Open Access Article
Open Access ArticlePropene-bridged cyanurate tetramers decorated on carbon nanosheets with antibacterial activity: insights from molecular modeling and in vitro studies†
Omnarayan
Agrawal‡
a,
Hitesh Kumar
Sharma‡
b,
Radhika
Chaurasia‡
a,
Gaganjyot Kaur
Bakshi
c,
Aakanksha
Agarwal
 a,
Mousumi
Sen
d,
Praveen
Mamidala
a,
Mousumi
Sen
d,
Praveen
Mamidala
 e,
R. K.
Dey
f,
Mukesh
Chourasia
*c and
Monalisa
Mukherjee
e,
R. K.
Dey
f,
Mukesh
Chourasia
*c and
Monalisa
Mukherjee
 *a
*a
aAmity Institute of Click Chemistry Research and Studies Amity University, Uttar Pradesh, Sector-125, Noida, U.P-201313, India. E-mail: mmukherjee@amity.edu; Tel: +91 98732 79964
bAmity Institute of Pharmacy, Amity University, Uttar Pradesh, Sector-125, Noida, U.P-201313, India
cCenter for Computational Biology and Bioinformatics, Amity Institute of Biotechnology, Amity University Uttar Pradesh, Noida 201301, India. E-mail: mchourasia@amity.edu; Tel: +91 9576161682
dAmity Institute of Applied Sciences, Amity University, Uttar Pradesh, Sector-125, Noida, U.P-201313, India
eLaboratory of Functional Genomics, Department of Biotechnology, Telangana University, Nizamabad-503322, India
fDepartment of Chemistry, Central University of Jharkhand, Ranchi-835205, Jharkhand, India
First published on 2nd May 2024
Abstract
The increasing antibiotic resistance owing to the limitations of current antibiotics accentuates the imperative need for a trailblazing antibacterial therapy. Their unique physicochemical attributes make propene-bridged cyanurate tetramers decorated on carbon nanosheets (CNHs) promising materials for biomedical applications. In this study, we synthesized CNHs using cyanuric acid as the dopant and glycerol and sulphuric acid as the precursors and investigated their antibacterial activity against prominent bacterial strains, Escherichia coli and Staphylococcus aureus. By conducting in vitro experiments, we assessed the antibacterial efficacy of these materials. Additionally, by performing in silico molecular docking analysis, we unraveled the intricate interactions between CNH materials and their targets in bacterial cells. These studies confirmed the profound antibacterial effectiveness of CNHs against both the bacterial strains. In addition, molecular dynamics simulation of total 3 μs revealed the interaction of CNHs with the enzymes GlmU of E. coli and MurE of S. aureus, which are responsible for bacterial cell wall synthesis, resulting in the inhibition of bacterial growth. These findings manifest the potential of the CNH as a promising antibacterial agent. The unique physicochemical properties of the CNH, coupled with antibacterial activity, substantiate their promising roles across an expansive array of biomedical applications, prominently inclining towards the combat against debilitating bacterial infections.
1. Introduction
Infectious maladies persist as a formidable public health predicament globally, with millions of fatalities and impairments caused by a myriad of pathogens. Conventional antibiotics exert their pharmacological effects by intricately modulating critical cellular processes, notably the inhibition of protein synthesis, impediment of cell wall turnover, and cessation of nucleic acid synthesis.1 Over time, the evolutionary process of pathogenic bacteria has led to the development of alarming levels of resistance to nearly all existing conventional antibiotics, thus emerging as a burgeoning concern in contemporary medicine. This antibiotic resistance is primarily attained via various mechanisms such as the manifestation of de novo genetic mutations and horizontal acquisition of resistant genes from divergent species, which adds to the difficulty of defeating these tough microbial foes.2 Combinatorial drug therapy has emerged as a powerful tool to overcome the prevailing issue. However, it surges the risks of adverse drug effects, potential for antagonistic interactions, and emergence of superinfections.3Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) are among the paramount agents of mortality worldwide, aggravating the burden of multi-drug resistance.4E. coli, a Gram-negative bacterium, colonizes the human gut and causes various diseases such as bacteremia, sepsis and various intestinal and extraintestinal infections, whereas, S. aureus a Gram-positive bacterium, is responsible for diverse infections such as skin and soft tissue infections, pneumonia, septicemia (blood poisoning), and meningitis.5,6 The rapid emergence of antibiotic resistance against these disease-causing pathogens stymied the effectiveness of antibiotics.7–9 Nanomaterials exhibit enhanced ability to permeate the membrane, a tendency to serve as efflux pump inhibitors, and effective antibacterial activity, and hence, they are less likely to induce bacterial resistance than the standard antibiotics.10 Carbon nanomaterials, including carbon nanotubes (CNTs), carbon nanofibers (CNFs), carbon nanohybrids, and graphene oxides (GOs), have been utilized in a variety of applications owing to their unique physical and chemical properties such as large surface area, high electrical conductivity, and tunable optical and mechanical properties.11–16The efficacy of carbon nanotubes (CNTs) in preventing microbial growth, however, depends on their concentration and the duration of therapy. Due to significant van der Waals interactions between nanotubes, raw CNTs are practically insoluble in any solvent. CNTs, being hydrophobic in nature, do not disperse in solutions and instead agglomerate, limiting their contact and interactions with microorganisms.17 Functionalization aids in the dispersion of CNTs; however, the cellular uptake of functionalized CNTs can activate immune cells and trigger subsequent immune responses. Graphene-based antibacterial agents have emerged as a promising solution for combating bacterial infections. However, the exact mechanism by which the graphene exerts its antibacterial activity remains controversial, warranting further investigation. Variability in antibacterial efficacy due to differences in the synthesis methods, size, shape, and functionalization of graphene materials further complicates their effective utilization.18 For graphene-based materials, it is crucial to evaluate their biosafety in mammals. The majority of current studies have shown a decrease in cell viability when mammals were exposed to graphene-based materials. Several challenges related to graphene toxicity need to be addressed, including toxicology standards, appropriate dosage, and combined toxicity.19 As a solution to the issue, the development of novel antibacterial agents is the need of the hour.
Nanohybrids have emerged as promising antibacterial agents due to their unique properties and enhanced antibacterial effects.20 The high surface-to-volume ratio of carbon nanomaterials facilitates efficient contact and interaction with bacterial cells, leading to improved antimicrobial efficacy.21
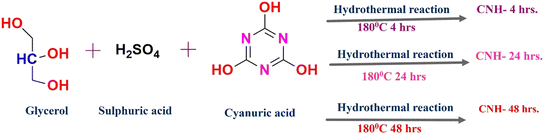
Herein, we report the antibacterial activity and mechanism of propene-bridged cyanurate tetramers decorated on carbon nanosheets (CNHs) towards Gram-negative Escherichia coli and Gram-positive Staphylococcus aureus. We successfully synthesized CNHs using cyanuric acid (CA) as the dopant, and glycerol and sulphuric acid as the precursors. CA plays a critical role in the formation of crystalline domains, in addition to oxygen and nitrogen doping. Crystalline domains comprising propene-bridged cyanurate tetramers (PBCTs) get embedded into 2D nanosheets. These crystalline domains exist both within and outside the 2D nanosheets, contributing to the nano and bulk regime of PBCT domains depending on the time of reaction (4, 24 and 48 h). These domains interact with the enzymes of the peptidoglycan biosynthesis pathway, effectively inhibiting or perturbing cell wall synthesis. Additionally, the PBCT domains nested within and outside the 2D nanosheets synergistically enhance the activity of the nanohybrid, thereby exerting antibacterial effects. The CNH plays two major roles: first, their layered structures envelop the bacteria, and second, the PBCT disrupts the integrity of the cell wall. The PBCT domains in CNH-4 show larger crystallite sizes due to the existence of bulk nanostructured material. However, PBCT domains in CNH-24 show the presence of a nano regime with the existence of more grain boundaries compared to CNH-4. Interestingly, nano regimes CNH-24 and CNH-48 exhibit a higher antibacterial activity than that of their bulk counterpart CNH-4. Plausibly, the bulk crystalline form CNH-4 with a lower number of grain boundaries favours charge transport in comparison to the nanocrystalline form CNH-24, which has a higher number of grain boundaries perturbing the charge transport mechanism. The lower the charge transport mechanism, the better the antibacterial activity. Through a suit of advanced characterization techniques, the inherent, structural, chemical, and morphological characteristics of the resulting CNH were revealed showing their crystalline nature, highly porous architecture, improved biocompatibility, enhanced charge separation, increased light absorption, sharp edge plane defects and ability to disrupt the integrity and function of bacterial cells, ultimately leading to their death.22 We conducted qualitative and quantitative in vitro antibacterial evaluation against these bacterial strains, utilizing the zone of inhibition and micro broth dilution assays. To further investigate the interaction of PBCTs with targets involved in the bacterial cell wall synthesis of E. coli and S. aureus, we conducted molecular docking and simulation studies. Moreover, to elucidate the bacteriostatic and bactericidal mechanisms of CNHs, we conducted a comprehensive time kill kinetics study along with SEM analysis. Our findings could pave the way for developing novel carbon-based materials with enhanced antibacterial properties, to tackle the growing predicament of antibiotic resistance.
2. Experimental section
2.1. Materials
Sulphuric acid, with a potency of 97%, was purchased from Thermo fisher Scientific India Pvt. Ltd., while glycerol, with a concentration of 99.5%, was received from Thermo fisher Scientific India Pvt. Cyanuric acid used in the experiment was procured from Tokyo Chemical Industry CO. Ltd., CNH-4, CNH-24 and CNH-48 were synthesized by a hydrothermal method and all other chemicals used throughout the experiments were of analytical grade or better quality. All the solutions have been designed and produced in accordance with the standard protocol that is described in the India Pharmacopoeia, 1996. Double deionized water was used in all the experiments. Escherichia coli (MTCC 1302, Gram negative) and Staphylococcus aureus (MTCC 96, Gram positive) were purchased from Microbial Type Culture Collection, CSIR-IMTech, Chandigarh, India.2.2. Methodology
2.3. CNH morphological and chemical characterization
CNHs were observed using a SEM (SU8010, Hitachi, Japan) and a HRTEM (JEOL, JEM-2100F) at an accelerating voltage of 200 kV. High-resolution transmission electron microscopy (HRTEM) was performed using a JEOL, JEM-2100F electron microscope. The sample preparation process involves drop-casting the sample dispersion material onto a copper grid coated with carbon, followed by drying at room temperature. Raman spectroscopy was performed at 100× magnification using a confocal micro-Raman LabRam HR spectrometer (Horiba Scientific) with backscattering geometry, a CCD detector, and a 532 nm Ar laser. Cu Kα radiation was utilized to record the powder X-ray diffraction (XRD) patterns in the 2θ range at 10–80 °C (D2 PHASER from Bruker). For surface analysis, XPS measurements were performed at the indus-2 beamline in RRCAT – Indore, using a pair of Si(111) crystals and a double-crystal monochromator (DCM). Fourier transform infrared (FT-IR) spectroscopy was performed using an FTIR (UPD) spectrometer in the 500–4000 cm−1 range.2.4. Isolation of mesenchymal stem cells (MSCs) from the bone marrow of BALB/c mice
A female BALB/c mouse weighing ∼26 g was kept under typical laboratory conditions (24–25 °C; alternating cycles of 12 h darkness and 12 h light) and fed with standard food pellets and water ad libitum. The mice were kept in cages made of polypropylene, and the bed of the caging was made of sterile rice husk. The experiments were conducted in accordance with the guidelines of the Institutional Animal Ethics Committee (IAEC), and utmost care was taken for handling mice according to the institutional guidelines. Ethical clearance was received from IAEC, INMAS, and DRDO (approval no. INM/IAEC/2019/05/Ext 01) before initiating the experiments. Bone marrow mesenchymal stem cells (MSCs) were isolated from the mouse. It was anesthetized using a concoction of ketamine and xylazine (20 mg kg−1 and 5 mg kg−1, respectively) and sacrificed according to the guidelines issued by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA). The hind limbs of the mice were amputated to harvest the bone marrow. Subsequently, the muscles surrounding the bone marrow were cleaned off and kept in a Petri dish with incomplete Dulbecco's modified Eagle medium (DMEM: Gibco). The cells in the bone marrow were flushed off using a syringe containing incomplete media and 2 micron syringe filter. The media containing the cells were then centrifuged at 300 g for 5 min. The pellet was then obtained, which was resuspended in low glucose DMEM media in a 25 cm2 flask. They were then placed in an incubator that was humidified and containing 5% carbon dioxide. For 3 days, the media was changed every 8 h to remove all hematopoietic stem cells (HSCs), which are suspended cells and retain only mesenchymal stem cells (MSCs), which are adhered cells. The cells matured into MSCs after 14 days and reached 80% confluency in a month. After washing for three days, the media was replaced every 5–6 days, until the color turned darker.2.5. Characterization of mesenchymal stem cells
Flow cytometry was performed to confirm MSCs using a positive marker (CD44) and a negative marker (CD19). The cells were centrifuged at 300 g for 5 min to obtain a pellet, which was then resuspended in 1200 μL PBS to be divided in four groups (300 μL each): Auto (no marker), CD44 (20 μL), CD19 (20 μL), and dual (40 μL) (CD19/CD44). Cells were incubated with the respective antibodies for 1–2 h and then centrifuged to obtain pellets. These pellets were then washed and resuspended with PBS (300 μL). A BD FACS Calibrur flow cytometer (BD Biosciences) instrument was used for cell fluorescence evaluation and data were analyzed using the FCS Express Flow Cytometry Software.2.6. Biocompatibility experiments
2.7. In vitro antibacterial analysis
2.8. Scanning electron microscopy of bacterial cells
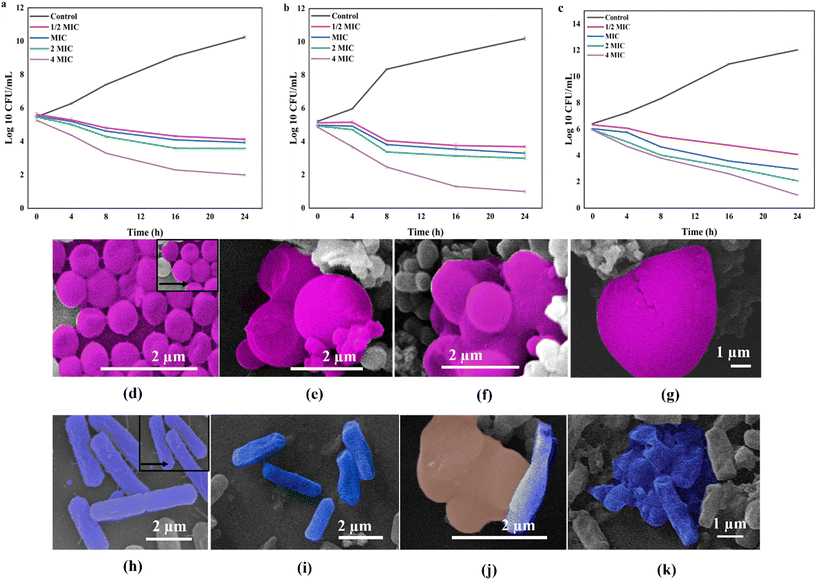
We investigated the ultrastructural changes induced by CNH-4, CNH-24 and CNH-48 treatments with E. coli and S. aureus cells by transmission electron microscopy (TEM). Bacterial cells were exposed to the respective compounds and subsequently fixed in a 2.5% glutaraldehyde solution in 0.1 M phosphate buffer saline to preserve their morphology. Following fixation, the samples were stained with 1% osmium tetroxide, which further enhanced the contrast for better visualization of cellular structures. To prepare the samples for microscopy, a dehydration process was carried out using a graded ethanol series (ranging from 50% to 100% ethanol). Finally, the samples were examined using a Zeiss Supra 55VP microscope (Oberkochen, Germany). The ultrastructural observations allowed us to gain insights into the specific cellular alterations caused by CNH-4, CNH-24 and CNH-48 treatment.2.9. In silico analyses of CNHs to investigate the antibacterial activity
Enzymes that are part of the cell wall biosynthesis pathways have been identified as highly promising targets for discovering novel inhibitors.27 To this end, we investigated how PBCT binds to the active sites of various enzymes of the cell wall biosynthesis pathway using molecular docking. The Glide module of Schrödinger suite 2023-1 was used to carry out the molecular docking studies.28 The crystal structures of target proteins involved in the cell membrane biosynthesis of both E. coli and S. aureus were obtained from the Protein Data Bank (PDB). The PDB ID for E. coli is GlmU (1HV9), MurC (2F00), MurD (1UAG), and MurG (1NLM), whereas for S. aureus, the PDB ID is GlmM (6GYZ), MurB (1HSK), MurE (4C13), and FemA (1LRZ). The Protein Preparation Wizard module was used to prepare the target structures, which were then optimized with the OPLS4 force field. This involved the addition of hydrogen atoms, assignment of bond orders and charge states, optimization of hydrogen bonds, and elimination of atomic clashes. Likewise, the LigPrep module was employed to prepare and optimize the structures of the substrate of selected enzymes and PBCTs. The protein active sites were defined by a grid box centered at the ligand binding site. In cases where no ligand was present, sitemap analysis was conducted on the proteins, and the site with the highest site score was chosen as the optimal cavity for docking. The Glide module was then utilized to dock the ligands onto the target proteins, generating ten poses in the standard precision (SP) mode. These poses were further subjected to the extra precision (XP) mode of docking.| ΔG = ΔGcomplex − (ΔGprotein + ΔGligand) |
3. Results and discussion
3.1. Characterization of CNHs
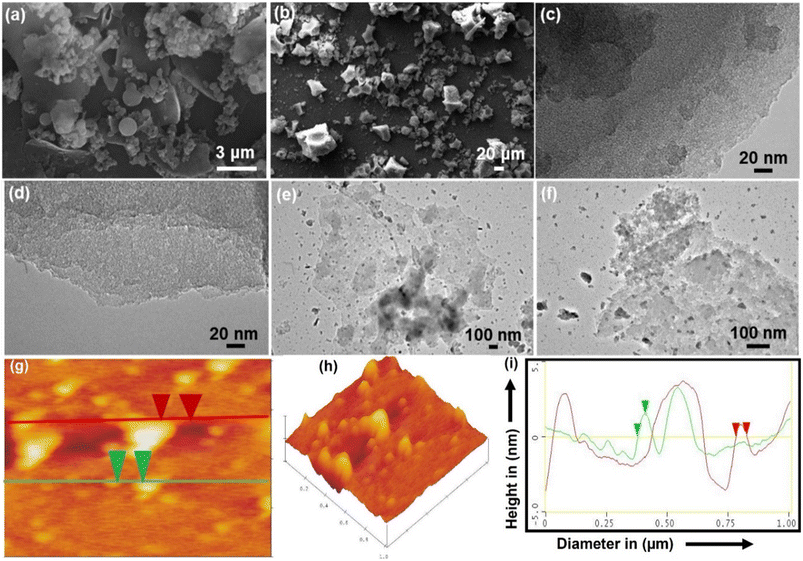
After hydrothermal treatment, the products CNH-4 and CNH-24 emerge as a cake of carbon that is malleable and easily pulverized after rinsing with a 50% solution of water and ethanol. As compared to CNH-24, CNH-4 looks like a lustrous carbon block, when smashed, integrates into crystalline powder. The SEM images of CNH-4 (Fig. 1a) exhibited folded structures of thin graphitic nanohybrids, and CNH-24 (Fig. 1b) showed the nanocrystalline structure of nanomaterials. The HRTEM image of CNH-4 (Fig. 1c and d) further confirmed the layered sheet-like structures, which suggested its bulk crystalline nature and CNH-24 (Fig. 1e and f) exhibited the nanocrystalline chunks embedded on the 2D sheet. The homogeneous dispersion of these nanocrystals on the laminated CNH, shown in the atomic force microscopic (AFM) images with an average height profile of 1.51 nm corresponding to the uniform distribution of these nanocrystals on the layered CNH. The AFM image of the CNH demonstrates the 2D view of CNH-4 (Fig. 1g), 3D view of CNH-4 (Fig. 1h) and height profile graph of CNH-4 (Fig. 1i) in which the green cap defines the height of CNH-4 as 1.51 nm and the red cap denotes the diameter of CNH-4 which is found to be 43.39 nm. After drying the CNH under vacuum at room temperature, XRD analysis was conducted, revealing a combination of major amorphous carbon phase with small crystalline domains (Fig. 2a). To test the hypothesis that temperature affects the degree of crystallinity of the precursor material, the CNH was subjected to oven drying, and XRD analysis was performed. The results were intriguing as the XRD pattern of CNH-24 (Fig. 2b) showed a cyclohexane monoclinic carbon phase at 45° and 56° having the (221) and (004) planes, respectively (JCPDS file 00-056-0159), and a graphene hexagonal phase at 26° degrees having the (002) plane that matches the standard data (JCPDS file 00–056-0159 is attached).30 The XRD analysis of CNH-4 indicated a broad peak at 23° 2θ, showing the amorphous nature along with some crystalline zones. When comparing CNH-24 with CNH-4, the XRD spectra exhibit numerous sharp peaks in CNH-4 featuring very narrow full width at half maximum (FWHM), indicating the existence of larger crystallite sizes. This phenomenon can be attributed to the presence of bulk nanostructured materials. This finding was consistent with the HRTEM images of CNH-4 presented in Fig. 1c and d.Raman spectroscopy is prevalently used to assess structural disorder in the CNH, which is based on a myriad of defects including intrinsic defects, edges,31 impurities, stacking defects, and grain structures, thereby providing a quantitative measure degree of disorder.32–34 Raman spectroscopy was performed in this study to examine the intrinsic vibrational differences between the as-synthesized products dependent on the time interval. The rate of imperfections in the CNH can be evaluated (Fig. 2c) by comparing the intensity of the Raman bands D and G (ID/IG). G-bands were visible in the Raman spectra of synthesized CNH-4 and CNH-24 at 1573 and 1575 cm−1, respectively. D-bands indicate the sp2 lattice structure, as they are prevalent in both materials.35 For CNH-4 and CNH-24, The D-band arises at 1347 and 1367 cm−1 respectively, indicating the presence of distorted structures in both materials. The intensity ratio of ID/IG decreased with the increase in time from 4 to 24 h [for CNH-4 (0.91) and CNH-24 (0.85)], and the ID/IG ratio declined because of the healing impact and the recovery of the sp2 characteristics. The FTIR spectra were used to figure out how precursor cyanuric acid grafted onto nanohybrids with thin sheets and how functional groups of CNH-4 and CNH-24 shifted (Fig. 2d). When we hydrothermally treated these CNHs, the N–H stretching peak gets wider at 3415 and 3404 cm−1 for CNH-4 and CNH-24, respectively. It insinuates that the molecule has distinct chemical phenolic domains that fluctuate the stability of the bonds. A small spike in the intensity of the peak at 1620 cm−1 for CNH-4 along with a slight blue transition of the peaks (at 2920 cm−1) in comparison to CNH-24 at 2915 cm−1 is indicative of the asymmetric CH2 stretching vibrations, which corroborate the reduction of oxygen functionalities and the restoration of the sp2 structure (π-conjugation). The spectra of CNH-4 and CNH-24 showing the presence of an oxygen-containing functional group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) at peak positions of 1706 and 1708 cm−1, respectively. Both CNH-4 and CNH-24 have a triazine fingerprint region in the range of 1400–1470 cm−1, which depict the chemical structure of the CNH, which were produced from cyanuric acid, having been preserved. After the hydrothermal reaction, new peaks at 1040 and 1042 cm−1 in the spectra of CNH-4 and CNH-24 portray that nitrogen was added to the carbon matrix of CNH. XPS evaluations were also carried out to provide additional insights into the chemical bonding states of CNH-4 and CNH-24 (Fig. 2e). The XPS survey spectra for both CNH-4 and CNH-24 over a wide range of binding energies (0–800 eV) show a predominant narrow C 1s peak at 285.3 eV, O 1s peak at 532.01 eV, and N 1s peak at 398.4 eV, showing the integration of nitrogen in the structure. At 167.3 eV, S 2p peaks were also observed.
O) at peak positions of 1706 and 1708 cm−1, respectively. Both CNH-4 and CNH-24 have a triazine fingerprint region in the range of 1400–1470 cm−1, which depict the chemical structure of the CNH, which were produced from cyanuric acid, having been preserved. After the hydrothermal reaction, new peaks at 1040 and 1042 cm−1 in the spectra of CNH-4 and CNH-24 portray that nitrogen was added to the carbon matrix of CNH. XPS evaluations were also carried out to provide additional insights into the chemical bonding states of CNH-4 and CNH-24 (Fig. 2e). The XPS survey spectra for both CNH-4 and CNH-24 over a wide range of binding energies (0–800 eV) show a predominant narrow C 1s peak at 285.3 eV, O 1s peak at 532.01 eV, and N 1s peak at 398.4 eV, showing the integration of nitrogen in the structure. At 167.3 eV, S 2p peaks were also observed.
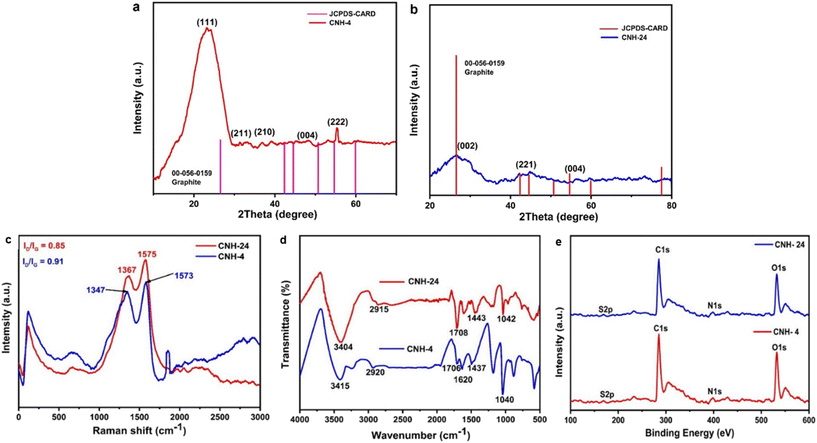 | ||
| Fig. 2 a) and b) X-ray diffraction pattern of CNH-4 and CNH-24. c) Raman spectra of CNH-4 and CNH-24. d) FTIR analysis of CNH-4 and CNH-24. e) High-resolution spectrum of C 1s, N 1s, O 1s and S 2p. | ||
3.2. Biocompatibility assay of CNH-4 and CNH-24
In Fig. 3a, it can be observed from the MTT assay that CNH-4 and CNH-24 were biocompatible with mesenchymal stem cells (MSC). Both CNH-4 and CNH-24 showed excellent biocompatibility till day 7, with CNH-24 being the most significant till day 7. Moreover, the increase in absorbance over the course of 1 week indicates the amelioration of metabolism of MSCs, suggesting possible MSC proliferation, which was further confirmed by the Cyquant assay. In Fig. 3b, the Cyquant cell proliferation assay shows a significant increase in cell proliferation with CNH-4. The amorphous nature of CNH-4 plausibly influenced better MSC proliferation in contrast to CNH-24.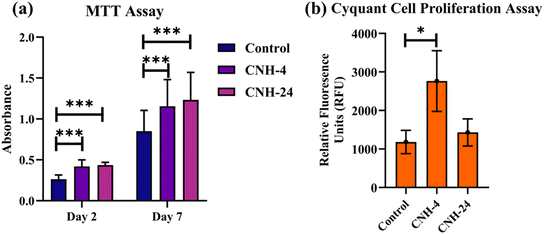 | ||
| Fig. 3 MTT assay (a) to evaluate the biocompatibility of CNH-4 and CNH-24 on mesenchymal stem cells. Cyquant cell proliferation assay (b) (* < 0.05, *** < 0.0007). | ||
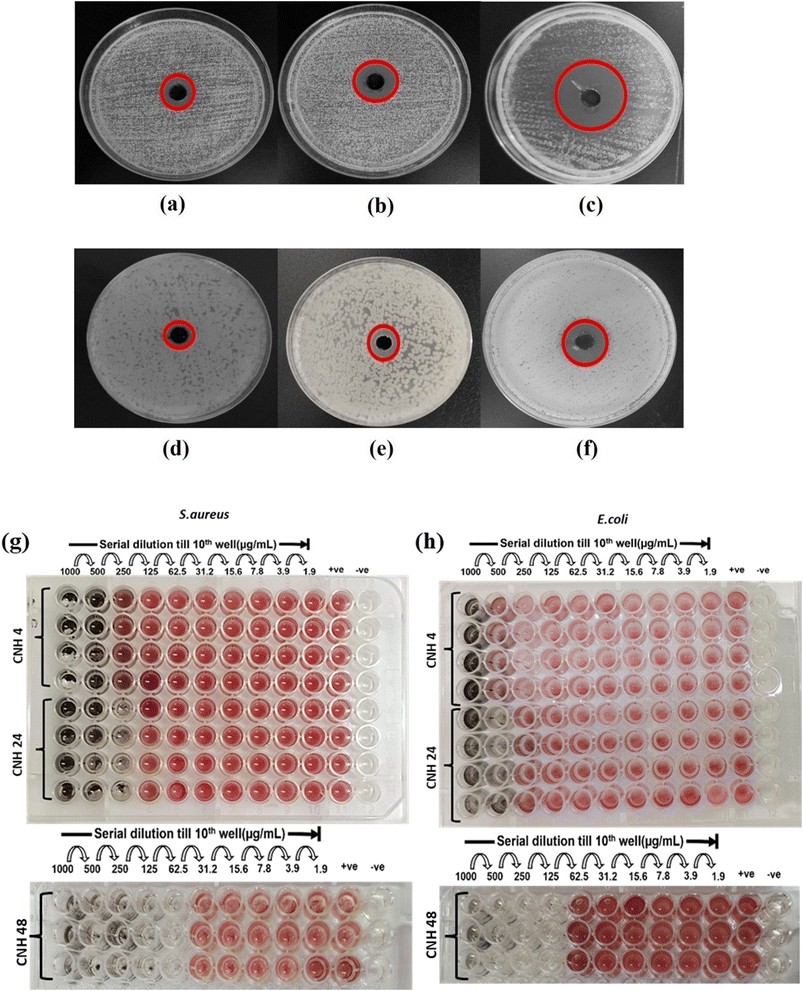
3.3. In vitro antibacterial analysis of CNH-4, CNH-24 and CNH-48
| Name of material | Zone of inhibition (mm) | Minimum inhibitory concentration (μg mL−1) | ||
|---|---|---|---|---|
| S. aureus | E. coli | S. aureus | E. coli | |
| CNH-4 | 16 | 8 | 500 | 1000 |
| CNH-24 | 20 | 12 | 250 | 500 |
| CNH-48 | 27 | 20 | 62.5 | 125 |
The observed morphological changes in the bacterial cells were consistent with the antibacterial activity of the CNH observed in the well diffusion and micro broth dilution assays. The SEM images provided further evidence that the CNH disrupted the bacterial cell membranes, leading to cell death. The ability of the CNHs to damage the bacterial cell membranes is probably due to their unique physical and chemical properties including their high surface area, large pore volume, and the presence of oxygen- and nitrogen-containing functional groups on their surface. These properties could enhance the interaction between the nanohybrids and bacterial cell membranes, leading to the disruption of membrane integrity and the leakage of cellular contents.
3.4. In silico studies
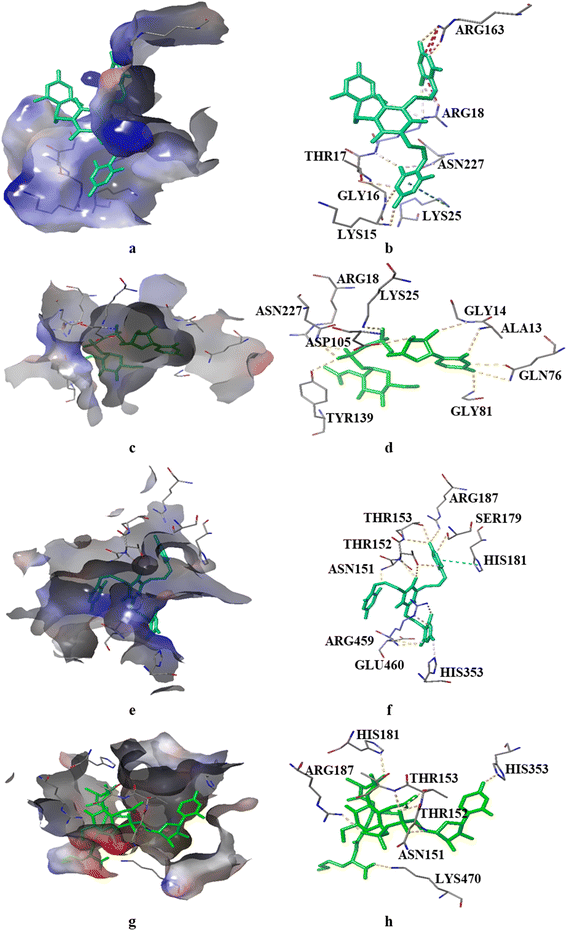
We investigated the binding potential of the PBCT with the active sites of various enzymes of the cell wall biosynthesis pathways by comparing it with the binding energy and interactions of their known substrate and inhibitor. Therefore, GlmU and MurC of E. coli and MurE of S. aureus were selected as the probable targets of the PBCT (ESI† Fig. S1 and S2 and Tables S1–S10). The lining of the active site of these enzymes is hydrophilic in nature and mostly composed of polar residues. The Glide XP docking results suggest that the binding of the PBCT with the active site of these enzymes is slightly less than the actual substrate. The GlmU enzyme shows binding energies of −7.18, −8.881 and −7.025 kcal mol−1 with the substrate (glucosamine-6-phosphate), inhibitor (N-acetylglucosamine-1-phosphate) and PBCT, respectively (Fig. S3†). The substrate (UDP-N-acetyl muramoyl-L-alanine), inhibitor (GW659893X), and PBCT of MurC show binding energies of −10.297, −5.351 and −7.827 kcal mol−1 respectively (Fig. S4†). The substrate (UDP-N-acetyl muramoyl-L-alanyl-D-glutamyl-L-lysine), inhibitor (fosfomycin) and PBCT are showing binding energies of −8.525, −5.800 and −7.255 kcal mol−1 respectively with MurE of S. aureus (Fig. S5†). The docking results indicate that the PBCT is a better binder than the known ligands of respective enzymes. We calculated the binding free energy (ΔG) of the complexes by MD simulation to confirm the protein–ligand affinity which was already predicted by the docking process. The MD trajectories were used in the calculation of ΔG. The MM-GBSA method was employed to calculate the combined values of polar and nonpolar solvation energies, as well as the total energies associated with electrostatic interactions, van der Waals forces and bonded interactions within all the complexes. According to the MM/GBSA-predicted binding energy, the GlmU-PBCT and GlmU-substrate complexes show −38.1 and −67.8 kcal mol−1, respectively. The MurC-PBCT and MurC-substrate complexes show −31.8 and −56.7 kcal mol−1, respectively and the MurE-PBCT and MurE-substrate complexes show −41.8 and −49.4 kcal mol−1 of ΔG, respectively. The docking and experimental results are well corroborated with the free binding energy calculation from the trajectories of MD simulations.Moreover, RMSD and conformational ensemble analysis suggest that the protein–ligand complexes are stable throughout the 500 ns simulations of GlmU and MurE (Fig. 6a and b and 7a and b). Similarly, the RMSF plot has not shown any significant mobility and flexibility per residue during the simulation at the binding site level upon PBCT binding (Fig. 6c and d and 7c and d). The conformational ensemble of all the four complexes contains representative conformations from the 4 clusters obtained from the clustering of production run trajectories (Fig. S6†). It is observed that conformations of all the complexes in the ensemble exhibit less structural diversity. Small conformational difference at the level of ligand in the binding pocket reveals differences in the interaction of the ligand with the protein. Three-dimensional interaction images were produced for the cluster conformation with the highest number of representatives for all the complexes (Fig. 8).
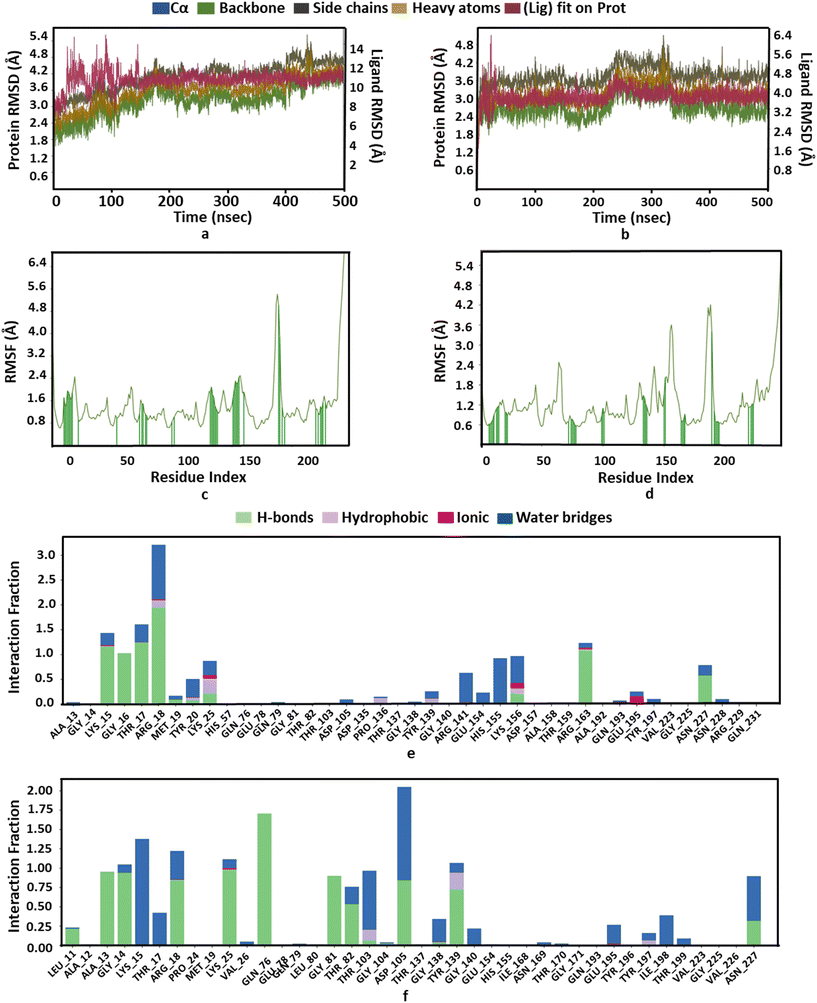 | ||
| Fig. 6 Structure and interaction analysis of MD simulation trajectories obtained for E. coli protein GlmU (PDB ID: 1HV9): RMSD plot of GlmU (a) with the PBCT and (b) with the substrate. A stable complex is formed without any significant conformational changes in the protein structure during 500 ns simulation. Backbone RMSF plot with interacting residues of GlmU (c) with the PBCT and (d) with the substrate. Bar graph for the interaction counts of GlmU (e) with the PBCT and (f) with the substrate. | ||
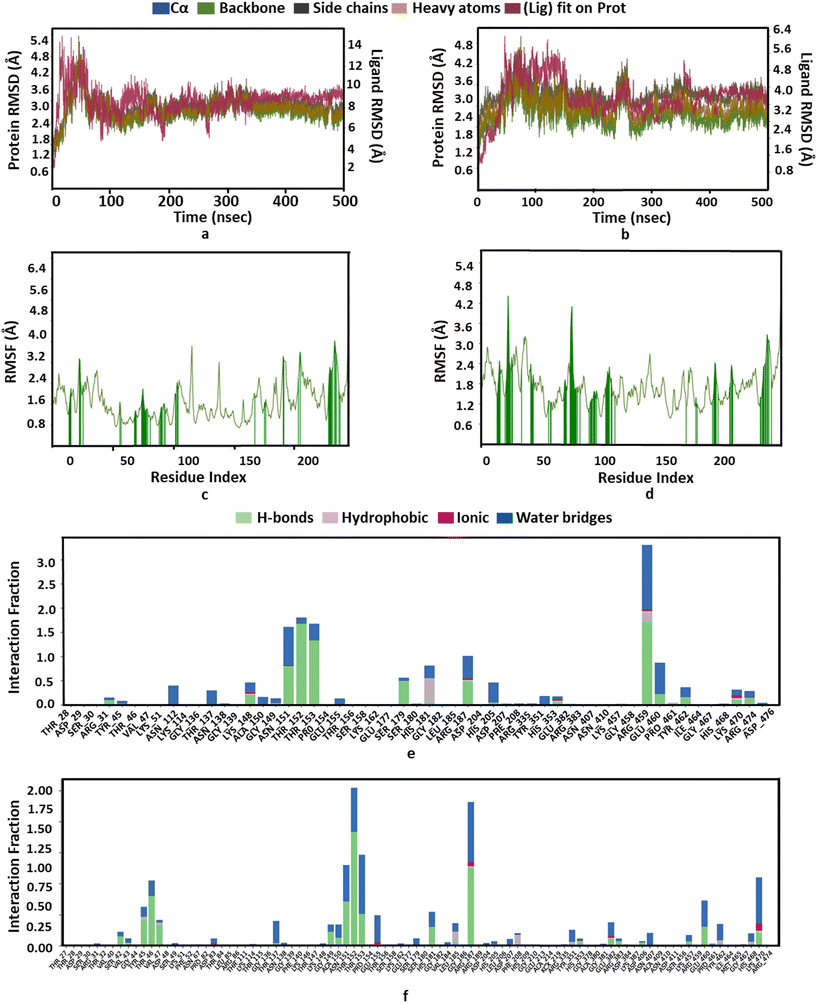 | ||
| Fig. 7 Structure and interaction analysis of MD simulation trajectories obtained for the S. aureus protein MurE (PDB ID: 4C13): RMSD plot of MurE (a) with the PBCT and (b) with the substrate. A stable complex is formed without any significant conformational changes in the protein structure during 500 ns simulation. Backbone RMSF plot with the interacting residues of MurE (c) with the PBCT and (d) with the substrate. Bar graph for the interaction counts of MurE (e) with the PBCT and (f) with the substrate. | ||
The oxygen atom of PBCT forms hydrogen bonds with the backbone of binding site residues GLY14, THR82, GLY225 and ALA13 of GlmU. The triazine ring of PBCT forms a π–π interaction with TYR103. ARG18, ARG141, HIS155 and LYS156 are interact with the PBCT through a water bridge. Moreover, the substrate also displays similar interactions with binding site residues. The hydroxyl and oxygen atoms of the substrate form hydrogen bonds with the backbone of GLY81, THR82, GLY14 and ALA13 and the sidechains of TYR103, ASP105, LYS25, GLN76, TYR139 and ALA85. The 2D interactions which are present in more than 30% of the trajectories are considered (Fig. S7†). The radius of gyration (Rg) was computed as a measure of the compactness of the receptor–ligand complexes. The analysis revealed that the Rg values of the protein complexes with the PBCT and substrate remained stable in about 17.6 ± 0.6 Å throughout the MD simulation frame of 500 ns (Fig. S8a and b†). Moreover, the PBCT and substrate appear to be stable at the GlmU active site as forming about 10 hydrogen bonds throughout the simulation (Fig. S8c and d†). To gain a better understanding of the bonding statistics and the efficacy of the compound with different proteins, a bar graph was generated to display the ligand interaction counts (Fig. 6e, f). The significant conformational changes were observed in the RMSD plot of the PBCT when complexed with MurC (Fig. S9†). Additionally, the Rg value of the PBCT-MurC complex is about 24.0 ± (+6.0 and −1.0) Å, which suggests that the PBCT is loosely packed in the binding site of MurC (Fig. S10†). Therefore, GlmU is predicted as the target of PBCTs rather than MurC of E. coli.
The PBCT in the complex with MurE showed hydrophobic interactions with HIS181 and ARG459; the oxygen atom of the PBCT is forming a network of hydrogen bonds with the backbone of THR152 and THR153 and sidechains of ARG187, ALA150, ASN151, HIS181, THR46, GLU382, ARG383, HIS468 and SER179. The triazine ring of PBCT shows π–π interaction with the sidechain of HIS205. Similarly, the substrate showed hydrophobic interactions with PHE208 and LEU185; hydrogen bond interactions with the backbone of TYR45, THR152 and VAL43 and sidechains of LYS457, ASP406, ASN151, ARG383, GLU382, SER179, HIS181 and SER42. Cation-π interactions are also observed with the sidechains of HIS353 and TYR45 during 500 ns MD simulations. A bar graph is generated to display the ligand interaction counts (Fig. 7e and f). The 2D interactions which are present in more than 30% of the trajectories are shown in Fig. S11.† The Rg value of the PBCT-MurE complex is stable at ∼24 Å after 250 ns of MD simulation (Fig. S12†).
4. Conclusion
Our study highlighted the antibacterial potential and underlying mechanism of CNHs against the Gram-negative bacterium Escherichia coli and the Gram-positive bacterium Staphylococcus aureus. In this synthesis, cyanuric acid plays a critical role in the formation of crystalline domains in addition to N and O doping. During the reaction, PBCT domains get embedded in the 2D nanosheet, resulting in the formation of CNHs. The increased time of hydrothermal treatment was responsible for the nucleation and growth of CNHs. The presence of these crystalline domains within and outside the 2D sheet contributes to both the nano and bulk aspects of PBCT domains in CNH-24 and CNH-4 respectively. Through their interaction with MurE and GlmU enzymes, the CNH effectively inhibits or disrupts cell wall synthesis in the bacteria. Additionally, the nestled domains within the CNH synergistically enhance the antibacterial effects by enveloping the bacteria with layered structures and utilizing the PBCT to undermine the integrity of cell membranes. Comparative analysis revealed that CNH-4, with crystalline domains in the bulk regime, exhibited a lower antibacterial activity than that of its nano regime counterpart, CNH-24. Extensive characterization techniques elucidated the intricate structural, chemical, and morphological attributes of CNH, underscoring their crystalline nature, excellent architecture, improved biocompatibility, enhanced charge separation, sharp edge plane defects, and capacity to disrupt the integrity and functionality of bacterial cells, ultimately leading to bacterial cell death. In vitro antibacterial evaluation assays validated the efficacy of CNHs against these bacterial strains. Molecular docking and simulation studies provided further insights into the interaction between the PBCT and targets involved in the bacterial cell wall synthesis in Escherichia coli and Staphylococcus aureus. The outcomes of our research hold great potential for the development of novel carbon-based hybrid materials endowed with enhanced antibacterial properties, addressing the exigent concern of antibiotic resistance.Abbreviations
| CA | Cyanuric acid |
| CFU | Colony forming unit |
| CNH | Propene-bridged cyanurate tetramers decorated on carbon nanosheets |
| CNH-4 | CNH 4 hours |
| CNH-24 | CNH 24 hours |
| CNH-48 | CNH 48 hours |
| CNT | Carbon Nano Tube |
| E. coli | Escherichia coli |
| GO | Graphene oxide |
| MD simulation | Molecular dynamics simulation |
| MIC | Minimum inhibitory concentration |
| PBCT | Propene-bridged cyanurate tetramer |
| RMSD | Root-mean-square deviation |
| RMSF | Root-mean-square fluctuation |
| S. aureus | Staphylococcus aureus |
| SP | Standard precision |
| XP | Extra precision |
Author contributions
OA performed all the synthesis, characterization; finalized the results; wrote the manuscript. HKS performed the in vitro studies, finalized the results; wrote the manuscript. RC contributed to the writing and editing of the manuscript, graphical abstract and figs. AA, MS and RKD contributed to literature survey. GKB and MC performed the in silico studies and wrote the manuscript. MM has conceptualized the work, contributed to writing the manuscript, proposed the title and mechanism for CNH formation. All authors have given approval to the final version of the manuscript. The authors confirm that all the figures/graphs/schemes/images/photos/ToC graphics that appear in the manuscript and ESI† file were created/taken by them.Conflicts of interest
There are no conflicts to declare.Acknowledgements
MM thanks the Department of Biotechnology (BT/PR21866/NNT/28/1145/2016), Department of Science and Technology-Science & Engineering Research Board (SERB)-(CRG/2023/001069) and University Grants Commission DAE CSR-(CRS/2021-22/04/642) for funding this work. MM and RC are also thankful to the Department of Science and Technology (DST/WOS-A/CS-106/2021) for their funds. MC thanks Council for Science and Technology, Uttar Pradesh (CST/Biotech/RS-1986/D1226) for funds. The authors would like to thank Amity University Uttar Pradesh for providing the research infrastructure.References
- M. A. Kohanski, D. J. Dwyer and J. J. Collins, How antibiotics kill bacteria: from targets to networks, Nat. Rev. Microbiol., 2010, 8, 423–435 CrossRef CAS PubMed.
- N. Händel, J. M. Schuurmans, Y. Feng, S. Brul and B. H. ter Kuile, Interaction between Mutations and Regulation of Gene Expression during Development of De Novo Antibiotic Resistance, Antimicrob. Agents Chemother., 2014, 58, 4371–4379 CrossRef PubMed.
- M. J. Rybak and B. J. McGrath, Combination Antimicrobial Therapy for Bacterial Infections, Drugs, 1996, 52, 390–405 CrossRef CAS PubMed.
- J. T. Poolman and A. S. Anderson, Escherichia coli and Staphylococcus aureus: leading bacterial pathogens of healthcare associated infections and bacteremia in older-age populations, Expert Rev. Vaccines, 2018, 17, 607–618 CrossRef CAS PubMed.
- C. L. Holmes, M. T. Anderson, H. L. T. Mobley and M. A. Bachman, Pathogenesis of Gram-negative bacteremia, Clin. Microbiol. Rev., 2021, 34, cmr.00234-20 CrossRef PubMed.
- A. P. Kulkarni, V. C. Nagvekar, B. Veeraraghavan, A. R. Warrier, D. Ts, J. Ahdal and R. Jain, Current perspectives on treatment of gram-positive infections in India: what is the way forward?, Interdiscip. Perspect. Infect. Dis., 2019, 7601847 CAS.
- B. A. Sheikh, B. A. Bhat and M. A. Mir, Antimicrobial resistance: new insights and therapeutic implications, Appl. Microbiol. Biotechnol., 2022, 106, 6427–6440 CrossRef CAS PubMed.
- E. Christaki, M. Marcou and A. Tofarides, Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence, J. Mol. Evol., 2020, 88, 26–40 CrossRef CAS PubMed.
- M. Frieri, K. Kumar and A. Boutin, Antibiotic resistance, J. Infect. Public Health, 2017, 10, 369–378 CrossRef PubMed.
- Q. Xin, H. Shah, A. Nawaz, W. Xie, M. Z. Akram, A. Batool, L. Tian, S. U. Jan, R. Boddula, B. Guo, Q. Liu and J. R. Gong, Antibacterial Carbon-Based Nanomaterials, Adv. Mater., 2019, 31, 1804838 CrossRef CAS PubMed.
- V. D. Punetha, S. Rana, H. J. Yoo, A. Chaurasia, J. T. McLeskey Jr, M. S. Ramasamy, N. G. Sahoo and J. W. Cho, Functionalization of carbon nanomaterials for advanced polymer nanocomposites: A comparison study between CNT and graphene, Prog. Polym. Sci., 2017, 67, 1–47 CrossRef CAS.
- M. Yousefi, M. Dadashpour, M. Hejazi, M. Hasanzadeh, B. Behnam, M. de la Guardia, N. Shadjou and A. Mokhtarzadeh, Anti-bacterial activity of graphene oxide as a new weapon nanomaterial to combat multidrug-resistance bacteria, Mater. Sci. Eng., C, 2017, 74, 568–581 CrossRef CAS PubMed.
- T. Govindaraju and M. B. Avinash, Two-dimensional nanoarchitectonics: organic and hybrid materials, Nanoscale, 2012, 4, 6102–6117 RSC.
- A. Dieter Schlüter, Mastering polymer chemistry in two dimensions, Commun. Chem., 2020, 3, 1–4 CrossRef PubMed.
- W. Wang, S. Chakrabarti, Z. Chen, Z. Yan, M. O. Tade, J. Zou and Q. Li, A novel bottom-up solvothermal synthesis of carbon nanosheets, J. Mater. Chem. A, 2014, 2, 2390–2396 RSC.
- M. A. Deshmukh, S.-J. Park, B.-C. Kang and T.-J. Ha, Carbon Nanohybrids for Advanced Electronic Applications, Phys. Status Solidi A, 2020, 217, 2000199 CrossRef CAS.
- C. D. Vecitis, K. R. Zodrow, S. Kang and M. Elimelech, Electronic-Structure-Dependent Bacterial Cytotoxicity of Single-Walled Carbon Nanotubes, ACS Nano, 2010, 4, 5471–5479 CrossRef CAS PubMed.
- H. Mohammed, A. Kumar, E. Bekyarova, Y. Al-Hadeethi, X. Zhang, M. Chen, M. S. Ansari, A. Cochis and L. Rimondini, Antimicrobial Mechanisms and Effectiveness of Graphene and Graphene-Functionalized Biomaterials. A Scope Review, Front. Bioeng. Biotechnol., 2020, 8, 465 CrossRef PubMed.
- P. Kumar, P. Huo, R. Zhang and B. Liu, Antibacterial Properties of Graphene-Based Nanomaterials, Nanomaterials, 2019, 9, 737 CrossRef CAS PubMed.
- M. K. A. Mohammed, D. S. Ahmed and M. R. Mohammad, Studying antimicrobial activity of carbon nanotubes decorated with metal-doped ZnO hybrid materials, Mater. Res. Express, 2019, 6, 055404 CrossRef CAS.
- M. Azizi-Lalabadi, H. Hashemi, J. Feng and S. M. Jafari, Carbon nanomaterials against pathogens; the antimicrobial activity of carbon nanotubes, graphene/graphene oxide, fullerenes, and their nanocomposites, Adv. Colloid Interface Sci., 2020, 284, 102250 CrossRef CAS PubMed.
- A. Chakraborty, P. Patni, D. Suhag, G. Saini, A. Singh, S. Chakrabarti and M. Mukherjee, N-doped carbon nanosheets with antibacterial activity: mechanistic insight, RSC Adv., 2015, 5, 23591–23598 RSC.
- M. Majood, A. Selvam, O. Agrawal, R. Chaurasia, S. Rawat, S. Mohanty and M. Mukherjee, Biogenic Carbon Quantum Dots as a Neoteric Inducer in the Game of Directing Chondrogenesis, ACS Appl. Mater. Interfaces, 2023, 15, 19997–20011 CrossRef CAS PubMed.
- S. Ali, M. Jalal, H. Ahmad, D. Sharma, A. Ahmad, K. Umar and H. Khan, Green Synthesis of Silver Nanoparticles from Camellia sinensis and Its Antimicrobial and Antibiofilm Effect against Clinical Isolates, Materials, 2022, 15, 6978 CrossRef CAS PubMed.
- H. K. Sharma, P. Gupta, D. Nagpal, M. Mukherjee, V. S. Parmar and V. Lather, Virtual screening and antimicrobial evaluation for identification of natural compounds as the prospective inhibitors of antibacterial drug resistance targets in Staphylococcus aureus, Fitoterapia, 2023, 168, 105554 CrossRef CAS PubMed.
- S. Sinha, V. K. Gupta, P. Kumar, R. Kumar, R. Joshi, A. Pal and M. P. Darokar, Usnic acid modifies MRSA drug resistance through down-regulation of proteins involved in peptidoglycan and fatty acid biosynthesis, FEBS Open Bio, 2019, 9, 2025–2040 CrossRef CAS PubMed.
- S. R. Prasad, P. Kumar, S. Mandal, A. Mohan, R. Chaurasia, A. Shrivastava, P. Nikhil, D. Aishwarya, P. Ramalingam, R. Gajbhiye, S. Singh, A. Dasgupta, M. Chourasia, V. Ravichandiran, P. Das and D. Mandal, Mechanistic insight into the role of mevalonate kinase by a natural fatty acid-mediated killing of Leishmania donovani, Sci. Rep., 2022, 12, 16453 CrossRef CAS PubMed.
- R. A. Friesner, R. B. Murphy, M. P. Repasky, L. L. Frye, J. R. Greenwood, T. A. Halgren, P. C. Sanschagrin and D. T. Mainz, Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes, J. Med. Chem., 2006, 49, 6177–6196 CrossRef CAS PubMed.
- Association for Computing Machinery New York NY United States, in Proceedings of the 2006 ACM/IEEE conference on Supercomputing - SC ‘06, ACM Press, New York, New York, USA.
- S. Sangam, A. Gupta, A. Shakeel, R. Bhattacharya, A. K. Sharma, D. Suhag, S. Chakrabarti, S. K. Garg, S. Chattopadhyay and B. Basu, Sustainable synthesis of single crystalline sulphur-doped graphene quantum dots for bioimaging and beyond, Green Chem., 2018, 20, 4245–4259 RSC.
- A. Agarwal, S. Jeevanandham, C. Kar, M. P. Rai, V. Kumar, H. B. Bohidar, S. Biswas and M. Mukherjee, Crystalline Domains Nested on Two-Dimensional Nanosheets as Heterogeneous Nanomachineries for the Sustainable Production of Bioactive Compounds from Chlorella sorokiniana, ACS Sustainable Chem. Eng., 2022, 10, 9732–9748 CrossRef CAS.
- K.-G. Zhou, F. Withers, Y. Cao, S. Hu, G. Yu and C. Casiraghi, Raman Modes of MoS2 Used as Fingerprint of van der Waals Interactions in 2-D Crystal-Based Heterostructures, ACS Nano, 2014, 8, 9914–9924 CrossRef CAS PubMed.
- K. Ganesan, S. Ghosh, N. G. Krishna, S. Ilango, M. Kamruddin and A. K. Tyagi, A comparative study on defect estimation using XPS and Raman spectroscopy in few layer nanographitic structures, Phys. Chem. Chem. Phys., 2016, 18, 22160–22167 RSC.
- A. C. Ferrari and D. M. Basko, Raman spectroscopy as a versatile tool for studying the properties of graphene, Nat. Nanotechnol., 2013, 8, 235–246 CrossRef CAS PubMed.
- W.-S. Tseng, C.-Y. Tseng and C.-T. Kuo, Effects of gas composition on highly efficient surface modification of multi-walled carbon nanotubes by cation treatment, Nanoscale Res. Lett., 2009, 4, 234–239 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lf00120f |
| ‡ O. A., H. K. S. and R. C. contributed equally as the first author. |
| This journal is © The Royal Society of Chemistry 2024 |