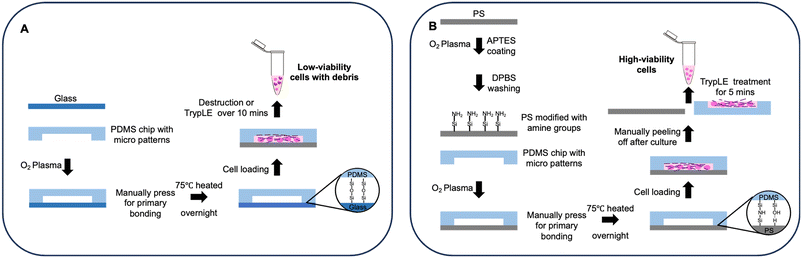 | ||
| Fig. 1 A: Process flow of conventional irreversible bonding devices and cell extraction; B: process flow of reversible bonding devices and rapid cell extraction. | ||
Experimental
APTES treatment on the substrate
Thermoplastics including a cyclic olefin copolymer (COC), PC, PMMA, and PS (Petri dish, #150318, ThermoFisher, USA) were used as substrates for bonding with PDMS, and were treated with oxygen plasma discharge in a 30 watt plasma cleaner (PDC-002 with 6′′ diameter × 6.5′′ length Pyrex chamber containing ambient air, Harrick Plasma, New York, USA) for 3 minutes, followed by treating with APTES ethanol dilutions for 10 minutes at ambient temperature in the chemical fume hood. Specifically, the concentration of APTES was set to 0.1% for cell culture and retrieval. The substrate was subsequently washed with Dulbecco's phosphate-buffered saline (DPBS, 14190235, ThermoFisher) or pure water until the APTES residue was removed, both of which were tested to depict the same performance (Fig. S1†). To completely remove the residual liquid, the cleaned substrates were placed on a hotplate at 75 °C for primary drying for 1 to 2 hours before fully drying out using a nitrogen gas spray gun.Microfluidic device design and fabrication
The microfluidic devices used in this study were made of PDMS based on the modification of a previous study (Fig. S2†).21 Soft lithography and replica molding were used to fabricate the device. Briefly, the lithography mask was designed using AutoCAD. A silicon wafer was then patterned with SU8-2050 photoresists (Microchem Corp., Westborough, USA) using standard photolithography techniques (ABM, USA). PDMS (Sylgard 184 Silicone Elastomer, Dow Corning, USA) was mixed at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of the elastomer to curing agent and then poured over the silicon mold. After degassing, the PDMS was cured on the mold for at least 2 hours at 80 °C for complete curing before removal from the master.22,23 The inlets and outlets of the PDMS chambers were punched using a PDMS puncher prior to substrate bonding. The PDMS slab to be bonded was cleaned with IPA, distilled water, and Scotch tape, and dried with an air spray gun. To form the reversible bonding between the PDMS slab and the treated substrate, these two components were both treated with oxygen plasma for 3 minutes, followed by immediately placing the PDMS slab surface in contact with the surface of the thermoplastic, and ensuring good sealing between the two surfaces by compression. Finally, the bonded device was incubated on a hotplate at 75 °C for at least 2 hours to enhance bonding, overnight incubation is also recommended.24 Over 75 °C heating may cause heat distortion of the PS substrate. Devices are sterilized by UV irradiation for at least 30 minutes before cell culture.
1 ratio of the elastomer to curing agent and then poured over the silicon mold. After degassing, the PDMS was cured on the mold for at least 2 hours at 80 °C for complete curing before removal from the master.22,23 The inlets and outlets of the PDMS chambers were punched using a PDMS puncher prior to substrate bonding. The PDMS slab to be bonded was cleaned with IPA, distilled water, and Scotch tape, and dried with an air spray gun. To form the reversible bonding between the PDMS slab and the treated substrate, these two components were both treated with oxygen plasma for 3 minutes, followed by immediately placing the PDMS slab surface in contact with the surface of the thermoplastic, and ensuring good sealing between the two surfaces by compression. Finally, the bonded device was incubated on a hotplate at 75 °C for at least 2 hours to enhance bonding, overnight incubation is also recommended.24 Over 75 °C heating may cause heat distortion of the PS substrate. Devices are sterilized by UV irradiation for at least 30 minutes before cell culture.
Surface characterization
The PS substrate was cut into small pieces, coated, mounted, and then subject to materials characterization of its functionalized surface by surface electron microscopy (SEM) (JSM-6320F, JEOL, JP), X-ray photoelectron spectroscopy (XPS) (PHI 5000 Versaprobe III, ULVAC-PHI, Japan), and atomic force microscopy (AFM) (Dimension ICON, Bruker, MA, USA). XPS analyses were conducted using a machine equipped with an aluminum X-ray source (mono-gun, 1486.6 eV) with a pass energy of 40 eV. The binding energy of C 1s (284.5 eV) was used as the reference. The resolution for the measurement of the binding energy was approximately 0.1 eV.Cell culture
For experiments, we used the U87 human glioblastoma cell line to test cell viability and tumor spheroid formation, and the HepG2 human hepatocellular cell line for cell extraction and separation, which were both cultured in Dulbecco's modified Eagle medium (DMEM, 11965-092, ThermoFisher) with 10% fetal bovine serum (FBS, A4766801, ThermoFisher).Human umbilical vein endothelial cells (ECs, CC-2519, Lonza) were used for the vascular network formation experiments, and were cultured in endothelial microvasculature growth medium (EGM-2MV BulletKit™, CC-3202, Lonza). Normal human lung fibroblasts (FBs) (NHLFs, CC-2512, Lonza) were cultured in fibroblast growth medium (FGM-2 BulletKit™, CC-3132, Lonza). To support vasculature formation in the device, FBs were transferred to the EGM-2 MV medium and cultured for at least 2 passages before proceeding on-chip co-culture with HUVECs.
Monocyte cell line THP-1 was used in cell retrieval experiments and cultured in RMPI-1640 (11875119, ThermoFisher) with 10% FBS. Cells were cultured on Petri dishes in a humidified incubator at 37 °C and 5% CO2 and grown up to 80% confluency for cell seeding experiments.
Gel preparation and cell seeding
Cell seeding gel was prepared following the established protocol for 3D cell culture.21 To generate the fibrin gel, the fibrinogen and the thrombin solutions were prepared separately. Briefly, the fibrinogen solution was prepared by dissolving 15 mg of bovine fibrinogen (F8630-1G, Sigma-Aldrich) in 2.5 ml of DPBS in a 37 °C water bath for 1 hour, followed by sterilization through a 0.22 μm filter. The thrombin stock solution was prepared by dissolving thrombin (T9549, Sigma-Aldrich) with 0.1% w/v BSA solution (B14, ThermoFisher) into a concentration of 100 U ml−1 and stored at −20 °C. Before use, the thrombin solution was diluted into 4 U ml−1 by adding the culture medium. For cell seeding, U87s, ECs, FBs, and HepG2s were trypsinized using TrypLE (#12605028, ThermoFisher) for 5 minutes, neutralized with culture medium, and then centrifuged for 3 minutes at 300 g. After aspirating the medium off, the cell pellets were resuspended with the diluted thrombin solution, which was then quickly mixed with the filtered fibrinogen solution. The cell-laden gel mixtures (6 million cells per ml of U87s for cell viability experiments; 3 million cells per ml of ECs and 1.5 million cells per ml of FBs for forming vascular networks on-chip; 3 million cells per ml of HepG2s for cell extraction and separation experiments) were then gently introduced into individual channels to polymerize for 15 minutes at room temperature within a humidified chamber. Finally, the corresponding channels were filled with cell culture medium by pipetting as the gel mixtures cross-linked. All the devices were incubated in a humidified incubator at 37 °C and 5% CO2, and the cell culture medium was removed and replenished every 24 hours.Cell viability calculation
Live and dead cells were distinctly stained using the LIVE/DEAD Cell Imaging Kit (488/570) (#R37601, Thermofisher, USA) for confocal microscopy. The subsequent calculation of cell viability was based on the formula below, incorporating the counts of live and dead cells as determined using ImageJ. | (1) |
Cell extraction
The microfluidic chip containing cells was carefully taken out from the incubator for cell extraction. For the irreversible chip, approximately 400 μL of TrypLE was added to the chip surface and the channel near to cells (Fig. 1A). For the reversible chip, approximately 200 μL of TrypLE was added to both the substrate and the PDMS chip after the PDMS slab was manually peeled off from the underlying substrate (Fig. 1B). An additional 200 μL of culture medium was then added to the chip after incubating at 37 °C, and the neutralized cell suspension was subsequently collected (Fig. 1A and B). The live cell counting was achieved using Countess II (#AMQAX1000, Thermofisher, USA) after adding trypan blue (#15250061, ThermoFisher, USA) to the cell suspension. For subculturing, the different extracted cell suspensions were transferred into two 60 mm Petri dishes containing 2 ml of DMEM medium respectively, and subsequently incubated for six days at 37 °C and 5% CO2 before imaging.To assess the efficacy of cell retrieval post-culture on the chip, we used ‘retrieval rates’ to quantify the proportion of cells retained after detachment of the PDMS slab. The retrieval rate of a given channel can be calculated by the following equation:
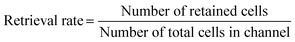 | (2) |
For instance, in calculating the retrieval rate of the 3D culture channel with the fibrin gel, the “number of retained cells” refers to the cells remaining attached to the PDMS slab after detachment, while the “number of total cells in the channel” refers to the total cells within the channel before the detachment of the slab and substrate. For calculating the retrieval rate of the 2D culture channel without fibrin gel, the “number of retained cells” refers to the cells adhering to the PS substrate after detachment, while the denominator is the same as above.
Burst pressure test
The bond strength of the interface was measured via a burst pressure test for PDMS to glass substrates (irreversible devices) and to APTES-treated PS dishes (reversible devices) after plasma treatment. For each test, the PDMS device (2 mm length, 1 mm width, and 50 μm height) was pressurized incrementally until the burst between PDMS and the substrate occurred. The burst pressure test setup consisted of a closed system in which a pneumatic pump with a digital manometer and the microfluidic chamber were interconnected using tubing. Measurements were conducted after the pressure reached equilibrium, and the final stable pressure prior to any PDMS–substrate burst was recorded respectively.Results and discussion
Potential aspects correlated with reversibility
Here, we define the “reversible bonding” between PDMS and substrates as their capacity to restore their pre-bonded stage, which requires two characteristics: the capability to re-bond together and to separate without significant deformations. The reversibility of our devices was investigated in the following five aspects: the nature of the substrates, the concentrations of APTES treatment, exposure to water immersion, the number of bond-detach events, and thermal aging of PDMS.Firstly, we explored the effect of different substrate and APTES concentrations on device reversibility (Fig. S3 and S4†). We have tested thermoplastics that are commonly applied in the microfluidics field, such as COC, PC, PMMA, and PS. The devices fabricated based on the listed four materials all demonstrated reversibility, as PDMS can be easily peeled off. The readouts of their burst pressure indicated a relatively limited number of chemical bonds contributing to their bonding with PDMS. Considering PS is the most commonly utilized in cell culture, it was thus chosen as the model material to expand the characterization and assessment.
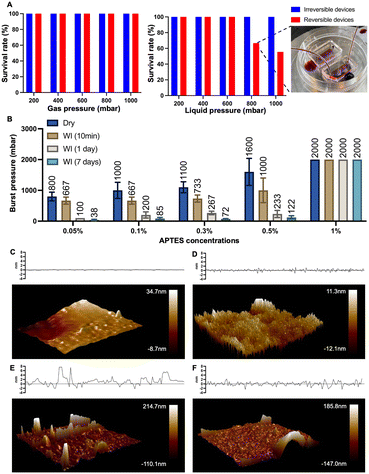
The bond formation was facilitated by APTES treatment at different concentrations. As we examined the APTES concentrations at 0.05%, 0.1%, 0.3%, 0.5%, and 1%, respectively, devices produced based on concentrations other than 1% demonstrated their reversibility by relatively low burst pressure resistance and negligible deformation after separation (Fig. 2 and S4†). At the same time, we observed that the resistance to burst increased as the concentration of APTES treatment increased (Fig. 2), which validates our philosophy of design that reducing the number of chemical bonds by decreasing the APTES concentration could thus lead to the reversible bonding between PDMS and the thermoplastic. Meanwhile, the effect of water immersion on reversible bonding was investigated, and will be discussed in detail through the change of surface morphology.
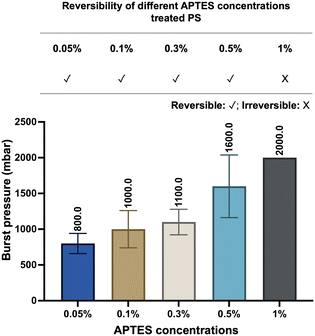 | ||
| Fig. 2 Reversibility and burst pressure of devices based on different concentrations of APTES-treated PS. The burst resistance of the devices increased with higher APTES concentrations (n = 6). | ||
A repetitive bond-detach event was subsequently tested to assess its effect on the reversibility by burst pressure tests (Fig. S5†). As the number of repetitions increases, the burst pressure resistance of the devices escalates, indicating not only an increase in the number of chemical bonds but also weakened reversibility. After three repetitions, the average value of the burst pressure of the device could reach 2000 mbar, which would be readily regarded as an irreversible device, exemplifying that the reversibility of our technique will remain until three bond-detach events. Still, given the main purpose of the technique is the application for cell retrieval after on-chip culture, multiple repetitions of bonding and detaching would rarely be encountered in empirical practice.
Since the thermal aging of PDMS has been demonstrated to influence the mechanical and chemical properties of PDMS,25,26 it is speculated to also affect the bonding of PDMS to the substrate. A straightforward experiment was conducted to compare the aged and non-aged PDMS: the aged PDMS was cured on a hotplate at 80 °C for 7 days, while the non-aged PDMS was cured for 2 hours at the same temperature. Both types of PDMS were then used to fabricate devices. The burst resistance of devices made with the aged PDMS showed no significant difference compared to those made with non-aged PDMS (Fig. S6†), revealing that the effect of thermal aging was negligible in this study.
In the following sections, with regard to the balance between pressure resistance and biocompatibility, we have selected a concentration of 0.1% for further investigation to provide a detailed account of the formation of chemical bonds and the feasibility of cell retrieval from the device.
Reduced APTES concentration facilitates reversible bonding between PS and PDMS
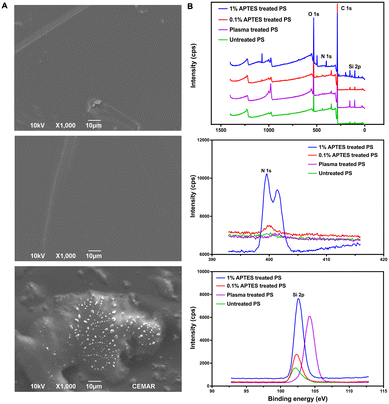
Oxygen plasma treatment is a typical and conventional bonding method for PDMS-based devices, enabling the irreversible bonding of PDMS-glass chips by activating the surface siloxane bonds of PDMS. Additionally, oxygen plasma followed by chemical treatment was also applied to other materials to enhance the bonding strength of PDMS-based devices.27 For instance, plasma-activated PDMS can bond to a PS substrate with a weak bonding strength of around 120 mbar. However, irreversible bonding can be achieved by treating the PS substrate with 1% APTES solution after oxygen plasma treatment.28,29To create a reversibly bonded device suitable for long-term cell culture and cell extraction, we used a 0.1% APTES ethanol solution to treat PS. This treatment resulted in the reversible bonding between the two surfaces, enabling long-term cell culture in the PDMS device, with the additional feature that the bonded surfaces can be easily separated by hand-peeling. SEM analysis was utilized to explore the bonding mechanism between PDMS and 0.1% v/v APTES treated-PS. The analysis confirmed the anticipated adhesion, which is shown in Fig. 3A. Fig. 3A top shows the untreated PS surface before the APTES treatment, and Fig. 3A middle displays the PS surface after being treated with APTES. According to SEM images, the PS surface maintained a smooth surface regardless of the APTES treatment, suggesting that there was no significant morphology change on the PS surface after oxygen plasma and low-concentration APTES treatment (Fig. 3A top and middle images). After peeling the PDMS slab off from the PS substrate, a few silicon residues along with partial deformation of the substrate were observed (Fig. 3A bottom images). These observations suggest the existence of covalent bonds between the PDMS and the treated PS, which have been induced by oxygen plasma and APTES treatment. The delamination-induced substrate deformation suggests the occurrence of chemical bonding between the PDMS and the treated PS substrate, rather than a simple stacking of the two materials. This bonding strategy evidently differs from the conventional PDMS-glass or APTES-treated irreversible chips, as there are only small silicone fragments remaining on the substrate after peeling, indicating that the limited number of chemical bonds between PDMS and treated PS results in the desired stable and reversible bonding for this device.
The effects of the 0.1% v/v APTES solution treatment are elucidated through the XPS survey spectrum, nitrogen (N 1s) fine spectrum, and silicon (Si 2p) fine spectrum, as depicted in Fig. 3B. By comparing the survey spectrum of the PS substrate treated with and without APTES, additional peaks at around 100 eV and 400 eV were found in the spectrum of APTES-treated PS, indicating the presence of additional chemical groups at the PS surface after treating with APTES solution (Fig. 3B top image). In the N 1s spectra (Fig. 3B middle image), peaks appeared at 400 eV for 1% and 0.1% APTES-treated PS surfaces, while the PS surfaces treated without APTES showed no peak. This indicates the successful incorporation of amino silane onto the PS surface. The amino functional groups form N–O bonds with the PDMS slab after plasma treatment, contributing to the bonding strength.30 The N 1s peak intensity of the PS surface treated with 1% APTES is much higher than that of the one treated with 0.1% APTES which showed a small peak at 400 eV. This suggests that the amino functional groups on the 0.1% APTES treated PS surface were fewer compared to the one treated with 1% APTES, resulting in a lower bonding strength that allowed it to be separated from the PDMS slab.
In the Si 2p spectra (Fig. 3B bottom image), the PS surfaces treated with APTES and the untreated PS surfaces showed peaks at 102.5 eV, indicating the presence of Si–O/Si–O–C bonds formed on the surface. Additionally, the plasma-treated PS exhibited a new peak at 104.2 eV, suggesting the presence of SiO2 on the PS surface.31 The Six+ on the untreated PS surface may originate from the manufacturing process. Upon reacting with the interface oxygen during the oxygen plasma treatment, a new layer of SiO2 is formed.31 This reaction results in the disappearance of the peak at 102.5 eV from the spectra of the plasma-treated PS surface. By comparing the peak areas of the plasma-treated PS surface with those treated with both oxygen plasma and APTES, the presence of these two corresponding components indicates successful silanization. The peak intensities of 102.5 eV on PS surfaces treated with different concentrations of APTES treatment revealed varying numbers of Si–O/Si–O–C bonds on the surfaces, which led to the different number of silicon bonds with PDMS afterward. The intensities of both N 1s and Si 2p peaks for the PS surfaces treated with 0.1% APTES solution were significantly lower than those for PS treated with 1% APTES solution. This observation demonstrates that a limited number of covalent bonds on PS surface resulted from low-concentration APTES treatment. Moreover, this reduced capability for covalent bond formation is crucial for reversible bonding.
Surface morphology revealing that the key of reversible bonding is water-weakened bonds
Having illustrated the effect of low-concentration APTES treatment on the PS surface, we sought to quantify the bonding strength and further explore the underlying mechanism responsible for the bonding strength variation when a lower concentration of APTES is used.We conducted two different pressure tests between conventional irreversible devices and reversible chips (0.1% APTES-treated) to demonstrate the bonding strength of the latter. In gas pressure tests, both types of chips showed comparable performance: both can withstand a gas pressure of up to 1000 mbar without any bursting (Fig. 4A). In fluid pressure tests, the pressure resistance threshold of reversible devices is 600 mbar, and the survival rate for the reversible devices was 66.7% at 800 mbar. Here the survival rate is defined as the proportion of chips that operate under this pressure without failure for at least half an hour. Upon increasing the liquid pressure to 1000 mbar, the survival rate for reversible bonding chips dropped to 55.6%. The reduction in pass rates under liquid pressure is likely due to the infiltration of water molecules to the PDMS–PS interface, which weakens the bonding strength. Thus, the water immersion experiments on the APTES-treated devices were performed to further quantify this reduction in burst resistance. Except for the 1% APTES-treated group, a notable reduction in burst resistance after water immersion was observed (Fig. 4B). All the pressures decreased to approximately 200 mbar after one day and to 100 mbar after seven days. Despite this reduction, the devices preserved adequate structural integrity to withstand the stress associated with medium changes during cell culture.
To explore the mechanism for this change in the bonding strength after the device has been used with aqueous reagents, we used AFM analysis to check for morphology transformations on surfaces under different conditions (Fig. 4C–F). A relatively flat surface profile was exhibited by the untreated PS surface (Fig. 4C). After treating the PS surface with APTES and prior to bonding, we observed a significant increase in PS surface roughness. The increase in surface roughness elevates the substrate's surface free energy, thereby enhancing its bonding characteristics when dry. We also noted that manual delamination of PDMS without prior water treatment led to a substantial amount of silicon residue on the PS surface (Fig. 4E). We performed the measurement again with APTES-treated PS that was bonded, treated with water to mimic the device contact with aqueous reagents such as buffer and media, and subsequently manually delaminated. This time the PS surface had few remaining silicon residues after delamination compared with the sample that did not undergo water treatment, and also had a reduced surface roughness (Fig. 3F). These results indicate that contact with water disrupts the bonding between the PDMS slab and PS substrate, thus easing the PDMS peeling process after cell culture. Overall, this water-weakened bonding strength is the basis of the reversible technique and makes hand peeling feasible.
Reversible devices demonstrating high biocompatibility by culturing multi-types of cells
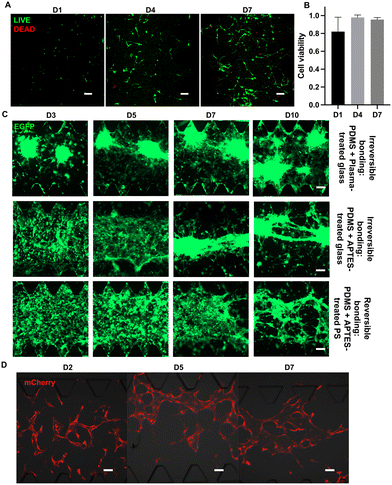
Reversible bonding is essential for retrieving cells from microfluidic devices for further analysis.29 Another basic requirement of reversible devices is biocompatibility, so that cells can live, propagate, and maintain their normal functions.After testing the physical performance of the reversible device, we next characterized the feasibility of this reversible device for cell culture. The U87 glioblastoma cell line was used to determine the biocompatibility of reversible chips and calculate cell viability. When seeded on the chip, U87 exhibited morphology characteristic of epithelial cells, mirroring the observed morphological attributes in conventional dish cultures.32 Notably, the cells developed to form spherical shapes over extended time, indicating their robust and sustained growth in the on-chip environment. On D1, D4, and D7, live/dead cell kits were applied to observe the growth status of U87 on the chip (Fig. 5A). At those selected time points, using confocal microscopy, we observed a large amount of green fluorescence indicating live cells, as well as a small amount of red fluorescence indicating dead cells. The calculated cell viabilities at selected time points are shown in Fig. 4B. The cell viability at D4 and D7 was nearly 100%, which indicates that the glioblastoma cells grew well on the reversible device. On D1, there was ∼80% on-chip cell viability due to cell damage during passaging.
Next, we tried to increase the seeding concentration of U87 cells and extend the on-chip culture period to more than 10 days. The U87 cells could form tumor spheroids and delay to form cell–cell connections between spheroids regardless of the bonding approach (Fig. 5C). The spheroid formation time on reversible devices is extended compared to that on irreversible devices, attributed to the differences in material properties such as stiffness and surface roughness of the different substrates or APTES treatment.33
Tumor cells are generally considered to be more robust to culture than other types of differentiated cells or primary cells. Hence, primary HUVECs and FBs were cultured in reversible devices to further assess the biocompatibility of this device with more delicate cell types. We seeded the HUVECs and FBs in different channels at their designated concentration. Notably, the HUVECs could form lumen-like structures after 5 days of on-chip culture (Fig. 5D), showing that reversible devices not only do not negatively affect the growth of HUVECs but also allow them to form functional lumen-like structures. In general, all seeded cells displayed expected growth rates on the reversible devices regardless of cell type, which validates the high biocompatibility of this reversible device.
Reversibly bonded devices enabling rapid and gentle cell extraction
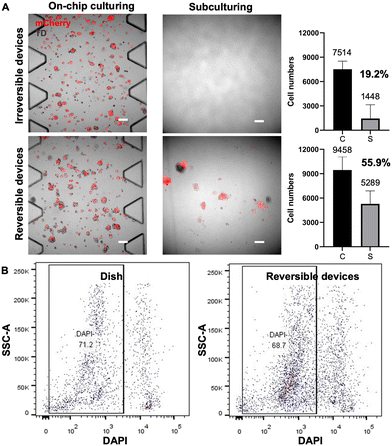
In addition to biocompatibility, the efficient and gentle extraction of cells from the device is also crucial for downstream cell profiling, such as FACS analysis, sorting and subculturing, as well as genomic sequencing. A gentle cell extraction process is necessary to minimize cell loss and avoid dramatically altering the cell state. We next demonstrated a simple method of retrieval of cells from this reversible device and assessed the damage to the cells during extraction.The HepG2-mCherry cells were seeded and cultured for 5 days on-chip. Before cell extraction, confocal images were taken of cells seeded and cultured on-chip, shown in the left part of Fig. 6A. After cell extraction, the retrieved cell suspensions were re-seeded onto a conventional culture dish for cell recovery, and confocal images were taken after six days of conventional cell culture on-dish. For the irreversible devices, trypsin was pushed into the device to release cells from the fibrin gel and flushed out for collection, whereas for the reversible devices, the PDMS was peeled off by hand, and then the cell-embedded side was subject to trypsinization. Peeling off the PDMS slab facilitates efficient access of trypsin to gel-embedded cells within the device, minimizing cellular damage during extraction and enabling comprehensive washing to maximize the collection of detached cells. Before cell extraction, fluorescent cells could be observed in both irreversible and reversible devices (Fig. 6A leftmost two panels). After collecting the cell suspensions from both devices, the cells were subcultured respectively for 6 days. No propagated cells recovered from the irreversible devices were observed on the culture dish, as shown by the microscope images of representative regions in Fig. 6A (upper right image). Presumably, because the retrieval process for the irreversible device was harsher and much less efficient, the cells retrieved were low in numbers and have lower viability. Consequently, no cells could be seen after 6 days of subculturing. In contrast, the cells recovered from the reversible devices showed fluorescence signals, indicating the successful subculturing of the retrieved cells (Fig. 6A, bottom right image).
Additionally, to determine the average number of live cells in retrieved suspensions, partial cell suspensions from both device types were stained with trypan blue. The resultant counts for live cells averaged 1448 ± 1698 for the irreversible devices and 5289 ± 1596 for the reversible devices, respectively (Fig. 6A, bar graphs). These results show that only 19.2% of cells were collected from the irreversible device by flushing and those retrieved cells were not capable of further propagation. In contrast, our reversible devices allowed for a significantly greater cell yield, with an extraction efficiency reaching 55.6%. These comparative results suggest that the cell suspensions retrieved from reversible devices yield a higher number of retrieved cells and ensure greater cell viability, offering advantages for subsequent experimental procedures.
Furthermore, we compared the damage caused by cell extraction from the reversible devices and normal culture dishes by DAPI staining flow cytometry. The viability of cells extracted from the dishes and reversible devices are similar, with live cell percentages recorded at 71.2% and 68.7%, respectively, as shown in Fig. 6B. The ability to extract an adequate number of cells from the PDMS microfluidic chips with satisfactory viability makes them suitable for downstream processes like single-cell sequencing.
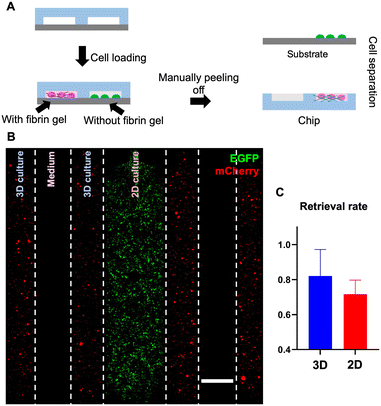
In addition to 3D cell culture where cells are embedded in matrix/gel, microfluidic devices are also often used for 2D cell culture.5,34 Due to the different conditions of 3D and 2D cell cultures, we wondered whether the cell retrieval rates from the device could differ depending on whether cells are embedded in gel or attached to the substrate surface.
To answer this question, we measured the cell retrieval rates for both scenarios using a multi-channel reversible device (Fig. 7A), in which 2 pairs of channels contain cells embedded in gel, interposed between each pair is a single channel allocated for the medium, and the channel in the center contains adherent cells cultured in 2D and attached to the bottom substrate. Specifically, in this 7-channel device, we introduced THP-1-EGFP cells into the center channel for 2D culture and placed HepG2-mCherry cells into the channels for 3D culture (Fig. 7B). During the culture, confocal images were taken to estimate the total number of cells in the channel; after 7 days of on-chip culture, we detached the PDMS slab from the PS substrate and retrieved cells adhering to the PDMS slab (embedded 3D culture scenario) and cells attached to the bottom substrate (2D culture scenario), respectively (Fig. S7†). Then, by counting the number of cells that were retrieved from each scenario, the retrieval rates for 3D and 2D culture can be calculated by respectively dividing over the total number of cells estimated while culturing on-chip (eqn (2)). For 3D culture (embedded), the retrieval rate was 82% and this rate was 72% for 2D culture (attached), indicating a high rate of retrieval for both cell culture scenarios (Fig. 7C).
Overall, this reversible bonding approach enabled streamlined on-chip cell extraction for both 3D and 2D microfluidic cell cultures.
Conclusions
In this paper, we developed an approach to achieve reversible PDMS–PS bonding by treating low-concentration APTES on PS, thus overcoming many limitations of live cell retrieval from PDMS-based microfluidic cell culture devices. This reversible device can withstand pressures exceeding 600 mbar in most cases, providing sufficient pressure resistance for practical use in many applications, and enabling long-term stable cell culture. We observed high cell viability for multiple cell types, including the glioblastoma cell line, hepatic cancer cell line, and endothelial cell line, which highlights the biocompatibility of these reversible microchips. Moreover, we demonstrated a platform for modeling the physiological tissue environment on-chip that allows rapid cell extraction, which is user-friendly and does not require specialized materials.This reversible bonding approach for microchips is an important addition to the microfluidics technology toolbox, enabling a wider range of on-chip cellular studies. The development of such a versatile and user-friendly approach for reversible bonding fabrication and efficient cell extraction could significantly impact biomedical applications of microfluidics, advancing further development in cell culture and organ-on-chip as well as the following studies such as drug discovery, disease modelling, and cellular interaction analysis.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization: X. F., X. L., A. R. W. Methodology: X. F., X. L., L. K. W. C., Z. W., Y. X., R. S. Data analysis: X. F., X. L. Supervision: X. L., A. R. W. Writing – original draft: X. F., X. L., A. R. W. Writing – review & editing: X. F., X. L., Z. W., A. R. W.Conflicts of interest
A. R. W., X. L., Z. W. and X. F. have filed a USPTO patent on the Reversibly-bonded Microfluidic Devices, application number 63/591113, filed 17 October 2023. All other authors declare that they have no competing interests.Acknowledgements
This study was funded by the Hong Kong Research Grants Council General Research Fund 16209820 (AW); the Center for Aging Science, The Hong Kong University of Science and Technology, Z1003 (AW); the Innovation and Technology Commission ITCPD/17-9 (AW). We would like to acknowledge the Nanosystem Fabrication Facility (CWB) of the HKUST for the device/system fabrication; we would like to acknowledge the technical support from SUSTech; SEM data were obtained using equipment maintained by the Center for Engineering Material and Reliability of Guangzhou HKUST Fok Ying Tung Research Institute, with assistance of Mr. Zhenyu Pan. For helpful discussion and suggestions, we thank all the members of the A. R. Wu research group for helpful discussions and administrative assistance during this project.References
- S. N. Bhatia and D. E. Ingber, Nat. Biotechnol., 2014, 32, 760–772 CrossRef CAS PubMed.
- S. Halldorsson, E. Lucumi, R. Gómez-Sjöberg and R. M. T. Fleming, Biosens. Bioelectron., 2015, 63, 218–231 CrossRef CAS PubMed.
- S. Lee, M. Chung, S.-R. Lee and N. L. Jeon, Biotechnol. Bioeng., 2020, 117, 748–762 CrossRef CAS PubMed.
- S. Jung, H. Jo, S. Hyung and N. L. Jeon, in Microfluidics and Biosensors in Cancer Research: Applications in Cancer Modeling and Theranostics, ed. D. Caballero, S. C. Kundu and R. L. Reis, Springer International Publishing, Cham, 2022, pp. 231–256, DOI:10.1007/978-3-031-04039-9_9.
- S. W. L. Lee, G. Adriani, E. Ceccarello, A. Pavesi, A. T. Tan, A. Bertoletti, R. D. Kamm and S. C. Wong, Front. Immunol., 2018, 9, 416 CrossRef PubMed.
- F. Akther, S. B. Yakob, N. T. Nguyen and H. T. Ta, Biosensors, 2020, 10, 182 CrossRef CAS PubMed.
- V. Sunkara, D.-K. Park, H. Hwang, R. Chantiwas, S. A. Soper and Y.-K. Cho, Lab Chip, 2011, 11, 962–965 RSC.
- S. Sonney, N. Shek and J. M. Moran-Mirabal, Biomicrofluidics, 2015, 9, 026501 CrossRef PubMed.
- J. Harris, H. Lee, B. Vahidi, C. Tu, D. Cribbs, C. Cotman and N. L. Jeon, J. Visualized Exp., 2007, 410, DOI:10.3791/410.
- S.-I. Funano, N. Ota and Y. Tanaka, Lab Chip, 2021, 21, 2244–2254 RSC.
- S. R. A. Kratz, C. Eilenberger, P. Schuller, B. Bachmann, S. Spitz, P. Ertl and M. Rothbauer, Sci. Rep., 2019, 9, 9287 CrossRef CAS PubMed.
- C. S. Thompson and A. R. Abate, Lab Chip, 2013, 13, 632–635 RSC.
- M. Chu, T. T. Nguyen, E. K. Lee, J. L. Morival and M. Khine, Lab Chip, 2017, 17, 267–273 RSC.
- X. Gong, X. Yi, K. Xiao, S. Li, R. Kodzius, J. Qin and W. Wen, Lab Chip, 2010, 10, 2622–2627 RSC.
- B. Jiang, A. White, W. Ou, S. Van Belleghem, S. Stewart, J. G. Shamul, S. O. Rahaman, J. P. Fisher and X. He, Bioact. Mater., 2022, 16, 346–358 CAS.
- C. Pérez-Sosa, A. B. Peñaherrera-Pazmiño, G. Rosero, N. Bourguignon, A. Aravelli, S. Bhansali, M. S. Pérez and B. Lerner, Micromachines, 2022, 13, 650 CrossRef PubMed.
- D. J. Teixeira Carvalho, L. Moroni and S. Giselbrecht, Nat. Rev. Mater., 2023, 8, 147–164 CrossRef.
- K.-Y. Song, H. Zhang, W.-J. Zhang and A. Teixeira, Microfluid. Nanofluid., 2018, 22, 135 CrossRef.
- E. A. Aeby, P. M. Misun, A. Hierlemann and O. Frey, Adv. Biosyst., 2018, 2, 1800054 CrossRef.
- L. S. Shiroma, M. H. O. Piazzetta, G. F. Duarte-Junior, W. K. T. Coltro, E. Carrilho, A. L. Gobbi and R. S. Lima, Sci. Rep., 2016, 6, 26032 CrossRef PubMed.
- S. Y. Tan, Q. Jing, Z. Leung, Y. Xu, L. K. W. Cheng, S. S. T. Tam and A. R. Wu, Lab Chip, 2022, 22, 3885–3897 RSC.
- D. Corning, Sylgard 184 silicone elastomer kit, https://www.dow.com/en-us/pdp.sylgard-184-silicone-elastomer-kit.01064291z.html#tech-content.
- I. D. Johnston, D. K. McCluskey, C. K. L. Tan and M. C. Tracey, J. Micromech. Microeng., 2014, 24, 035017 CrossRef CAS.
- PDMS Bonding, https://harrickplasma.com/pdms-bonding/.
- Z. Brounstein, J. Zhao, D. Geller, N. Gupta and A. Labouriau, Polymers, 2021, 13, 3125 CrossRef CAS PubMed.
- D. T. Eddington, J. P. Puccinelli and D. J. Beebe, Sens. Actuators, B, 2006, 114, 170–172 CrossRef CAS.
- T. H. N. Dinh, H. H. Cao, F. S. Hamdi, M. Couty, E. Martincic, M. Woytasik and E. Dufour-Gergam, Microfluid. Nanofluid., 2015, 19, 751–756 CrossRef CAS.
- R. Sivakumar and N. Y. Lee, Analyst, 2020, 145, 4096–4110 RSC.
- Y. Zhang, K. Sun, Y. Xie, K. Liang, J. Zhang and Y. Fan, Rev. Sci. Instrum., 2023, 94, 061501 CrossRef CAS PubMed.
- A. Borók, K. Laboda and A. Bonyár, Biosensors, 2021, 11, 292 CrossRef PubMed.
- A. Kaur, P. Chahal and T. Hogan, IEEE Electron Device Lett., 2016, 37, 142–145 CAS.
- ATCC, U-87 MG - HTB-14 | ATCC, https://www.atcc.org/products/htb-14.
- A. Ross, Z. Jiang, M. Bastmeyer and J. Lahann, Small, 2012, 8, 336–355 CrossRef CAS PubMed.
- L. de Haan, J. Suijker, R. van Roey, N. Berges, E. Petrova, K. Queiroz, W. Strijker, T. Olivier, O. Poeschke, S. Garg and L. J. van den Broek, Int. J. Mol. Sci., 2021, 22, 8234 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc01019h |
| This journal is © The Royal Society of Chemistry 2024 |