Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Targeted release of live probiotics from alginate-based nanofibers in a simulated gastrointestinal tract†
Emily
Diep
 and
Jessica D.
Schiffman
and
Jessica D.
Schiffman
 *
*
Department of Chemical Engineering, University of Massachusetts Amherst, Amherst, Massachusetts 01003-9303, USA. E-mail: schiffman@umass.edu
First published on 21st May 2024
Abstract
Gut bacteria influence human health by digesting nutrients, modulating immune responses, and communicating with the nervous system. Orally delivered probiotics must survive the harsh environment of stomach acid to reach the gut microbiome and incur a health benefit. Here, the probiotic Lactococcus lactis was encapsulated via coaxial electrospinning into alginate-based nanofibers containing the antacid calcium carbonate. Though the high molecular weight polyethylene oxide was used to facilitate fiber formation, crosslinking the nanofibers in an aqueous solution allowed the polyethylene oxide to diffuse out of the nanofiber to form a safe for oral consumption formulation. The antacid protected the encapsulated living bacteria against acidic insults; thus, bacteria remained viable and encapsulated during submersion in a simulated stomach model. After transfer to the intestinal phase, up to 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 viable probiotic cells were released per gram of nanofibers demonstrating the pH-dependent delivery of the electrospun alginate nanofibers.
000 viable probiotic cells were released per gram of nanofibers demonstrating the pH-dependent delivery of the electrospun alginate nanofibers.
Introduction
In the gut, a community of bacteria influences human health by converting food into nutrients, defending the body against pathogenic infection, and communicating with the immune and nervous systems.1–3 Researchers have found that a balanced and diverse community is key to the regulation of immune responses.4,5 Thus, the delivery of bacteria through probiotic supplements can be used to modulate the gut community to produce biotherapeutic effects.6,7 Bacteria cells can be freeze-dried to increase their shelf-life, while also forming them into a powder that can be incorporated into an oral supplement.8,9 While the processing conditions, low temperature, and pressure used during free drying can be harmful to cells, the encapsulation of bacteria within materials like, proteins, carbohydrates, or polymers protects the cells from damage during processing. Other technologies – spray drying, emulsions, microfluidics, 3D printing, extrusion, etc. – have also been used to encapsulate bacteria in a variety of polymers to improve cell viability during processing, storage, and usage.6,10–13Recently, electrospinning has been used to encapsulate bacteria cells into multifunctional polymer nanofibers.7,14,15 The high surface area-to-volume matrix makes electrospun nanofiber scaffolds ideal for rapid release from oral dosages, such as oral probiotic supplements. Encapsulated bacteria remain viable for weeks at room temperature and months under refrigeration while encapsulated in the nanofibers.16 Various bacterial strains have been encapsulated using electrospinning, primarily using biocompatible, synthetic polymers like polyethylene oxide and polyvinyl alcohol.16–20 While synthetic polymers can be easily electrospun, natural polymers provide biocompatibility and additional functional benefits.
For example, the natural polymer alginate, composed of 1,4-linked α-L-guluronic acid and β-D-mannuronic acid groups, can be ionically crosslinked with biocompatible calcium ions and has pH-dependent behavior for the targeted delivery of cargo into the intestines.21–24 In acidic environments, protonated alginate chains are held together by hydrogen bonding to maintain the encapsulation mechanism. In higher pH environments, like that of the intestines where the gut bacteria reside, the alginate chains are deprotonated and repel each other which causes the structure to swell and release encapsulants. Though the formation of alginate nanofibers from aqueous solutions has been challenging, the use of biocompatible carrier polymers and Food and Drug Administration (FDA)-approved surfactants has been shown to facilitate nanofiber formation and safely encapsulate viable bacteria.25–27
In this work, probiotic-loaded nanofibers were coaxially electrospun, crosslinked, and used to deliver live bacteria cells into a simulated gastrointestinal tract. Coaxial electrospinning allowed high concentrations of the probiotic Lactococcus lactis to be drawn into a precursor solution containing alginate and antacid. Crosslinking methods, which were previously explored for alginate solutions electrospun using single-nozzle electrospinning, were used to chemically stabilize the coaxial fibers while maintaining the viability of encapsulated bacteria. Crosslinking in aqueous solution would also allow the carrier polymers and surfactants required for electrospinning to diffuse out of the nanofiber forming a safe, oral delivery system. The pH-dependent behavior of these crosslinked alginate nanofibers carries bacteria through the stomach and releases them to the bacterial gut community located in the intestines. For the first time, it was demonstrated that for the probiotics to survive in the harsh stomach acid environment, an antacid, calcium carbonate, was needed in the nanofiber formulation.
Experimental
Materials
All chemicals were used as received without further purification. Low-viscosity alginic acid sodium salt from brown algae (SA, ν = 4–12 cP, 1 wt% in H2O at 25 °C, MW = 58.9 kDa, M/G ratio = 2.65)28 was purchased from Sigma Aldrich (St Louis, MO). Brain heart infusion (BHI), calcium carbonate (CaCO3), calcium chloride (CaCl2), glutaraldehyde, poly(ethylene oxide) (PEO, Mw = 600 kDa), polysorbate 80 (PS80), potassium phosphate (KH2PO4), sodium chloride (NaCl), sodium phosphate dibasic (Na2HPO4) were purchased from Sigma Aldrich (St Louis, MO). Agar, hydrochloric acid (HCl, percent purity: 36.5–38.0%), potassium chloride (KCl), and sodium hydroxide (NaOH) were purchased from Fisher Scientific (Hampton, NH). Deionized (DI) water was obtained from a Barnstead Nanopure Infinity water purification system (Thermo Fisher Scientific, Waltham, MA).Preparation of Lactococcus lactis
Lactococcus lactis (L. lactis) 11454 was grown overnight (16 h) in BHI media (37 g L−1 in DI water autoclaved at 120 °C, 1 bar, for 15 min) in an incubator (37 °C) on a shake plate (250 rpm). Centrifugation (2500g at 25 °C, Sorvall™ ST 40R, Thermofisher Scientific, Waltham, MA) for 5 min was used to pelletize the cells. Pellets were washed thrice with phosphate buffered saline (PBS, 8 g L−1 NaCl, 0.2 g L−1 KCl, 1.44 g L−1 Na2HPO4, 0.24 g L−1 KH2PO4 in DI water, adjusted to pH 7.4 using HCl) before resuspension in sterile DI water.Preparation of coaxially electrospun L. lactis in alginate-based nanofibers
The core precursor solution was made by dissolving 5 wt% of SA in sterile DI water. The SA solution was mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with the bacteria suspension via solution rotation (20 rpm) in a Roto-Therm™ Incubated Tube Rotator (Benchmark Scientific, Sayreville, NJ) for 1 h before electrospinning. The final core precursor solution contained 2.5 wt% SA with 5 × 109 colony forming units (CFU) mL−1. To create the shell precursor solution, SA and PEO were dissolved in sterile DI water, respectively, and mixed via rotation for 24 h. SA, PEO, PS80, and CaCO3 were then rotated together for 24 h to create a final concentration of 2.5/1.5/3/2 wt% SA/PEO/PS80/CaCO3.
1 ratio with the bacteria suspension via solution rotation (20 rpm) in a Roto-Therm™ Incubated Tube Rotator (Benchmark Scientific, Sayreville, NJ) for 1 h before electrospinning. The final core precursor solution contained 2.5 wt% SA with 5 × 109 colony forming units (CFU) mL−1. To create the shell precursor solution, SA and PEO were dissolved in sterile DI water, respectively, and mixed via rotation for 24 h. SA, PEO, PS80, and CaCO3 were then rotated together for 24 h to create a final concentration of 2.5/1.5/3/2 wt% SA/PEO/PS80/CaCO3.
Electrospinning was conducted in an environmental box kept at 23 °C and 20–30% relative humidity. Both core and shell solutions were loaded into 5 mL syringes and connected to a homemade coaxial needle with a blunt 20-gauge inner needle and a blunt 14-gauge outer needle. Three core/shell flow rate combinations (0.35/0.70 mL h−1, 0.70/070 mL h−1, and 0.70/0.35 mL h−1) were used in this study. Horizontal syringe pumps advanced the solutions from the syringes to specified rates. Aluminum covered copper collector plate (15 cm × 15 cm × 0.32 cm) was used for sample collection. A high voltage supply connected to the coaxial needle and the collector plate via alligator clips which applied 17.5 kV over a tip-to-collector distance of 18 cm.
Crosslinking of L. lactis-loaded, alginate-based nanofibers
Crosslinking solutions were sterilized in Stericup® Quick Release Vacuum Driven Disposable Filtration System, (MiliporeSigma, Burlington, MA) before use. After electrospinning, as-spun nanofibers were removed from aluminum foil, crosslinked in either 2 wt% CaCl2 in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 glycerol
1 glycerol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water for 1 min (CLNF-G) or 2 wt% CaCl2 in DI water (CLNF-W), and rinsed in sterile DI water for 1 min to remove excess calcium ions. Sample names and crosslinking methods are outlined in Table 1.
water for 1 min (CLNF-G) or 2 wt% CaCl2 in DI water (CLNF-W), and rinsed in sterile DI water for 1 min to remove excess calcium ions. Sample names and crosslinking methods are outlined in Table 1.
| Name | Crosslinking solution | Rinsing |
|---|---|---|
| As-spun | n/a | n/a |
| CLNF-G | 1 min in 2% CaCl2 in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 glycerol 1 glycerol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water water |
1 min in water |
| CLNF-W | 1 min in 2% CaCl2 in water | 1 min in water |
Scanning electron microscopy of bacteria cells and electrospun nanofibers
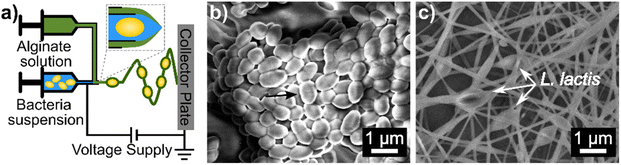
For SEM imaging of bacteria cells, overnight cultures of L. lactis in BHI media were incubated. The bacterial suspension was adjusted to OD600 = 0.3–0.4 by diluting with BHI media, inoculated onto filter paper (10 μL), and dried at room temperature for 1 h. The bacteria-loaded filter paper was fixed using 2.5 w/v% of glutaraldehyde in PBS for 2 h at room temperature29 and then rinsed in DI water twice. After fixation, samples were dehydrated in 30%, 50%, 75%, 85%, 95%, and 100% alcohol–water gradients for 5 min in each solution.30 Samples were oven-dried at 70 °C before being mounted to SEM stubs with carbon tabs for analysis.Bacteria and nanofiber samples were sputter-coated (Cressington208 Sputter Coater, Watford, UK) with 3 nm of platinum before imaging with FEI Magellan 400 XHR-SEM (ThermoFisher Scientific, Hillsboro, OR). The diameters of 50 random fibers from five micrographs were measured using ImageJ 1.52a software (National Institutes of Health, Bethesda, MD) to determine the average fiber diameter.31 Fiber diameter was measured as consistent widths across fibers excluding bulges that might have been bacterial cells.
Thermogravimetric analysis (TGA) of L. lactis-loaded nanofibers
For characterization, nanofibers using the 0.35/0.70 mL h−1 core/shell flow rate were utilized to maximize sample production per volume of precursor solutions. As-spun nanofibers and crosslinked nanofiber samples (5–10 mg) were weighed while heating from 20–600 °C at a scan rate of 10 °C min−1 in nitrogen gas (25 mL min−1) using Q50 Thermogravimetric Analyzer (TA Instruments, New Castle, DE).32,33 Mass loss per step was determined as the difference is mass (%) between local minima using first derivative thermograms.Bacterial loading tests
The bacterial loading within as-spun nanofibers was determined by dissolved nanofibers in DI water and performing single plate-serial dilution spotting (SP-SDS) drop plate method on BHI agar plates.34 For crosslinked samples, additional steps of suspending in BHI, vertexing (1 min), and incubating (37 °C, 250 rpm, 1 h) were used to release bacteria from the nanofibers prior to serial dilution. The plates were incubated for 16 h to allow cells to develop into visible colonies which were used to directly calculate the number of colony forming units (CFU) per g of as-spun nanofiber mat (XP204 Analytical Balance, Mettler Toledo, Vernon Hills, IL).Release of L. lactis into a simulated gastrointestinal tract (GIT)
The gastrointestinal tract (GIT) model used in this study consists of the nanofiber mats being incubated in a solution that mimicked the “stomach” followed by incubation in a solution that mimicked the “intestines”. Specifically, L. lactis was coaxially electrospun into alginate-based nanofibers, as previously described. As-spun fibers (5–10 mg) were crosslinked as previously described and outlined in Table 1. Next, the nanofiber mats were sequentially incubated at 37 °C for 2 h in the solution that mimicked the (1) stomach then (2) the intestines. To prepare the acidic stomach solution (2 mL), HCl was added dropwise to DI water until a pH value of 2.5 was obtained. The neutral intestinal stage (2 mL) was prepared by dissolving NaOH in water and diluting with DI water until a pH value of 7.0 was obtained.35 Each solution was freshly prepared before each experimental replicate. The nanofiber mats were blotted on the sides of the wells to remove excess solution before transfer to the next stage. At predetermined time points, the simulated stomach solution or the simulated intestinal solution was sampled (20 μL) and plated on BHI agar. After plates were incubated for 24 h, colony counts were used to determine the number of viable bacteria cells released per gram of as-spun nanofiber throughout the model. The pH value of the simulated stomach phase and intestinal phase was also measured using the Fisherbrand™ accumet™ Benchtop pH meter (Hampton, NH) after samples had been removed from each stage.Statistical analysis
Significant differences between samples were determined using one-way ANOVA with Tukey post-hoc test on OriginPro 2022 (OriginLab, Northampton, MA) between multiple samples. Fiber diameters are expressed as mean ± standard deviation. Bacteria loading and pH values of the GIT model are reported as mean ± standard error. Standard deviation and standard error are represented as error bars in figures. Statistical significance was set to p < 0.05.Results and discussion
Coaxial electrospinning of L. lactis in alginate-based nanofibers
Here, we report the encapsulation of L. lactis into alginate nanofibers with CaCO3 antacid using coaxial electrospinning. A schematic of the coaxial electrospinning setup, as well as SEM micrographs of free and electrospun L. lactis are included in Fig. 1. A concentrated suspension of bacteria mixed with alginate solution was loaded into the inner needle, which was electrospun at the same time as the alginate/antacid precursor solution that was loaded into the outer needle. Flowing alginate in the core needle helped to reduce interfacial tension variance that can occur where both solutions meet at the coaxial needle tip.36,37 “Electrospinnable” solutions are preferred for the shell solution in coaxial electrospinning to carry along the core solution.36,38 Specifically, our shell solution consisted of a 2.5/1.5/3% mixture of SA/PEO/PS80 which was selected because it formed uniform nanofibers when electrospun using a single nozzle set up in a previous study.24For the first time, 2% CaCO3 was mixed into the shell solution to improve the acid survivability of encapsulated bacteria. Varying coaxial flow rates of 0.35/0.70, 0.70/0.70, and 0.70/0.35 mL h−1 produced statistically different average fiber diameters of 220.43 ± 83.76 nm, 120.39 ± 39.34 nm, and 137.61 ± 37.14 nm, respectively. The 0.35/0.70 mL h−1 flow rate system may maximize the flow of the more “electrospinnable” outer solution relative to the core solution during electrospinning allowing more polymer to be drawn into the nanofiber. Unless otherwise stated, the 0.35/0.70 mL h−1 core/shell flow rate was used throughout the remainder of this study.
Chemical composition of coaxially as-spun and crosslinked nanofibers
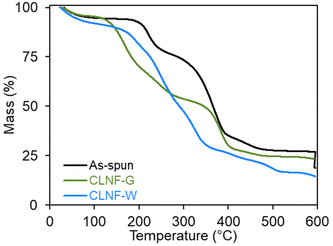
Crosslinking alginate with calcium ions improves the chemical stability of alginate structures in different environments, including aqueous solutions. For alginate nanofibers, different co-solvent systems have been studied to prevent the dissolution of the alginate chains from the nanofiber into aqueous crosslinking solution.24,39 In this study, we explored crosslinking the fibers with CaCl2 in either a water/glycerol mixture (CLNF-G) or water (CLNF-W) solution. Based on previous studies,24 these crosslinking methods may cause diffusion of the water-soluble carrier polymer, PEO, and/or the surfactant, PS80, out of the nanofiber and into the aqueous-based crosslinking solutions.TGA was used to determine the mass composition of the as-spun nanofibers. Bacteria, alginate, and glycerol degrade at lower temperatures (<300 °C)40–42 compared to the degradation temperatures of PEO and PS80 (>300 °C) as shown in Fig. S1.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33,43 As-spun nanofibers displayed two major degradation steps indicative of alginate/bacteria and PEO/PS80 within the nanofiber (Fig. 2). Though TGA cannot distinguish between the degradation of alginate from bacteria or PEO from PS80 due to similar degradation temperature ranges, the composition of the as-spun nanofibers was determined to be 17.7% alginate/bacteria and 42.7% PEO/PS80 based on mass loss in each temperature range with 33.8% of residue commonly associated with organic materials.
33,43 As-spun nanofibers displayed two major degradation steps indicative of alginate/bacteria and PEO/PS80 within the nanofiber (Fig. 2). Though TGA cannot distinguish between the degradation of alginate from bacteria or PEO from PS80 due to similar degradation temperature ranges, the composition of the as-spun nanofibers was determined to be 17.7% alginate/bacteria and 42.7% PEO/PS80 based on mass loss in each temperature range with 33.8% of residue commonly associated with organic materials.
The thermogram of CLNF-G contained two degradation steps similar to thermograms of as-spun nanofibers; however, more mass loss occurred in the lower temperature range due to the additional presence of glycerol. In alginate materials, glycerol forms hydrogen bonding between alginate chains which may hinder the diffusion of PEO and PS80 out of the fiber during crosslinking.24,42 The thermogram indicates the CLNF-G nanofiber composition is 42.6% alginate/glycerol/bacteria and 25.7% PEO/PS80.
Alternatively, CLNF-W only showed one degradation step in the lower temperature range indicating only the presence of bacteria and alginate. In the aqueous crosslinking system, alginate materials swell due to the pH of the solution (10.3) allowing water-soluble PEO and PS80 to diffuse out of the nanofiber and into the crosslinking solution.24 While PEO is biocompatible, high molecular weights are not suitable for renal clearance in the body.44 Thus, the removal of the carrier polymer from the nanofiber is beneficial for the development of oral formulations.
Bacteria loading in crosslinked, coaxially electrospun nanofibers
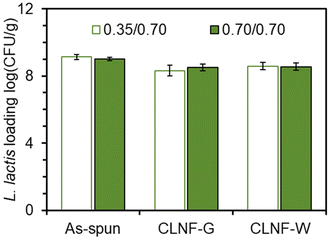
To study the effects of core/shell flow rate on bacteria loading, the flow rate systems 0.35/0.70 mL h−1 and 0.70/0.70 mL h−1 were tested. Bacteria loading in the as-spun nanofibers using these flow rates were statistically equivalent with over 109 CFU g−1 encapsulated cells (Fig. 3 & Table S1†). These studies were conducted with the 035/0.70 mL h−1 core/shell flow rates. The glycerol/water and pure water as the crosslinking solvent enabled the encapsulated bacteria to survive the crosslinking process. The bacteria loading after crosslinking was found to be statistically equivalent to as-spun loading which could allow between 1.33 × 108 and 3.79 × 108 CFU per g of electrospun mat to be delivered depending on the flow rate and crosslinking method used. In the gastrointestinal tract, adequate amounts of released probiotics are necessary to incur biotherapeutic effects.2,45Release of L. lactis from nanofibers in a simulated gastrointestinal tract
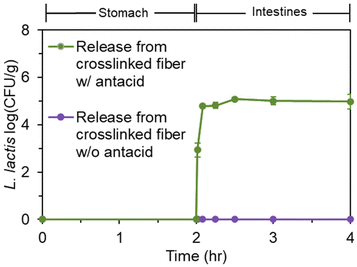
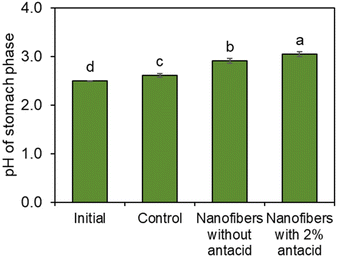
Next, a gastrointestinal tract model35 was used to highlight the pH-dependent behavior of the bacteria-loaded, electrospun alginate nanofibers. Initially, electrospun L. lactis without antacid was subjected to the simulated gastrointestinal tract. However, viable bacteria were not detected in the stomach phase or the intestinal phase. Without antacid, protons can penetrate the alginate encapsulation to deactivate bacteria.Thus, an antacid was formulated into the shell solution prior to electrospinning. Again, no viable bacteria were detected in the stomach phase after two hours of submersion of the fibers containing both L. lactis and antacid (Fig. 4). However, as desired, over 105 CFU was released per g of mat after transfer into the intestinal phase. The concentration of bacteria remained steady for the remaining duration in the intestinal tract. This behavior is consistent with our previous study24 on the pH-dependent behavior of alginate nanofibers, which is stable in acidic environments and swells in higher pH environments. In a study by Zhang et al.,46 alginate microgels containing antacid maintained a neutral pH during submersion in an acidic environment. This suggested the antacid could neutralize protons that penetrate the alginate structure and reduce damage to encapsulated bacterial cells.47 The pH value of the stomach phase was measured after submersion of bacteria-loaded, alginate-based nanofibers electrospun with and without CaCO3 to determine the effects of the antacid-loaded nanofibers on the pH of the stomach (Fig. 5). It was determined that the initial pH of the stomach (2.5 ± 0.0), the pH after treatment with nanofibers without antacid (2.9 ± 0.1), and the pH after treatment with fibers containing antacid (3.0 ± 0.0) were all statistically different from each other. The increase in pH can be attributed to both the presence of the fiber and antacid delivered from the electrospun nanofibers that likely dissolved into the stomach phase.47 Overall, these pH values are well within the normal pH range of the stomach (2.5–3.0), and the antacid enabled the survivability of encapsulated bacteria in the stomach.
Conclusion
Encapsulation systems can protect probiotics against the harsh, acidic environment of the stomach and deliver probiotics to the gut community in the intestines to influence human health. In this study, a combination of alginate and the antacid, CaCO3, were used to encapsulate probiotic bacteria for targeted delivery in a simulated gastrointestinal tract. Coaxial electrospinning enabled the encapsulation of high loadings of the probiotic L. lactis into alginate-based nanofibers. While the carrier polymer, polyethylene oxide, and a surfactant, polysorbate 80, helped facilitate alginate nanofiber formation, the high molecular weight of PEO is not ideal for ingestion. The nanofiber composition was altered during crosslinking of the alginate chains in an aqueous solution. TGA revealed the diffusion of PEO and PS80 out of the nanofibers leaving behind a calcium alginate nanofiber. The aqueous crosslinking systems also supported the viability of encapsulated bacteria.Without antacid, no viable bacteria were detected in the simulated gastrointestinal solutions. The formulation of 2% calcium carbonate (antacid) into the outer precursor solution proved imperative for the survival of the encapsulated L. lactis in a simulated gastrointestinal tract. Adding the antacid did not impact bacterial loading within the nanofibers. No viable bacteria were released in the simulated stomach phase due to hydrogen bonding between protonated alginate chains whereas encapsulated bacteria were released into the simulated intestinal phase due to swelling of the alginate nanofibers in higher pH environments. After the delivery to the gut community, the probiotics can promote various health benefits. While this work exemplifies one application of electrospun probiotics, it may help to motivate future research of beneficial bacteria-loaded nanofibers.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
E. D. acknowledges support from the Spaulding Smith Fellowship and the PPG Fellowship. We acknowledge the facilities at the W. M. Keck Center for Electron Microscopy.References
- S. K. Dash, A. N. Spreen and B. M. Ley, Health Benefits of Probiotics, BL Publications, 2000 Search PubMed.
- FAO/WHO, Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria, Cordoba, Argentina, 2001 Search PubMed.
- K. Fenster, B. Freeburg, C. Hollard, C. Wong, R. Rønhave Laursen and A. C. Ouwehand, Microorganisms, 2019, 7, 83 CrossRef CAS PubMed.
- J. L. Round and S. K. Mazmanian, Nat. Rev. Immunol., 2009, 9, 313–323 CrossRef CAS PubMed.
- C. Petersen and J. L. Round, Cell. Microbiol., 2014, 16, 1024–1033 CrossRef CAS PubMed.
- A. K. Anal and H. Singh, Trends Food Sci. Technol., 2007, 18, 240–251 CrossRef CAS.
- L. Deng and H. Zhang, ES Food Agrofor., 2020, 2, 3–12 Search PubMed.
- F. Fonseca, S. Cenard and S. Passot, in Cryopreservation and Freeze-Drying Protocols, ed. W. F. Wolkers and H. Oldenhof, Springer, New York, NY, 2015, pp. 477–488 Search PubMed.
- W. Savedboworn, K. Teawsomboonkit, S. Surichay, W. Riansa-ngawong, S. Rittisak, R. Charoen and K. Phattayakorn, Food Sci. Biotechnol., 2019, 28, 795–805 CrossRef CAS PubMed.
- S. Moumita, B. Das, U. Hasan and R. Jayabalan, LWT – Food Sci. Technol., 2018, 96, 127–132 CrossRef CAS.
- P. Wongkongkatep, K. Manopwisedjaroen, P. Tiposoth, S. Archakunakorn, T. Pongtharangkul, M. Suphantharika, K. Honda, I. Hamachi and J. Wongkongkatep, Langmuir, 2012, 28, 5729–5736 CrossRef CAS PubMed.
- T. Alkayyali, T. Cameron, B. Haltli, R. G. Kerr and A. Ahmadi, Anal. Chim. Acta, 2019, 1053, 1–21 CrossRef CAS PubMed.
- E. Ning, G. Turnbull, J. Clarke, F. Picard, P. Riches, M. Vendrell, D. Graham, A. W. Wark, K. Faulds and W. Shu, Biofabrication, 2019, 11, 045018 CrossRef CAS PubMed.
- E. Diep and J. D. Schiffman, ACS Appl. Bio Mater., 2023, 6, 951–964 CrossRef CAS PubMed.
- A. C. Mendes and I. S. Chronakis, Food Hydrocolloids, 2021, 117, 106688 CrossRef CAS.
- A. López-Rubio, E. Sanchez, Y. Sanz and J. M. Lagaron, Biomacromolecules, 2009, 10, 2823–2829 CrossRef PubMed.
- M. Kurečič, T. Rijavec, S. Hribernik, A. Lapanje, K. S. Kleinschek and U. Maver, Nanomedicine, 2018, 13, 1583–1594 CrossRef PubMed.
- W. Salalha, J. Kuhn, Y. Dror and E. Zussman, Nanotechnology, 2006, 17, 4675–4681 CrossRef CAS PubMed.
- Š. Zupančič, K. Škrlec, P. Kocbek, J. Kristl and A. Berlec, Pharmaceutics, 2019, 11, 483 CrossRef PubMed.
- K. Škrlec, Š. Zupančič, S. Prpar Mihevc, P. Kocbek, J. Kristl and A. Berlec, Eur. J. Pharm. Sci., 2019, 136, 108–119 Search PubMed.
- K. Y. Lee and D. J. Mooney, Prog. Polym. Sci., 2012, 37, 106–126 CrossRef CAS PubMed.
- M. George and T. E. Abraham, J. Controlled Release, 2006, 114, 1–14 CrossRef CAS PubMed.
- S. Ahirrao, P. Gide, B. Shrivastav and P. Sharma, Part. Sci. Technol., 2014, 32, 105–111 CrossRef CAS.
- E. Diep and J. D. Schiffman, Biomacromolecules, 2023, 24, 2908–2917 CrossRef PubMed.
- C. A. Bonino, K. Efimenko, S. I. Jeong, M. D. Krebs, E. Alsberg and S. A. Khan, Small, 2012, 8, 1928–1936 CrossRef CAS PubMed.
- E. Diep and J. D. Schiffman, Biomater. Sci., 2021, 9, 4364–4373 RSC.
- M. T. Yilmaz, O. Taylan, C. Y. Karakas and E. Dertli, Carbohydr. Polym., 2020, 244, 116447 CrossRef CAS PubMed.
- M. Marounek, Z. Volek, T. Taubner, D. Dušková and M. Czauderna, bioRxiv, 2020, 239335.
- Y. Chao and T. Zhang, Appl. Microbiol. Biotechnol., 2011, 92, 381–392 CrossRef CAS PubMed.
- M. Becce, A. Klöckner, S. G. Higgins, J. Penders, D. Hachim, C. J. Bashor, A. M. Edwards and M. M. Stevens, J. Mater. Chem. B, 2021, 9, 4906–4914 RSC.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- P. Kianfar, A. Vitale, S. Dalle Vacche and R. Bongiovanni, Carbohydr. Polym., 2019, 217, 144–151 CrossRef CAS.
- T. Çaykara, S. Demirci, M. S. Eroğlu and O. Güven, Polymer, 2005, 46, 10750–10757 CrossRef.
- P. Thomas, A. C. Sekhar, R. Upreti, M. M. Mujawar and S. S. Pasha, Biotechnol. Rep., 2015, 8, 45–55 CrossRef PubMed.
- L. Mei, F. He, R.-Q. Zhou, C.-D. Wu, R. Liang, R. Xie, X.-J. Ju, W. Wang and L.-Y. Chu, ACS Appl. Mater. Interfaces, 2014, 6, 5962–5970 CrossRef CAS PubMed.
- P. Rathore and J. D. Schiffman, ACS Appl. Mater. Interfaces, 2021, 13, 48–66 CrossRef CAS PubMed.
- A. K. Moghe and B. S. Gupta, Polym. Rev., 2008, 48, 353–377 CrossRef CAS.
- D. Han and A. Steckl, ChemPlusChem, 2019, 84, 1453–1497 CrossRef CAS PubMed.
- C. A. Bonino, M. D. Krebs, C. D. Saquing, S. I. Jeong, K. L. Shearer, E. Alsberg and S. A. Khan, Carbohydr. Polym., 2011, 85, 111–119 CrossRef CAS.
- K. Feng, M.-Y. Zhai, Y. Zhang, R. J. Linhardt, M.-H. Zong, L. Li and H. Wu, J. Agric. Food Chem., 2018, 66, 10890–10897 CrossRef CAS PubMed.
- Md. S. Islam and M. R. Karim, Colloids Surf., A, 2010, 366, 135–140 CrossRef CAS.
- C. Gao, E. Pollet and L. Avérous, Food Hydrocolloids, 2017, 63, 414–420 CrossRef CAS.
- R. S. K. Kishore, A. Pappenberger, I. B. Dauphin, A. Ross, B. Buergi, A. Staempfli and H.-C. Mahler, J. Pharm. Sci., 2011, 100, 721–731 CrossRef CAS PubMed.
- J. J. F. Verhoef and T. J. Anchordoquy, Drug Delivery Transl. Res., 2013, 3, 499–503 CrossRef CAS PubMed.
- K. Qiu, Y. Huang and A. C. Anselmo, Cell. Mol. Bioeng., 2021, 14, 487–499 CrossRef CAS.
- Z. Zhang, R. Zhang, Q. Sun, Y. Park and D. J. McClements, Food Hydrocolloids, 2017, 65, 198–205 CrossRef CAS.
- M. Gu, Z. Zhang, C. Pan, T. R. Goulette, R. Zhang, G. Hendricks, D. J. McClements and H. Xiao, Food Hydrocolloids, 2019, 91, 283–289 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lp00023d |
| This journal is © The Royal Society of Chemistry 2024 |