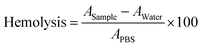Open Access Article
Open Access ArticlePoly(L-glutamic acid) augments the transfection performance of lipophilic polycations by overcoming tradeoffs among cytotoxicity, pDNA delivery efficiency, and serum stability†
Ram Prasad
Sekar
a,
Jessica L.
Lawson
b,
Aryelle R. E.
Wright
c,
Caleb
McGrath
c,
Cesar
Schadeck
b,
Praveen
Kumar
 e,
Jian Wei
Tay
d,
Joseph
Dragavon
d and
Ramya
Kumar
*a
e,
Jian Wei
Tay
d,
Joseph
Dragavon
d and
Ramya
Kumar
*a
aChemical and Biological Engineering, Colorado School of Mines, Golden, CO 80401, USA. E-mail: ramyakumar@mines.edu
bMaterials Science, Colorado School of Mines, Golden, CO 80401, USA
cQuantitative Biosciences and Engineering, Colorado School of Mines, Golden, CO 80401, USA
dBioFrontiers Institute, University of Colorado, Boulder, CO 80303, USA
eShared Instrumentation Facility, Colorado School of Mines, Golden, CO, USA
First published on 30th April 2024
Abstract
Polycations are scalable and affordable nanocarriers for delivering therapeutic nucleic acids. Yet, cationicity-dependent tradeoffs between nucleic acid delivery efficiency, cytotoxicity, and serum stability hinder clinical translation. Typically, the most efficient polycationic vehicles also tend to be the most toxic. For lipophilic polycations—which recruit hydrophobic interactions in addition to electrostatic interactions to bind and deliver nucleic acids—extensive chemical or architectural modifications sometimes fail to resolve intractable toxicity—efficiency tradeoffs. Here, we employ a facile post-synthetic polyplex surface modification strategy wherein poly(L-glutamic acid) (PGA) rescues toxicity, inhibits hemolysis, and prevents serum inhibition of lipophilic polycation-mediated plasmid (pDNA) delivery. Importantly, the sequence in which polycations, pDNA, and PGA are combined dictates pDNA conformations and spatial distribution. Circular dichroism spectroscopy reveals that PGA must be added last to polyplexes assembled from lipophilic polycations and pDNA; else, PGA will disrupt polycation-mediated pDNA condensation. Although PGA did not mitigate toxicity caused by hydrophilic PEI-based polycations, PGA tripled the population of transfected viable cells for lipophilic polycations. Non-specific adsorption of serum proteins abrogated pDNA delivery mediated by lipophilic polycations; however, PGA-coated polyplexes proved more serum-tolerant than uncoated polyplexes. Despite lower cellular uptake than uncoated polyplexes, PGA-coated polyplexes were imported into nuclei at higher rates. PGA also silenced the hemolytic activity of lipophilic polycations. Our work provides fundamental insights into how polyanionic coatings such as PGA transform intermolecular interactions between lipophilic polycations, nucleic acids, and serum proteins, and facilitate gentle yet efficient transgene delivery.
Introduction
Nucleic acids are transforming therapeutic landscapes, not only for relatively rare inherited disorders such as Duchenne's muscular dystrophy, but also for more prevalent diseases such as diabetes, hypercholesterolemia, or cardiovascular diseases.1–3 Anionic, bulky, and hydrophilic, nucleic acids struggle to cross cell membranes and enter cellular targets, making engineered viral vectors indispensable. Although viral vectors are safe, efficient, and tissue-specific,4,5 they are prohibitively expensive to manufacture at scale, curtailing their utility in highly prevalent diseases where patient populations may span hundreds of millions.6–8 To overcome scale-up bottlenecks, synthetic nanocarriers such as lipid nanoparticles, polypeptides, polymers, or inorganic nanoparticles have been extensively investigated for the intracellular delivery of messenger RNA, plasmids (pDNA) or ribonucleoprotein payloads.9–12 Polymers, in particular, can be precisely tailored to fulfill design requirements imposed by diverse nucleic acid payloads and cellular targets.13,14Positive charge centers, typically installed as ionizable primary, secondary, or tertiary amines, are ubiquitous design motifs. Polycations exploit electrostatic interactions to bind and condense nucleic acids into polyplexes. Subsequently, polycations must safeguard their payloads from degradation, improvise cellular uptake, and navigate endolysosomal interrogation.15 Long considered indispensable for polymer-mediated nucleic acid delivery, cationicity is also a double-edged sword. For instance, non-phagocytic cells internalize cationic nanocarriers at higher rates than anionic ones.16,17 But polycations disrupt the integrity of cell membranes, depolarize mitochondria, and accelerate autophagy-mediated degradation of their nucleic acid payloads.16,18 To juggle the seemingly conflicting demands of efficient nucleic acid delivery and the preservation of cellular viability and phenotype, several researchers modulated the distribution of positive charges along polymer backbones.19,20 Others employed charge conversion,21–23 fluorination,24–27 aromatic residues,28–32 or other lipophilic functionalities.33,34 Despite these time-consuming and iterative chemical or architectural modifications to promising polycations, tradeoffs between delivery efficiency and cytotoxicity still persist.35,36
Cytotoxicity apart, polycations are also susceptible to non-specific protein adsorption,37 which triggers rapid polyplex clearance by the reticuloendothelial system.38,39 Cationicity aggravates the deleterious effects of non-specific protein adsorption40 especially since plasma proteins are predominantly negatively charged.41 Even during ex vivo gene delivery, non-specific adsorption of serum proteins inhibits the cellular uptake of polyplexes.42 Cationic polyplexes are generally serum-intolerant, requiring either serum-free or reduced serum media formulations such as OptiMEM. However, serum starvation reduces transgene expression in clinically valuable cell types, requiring us to engineer serum-compatible polycations that transfect diverse cellular targets.43 Recognizing that careful control over cationicity is essential to resolve tradeoffs among nucleic acid delivery efficiency, toxicity, and serum compatibility, researchers incorporated anionic moieties within polycations, creating amphoteric polymers.44,45 Others employed electrostatic self-assembly rather than covalent conjugation to append anionic moieties that mask polyplex cationicity.46 In this “layer-by-layer” approach, anionic glycosaminoglycans47 such as heparin,48 or hyaluronic acid,49–52 polypeptides,53 or other polyanions are added to enhance polyplex interfacial properties.
Prior work on polyanionic surface modifiers focused overwhelmingly on poly-ethylene-imine (PEI), a hydrophilic (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of −0.97
P of −0.97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 54,55) polycation and various polyanionic surface modifiers suppressed non-specific protein adsorption by PEI polyplexes.53 In these studies, polyplex stoichiometry (the C/N/P ratio or the molar ratio of carboxylates in polyanions to amines in polycations to phosphate groups in nucleic acids) defined the magnitude and polarity of polyplex surface charge. For instance, Pack and colleagues found that if PGA incorporation exceeded a critical C/N/P threshold, PEI polyplexes became anionic,17 causing them to be internalized via clathrin-mediated rather than caveolin-mediated endocytosis pathways. Clathrin-trafficked polyplexes would subsequently suffer entrapment within lysosomes, ultimately sabotaging their transfection efficiency. Similarly, for PEI polyplexes coated with poly(ethylene-alt-maleic acid), Pearson and colleagues employed N/P ratios as high as 30 to maintain polyplex cationicity.38 Previous studies explored the roles of polyanion pKa, polyplex stoichiometry, architectural and chemical modifications (such as acetylation) of PEI, focusing almost exclusively on hydrophilic polycations such as PEI rather than lipophilic polycations.56–61 Lipophilic polycations effect highly efficient transfection even at N/P ratios as low as 1, underscoring the importance of non-electrostatic pDNA binding interactions, particularly hydrophobicity.62–65 Because lipophilic polycations are less reliant on electrostatic interactions to bind and deliver pDNA,66 it is unclear whether electrostatically appended polyanionic coatings will conformally coat polyplex surfaces or substantially improve transfection outcomes. In this work, we demonstrate that surface modification with poly(L-glutamic acid) (PGA) helps lipophilic polycations navigate toxicity–efficiency tradeoffs, prevents polycation-induced hemolysis, and promotes plasmid (pDNA) delivery in serum-containing media.
54,55) polycation and various polyanionic surface modifiers suppressed non-specific protein adsorption by PEI polyplexes.53 In these studies, polyplex stoichiometry (the C/N/P ratio or the molar ratio of carboxylates in polyanions to amines in polycations to phosphate groups in nucleic acids) defined the magnitude and polarity of polyplex surface charge. For instance, Pack and colleagues found that if PGA incorporation exceeded a critical C/N/P threshold, PEI polyplexes became anionic,17 causing them to be internalized via clathrin-mediated rather than caveolin-mediated endocytosis pathways. Clathrin-trafficked polyplexes would subsequently suffer entrapment within lysosomes, ultimately sabotaging their transfection efficiency. Similarly, for PEI polyplexes coated with poly(ethylene-alt-maleic acid), Pearson and colleagues employed N/P ratios as high as 30 to maintain polyplex cationicity.38 Previous studies explored the roles of polyanion pKa, polyplex stoichiometry, architectural and chemical modifications (such as acetylation) of PEI, focusing almost exclusively on hydrophilic polycations such as PEI rather than lipophilic polycations.56–61 Lipophilic polycations effect highly efficient transfection even at N/P ratios as low as 1, underscoring the importance of non-electrostatic pDNA binding interactions, particularly hydrophobicity.62–65 Because lipophilic polycations are less reliant on electrostatic interactions to bind and deliver pDNA,66 it is unclear whether electrostatically appended polyanionic coatings will conformally coat polyplex surfaces or substantially improve transfection outcomes. In this work, we demonstrate that surface modification with poly(L-glutamic acid) (PGA) helps lipophilic polycations navigate toxicity–efficiency tradeoffs, prevents polycation-induced hemolysis, and promotes plasmid (pDNA) delivery in serum-containing media.
Owing to its dense negative charge, hydrophilicity, and biodegradability, poly(L-glutamic acid) or PGA is an attractive stealth coating for nanocarriers.67–70 In a head-to head comparison between PGA, alginate, heparan, and poly(aspartic acid), PGA was most effective in inhibiting the non-specific adsorption of serum proteins.71 Hammond and coworkers reported that PGA-coated nanoparticles bind strongly and specifically to ovarian cancer cells67,72 and established that PGA reconfigures intracellular trafficking pathways to favor endosome-evading caveolar pathways over clathrin-mediated pathways.67,72 We chose PGA to augment the interfacial properties and transfection performance of lipophilic polycations due to its anti-inflammatory properties, endosome-evading intracellular trafficking patterns, and potential for receptor-mediated uptake.73,74
We selected p(DIPAEMA56-st-HEMA48) (DIP50H50) as a model lipophilic polycation (Fig. 1), since toxicity–efficiency tradeoffs undermine its utility as a pDNA carrier.75,76 Although this contribution focuses on DIP50H50 and PGA as a model system, our findings will be broadly valuable for other combinations of lipophilic polycations and polyanionic surface modifiers. Our paper informs the design of ternary polyplexes by contributing three fundamentally valuable insights. First, we thoroughly investigated the sequence of addition of the lipophilic polycations (DIP50H50), pDNA payloads, and PGA during the assembly of ternary polyplexes—a variable that has not been explored previously—and found that the addition sequence governed pDNA spatial distribution and conformations. Second, although PGA mitigated toxicity–efficiency tradeoffs for DIP50H50, the toxicity of PEI-based polyplexes remained unaffected. Third, we employed quantitative confocal microscopy to map differences in nuclear localization between uncoated and PGA-coated polyplexes. Our work illuminates binding interactions between polycations, nucleic acids, and serum proteins, facilitating rational design of surface modification strategies for lipophilic polycations.
Results and discussion
Here, we demonstrate that PGA mitigates tradeoffs between cytotoxicity, pDNA delivery efficiency, and serum stability for polyplexes formed from the lipophilic polycation, DIP50H50. First, CD spectroscopy identified the optimal sequence of addition of DIP50H50, pDNA, and PGA to mask cationicity without triggering pDNA leakage. Subsequently, we performed pDNA delivery assays, establishing that PGA is a highly effective surface modifier that triples the population of transgene-expressing cells arising from DIP50H50-mediated transfection. PGA also assists DIP50H50 in forming stable polyplexes in serum-containing media, potentiating serum-tolerant transfection. Confocal analysis revealed that, despite lower cellular uptake, PGA-coated polyplexes have a higher propensity to occupy nuclear interiors than uncoated polyplexes. Finally, hemolysis assays demonstrated that PGA augments the hemocompatibility of DIP50H50 polyplexes.Addition sequence dictates polycation–pDNA binding configurations
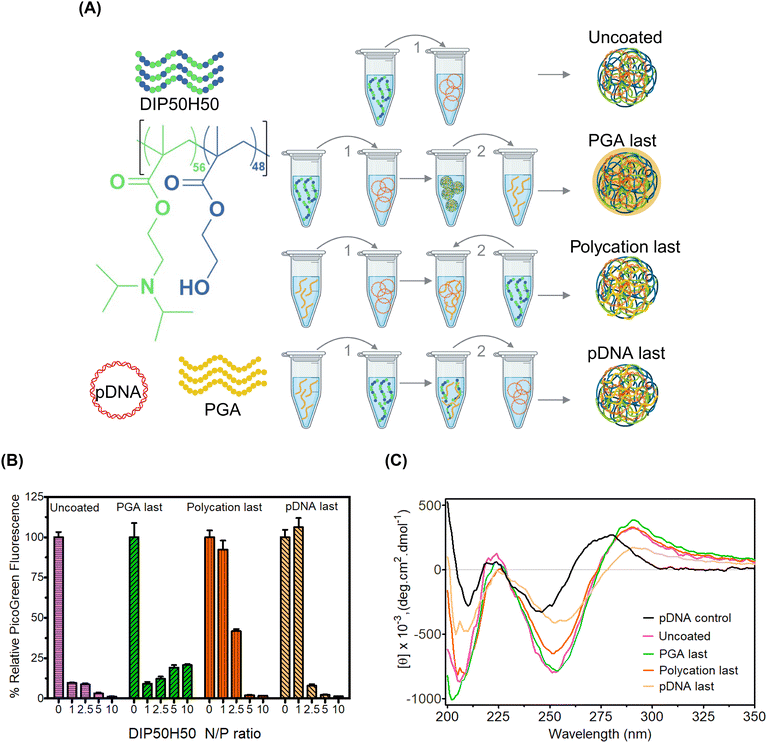
Our rationale is that for PGA to overcome tradeoffs among pDNA delivery efficiency, toxicity, and serum stability without perturbing DIP50H50–pDNA binding interactions, the addition sequence must be optimized first. PGA molecules must occupy polyplex exteriors instead of competing with pDNA for polycation binding sites in polyplex cores. The sequence of addition of polycations and pDNA modulates exerts dramatic effects on polyplex size distribution and encapsulation efficiency.77,78 However, we do not understand how the sequence in which polycations, pDNA, and PGA are combined (Fig. 1) impacts polyplex properties and performance. We tested four addition sequences (Fig. 1A): (1) for uncoated polyplexes, we added the polycation, DIP50H50 to pDNA and realized desired N/P ratios (molar ratio of protonatable amines in DIP50H50 to phosphates in pDNA), (2) for “PGA last” polyplexes, we added PGA to uncoated polyplexes, (3) for “polycation last” polyplexes, we added DIP50H50 to a mixture of PGA and pDNA, (4) for “pDNA last” polyplexes, we added the pDNA payload to a mixture of DIP50H50 and PGA. We kept the C/N ratio (molar ratios of carboxylates in PGA to amines in DIP50H50) fixed at 1 for sequences (2)–(4). During preliminary experiments, we discovered that a C/N ratio of 1 was sufficient to realize polyplex charge reversal from cationic to anionic, and that higher C/N ratios did not offer any benefits.For all three addition sequences, PGA reverses the polarity of polyplex surface charge from positive to negative, confirming that PGA effectively masks cationicity (Fig. S2(a) in the ESI†). Further, the addition sequence had no impact whatsoever on polyplex size distributions (Fig. S2(b) in the ESI†). Using dye exclusion assays we asked whether the addition sequence influenced pDNA encapsulation efficiency (Fig. 1B). In PicoGreen assays, fluorescence quenching estimates the strength of DIP50H50−pDNA binding. The fluorescence intensity of uncoated polyplexes decreased steeply upon complexation with DIP50H50; strong DIP50H50−pDNA binding was observed at N/P ratios as low as 1. In PGA last polyplexes, results mirrored that of the uncoated polyplexes at an N/P value of 1. However at this N/P value, polycation last and pDNA last polyplexes exhibited four- to eight-fold higher fluorescence intensities than uncoated polyplexes. However, a large excess of polycation counteracted the destabilizing influence of PGA in polycation last and pDNA last polyplexes. For instance, the fluorescence intensity at the highest N/P ratios (N/P of 5 and 10) in the polycation last and pDNA last polyplexes was on par with values from the uncoated and the PGA last polyplexes.
To deepen our understanding of how PGA addition sequence impacts DIP50H50–pDNA binding, we performed circular dichroism (CD) spectroscopy, a highly sensitive tool that detects helicity changes as pDNA is condensed. CD tracked pDNA conformational changes as a function of addition sequence (Fig. 1C). Neither DIP50H50 nor PGA generated CD signals above 220 nm (Fig. S3 in the ESI†), so any CD signal above 220 nm is from pDNA alone. CD spectra were normalized with respect to relative pDNA concentration, which was determined from the absorbance maximum (between 250–275 nm). Uncomplexed pDNA exhibited a positive band around 280 nm and a negative band around 250 nm, consistent with its native B-type helical conformation. When DIP50H50 is added to pDNA, CD spectra shifted to the right and the molar ellipticity increased. This increased ellipticity is consistent with polycation-induced changes in pDNA conformation, wherein DIP50H50 collapses the native B-type helical structure of pDNA, triggering a conformational shift to C-type structures.53
CD spectra for PGA last polyplexes and uncoated polyplexes were near-identical, suggesting the PGA forms an anionic coating around polyplexes without perturbing DIP50H50–pDNA interactions in polyplex cores. As for polycation last polyplexes, the ellipticity of the negative band at 250 nm was lower than uncoated and PGA last polyplexes. We attribute this reduced ellipticity to competition between pDNA and PGA for binding sites on DIP50H50. In polycation last polyplexes, pDNA has fewer opportunities to complex with DIP50H50, hindering pDNA condensation and depressing ellipticity. Finally, the CD spectra of pDNA last polyplexes closely resembled uncomplexed pDNA. In pDNA last polyplexes, polycations bind PGA preferentially over pDNA, leaving fewer polycation binding sites available for pDNA payloads. Unencumbered by polycations, uncondensed pDNA occupies the polyplex exterior, adopting conformations nearly identical to that of uncomplexed pDNA. Previously, CD spectroscopy tracked the unfolding of -helices in PGA and identified an isodichroic point at 204 nm, where folded and unfolded PGA have the same CD signal.79 This 204 nm signal is far more prominent in polyplexes where PGA is added last compared to those where polycation or pDNA is added last. Cumulatively, our data suggests that the sequence of addition influences the organization of pDNA and PGA within the polyplexes.
The order in which polycations, PGA, and pDNA are combined governs how tightly DIP50H50 binds pDNA payloads and the extent to which PGA interferes with DIP50H50–pDNA complexation. PGA will be most effective as a surface modifier when the PGA last addition sequence is employed; in other sequences, PGA weakens DIP50H50–pDNA binding.
PGA resolves cationicity-dependent tradeoffs between pDNA delivery efficiency and cytotoxicity
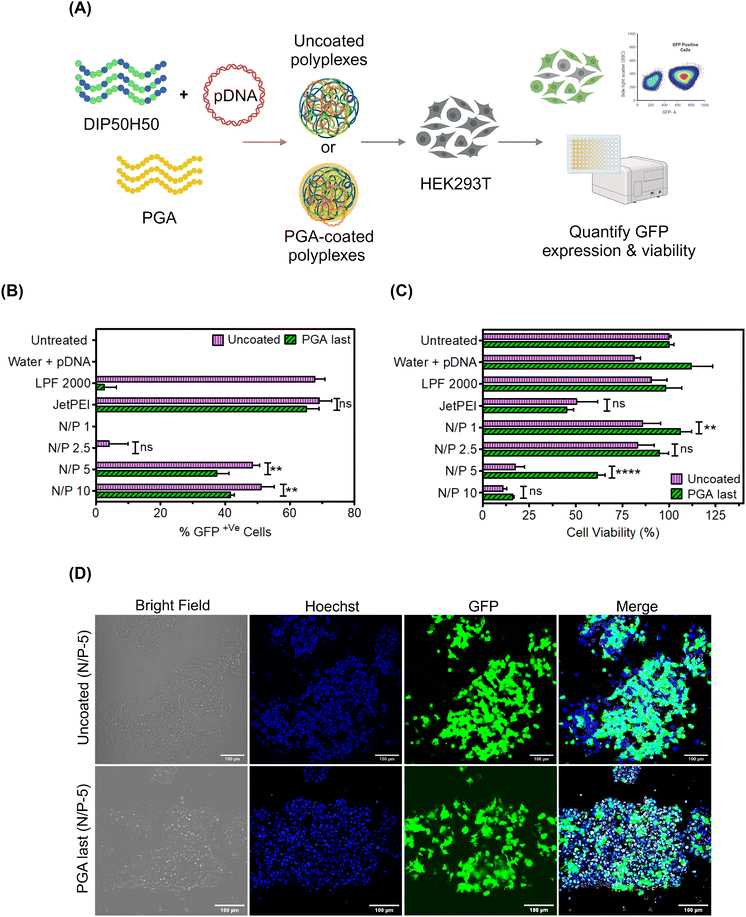
Based on CD spectroscopy results, we decided to focus on PGA last polyplexes in the rest of the paper; results from other addition sequences are furnished in the ESI (Fig. S4 and S8–S10†) Having identified PGA last as the optimal addition sequence to modify polyplex surfaces without triggering pDNA leakage, we next asked whether PGA helps DIP50H50 overcome toxicity–efficiency tradeoffs (Fig. 2). We formulated uncoated and PGA-coated DIP50H50 polyplexes with GFP-encoding pDNA and compared transfection efficiency and cell viability (Fig. 2A). Lipid- and PEI-based commercial reagents for pDNA delivery, Lipofectamine 2000 and JetPEI, were employed as positive controls. Although uncoated polyplexes exhibited marginally higher transfection efficiency than PGA-coated polyplexes (Fig. 2B and D), cell viability was three-fold higher for PGA-coated polyplexes (Fig. 2C).The percentage of GFP+ cells—the fraction of viable cells expressing GFP—can be a misleading measure of transfection efficiency. Since we assign equal priority to cellular viability and transfection efficiency, the number of GFP+ cells is a more useful metric than either the percentage of GFP+ cells or cell viability alone. PGA-coated polyplexes exhibited lower transfection efficiency (by 10–15%) and higher viability (by 10–35%) than uncoated polyplexes. PGA tripled the population of GFP+ cells (Fig. S5 in the ESI†) relative to uncoated polyplexes and helped DIP50H50 mediate gentle yet efficient pDNA delivery. From our correlation analysis (Fig. S6 and Table S1 in the ESI†), we observed a strong negative correlation (−0.935 ± 0.55) between cell viability and transfection efficiency in uncoated polyplexes but PGA coating weakened this correlation considerably (−0.84 ± 0.13). We concluded that surface modification with PGA mitigates cationicity-dependent tradeoffs between cytotoxicity and delivery efficiency for lipophilic polycations like DIP50H50.
Interestingly, PGA exerted divergent effects on the transfection performance of lipophilic (DIP50H50, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of 6.475) and hydrophilic polycations (JetPEI, log
P of 6.475) and hydrophilic polycations (JetPEI, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of −0.9754,55). While PGA blunted the toxicity of DIP50H50, JetPEI was largely indifferent to PGA, exhibiting similar toxicity and transgene expression levels with or without PGA. We measured the reduction in ζ-potential for JetPEI and DIP50H50 polyplexes after PGA coating (Fig. S7(a) in the ESI†), and observed striking differences. PGA transformed the surface character of DIP50H50 polyplexes from moderately cationic (+17 ± 1 mV) to strongly anionic (−30 ± 4 mV), suggesting that PGA conformally covered polyplex surfaces. In contrast, PGA did not reduce the surface charge of JetPEI polyplexes (+27 ± 3 mV to −2.5 ± 15 mV) to the same extent as it did with DIP50H50, suggesting that the surface coverage of JetPEI polyplexes by PGA was patchy and incomplete. We attribute the contrasting effects of PGA on the transfection performance DIP50H50 and JetPEI to incomplete surface coverage of JetPEI polyplexes by PGA. Overall, PGA proved to be a more effective surface modifier for polyplexes formed from lipophilic polycations than hydrophilic polycations.
P of −0.9754,55). While PGA blunted the toxicity of DIP50H50, JetPEI was largely indifferent to PGA, exhibiting similar toxicity and transgene expression levels with or without PGA. We measured the reduction in ζ-potential for JetPEI and DIP50H50 polyplexes after PGA coating (Fig. S7(a) in the ESI†), and observed striking differences. PGA transformed the surface character of DIP50H50 polyplexes from moderately cationic (+17 ± 1 mV) to strongly anionic (−30 ± 4 mV), suggesting that PGA conformally covered polyplex surfaces. In contrast, PGA did not reduce the surface charge of JetPEI polyplexes (+27 ± 3 mV to −2.5 ± 15 mV) to the same extent as it did with DIP50H50, suggesting that the surface coverage of JetPEI polyplexes by PGA was patchy and incomplete. We attribute the contrasting effects of PGA on the transfection performance DIP50H50 and JetPEI to incomplete surface coverage of JetPEI polyplexes by PGA. Overall, PGA proved to be a more effective surface modifier for polyplexes formed from lipophilic polycations than hydrophilic polycations.
PGA rescues polyplexes from serum-induced aggregation without exacerbating serum-mediated pDNA unbinding
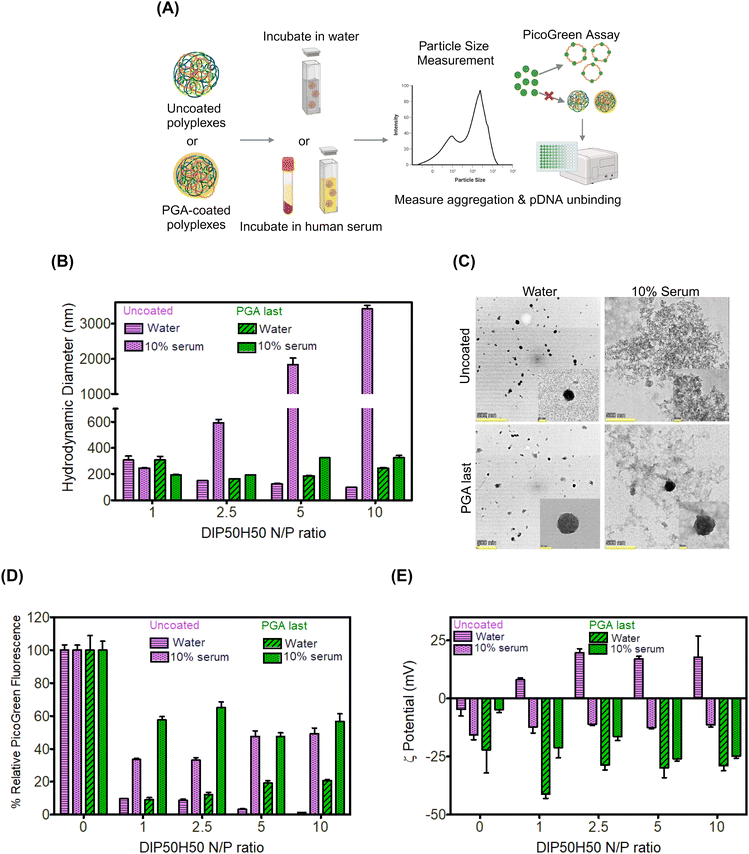
We anticipated that PGA-coated DIP50H50 polyplexes will resist aggregation in serum-containing media (Fig. 3A), potentiating serum-tolerant transfection. Since serum proteins may displace pDNA from polyplexes by competing with pDNA for binding sites on DIP50H50, we tested whether PGA-coated polyplexes were more susceptible to serum-mediated pDNA release than uncoated polyplexes. Uncoated polyplexes and three sets of PGA-coated polyplexes were incubated in either a 10% or 50% solution of serum extracted from whole human blood. Size distributions, electrokinetic analyses, and pDNA encapsulation efficiencies for all addition sequences and serum concentrations are furnished in Fig. S8–S10 in the ESI,† respectively.We measured the size distributions of uncoated and PGA-coated polyplexes in water and in serum using dynamic light scattering (DLS). In water, the hydrodynamic diameters of uncoated and PGA-coated polyplexes were comparable (<300 nm). In human serum, uncoated polyplexes aggregated severely, with hydrodynamic diameters approaching 7–8 × their values in water (Fig. 3B). Further, serum-induced aggregation of uncoated polyplexes was strongly N/P-dependent with the highest sizes (diameters approaching 3 μm) observed for N/P 10 polyplexes. In contrast, the hydrodynamic diameters of PGA-coated polyplexes were nearly identical in water and in serum (Fig. 3B).
To substantiate conclusions from DLS, we performed transmission electron microscopy (TEM) of uncoated and PGA-coated polyplexes (N/P ratio of 5) in water and in 10% serum. Both uncoated and PGA-coated polyplexes formed well-defined sub-100 nm assemblies in water but micron-scale aggregates appeared prominently when uncoated polyplexes were incubated in serum (Fig. 3C). Consistent with observations from DLS, PGA-coated polyplexes exhibited comparable size distributions in water and in serum in TEM images. Serum exposure transformed uncoated polyplexes from cationic to anionic (Fig. 3D). PGA-coated polyplexes were already negatively charged in water; anionic serum proteins may have displaced PGA from polyplex surfaces, modifying the magnitude of negative charge.
Serum-triggered pDNA release was comparable across PGA-coated and uncoated polyplexes (Fig. 3C), suggesting that PGA does not exacerbate competitive unbinding of pDNA in serum. Physicochemical characterization of polyplex–serum interactions established that PGA augments polyplex colloidal stability in serum without aggravating pDNA unbinding.
Despite depressing cellular uptake, PGA helps lipophilic polycations mediate pDNA delivery in serum-containing media
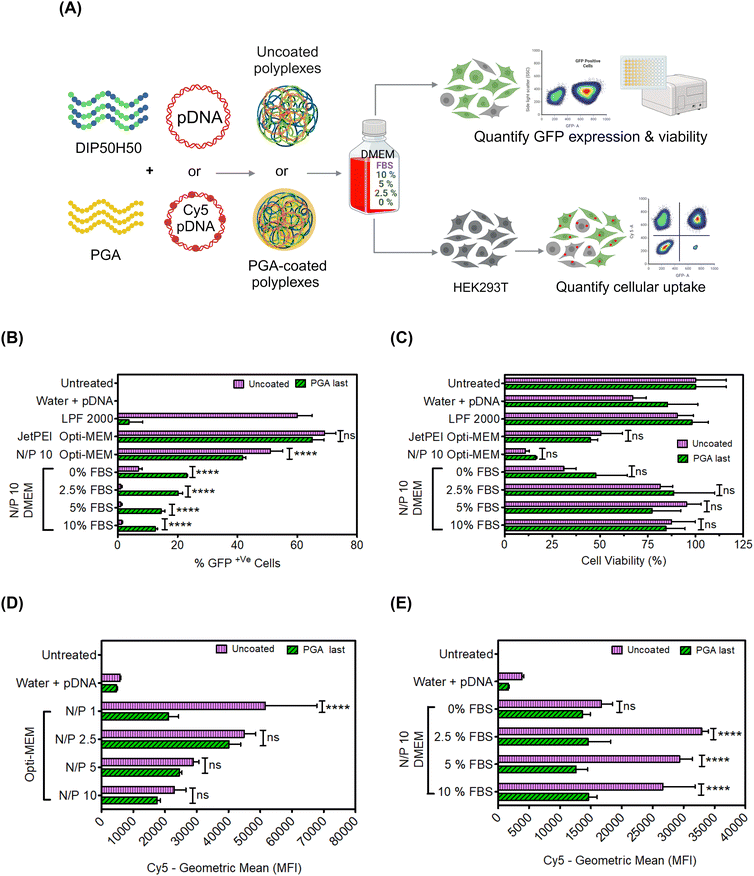
After verifying that PGA prevented polyplex aggregation in 10% serum without exacerbating pDNA unbinding, we evaluated pDNA delivery efficiency of PGA-coated polyplexes in serum-containing cell culture media. Serum proteins inhibit polycation-mediated pDNA delivery, forcing experimenters to perform transfection in reduced serum media (e.g., proprietary formulations such as OptiMEM) or in serum-free media.80 We compared five media formulations (Fig. 4A): (1) OptiMEM, a proprietary reduced-serum formulation that is ubiquitously used in polycation-mediated delivery, (2) serum-free DMEM without fetal bovine serum (FBS) and (3–5) DMEM with FBS supplementation of 2.5, 5, or 10% v/v. We compared the transfection efficiency of uncoated and PGA-coated polyplexes in each of the above five media formulations.PGA-coated polyplexes fared worse than uncoated polyplexes (Fig. 4B) in OptiMEM but performed better in DMEM (across all serum concentration levels). Serum concentration levels as low as 2.5% completely abolished the transfection of uncoated polyplexes but only halved the transfection efficiency of PGA-coated polyplexes. At higher serum concentration levels (5% and 10%), the gulf in transfection performance between uncoated and PGA-coated polyplexes grew wider.
Interestingly, the transfection efficiency of uncoated polyplexes was lower in serum-free DMEM (0% FBS) than in OptiMEM. We attribute this result to differences in glucose concentration between OptiMEM and DMEM; DMEM contains more glucose (which has previously been shown to impact cell metabolism81) than OptiMEM.82 However, DMEM may lack other biomolecules found in OptiMEM; OptiMEM is a proprietary formulation, whose components remain a matter of conjecture, preventing us from digging deeper into this result. Another interesting albeit unexpected result was that uncoated polyplexes underperform PGA-coated polyplexes in serum-free DMEM (0% FBS) but exhibit higher transfection efficiencies in the presence of FBS. We speculate that serum proteins from FBS exert divergent effects on uncoated and PGA-coated polyplexes, with some adsorbed serum proteins hindering transfection and others promoting uptake and transfection. To fully understand how serum proteins from FBS or OptiMEM components interact with uncoated and PGA-coated polyplexes, we must characterize the composition of protein coronae formed around polyplexes.83–85 Protein corona investigations of polyplexes are expensive and challenging, requiring careful sample preparation and extensive method development.86,87 We opted to pursue proteomics investigations in a future study.
Cell viability was much higher in serum-containing media than in OptiMEM (Fig. 4C), particularly for PGA-coated polyplexes (Fig. S11 in the ESI†). By potentiating rescuing cells from toxicity associated with serum starvation, PGA delays the onset of cellular stress responses that inhibit GFP expression.88–90
Next, we compared cellular internalization for PGA-coated and uncoated polyplexes in diverse media formulations (reduced serum OptiMEM and DMEM with serum concentration levels spanning 0–10% v/v). We labeled pDNA with Cy5 fluorophores,76 and measured Cy5 fluorescence intensity in OptiMEM and DMEM (Fig. 4D and E respectively). In OptiMEM, PGA-coated polyplexes were taken up by cells at lower levels than uncoated polyplexes. In the presence of serum, PGA-coated polyplexes are smaller than uncoated polyplexes, which in turn lowers their sedimentation velocity and lowers the frequency of cell–polyplex contact.91 Despite lower cellular uptake, PGA-coated polyplexes displayed higher transfection efficiency than uncoated polyplexes in serum-containing media (Fig. 4D and E). Cellular uptake, therefore, cannot explain PGA-mediated enhancements in transfection efficiency in serum-containing media. To verify whether PGA-coated polyplexes compensate for depressed cellular uptake by routing polyplexes along intracellular trajectories culminating in nuclear import, we performed confocal microscopy.
PGA coating promotes nuclear import of pDNA
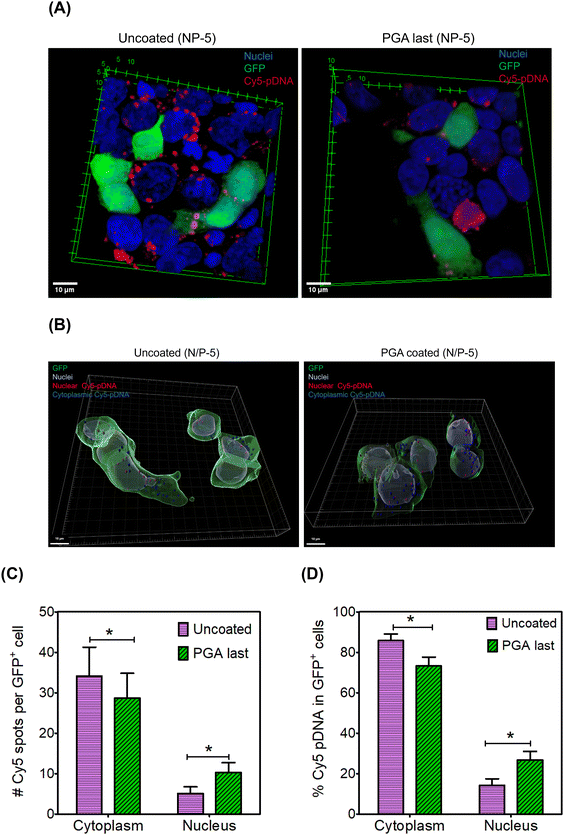
We hypothesized that PGA will reroute polyplexes along intracellular itineraries that promote nuclear import. To test this hypothesis, we mapped the intracellular distribution of Cy5-labeled polyplexes using confocal microscopy. We formulated uncoated and PGA-coated polyplexes with Cy5-labeled pDNA at an N/P ratio of 5. Twenty-four hours after polyplex administration, we fixed and stained cells with Hoechst (Fig. 5A). Interestingly, Cy5 intensity was noticeably lower in the PGA-coated condition than in the uncoated group (Fig. S12 and S13 in the ESI†), corroborating uptake measurements (Fig. 4D).Internalized polyplexes were classified as cytoplasmic (cyan) and nuclear (white) based on the spatial location of Cy5 signals (Fig. 5B). Thereafter, we enumerated polyplexes (Fig. 5C) and computed the fraction of pDNA partitioning between cytoplasmic and nuclear regions (Fig. 5D). Among GFP+ cells, pDNA is far more likely to accumulate within the nuclear periphery when polyplexes are modified with PGA (Fig. 5D). Although uncoated polyplexes are internalized more efficiently than their PGA-coated counterparts, nuclear import was significantly higher for PGA-coated polyplexes. PGA might be rerouting polyplexes along caveolar rather than clathrin-mediated pathways, consistent with reports by17 Consequently, polyplexes trafficked along caveolar pathways (PGA-coated) enjoy a higher likelihood of nuclear import than clathrin-trafficked polyplexes (uncoated). Although we found fewer polyplexes per cell in the PGA-coated condition than in the uncoated condition, PGA-coated polyplexes were more likely to inhabit nuclear regions than uncoated polyplexes. One limitation of our study is that fluorescence labeling is poorly suited for accurate quantification of pDNA within nuclei. This is because fluorescent labeling alters the hydrophobicity of pDNA, potentially impacting their intracellular trajectories. In future work, we will compare nuclear import using label-free techniques such as the qPCR method developed by Szoka et al.92
PGA silences the hemolytic activity of polyplexes
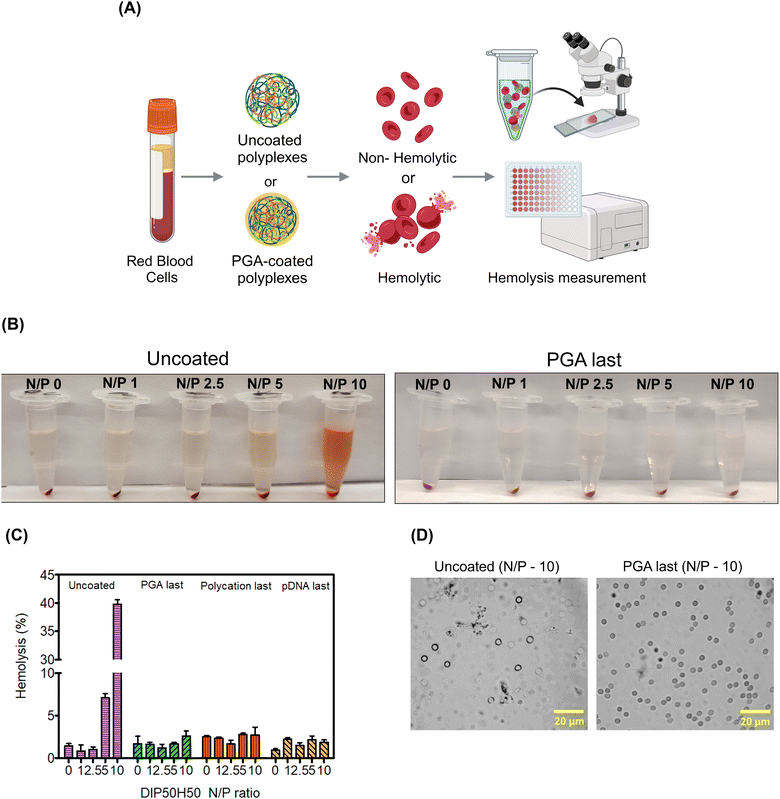
Since red blood cells (RBCs) are the most abundant cell type in the blood stream,93 studying RBC lysis will illuminate the role of PGA in augmenting polyplex hemocompatibility.94 Polyplex-induced toxicity in HEK293T cells does not predict hemolysis since RBCs possess distinct cellular characteristics. For instance, RBC plasma membranes are unusually thin to facilitate oxygen uptake, RBCs are also smaller and more discoid, and lack organelles that may initiate cellular repair following polycation-mediated lysis.93,95We determined whether PGA improves RBCs’ tolerance to polyplexes. We harvested RBCs from healthy human donors and exposed them to uncoated and PGA-coated polyplexes. Then, we measured RBC lysis as a function of N/P ratio and surface treatment (Fig. 6A). To quantify hemolysis, we measured the absorbance of hemoglobin liberated from lysed cells into the supernatant. For RBCs treated with uncoated polyplexes (Fig. 6B), the pinkish red colour in the supernatant intensified progressively with increasing N/P ratio, confirming that uncoated polyplexes are detrimental to RBC health. On the other hand, PGA-coated polyplexes maintained a transparent colorless supernatant even at the highest N/P ratio tested (N/P = 10), suggesting that PGA protected RBCs from polycation-triggered lysis. Plate reader measurements (Fig. 6C) confirmed visual observation, with the highest absorbance detected for uncoated polyplexes (N/P = 10). Interestingly, whether PGA protects RBCs from polyplex-induced hemolysis is independent of addition sequence, with PGA silencing the hemolytic activity of DIP50H50 for all three PGA-coated polyplexes (PGA last, polycation last and pDNA last).
PGA effectively inhibits lipophilic polycations from triggering hemolysis, potentially augmenting polyplex hemocompatibility. In future work we will study polyplex interactions with platelets and white blood cells, and elucidate how PGA impacts complement activation, platelet aggregation, and blood clotting.
Conclusions
Our study demonstrates the utility of polyplex surface modification in navigating tradeoffs among toxicity, transfection efficiency, and serum stability during polycation-mediated gene delivery. Instead of pursuing chemical and architectural modifications of a promising lipophilic polycation for pDNA delivery (DIP50H50), we augmented cytocompatibility and serum stability through post-synthetic surface modification with PGA. The conformations adopted by pDNA payloads and the degree to which polycations condensed pDNA was highly sensitive to the sequence of addition of PGA, polycations, and pDNA—a variable that to the best of our knowledge has never been studied before. Although PGA dramatically expanded the population of transfected cells in the DIP50H50 treatment condition, PGA had no impact on JetPEI-induced toxicity. Interestingly, PGA-coated polyplexes lagged uncoated polyplexes in cellular uptake but still exhibited higher pDNA delivery efficiency in serum-containing media. This suggested that intracellular transport, rather than cellular uptake, is the key delivery bottleneck. To understand differences in the intracellular fates of uncoated and PGA-coated polyplexes, we studied how each polyplexes partition between nuclear and cytoplasmic regions using quantitative confocal microscopy, and found that PGA-coated polyplexes were more likely to undergo nuclear import than uncoated polyplexes. Uncoated polyplexes triggered lysis in almost 50% of red blood cells, but PGA inhibited hemolysis by masking polyplex cationicity. PGA also shielded polyplexes from non-specific protein adsorption and aggregation in serum-containing media. By carefully characterizing binding interactions between polycations, pDNA payloads, serum proteins, and cells, our paper provides a useful framework to rationally design polyplex surface modification protocols. In the future, we foresee that the protein corona and cell type specificity of polyplexes can be rationally tuned by individually tailoring polyanionic coating protocols for each polycation.Experimental section
Materials
2-(diisopropylamino)ethyl methacrylate (DIPAEMA), 2-hydroxyethyl methacrylate (HEMA), 4-cyano-4-[(ethylsulfanylthiocarbonyl)sulfanyl]pentanoic acid (CEP), 4,4′-azobis(4-cyano-valeric acid) (V501), dimethyl formamide (DMF) were purchased from Sigma Aldrich (St Louis, MO). Poly-L-glutamic acid sodium salt (PGA) (MW 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–100
000–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) was purchased from Biosynth. The plasmid, pZsgreen N-1 (4708 bp), was obtained from Aldevron (Fargo, ND) and the stock solution was further diluted in ultrapure water. DMEM (Dulbecco's modified Eagle's medium), fetal bovine serum, antibiotic-antimycotic (100×), Trypsin-EDTA (0.25%), phenol red, Ultrapure™ DNase/RNase-free distilled water, phosphate-buffered saline (PBS, 1×), Opti-MEM™ I reduced serum medium, FluoroBrite,™ DMEM, Hoechst 33342 solution (20 mM), trypan blue solution, 0.4%, and Lipofectamine™ 2000 were purchased from Thermo Fisher (Waltham, MA), jetPEI®, HTS DNA (catalog #89129-916) was purchased from Polyplus. Pierce™ 16% formaldehyde (w/v), methanol-free, ProLong™ glass antifade, were purchased from Invitogen, Thermo Fisher (Waltham, MA). Label IT® nucleic acid labeling kit was purchased from Cy5®, Mirus bio (Madison, WI). Calcein Deep Red™ AM ester was procured from ATT Bioquest (Pleasanton, CA). Human serum and red blood cells (RBCs) were shared by Dr Nanette Boyle's lab at the Colorado School of Mines. Dialysis membrane tubing (MWCO 3 kDa) was purchased from Spectrum® Laboratories (Reseda, CA).
000 g mol−1) was purchased from Biosynth. The plasmid, pZsgreen N-1 (4708 bp), was obtained from Aldevron (Fargo, ND) and the stock solution was further diluted in ultrapure water. DMEM (Dulbecco's modified Eagle's medium), fetal bovine serum, antibiotic-antimycotic (100×), Trypsin-EDTA (0.25%), phenol red, Ultrapure™ DNase/RNase-free distilled water, phosphate-buffered saline (PBS, 1×), Opti-MEM™ I reduced serum medium, FluoroBrite,™ DMEM, Hoechst 33342 solution (20 mM), trypan blue solution, 0.4%, and Lipofectamine™ 2000 were purchased from Thermo Fisher (Waltham, MA), jetPEI®, HTS DNA (catalog #89129-916) was purchased from Polyplus. Pierce™ 16% formaldehyde (w/v), methanol-free, ProLong™ glass antifade, were purchased from Invitogen, Thermo Fisher (Waltham, MA). Label IT® nucleic acid labeling kit was purchased from Cy5®, Mirus bio (Madison, WI). Calcein Deep Red™ AM ester was procured from ATT Bioquest (Pleasanton, CA). Human serum and red blood cells (RBCs) were shared by Dr Nanette Boyle's lab at the Colorado School of Mines. Dialysis membrane tubing (MWCO 3 kDa) was purchased from Spectrum® Laboratories (Reseda, CA).
Polymer synthesis and characterization
p(DIPAEMA56-st-HEMA48) was synthesized using RAFT polymerization.75 Briefly, DIPAEMA (0.0025 mol), HEMA (0.0025 mol), CEP (0.00007140 mol) and V-501 (0.000007140 mol) were weighed, dissolved in DMF (4 mL) and transferred into scintillation glass vials (20 mL). The reaction mixture was degassed by bubbling nitrogen through the solution for 30 min, heated to 80 °C, and magnetically stirred overnight under nitrogen environment. After 18 h, the reaction mixture was quenched with liquid nitrogen followed by addition of 1 M HCL to dissolve precipitated polymer. The product was purified using dialysis in acidified milliQ water over 5 days, lyophilized, and stored at 4 °C. Purifiedpolymer was designated as DIP50H50. Molecular weight distribution was determined by size exclusion chromatography (Agilent, Santa Clara, CA) with multi-angle light scattering (SEC-MALS) using refractive index and multiple-angle light-scattering detectors (Wyatt, Santa Barbara, CA). 1H NMR spectra were recorded using a JEOL ECA 500 instrument (JEOL, Peabody, MA). Polymer ζ-potential was measured using Brookhaven NanoBrook-90 Plus PALS (Brookhaven Instruments, Holtsville, NY). Characterization data is furnished in Fig. S1 in the ESI.†Polyplex formulation
To obtain DIP50H50 stock solutions, 4.95 mg of DIP50H50 was dissolved in 1 mL of ultrapure water and sterile-filtered. pDNA stock solutions were prepared at 1.04 mg mL−1. Polyplexes were formed by adding equal volumes of diluted polymer stock solutions (diluted to realize targeted N/P ratios) to a 50 ng μL−1 pDNA solution. After gentle pipette-mixing, polyplexes were incubated at 23 °C for 45 min. Polyplexes were further diluted with ultrapure water prior to dye exclusion, dynamic light scattering, and electrokinetic characterization.A stock solution of PGA (1 mg mL−1) was prepared and sterile-filtered, and diluted to achieve targeted C/N/P ratios. Three PGA coating methods were compared (Fig. 1). In PGA last, PGA was added to pre-made polyplexes and incubated for 45 min. In polycation last, PGA was added to pZsGreen and incubated for 10 min before adding DIP50H50, and incubating for 45 min. In pDNA last, PGA was added to DIP50H50 and incubated for 10 min. Then pZsGreen was added and ternary polyplexes incubated for an additional 45 min. Ternary polyplexes were further diluted with ultrapure water before physical characterization (dye exclusion, DLS, electrokinetic characterization).
Polyplex characterization
To determine whether PGA coating attenuated DIP50H50–pDNA binding, we performed PicoGreen assays. Unbound pDNA will bind PicoGreen, generating a fluorescence signal whereas polymer-bound pDNA will be inaccessible to the PicoGreen dye. We expect the reduction in fluorescence signal to scale with the strength of DIP50H50–pDNA binding. Uncoated or PGA-coated polyplexes were prepared at a pDNA concentration of 50 ng μL−1 and further diluted with ultrapure water. Consistent with the manufacturer's protocol, PicoGreen was diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 ratio with ultrapure water. Equal volumes of diluted PicoGreen and polyplexes were mixed and incubated at 23 °C for 30 min. Samples were transferred into black bottom 96 well plates. The fluorescence intensity was measured with monochromator settings of excitation/emission 485/520 nm. Particle size and ζ-potential of polyplexes were analyzed by using Brookhaven NanoBrook 90Plus PALS. The samples were transferred into plastic disposable cuvettes and three measurements performed. The mean diameter, PDI, and ζ-potential values were averaged and reported.
200 ratio with ultrapure water. Equal volumes of diluted PicoGreen and polyplexes were mixed and incubated at 23 °C for 30 min. Samples were transferred into black bottom 96 well plates. The fluorescence intensity was measured with monochromator settings of excitation/emission 485/520 nm. Particle size and ζ-potential of polyplexes were analyzed by using Brookhaven NanoBrook 90Plus PALS. The samples were transferred into plastic disposable cuvettes and three measurements performed. The mean diameter, PDI, and ζ-potential values were averaged and reported.
Cellular assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 24-well plates. The cells were then incubated for 24 h at 37 °C and 5% CO2. Polyplexes were formed at 4 N/P ratios (0, 1, 2.5 & 10) in sterile conditions and incubated for 45 min. Positive controls, jetPEI and Lipofectamine 2000 (LPF 2000) were used in accordance with the manufacturer's directions. Polyplexes (uncoated and PGA-coated) and jetPEI controls were diluted in OptiMEM and added to the HEK293T cells and incubated at 37 °C and 5% CO2 for 4 h. The concentration of pDNA was fixed as 1 μg per well across all treatment conditions. After 4 h of incubation, 1 mL of DMEM media was added to polyplex and JetPEI sample groups and incubated for 24 h at 37 °C and 5% CO2. At the 24 h time point, the media was replaced with fresh DMEM media. After 48 h, the cells were trypsinized, centrifuged at 1000 rpm for 5 min and pellets were washed with PBS. The cells were stained with Calcein Red viability dye (400 nM) in PBS for 30 min at 23 °C. The cells were transferred into FACS tubes and GFP expression was measured using flow cytometry (Cytek, Fremont, CA). Gating strategies are furnished in Fig. S15 and S16 in the ESI.† Transfection efficiency was computed by determining the proportion of GFP+ cells in each treatment group.
000 cells per well in 24-well plates. The cells were then incubated for 24 h at 37 °C and 5% CO2. Polyplexes were formed at 4 N/P ratios (0, 1, 2.5 & 10) in sterile conditions and incubated for 45 min. Positive controls, jetPEI and Lipofectamine 2000 (LPF 2000) were used in accordance with the manufacturer's directions. Polyplexes (uncoated and PGA-coated) and jetPEI controls were diluted in OptiMEM and added to the HEK293T cells and incubated at 37 °C and 5% CO2 for 4 h. The concentration of pDNA was fixed as 1 μg per well across all treatment conditions. After 4 h of incubation, 1 mL of DMEM media was added to polyplex and JetPEI sample groups and incubated for 24 h at 37 °C and 5% CO2. At the 24 h time point, the media was replaced with fresh DMEM media. After 48 h, the cells were trypsinized, centrifuged at 1000 rpm for 5 min and pellets were washed with PBS. The cells were stained with Calcein Red viability dye (400 nM) in PBS for 30 min at 23 °C. The cells were transferred into FACS tubes and GFP expression was measured using flow cytometry (Cytek, Fremont, CA). Gating strategies are furnished in Fig. S15 and S16 in the ESI.† Transfection efficiency was computed by determining the proportion of GFP+ cells in each treatment group.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) were seeded in black 24-well glass bottomed plates and incubated for 24 h at 37 °C and 5% CO2. Transfection experiments were conducted as described above. Twenty-four hours after pDNA delivery, cells were washed with PBS (1×) and stained with Lysobrite Red and incubated for 30 min in dark and washed with cold PBS (3 × 5min). Cells were counter-stained with Hoechst 3342, incubated for 20 min, washed with cold PBS (3 × 5 min), and fixed with 4% v/v paraformaldehyde. Cells were imaged under laser scanning confocal microscopy (Nikon A1R) with the following laser emission filter combinations for each fluorophore: Hoescht 3342 - 405 nm laser, Chroma ET450/50m emission filter, GFP - 488 nm laser, Chroma ET525/50m emission filter, Lysobrite Red - 561 nm laser, Chroma 600/50m emission filter, Cy5 - 640 nm laser, Chroma ET685/70m emission filter. The images were captured using 20 × 0.75 NA Plan Apo Lambda (air) and 60× 1.4 NA Plan Apo Lambda (Oil) objective lenses.
000 cells per well) were seeded in black 24-well glass bottomed plates and incubated for 24 h at 37 °C and 5% CO2. Transfection experiments were conducted as described above. Twenty-four hours after pDNA delivery, cells were washed with PBS (1×) and stained with Lysobrite Red and incubated for 30 min in dark and washed with cold PBS (3 × 5min). Cells were counter-stained with Hoechst 3342, incubated for 20 min, washed with cold PBS (3 × 5 min), and fixed with 4% v/v paraformaldehyde. Cells were imaged under laser scanning confocal microscopy (Nikon A1R) with the following laser emission filter combinations for each fluorophore: Hoescht 3342 - 405 nm laser, Chroma ET450/50m emission filter, GFP - 488 nm laser, Chroma ET525/50m emission filter, Lysobrite Red - 561 nm laser, Chroma 600/50m emission filter, Cy5 - 640 nm laser, Chroma ET685/70m emission filter. The images were captured using 20 × 0.75 NA Plan Apo Lambda (air) and 60× 1.4 NA Plan Apo Lambda (Oil) objective lenses.
The images were analyzed using the Imaris software (Bitplane, version 10.0.0). An initial surface was generated using the Cell (Nucleus and Cytoplasm) tool in Imaris. The DAPI channel was used to segment the nuclei and the GFP channel was used to identify cell bodies. We set a nucleus diameter of 5 μm and a smoothing filter width of 0.5 μm was used for both channels. Intensity thresholds were chosen visually. Nuclei in non-GFP labelled cells were erased. We then manually corrected the surfaces generated by this tool to obtain individual surfaces for the nucleus and cell body for each cell. Spots were then identified using the Spot tool, with an estimated spot size of 0.866. Point spread function elongation along the z-axis was estimated at 2 μm. The spots were then filtered by the “quality” metric, which was set visually between 9.43–16.9.
Finally, the spots were filtered for individual cells and nuclei using the distance to the surface. Spots were initially filtered to identify spots within individual cell bodies, then within individual nuclei (distance to the corresponding surface was set to below 0 μm in the corresponding filters). The total number of spots within the cell body only and within the nucleus was then tallied.
Conflicts of interest
The authors do not have any conflicts of interest to disclose.Acknowledgements
Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award number 1 R21 EB034464-01. J. L. L. acknowledges support from the National Science Foundation Graduate Research Fellowship Program. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. 2137099. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. Authors are grateful to Prof. Kevin Cash and his group for access to a plate reader. Authors acknowledge Prof. Nanette Boyle and her group for sharing human blood samples. Confocal imaging was performed at the BioFrontiers Institute Advanced Light Microscopy Core (RRID: SCR018302). Laser scanning confocal microscopy was performed on an Nikon A1R microscope supported by NIST-CU Cooperative Agreement award number 70NANB15H226. The analysis workstation and the software package Imaris were supported by NIH 1S10RR026680. TEM was performed in the following core facility, which is a part of Colorado School of Mines’ Shared Instrumentation Facility (Electron Microscopy: RRID:SCR022048). Schematics were created using https://biorender.com.References
- J. Doudna, The Promise and Challenge of Therapeutic Genome Editing, Nature, 2020, 578, 229–236 CrossRef CAS PubMed.
- M. Eisenstein, Gene Therapies Close in on a Cure for Sickle-Cell Disease, Nature, 2021, 596, S2–S4 CrossRef CAS.
- X. M. Anguela and K. A. High, Entering the Modern Era of Gene Therapy, Annu. Rev. Med., 2019, 70, 273–288 CrossRef CAS PubMed.
- J. T. Bulcha, Y. Wang, H. Ma, P. W. L. Tai and G. Gao, Viral Vector Platforms within the Gene Therapy Landscape, Signal Transduction Targeted Ther., 2021, 6, 1–24 CrossRef PubMed.
- A. Pupo, A. Fernández, S. H. Low, A. François, L. Suárez-Amarán and R. J. Samulski, AAV Vectors: The Rubik's Cube of Human Gene Therapy, Mol. Ther., 2022, 30, 3515–3541 CrossRef CAS PubMed.
- Editorial, Gene Therapy at the Crossroads, Nat. Biotechnol., 2022, 40, 621–621 CrossRef PubMed.
- J. F. Wright, AAV Vector Manufacturing Process Design and Scalability - Bending the Trajectory to Address Vector-Associated Immunotoxicities, Mol. Ther., 2022, 30, 2119–2121 CrossRef CAS PubMed.
- C. C. D. Joe, N. Chopra, P. Nestola, J. Niemann and A. D. Douglas, Rapid-Response Manufacturing of Adenovirus-Vectored Vaccines, Nat. Biotechnol., 2023, 41, 314–316 CrossRef CAS PubMed.
- M. Durymanov and J. Reineke, Non-Viral Delivery of Nucleic Acids: Insight into Mechanisms of Overcoming Intracellular Barriers, Front. Pharmacol., 2018, 9, 971 CrossRef PubMed.
- J. Li, J. J. Raise, M. He, R. Das and N. Murthy, Non-Viral Strategies for Delivering Genome Editing Enzymes, Adv. Drug Delivery Rev., 2021, 168, 99–117 CrossRef CAS PubMed.
- J. van Haasteren, J. Li, O. J. Scheideler, N. Murthy and D. Schaffer, The Delivery Challenge: Fulfilling the Promise of Therapeutic Genome Editing, Nat. Biotechnol., 2020, 38, 845–855 CrossRef CAS PubMed.
- J. O. Ahern, I. Lara-Saez, D. Zhou, R. Murillas, J. Bonafont, A. Mencia, M. Garcia, D. Manzanares, J. Lynch, R. Foley, Q. Xu, A. Sigen, F. Larcher and W. Wang, Non-Viral Delivery of CRISPR-Cas9 Complexes for Targeted Gene Editing via a Polymer Delivery System, Gene Ther., 2022, 29, 157–170 CrossRef PubMed.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. V. Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Polymeric Delivery of Therapeutic Nucleic Acids, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- A. I. S. van den Berg, C.-O. Yun, R. M. Schiffelers and W. E. Hennink, Polymeric Delivery Systems for Nucleic Acid Therapeutics: Approaching the Clinic, J. Controlled Release, 2021, 331, 121–141 CrossRef CAS PubMed.
- A. Verma and F. Stellacci, Effect of Surface Properties on Nanoparticle–Cell Interactions, Small, 2010, 6, 12–21 CrossRef CAS PubMed.
- E. Fröhlich, The Role of Surface Charge in Cellular Uptake and Cytotoxicity of Medical Nanoparticles, Int. J. Nanomed., 2012, 7, 5577–5591 CrossRef PubMed.
- L. Mott, C. Akers and D. W. Pack, Effect of Polyplex Surface Charge on Cellular Internalization and Intracellular Trafficking, J. Drug Delivery Sci. Technol., 2023, 84, 104465 CrossRef CAS.
- L. Parhamifar, H. Andersen, L. Wu, A. Hall, D. Hudzech and S. M. Moghimi, Advances in Genetics, Academic Press, 2014, vol. 88, pp. 353–398 Search PubMed.
- J. S. Correia, S. Mirón-Barroso, C. Hutchings, S. Ottaviani, B. Somuncuoğlu, L. Castellano, A. E. Porter, J. Krell and T. K. Georgiou, How Does the Polymer Architecture and Position of Cationic Charges Affect Cell Viability?, Polym. Chem., 2023, 14, 303–317 RSC.
- A. M. Weiss, M. A. I. Lopez, B. W. Rawe, S. Manna, Q. Chen, E. J. Mulder, S. J. Rowan and A. P. Esser-Kahn, Understanding How Cationic Polymers’ Properties Inform Toxic or Immunogenic Responses via Parametric Analysis, Macromolecules, 2023, 56, 7286–7299 CrossRef CAS PubMed.
- K. Ryu, M. K. Lee, J. Park and T.-i. Kim, pH-Responsive Charge-Conversional Poly(ethylene imine)-Poly(l-lysine)-Poly(l-glutamic acid) with Self-Assembly and Endosome Buffering Ability for Gene Delivery Systems, ACS Appl. Bio Mater., 2018, 1, 1496–1504 CrossRef CAS PubMed.
- K. Dutta, R. Das, J. Medeiros, P. Kanjilal and S. Thayumanavan, Charge-Conversion Strategies for Nucleic Acid Delivery, Adv. Funct. Mater., 2021, 31, 2011103 CrossRef CAS PubMed.
- R. W. Lewis, A. Muralidharan, B. Klemm, P. E. Boukany and R. Eelkema, Nucleophile responsive charge-reversing polycations for pDNA transfection, Polym. Chem., 2023, 14, 1591–1601 RSC.
- G. Chen, K. Wang, Q. Hu, L. Ding, F. Yu, Z. Zhou, Y. Zhou, J. Li, M. Sun and D. Oupický, Combining Fluorination and Bioreducibility for Improved siRNA Polyplex Delivery, ACS Appl. Mater. Interfaces, 2017, 9, 4457–4466 CrossRef CAS PubMed.
- Y. Jin, W. Yu, W. Zhang, C. Wang, Y. Liu, W.-E. Yuan and Y. Feng, A novel fluorinated polyethyleneimine with microRNA-942-5p-sponges polyplex gene delivery system for non-small-cell lung cancer therapy, J. Colloid Interface Sci., 2023, 648, 287–298 CrossRef CAS PubMed.
- X. Guo, Z. Yuan, Y. Xu, M. Wei, Z. Fang and W.-E. Yuan, A fluorinated low-molecular-weight PEI/HIF-1α shRNA polyplex system for hemangioma therapy, Biomater. Sci., 2020, 8, 2129–2142 RSC.
- Z. Yuan, X. Guo, M. Wei, Y. Xu, Z. Fang, Y. Feng and W.-E. Yuan, Novel fluorinated polycationic delivery of anti-VEGF siRNA for tumor therapy, NPG Asia Mater., 2020, 12, 34 CrossRef CAS.
- Z. Liu, Z. Zhang, C. Zhou and Y. Jiao, Hydrophobic modifications of cationic polymers for gene delivery, Prog. Polym. Sci., 2010, 35, 1144–1162 CrossRef CAS.
- W.-J. Yi, X.-C. Yu, B. Wang, J. Zhang, Q.-Y. Yu, X.-D. Zhou and X.-Q. Yu, TACN-based oligomers with aromatic backbones for efficient nucleic acid delivery, Chem. Commun., 2014, 50, 6454–6457 RSC.
- Y. Liu, L. Zhou, X. Xu, Z. Cheng, Y. Chen, X.-A. Mei, N. Zheng, C. Zhang and Y. Bai, Combination of Backbone Rigidity and Richness in Aryl Structures Enables Direct Membrane Translocation of Polymer Scaffolds for Efficient Gene Delivery, Biomacromolecules, 2023, 24, 5698–5706 CrossRef CAS PubMed.
- X. Nie, Z. Zhang, C.-H. Wang, Y.-S. Fan, Q.-Y. Meng and Y.-Z. You, Interactions in DNA Condensation: An Important Factor for Improving the Efficacy of Gene Transfection, Bioconjugate Chem., 2019, 30, 284–292 CrossRef CAS PubMed.
- C.-R. Luan, Y.-H. Liu, J. Zhang, Q.-Y. Yu, Z. Huang, B. Wang and X.-Q. Yu, Low Molecular Weight Oligomers with Aromatic Backbone as Efficient Nonviral Gene Vectors, ACS Appl. Mater. Interfaces, 2016, 8, 10743–10751 CrossRef CAS PubMed.
- E. Ben-Akiva, J. Karlsson, S. Hemmati, H. Yu, S. Y. Tzeng, D. M. Pardoll and J. J. Green, Biodegradable lipophilic polymeric mRNA nanoparticles for ligand-free targeting of splenic dendritic cells for cancer vaccination, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2301606120 CrossRef PubMed.
- Y. Wu, A. E. Smith and T. M. Reineke, Lipophilic Polycation Vehicles Display High Plasmid DNA Delivery to Multiple Cell Types, Bioconjugate Chem., 2017, 28, 2035–2040 CrossRef CAS PubMed.
- J. S. Correia, S. Mirón-Barroso, C. Hutchings, S. Ottaviani, B. Somuncuoğlu, L. Castellano, A. E. Porter, J. Krell and T. K. Georgiou, How Does the Polymer Architecture and Position of Cationic Charges Affect Cell Viability?, Polym. Chem., 2023, 14, 303–317 RSC.
- B. D. Monnery, M. Wright, R. Cavill, R. Hoogenboom, S. Shaunak, J. H. G. Steinke and M. Thanou, Cytotoxicity of Polycations: Relationship of Molecular Weight and the Hydrolytic Theory of the Mechanism of Toxicity, Int. J. Pharm., 2017, 521, 249–258 CrossRef CAS PubMed.
- L. Cui, L. Kudsiova, F. Campbell, D. J. Barlow, H. C. Hailes, A. B. Tabor and M. J. Lawrence, Understanding and optimising the transfection of lipopolyplexes formulated in saline: the effects of peptide and serum, Biomater. Sci., 2023, 11, 3335–3353 RSC.
- A. Chakraborty, J. J. Martín Lasola, N. Truong and R. M. Pearson, Serum-Independent Nonviral Gene Delivery to Innate and Adaptive Immune Cells Using Immunoplexes, ACS Appl. Bio Mater., 2020, 3, 6263–6272 CrossRef CAS PubMed.
- L. F. Ferreira, A. S. Picco, F. E. Galdino, L. J. C. Albuquerque, J.-F. Berret and M. B. Cardoso, Nanoparticle–Protein Interaction: Demystifying the Correlation between Protein Corona and Aggregation Phenomena, ACS Appl. Mater. Interfaces, 2022, 14, 28559–28569 CrossRef CAS PubMed.
- K. Dutta, R. Das, J. Medeiros, P. Kanjilal and S. Thayumanavan, Charge-Conversion Strategies for Nucleic Acid Delivery, Adv. Funct. Mater., 2021, 31, 2011103 CrossRef CAS PubMed.
- L. Rügheimer, P. Hansell and M. Wolgast, Determination of the Charge of the Plasma Proteins and Consequent Donnan Equilibrium across the Capillary Barriers in the Rat Microvasculature, Acta Physiol., 2008, 194, 335–339 CrossRef PubMed.
- K. C. Remant Bahadur, B. Thapa and P. Xu, Design of Serum Compatible Tetrary Complexes for Gene Delivery, Macromol. Biosci., 2012, 12, 637–646 CrossRef PubMed.
- M.-u. Rashid and K. M. Coombs, Serum-reduced Media Impacts on Cell Viability and Protein Expression in Human Lung Epithelial Cells, J. Cell. Physiol., 2019, 234, 7718–7724 CrossRef CAS PubMed.
- T. Ito, N. Iida-Tanaka, T. Niidome, T. Kawano, K. Kubo, K. Yoshikawa, T. Sato, Z. Yang and Y. Koyama, Hyaluronic Acid and Its Derivative as a Multi-Functional Gene Expression Enhancer: Protection from Non-Specific Interactions, Adhesion to Targeted Cells, and Transcriptional Activation, J. Controlled Release, 2006, 112, 382–388 CrossRef CAS PubMed.
- S. Mahor, B. C. Dash, S. O'Connor and A. Pandit, Mannosylated Polyethyleneimine–Hyaluronan Nanohybrids for Targeted Gene Delivery to Macrophage-Like Cell Lines, Bioconjugate Chem., 2012, 23, 1138–1148 CrossRef CAS PubMed.
- N. Boehnke, K. J. Dolph, V. M. Juarez, J. M. Lanoha and P. T. Hammond, Electrostatic Conjugation of Nanoparticle Surfaces with Functional Peptide Motifs, Bioconjugate Chem., 2020, 31, 2211–2219 CrossRef CAS PubMed.
- K. Hagiwara, M. Nakata, Y. Koyama and T. Sato, The effects of coating pDNA/chitosan complexes with chondroitin sulfate on physicochemical characteristics and cell transfection, Biomaterials, 2012, 33, 7251–7260 CrossRef CAS PubMed.
- W. S. Boyle, K. Senger, J. Tolar and T. M. Reineke, Heparin Enhances Transfection in Concert with a Trehalose-Based Polycation with Challenging Cell Types, Biomacromolecules, 2017, 18, 56–67 CrossRef CAS PubMed.
- S.-H. Cho, Y.-W. Noh, M. Y. Cho and Y. T. Lim, An Electrostatically Self-Assembled Ternary Nanocomplex as a Non-Viral Vector for the Delivery of Plasmid DNA into Human Adipose-Derived Stem Cells, Molecules, 2016, 21, 572 CrossRef PubMed.
- Y. He, G. Cheng, L. Xie, Y. Nie, B. He and Z. Gu, Polyethyleneimine/DNA Polyplexes with Reduction-Sensitive Hyaluronic Acid Derivatives Shielding for Targeted Gene Delivery, Biomaterials, 2013, 34, 1235–1245 CrossRef CAS PubMed.
- Y. Liu, X. Dai, B. Yu, M. Chen, N. Zhao and F.-J. Xu, pH-Responsive hyaluronic acid-cloaked polycation/gold nanohybrids for tumor-targeted synergistic photothermal/gene therapy, Biomater. Sci., 2022, 10, 2618–2627 RSC.
- K. Suzuki, Y. Yoshizaki, K. Horii, N. Murase, A. Kuzuya and Y. Ohya, Preparation of hyaluronic acid-coated polymeric micelles for nasal vaccine delivery, Biomater. Sci., 2022, 10, 1920–1928 RSC.
- C. Wang, M. Feng, J. Deng, Y. Zhao, X. Zeng, L. Han, S. Pan and C. Wu, Poly(α-Glutamic Acid) Combined with Polycation as Serum-Resistant Carriers for Gene Delivery, Int. J. Pharm., 2010, 398, 237–245 CrossRef CAS PubMed.
- Y. Li, T. Zhao, C. Wang, Z. Lin, G. Huang, B. D. Sumer and J. Gao, Molecular basis of cooperativity in pH-triggered supramolecular self-assembly, Nat. Commun., 2016, 7, 13214 CrossRef PubMed.
- C. N. Lungu, M. V. Diudea, M. V. Putz and I. P. Grudziński, Linear and Branched PEIs (Polyethylenimines) and Their Property Space, Int. J. Mol. Sci., 2016, 17, 555 CrossRef PubMed.
- M. B. Parmar, K. C. Remant Bahadur, R. Löbenberg and H. Uludağ, Additive Polyplexes to Undertake siRNA Therapy against CDC20 and Survivin in Breast Cancer Cells, Biomacromolecules, 2018, 19, 4193–4206 CrossRef CAS PubMed.
- C. Wang, X. Luo, Y. Zhao, L. Han, X. Zeng, M. Feng, S. Pan, H. Peng and C. Wu, Influence of the Polyanion on the Physico-Chemical Properties and Biological Activities of Polyanion/DNA/Polycation Ternary Polyplexes, Acta Biomater., 2012, 8, 3014–3026 CrossRef CAS PubMed.
- W. Tansey, S. Ke, X. Y. Cao, M. J. Pasuelo, S. Wallace and C. Li, Synthesis and Characterization of Branched Poly(l-Glutamic Acid) as a Biodegradable Drug Carrier, J. Controlled Release, 2004, 94, 39–51 CrossRef CAS PubMed.
- Y. Pu, S. Chang, H. Yuan, G. Wang, B. He and Z. Gu, The Anti-Tumor Efficiency of Poly(L-glutamic Acid) Dendrimers with Polyhedral Oligomeric Silsesquioxane Cores, Biomaterials, 2013, 34, 3658–3666 CrossRef CAS PubMed.
- M. Ahmed and R. Narain, Cell Line Dependent Uptake and Transfection Efficiencies of PEI–Anionic Glycopolymer Systems, Biomaterials, 2013, 34, 4368–4376 CrossRef CAS PubMed.
- M.-I. Syga, E. Nicolì, E. Kohler and V. P. Shastri, Albumin Incorporation in Polyethylenimine–DNA Polyplexes Influences Transfection Efficiency, Biomacromolecules, 2016, 17, 200–207 CrossRef CAS PubMed.
- J. Shi, Y. Zhang, B. Ma, H. Yong, D. Che, C. Pan, W. He, D. Zhou and M. Li, Enhancing the Gene Transfection of Poly(β-Amino Ester)/DNA Polyplexes by Modular Manipulation of Amphiphilicity, ACS Appl. Mater. Interfaces, 2023, 15, 42130–42138 CrossRef CAS PubMed.
- P. Klemm, J. I. Solomun, M. Rodewald, M. T. Kuchenbrod, V. G. Hänsch, F. Richter, J. Popp, C. Hertweck, S. Hoeppener, C. Bonduelle, S. Lecommandoux, A. Traeger and S. Schubert, Efficient Gene Delivery of Tailored Amphiphilic Polypeptides by Polyplex Surfing, Biomacromolecules, 2022, 23, 4718–4733 CrossRef CAS PubMed.
- J. M. Horn and A. C. Obermeyer, Genetic and Covalent Protein Modification Strategies to Facilitate Intracellular Delivery, Biomacromolecules, 2021, 22, 4883–4904 CrossRef CAS PubMed.
- Y. Feng, Z. Guo, J. Chen, S. Zhang, J. Wu, H. Tian and X. Chen, Cationic Polymer Synergizing with a Disulfide-Containing Enhancer Achieved Efficient Nucleic Acid and Protein Delivery, Biomater. Sci., 2022, 10, 6230–6243 RSC.
- Z. Zhang, N. Qiu, S. Wu, X. Liu, Z. Zhou, J. Tang, Y. Liu, R. Zhou and Y. Shen, Dose-Independent Transfection of Hydrophobized Polyplexes, Adv. Mater., 2021, 33, 2102219 CrossRef CAS PubMed.
- S. Vicente-Ruiz, A. Armiñán, K. Maso, E. Gallon, O. Zagorodko, J. Movellan, F. Rodríguez-Otormín, M. Baues, J.-N. May, F. De Lorenzi, T. Lammers and M. J. Vicent, Poly-l-Glutamic Acid Modification Modulates the Bio-Nano Interface of a Therapeutic Anti-IGF-1R Antibody in Prostate Cancer, Biomaterials, 2023, 301, 122280 CrossRef CAS PubMed.
- T. F. Martens, K. Remaut, H. Deschout, J. F. J. Engbersen, W. E. Hennink, M. J. van Steenbergen, J. Demeester, S. C. De Smedt and K. Braeckmans, Coating Nanocarriers with Hyaluronic Acid Facilitates Intravitreal Drug Delivery for Retinal Gene Therapy, J. Controlled Release, 2015, 202, 83–92 CrossRef CAS PubMed.
- L. Rose, H. M. Aliabadi and H. Uludağ, Gelatin Coating to Stabilize the Transfection Ability of Nucleic Acid Polyplexes, Acta Biomater., 2013, 9, 7429–7438 CrossRef CAS PubMed.
- J. Chen, Z. Guo, Z. Jiao, L. Lin, C. Xu, H. Tian and X. Chen, Poly (l-glutamic acid)-based zwitterionic polymer in a charge conversional shielding system for gene therapy of malignant tumors, ACS Appl. Mater. Interfaces, 2020, 12, 19295–19306 CrossRef CAS PubMed.
- N. Boehnke, S. Correa, L. Hao, W. Wang, J. P. Straehla, S. N. Bhatia and P. T. Hammond, Theranostic Layer-by-Layer Nanoparticles for Simultaneous Tumor Detection and Gene Silencing, Angew. Chem., 2020, 132, 2798–2805 CrossRef.
- S. Correa, N. Boehnke, A. E. Barberio, E. Deiss-Yehiely, A. Shi, B. Oberlton, S. G. Smith, I. Zervantonakis, E. C. Dreaden and P. T. Hammond, Tuning Nanoparticle Interactions with Ovarian Cancer through Layer-by-Layer Modification of Surface Chemistry, ACS Nano, 2020, 14, 2224–2237 CrossRef CAS PubMed.
- T. Kurosaki, T. Kitahara, S. Kawakami, Y. Higuchi, A. Yamaguchi, H. Nakagawa, Y. Kodama, T. Hamamoto, M. Hashida and H. Sasaki, γ-Polyglutamic Acid-Coated Vectors for Effective and Safe Gene Therapy, J. Controlled Release, 2010, 142, 404–410 CrossRef CAS PubMed.
- R. Wang, K. Guo, W. Zhang, Y. He, K. Yang, Q. Chen, L. Yang, Z. Di, J. Qiu, P. Lei, Y. Gu, Z. Luo, X. Xu, Z. Xu, X. Feng, S. Li, Z. Yu and H. Xu, Poly-γ-Glutamic Acid Microgel-Encapsulated Probiotics with Gastric Acid Resistance and Smart Inflammatory Factor Targeted Delivery Performance to Ameliorate Colitis, Adv. Funct. Mater., 2022, 32, 2113034 CrossRef CAS.
- R. Kumar, N. Le, Z. Tan, M. E. Brown, S. Jiang and T. M. Reineke, Efficient Polymer-Mediated Delivery of Gene-Editing Ribonucleoprotein Payloads through Combinatorial Design, Parallelized Experimentation, and Machine Learning, ACS Nano, 2020, 14, 17626–17639 CrossRef CAS PubMed.
- R. Kumar, N. Le, F. Oviedo, M. E. Brown and T. M. Reineke, Combinatorial Polycation Synthesis and Causal Machine Learning Reveal Divergent Polymer Design Rules for Effective pDNA and Ribonucleoprotein Delivery, JACS Au, 2022, 2, 428–442 CrossRef CAS PubMed.
- S. K. Cho, C. Dang, X. Wang, R. Ragan and Y. J. Kwon, Mixing-Sequence-Dependent Nucleic Acid Complexation and Gene Transfer Efficiency by Polyethylenimine, Biomater. Sci., 2015, 3, 1124–1133 RSC.
- C. Malloggi, D. Pezzoli, L. Magagnin, L. D. Nardo, D. Mantovani, E. Tallarita and G. Candiani, Comparative Evaluation and Optimization of Off-the-Shelf Cationic Polymers for Gene Delivery Purposes, Polym. Chem., 2015, 6, 6325–6339 RSC.
- L. Mendonça and F. Hache, Nanosecond T-Jump Experiment in Poly(Glutamic Acid): A Circular Dichroism Study, Int. J. Mol. Sci., 2012, 13, 2239–2248 CrossRef PubMed.
- C. V. Bruggen, D. Punihaole, A. R. Keith, A. J. Schmitz, J. Tolar, R. R. Frontiera and T. M. Reineke, Quinine Copolymer Reporters Promote Efficient Intracellular DNA Delivery and Illuminate a Protein-Induced Unpackaging Mechanism, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 32919–32928 CrossRef PubMed.
- S. M. Baumgartner-Parzer, L. Wagner, M. Pettermann, J. Grillari, A. Gessl and W. Waldhäusl, High-Glucose–Triggered Apoptosis in Cultured Endothelial Cells, Diabetes, 1995, 44, 1323–1327 CrossRef CAS PubMed.
- A. T. L. Young, R. B. Moore, A. G. Murray, J. C. Mullen and J. R. T. Lakey, Assessment of Different Transfection Parameters in Efficiency Optimization, Cell Transplant., 2004, 13, 179–185 CrossRef PubMed.
- N. Hartl, B. Gabold, F. Adams, P. Uhl, S. Oerter, S. Gätzner, M. Metzger, A.-C. König, S. M. Hauck, A. Appelt-Menzel, W. Mier, G. Fricker and O. M. Merkel, Overcoming the Blood-Brain Barrier? - Prediction of Blood-Brain Permeability of Hydrophobically Modified Polyethylenimine Polyplexes for siRNA Delivery into the Brain with in Vitro and in Vivo Models, J. Controlled Release, 2023, 360, 613–629 CrossRef CAS PubMed.
- D. Zhu, H. Yan, Z. Zhou, J. Tang, X. Liu, R. Hartmann, W. J. Parak, N. Feliu and Y. Shen, Detailed Investigation on How the Protein Corona Modulates the Physicochemical Properties and Gene Delivery of Polyethylenimine (PEI) Polyplexes, Biomater. Sci., 2018, 6, 1800–1817 RSC.
- D. Zhu, H. Yan, Z. Zhou, J. Tang, X. Liu, R. Hartmann, W. J. Parak, Y. Shen and N. Feliu, Influence of the Modulation of the Protein Corona on Gene Expression Using Polyethylenimine (PEI) Polyplexes as Delivery Vehicle, Adv. Healthcare Mater., 2021, 10, 2100125 CrossRef CAS PubMed.
- D. Maiolo, J. Colombo, J. Beretta, C. Malloggi, G. Candiani and F. Baldelli Bombelli, The Polyplex, Protein Corona, Cell Interplay: Tips and Drawbacks, Colloids Surf., B, 2018, 168, 60–67 CrossRef CAS PubMed.
- N. Hartl, D. C. Jürgens, S. Carneiro, A.-C. König, X. Xiao, R. Liu, S. M. Hauck and O. M. Merkel, Protein Corona Investigations of Polyplexes with Varying Hydrophobicity – From Method Development to in Vitro Studies, Int. J. Pharm., 2023, 643, 123257 CrossRef CAS PubMed.
- A. Hamann, K. Broad, A. Nguyen and A. K. Pannier, Mechanisms of unprimed and dexamethasone-primed nonviral gene delivery to human mesenchymal stem cells, Biotechnol. Bioeng., 2019, 116, 427–443 CrossRef CAS PubMed.
- A. Hamann, T. Kozisek, K. Broad and A. K. Pannier, Glucocorticoid Priming of Nonviral Gene Delivery to hMSCs Increases Transfection by Reducing Induced Stresses, Mol. Ther. – Methods Clin. Dev., 2020, 18, 713–722 CrossRef CAS PubMed.
- T. M. Martin, B. J. Wysocki, J. P. Beyersdorf, T. A. Wysocki and A. K. Pannier, Integrating mitosis, toxicity, and transgene expression in a telecommunications packet-switched network model of lipoplex-mediated gene delivery, Biotechnol. Bioeng., 2014, 111, 1659–1671 CrossRef CAS PubMed.
- Z. Tan, Y. Jiang, M. S. Ganewatta, R. Kumar, A. Keith, K. Twaroski, T. Pengo, J. Tolar, T. P. Lodge and T. M. Reineke, Block Polymer Micelles Enable CRISPR/Cas9 Ribonucleoprotein Delivery: Physicochemical Properties Affect Packaging Mechanisms and Gene Editing Efficiency, Macromolecules, 2019, 52, 8197–8206 CrossRef CAS.
- R. N. Cohen, M. A. van der Aa, N. Macaraeg, A. P. Lee and F. C. Szoka, Quantification of Plasmid DNA Copies in the Nucleus after Lipoplex and Polyplex Transfection, J. Controlled Release, 2009, 135, 166–174 CrossRef CAS PubMed.
- V. Pretini, M. H. Koenen, L. Kaestner, M. H. A. M. Fens, R. M. Schiffelers, M. Bartels and R. Van Wijk, Red Blood Cells: Chasing Interactions, Front. Physiol., 2019, 10, 945 CrossRef PubMed.
- H. R. Phillips, Z. P. Tolstyka, B. C. Hall, J. K. Hexum, P. B. Hackett and T. M. Reineke, Glycopolycation–DNA Polyplex Formulation N/P Ratio Affects Stability, Hemocompatibility, and in Vivo Biodistribution, Biomacromolecules, 2019, 20, 1530–1544 CrossRef CAS PubMed.
- V. Heinrich, K. Ritchie, N. Mohandas and E. Evans, Elastic Thickness Compressibilty of the Red Cell Membrane, Biophys. J., 2001, 81, 1452–1463 CrossRef CAS PubMed.
- I. Sovadinova, E. F. Palermo, R. Huang, L. M. Thoma and K. Kuroda, Mechanism of Polymer-Induced Hemolysis: Nanosized Pore Formation and Osmotic Lysis, Biomacromolecules, 2011, 12, 260–268 CrossRef CAS PubMed.
- A. P. Francis and A. Jayakrishnan, Conjugating Doxorubicin to Polymannose: A New Strategy for Target Specific Delivery to Lung Cancer Cells, J. Biomater. Sci., Polym. Ed., 2019, 30, 1471–1488 CrossRef CAS PubMed.
- V. Bulmus, A New pH-responsive and Glutathione-Reactive, Endosomal Membrane-Disruptive Polymeric Carrier for Intracellular Delivery of Biomolecular Drugs, J. Controlled Release, 2003, 93, 105–120 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lp00085d |
| This journal is © The Royal Society of Chemistry 2024 |