Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Recent advancement in the detection of potential cancer biomarkers using the nanomaterial integrated electrochemical sensing technique: a detailed review
Hema
Bhardwaj
a,
Archana
a,
Ashab
Noumani
a,
Jayendra Kumar
Himanshu
ab,
Shreeti
Chakravorty
a and
Pratima R.
Solanki
 *a
*a
aNano-Bio Laboratory, Special Centre for Nano Science (SCNS), Jawaharlal Nehru University, Delhi 110067, India. E-mail: pratimarsolanki@gmail.com; partima@mail.jnu.ac.in
bDepartment of Biotechnology, School of Life Sciences, Mahatma Gandhi Central University, Motihari, Bihar 845401, India
First published on 4th January 2024
Abstract
Cancer is a leading cause of morbidity and mortality worldwide but early diagnosis, management or screening and treatment of cancer can significantly improve the survival rate of cancer patients. Oral cancer is the sixth most common cancer leading to approx. one-third of deaths worldwide. Oral cancer poses a serious threat due to its soaring case-fatality rate and metastatic characteristics and it is a type of head and neck cancer where cancerous tissue growth is located in the oral cavity. Also, lung cancer is the second most common cancer in the world with a five-year survival rate of only 15%. However, conventional techniques are available for cancer detection but these techniques are time consuming and involve a pretreatment process and sophisticated instruments. Therefore, technological advancement in the area of electrochemical sensors with miniaturization of point-of-care devices including nanomaterials has shown significant enhancement in overall sensing performance for cancer detection. Non-invasive diagnosis of cancer reduces the death risk of the disease or increases survival rate. Therefore, in this review article, the background of cancer, definition of the problem related to cancer, available conventional techniques for diagnosis and recent progress in the development of electrochemical sensors for cancer detection using nanomaterials are summarized. This review emphasizes on the use of an electrochemical sensing technique that can provide simpler, faster, non-invasive and ultraprecise detection of cancer biomarkers. Additionally, this review focuses on the recent literature on cancer detection and advancement in the diagnosis and treatment together with future perspectives on further advancements.
1. Introduction
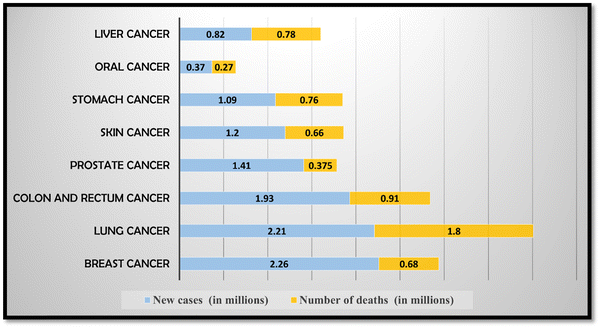
According to the National Cancer Institute, cancer refers to a complex group of diseases characterized by the uncontrolled growth and division of abnormal cells.1 These abnormal cells have the potential to invade and infiltrate nearby tissues and organs, interfering with their normal functioning, and can affect any organ or part of the body, and lead to cancer.2 The development of cancer typically begins with genetic mutations or changes in the DNA of cells. When cancer cells divide and multiply, they form a mass of tissue called a tumor. Tumors can be classified as benign or malignant. Benign tumors are non-cancerous and generally do not spread to other parts of the body. They are usually localized and can often be surgically removed without causing significant harm. On the other hand, malignant tumors are cancerous and have the ability to invade the surrounding tissues and spread to distant sites in the body through the lymphatic system or bloodstream.3 This process is known as metastasis. There are over 100 types of cancer that have been identified, each with its own unique characteristics, treatment approaches, and prognosis. Some common types of cancer include breast cancer, lung cancer, colorectal cancer, prostate cancer, skin cancer, and ovarian cancer, among many others.4 Diagnosing cancer involves various methods such as imaging tests (e.g., X-rays, CT scans, MRI), laboratory tests (e.g., blood tests, tumor marker tests), and invasive procedures (e.g., biopsies) to analyze tissue samples for the presence of cancer cells.5–9 Common treatment modalities include surgery, radiation therapy, chemotherapy, targeted therapy, immunotherapy, and hormone therapy. In many cases, a combination of these treatments may be used to achieve the best possible outcome.10It is important to note that significant advancements have been made in cancer research and treatment over the years, leading to improved outcomes and increased survival rates for many types of cancer. Early detection, timely intervention, and access to quality healthcare services play crucial roles in improving prognosis and quality of life for individuals diagnosed with cancer.11 The recent worldwide incidence of major types of cancer with new cancer cases as well as deaths is depicted in Fig. 1. In 2020, there were an estimated approximately 18 million new cancer cases and around 10 million cancer deaths worldwide.
The second-leading cause of mortality in India and the United States is cancer, which represents a serious global public health concern. Sadly, the 2019 coronavirus disease (COVID-19) pandemic has had a negative impact on cancer diagnosis and therapy. Cancer care has been delayed as a result of a number of issues, such as temporary closure of healthcare institutions, changes in employment and health insurance, and worries about COVID-19 exposure. The COVID-19 outbreak's peak in the middle of 2020 had the most effect, and the healthcare system has not yet fully recovered.1,12 The Massachusetts General Hospital's surgical oncology operations provide an illustration of the scope of the problem. These operations were only at 72% of the levels recorded in 2019 in the second half of 2020. Additionally, they only got to 84% in 2021, which was the slowest recovery rate among all surgical specialties. Potential effects of the delay in cancer detection and treatment include a rise in advanced-stage disease and fatality rates. Due to the delay in gathering complete data on cancer incidence and mortality, which normally takes two to three years, these harmful consequences may manifest gradually over time and will take several years to accurately assess at the population level. The pandemic disproportionate effect on communities of color is one factor that has received considerable attention. This inequality is a consequence of both direct and indirect effects of a number of things, including uneven socioeconomic situations, systemic inequalities, and restricted access to healthcare services. The detrimental consequences in these areas emphasize the urgent need for targeted measures and assistance to lessen the uneven impact of the epidemic on cancer treatment.13–15
According to a cancer survey, in 2018, 18.1 million cancer cases were diagnosed and 9.6 million cancer related mortalities were reported. Of all cancers, lung cancer has the highest mortality rate, followed by oral cancer, stomach cancer, liver cancer and colorectal cancer.16 One of the major issues leading to cancer fatalities is late and incorrect diagnosis of cancer cell and improper treatment.17 According to analysis and research conducted by the World Health Organization, 80% or more of cancer patients can be successfully treated if they are diagnosed when the disease is still in its early stages.18 Therefore, in view of the highest cancer cases and deaths of lung and oral cancer, this review article is focused on these two types of cancer, i.e. oral and lung cancer. Table 1 provides a brief summary of the type of cancer, and estimated new cancer cases and deaths that occurred worldwide as well as in India due to oral and lung cancer.
| Cancer site | Cancer type | Estimated new cases in India | Estimated death in India | Estimated new cases in the world | Estimated death in the world |
|---|---|---|---|---|---|
| Oral cavity and pharynx | Tongue | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 220 220 |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 350 350 |
120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 140 140 |
54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680 680 |
| Mouth | 114![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 230 230 |
422![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440 440 |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 230 230 |
|
| Pharynx | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
8500 | 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
|
| Other oral cavity | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
6300 | 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
|
| Respiratory system | Larynx | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
5500 | 166![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| Lung and bronchus | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 067 067![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
669![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 210 210![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 376 376![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
|
| Other respiratory organs | 5000 | 2500 | 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
In the following sections, detailed information about oral and lung cancer including biology of cancer development, conventional techniques for diagnosis and the recent progress in the detection of oral and lung cancer using electrochemical sensors as well as future perspectives related to the monitoring or prognosis of oral or lung cancer is provided.
Oral cancer
Head and neck squamous cell carcinoma (HNSCC) is a type of cancer that arises from the squamous cells lining the mucosal surfaces of the head and neck region. It is a significant health concern worldwide, with a substantial impact on morbidity and mortality. Squamous cell carcinoma accounted for 85% of cases in the current study, and confirmation of the diagnosis is typically made at an advanced stage of the tumor, which lowers patient survival. The head and neck region includes the oral cavity (mouth), oropharynx (tonsils, base of the tongue), larynx (voice box), and hypopharynx (lower part of the throat).19 Oral cancer is a significant malignancy that ranks sixth as the most common cancer globally.20 Oral cancer poses a serious health challenge to the nations undergoing economic transition. In India, around 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new cases and 52
000 new cases and 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths are reported annually, which is approximately one-fourth of global incidences.21 The increasing cases of oral cancer are the most important concern for community health as it oral cancer is one of the common types of cancer in India. In the year 2012, the incidence of cancer of the oral cavity and lip reached approximately 300
000 deaths are reported annually, which is approximately one-fourth of global incidences.21 The increasing cases of oral cancer are the most important concern for community health as it oral cancer is one of the common types of cancer in India. In the year 2012, the incidence of cancer of the oral cavity and lip reached approximately 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cases, representing 2.1% of the total cancer burden; on the other hand, the reported cancer incidence in India 2023 was estimated to be 1.9 to 2.0 million, whereas the real incidence is 1.5 to 3 times higher than the reported cases.22 Regrettably, the mortality rate associated with this disease is equally alarming, resulting in the loss of 145
000 cases, representing 2.1% of the total cancer burden; on the other hand, the reported cancer incidence in India 2023 was estimated to be 1.9 to 2.0 million, whereas the real incidence is 1.5 to 3 times higher than the reported cases.22 Regrettably, the mortality rate associated with this disease is equally alarming, resulting in the loss of 145![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 lives. Pharyngeal and laryngeal tumors may be treated with radiation and CRT, but oral cavity cancers are often treated with surgery. Understanding the scientific aspects and implications of oral cancer is crucial for addressing this public health issue effectively.23 Cancer of the oral cavity and lip primarily encompasses tumors that arise from the epithelial lining of the mouth, including the lips, tongue, gums, and oral mucosa. The development of oral cancer is influenced by various risk factors, including tobacco and alcohol use, betel quid chewing, poor oral hygiene, viral infections such as human papillomavirus (HPV) infection, and genetic predisposition. These risk factors contribute to the accumulation of genetic mutations and alterations in key cellular signaling pathways, leading to uncontrolled cell growth and tumor formation. The scientific community has made significant progress in understanding the molecular mechanisms underlying oral cancer. Studies have identified specific genetic mutations, such as alterations in tumor suppressor genes (e.g., TP53, CDKN2A) and oncogenes (e.g., EGFR, KRAS), which play critical roles in the initiation and progression of oral cancer. Nowadays there are a lot of studies that have been done and some of the recent research is briefly discussed in this review article. One example is a 2023 study in which Balakittnen et al. introduced noncoding RNAs in an oral cancer. Due to their participation in almost all biological processes, it can be said that this group of biomarkers is among the most promising ones.24 With regard to early detection of cancer, scientists have developed a deep learning concept. In 2023, Huang et al. were the first to use preprocessing techniques such as data augmentation, gamma correction, and noise reduction to improve the raw image quality and boost their quantity to supply enough data for convolutional neural network training. They used ISSA (an improved version of the squirrel search algorithm) to select the network weights optimally and deliver greater accuracy.25 By utilizing artificial intelligence, Rawi et al. offered a cutting-edge technological advancement in the management of oral cancer in the year 2022.26
000 lives. Pharyngeal and laryngeal tumors may be treated with radiation and CRT, but oral cavity cancers are often treated with surgery. Understanding the scientific aspects and implications of oral cancer is crucial for addressing this public health issue effectively.23 Cancer of the oral cavity and lip primarily encompasses tumors that arise from the epithelial lining of the mouth, including the lips, tongue, gums, and oral mucosa. The development of oral cancer is influenced by various risk factors, including tobacco and alcohol use, betel quid chewing, poor oral hygiene, viral infections such as human papillomavirus (HPV) infection, and genetic predisposition. These risk factors contribute to the accumulation of genetic mutations and alterations in key cellular signaling pathways, leading to uncontrolled cell growth and tumor formation. The scientific community has made significant progress in understanding the molecular mechanisms underlying oral cancer. Studies have identified specific genetic mutations, such as alterations in tumor suppressor genes (e.g., TP53, CDKN2A) and oncogenes (e.g., EGFR, KRAS), which play critical roles in the initiation and progression of oral cancer. Nowadays there are a lot of studies that have been done and some of the recent research is briefly discussed in this review article. One example is a 2023 study in which Balakittnen et al. introduced noncoding RNAs in an oral cancer. Due to their participation in almost all biological processes, it can be said that this group of biomarkers is among the most promising ones.24 With regard to early detection of cancer, scientists have developed a deep learning concept. In 2023, Huang et al. were the first to use preprocessing techniques such as data augmentation, gamma correction, and noise reduction to improve the raw image quality and boost their quantity to supply enough data for convolutional neural network training. They used ISSA (an improved version of the squirrel search algorithm) to select the network weights optimally and deliver greater accuracy.25 By utilizing artificial intelligence, Rawi et al. offered a cutting-edge technological advancement in the management of oral cancer in the year 2022.26
Lung cancer
Throughout the world, lung cancer is one of the leading causes of death.27 Surgery, radiation therapy, chemotherapy, and targeted medication therapy are therapeutic options for treating lung cancer.28 Medical management is frequently linked to the emergence of treatment resistance that results in relapse. Due to its manageable safety profile, persistent therapeutic response due to immunological memory development, and effectiveness across a wide patient population, immunotherapy is significantly changing how cancer is treated.29 Different tumor-specific vaccination strategies are gaining ground in the treatment of lung cancer.28 This review discusses the recent developments in the clinical studies of lung cancer and the related challenges.Globally, cancer incidence and death are rising, with lung cancer being the most commonly diagnosed form of cancer (11.6% of the total cases).30 Also, it is found that there was a roughly 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 236
000 and 236![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 740 and 2.2 million cases and new cases of lung cancer along with 60
740 and 2.2 million cases and new cases of lung cancer along with 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 130
000, 130![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 180 and 1.8 million fatalities, respectively were observed. With a share of 18.4% of all cancer-related fatalities worldwide, this malignancy maintains its position as the main cause of cancer-related deaths.31 As a result, it places heavy societal and economic burden. Approximately 80% of lung cancer fatalities are primarily caused by tobacco use. Exposure to substances like radon and asbestos, and continuous and cumulative contact with airborne contaminants, particularly emissions containing polycyclic aromatic hydrocarbons (PAH), are additional risk factors for lung cancer. Lung cancer history, either personal or familial, is also acknowledged as a risk factor.32
180 and 1.8 million fatalities, respectively were observed. With a share of 18.4% of all cancer-related fatalities worldwide, this malignancy maintains its position as the main cause of cancer-related deaths.31 As a result, it places heavy societal and economic burden. Approximately 80% of lung cancer fatalities are primarily caused by tobacco use. Exposure to substances like radon and asbestos, and continuous and cumulative contact with airborne contaminants, particularly emissions containing polycyclic aromatic hydrocarbons (PAH), are additional risk factors for lung cancer. Lung cancer history, either personal or familial, is also acknowledged as a risk factor.32
While both non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) are treated using a variety of therapeutic modalities, including surgery, chemotherapy, and radiation, there is an urgent need for the development of effective methods to either cure or effectively manage lung cancer, particularly at advanced stages. Due to the lack of a reliable platform for early-stage identification and the delayed onset of symptoms during disease development, which restricts therapy options and overall survival, the prognosis of NSCLC is particularly difficult. Low-dose computed tomography (LDCT), the current gold standard for lung cancer screening in patients, is used. However, LDCT has issues like false-positive identification, radiation risk, early detection sensitivity, and the lack of funding for effective CT-based screening programs. Only 5% of the recommended 15 million high-risk persons in the United States have received LDCT screening. Despite the potential benefits of early detection in facilitating tumor removal, treatment effectiveness, and favourable outcomes, the refractory nature of lung cancer is largely due to the lack of a suitable screening platform, as well as the disease's metastatic nature, genetic diversity, and limited responsiveness to late-stage chemotherapy.28
However, for locally advanced and metastatic lung malignancies, chemotherapy and radiation therapy regimens, including neoadjuvant and/or adjuvant methods, are advised. These therapies, however, have limited overall survival rates and frequently have negative side effects. Notably, targeted treatments in combination with chemotherapy have become standard procedures for NSCLC patients carrying actionable oncogenic changes such as driver mutations and fusions/rearrangements. In some circumstances, these targeted medicines have shown promise in improving progression-free survival (PFS) and overall survival. It is vital to understand that targeted therapies have different side effect profiles from standard chemotherapy and might not always produce long-lasting therapeutic results.28
2. Development of cancer and problem definition
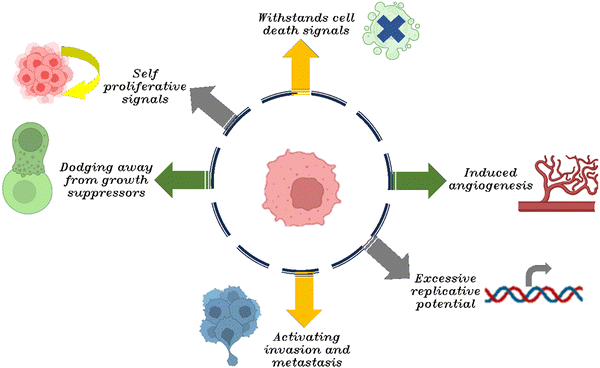
Tumorigenesis is a multifaceted process, and the metastatic behavior of malignant neoplasms represents both hallmark and a significant challenge in oncology. As neoplastic progression occurs, cells from the primary neoplasm infiltrate the surrounding healthy tissues, migrate to distal sites, and establish secondary colonies. It is estimated that metastatic events account for approximately 90% of cancer-related fatalities.33,34 Also, the neoplastic cells demonstrate a dynamic shift between epithelial and mesenchymal phenotypes.35,36 As cancer predominantly originates in epithelial tissue, with roughly account 80 to 90% of all cancer cases that being characterized as carcinomas. In 2000, Hanahan and Weinberg delineated six hallmark capabilities that typify the majority of cancers, if not all. Fig. 2 illustrates the six characteristic features commonly exhibited by cancer cells. These hallmarks encompass sustaining self-proliferative signals, dodging away from growth suppressors, withstanding cell death signals, inducing angiogenesis, excessive replicative potential, and activating invasion and metastasis.37During tumorigenesis, cancer cells are subjected to an array of stresses, stemming from dysregulated proliferative signals driven by oncogenes and a substantial mutational load. While these signals would typically initiate apoptosis in normal cells, malignancies have evolved ways to bypass these death cues, facilitating progression to more advanced and aggressive cancer stages.
3. Conventional techniques used for cancer biomarker detection
3.1 Histopathology
Cancer incidence and mortality have dramatically increased and cancer is a notable cause of death worldwide in recent decades.38,39 Unfortunate incidents of cancer are a result of changes in lifestyle as there is a dynamic global change in geographical and economic factors. The factors which are responsible for cancer include obesity, alcohol intake, lack of physical activity, hormonal imbalance, and smoking. Also, there is a strong relationship between enhanced risk of cancer and mental health ailments. Therefore, change in the modern lifestyle adversely impacts the human health that lead to cause several health issues including cancer. Also, in the view of elevation of cancer cases, recent progressive advancement also being achieved towards the diagnosis of diseases and monitoring of human health. Thereby, advanced screening facilities also increased for rapid diagnoses of cancer cases and these facilities are available not only in the urban areas but also in the rural region. Since 2020, awareness about the cancer prevention steps and advanced treatment strategies have assisted the people to increase the likelihood of fighting against the diseases.40,41 As a result, cancer diagnosis is absolutely essential. Cancer can be diagnosed using a variety of techniques, such as biochemical analysis, surgery, diagnostic medical imaging (DMI), clinical tests, etc. The most popular diagnostic method used by experts is image diagnosis such as ultrasound, magnetic resonance imaging, nuclide, X-ray computed tomography (CT), X-ray, differential contrast, and histopathology image analysis. Histopathology image analysis involves examining the biopsy tissue under a microscope to enable specialists to clearly see the details of the cell characteristics in tissue.42,43Histopathological detection techniques are considered to be the best for determining the type of cancer.44 Histopathology, which derives from the Greek terms histos (tissue), pathos (sickness/suffering), and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) y (−log
y (−log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ia), is the study of changes in any tissue, whether it be human, animal, or plant, connected with a disease or disorder. In a medical setting, sampling occurs either during surgery or during an autopsy, and the tissue is then processed for microscopic study.45 The examination of a slide for research and diagnostic purpose is known as histopathology. It is a field of study that only relies on microscopic interpretation and analysis. For an accurate diagnosis to be made, processing procedures, suitable fixation and suitable biopsy technique, staining, and sufficient sectioning are crucial. It is crucial to distinguish the structural and morphological characteristics of tissue components for a complete diagnosis.46 The majority of tissue-based diagnostics require staining. Haematoxylin and eosin (H&E) staining is a staining technique that is often used in histology labs.47 In this procedure, different tissue components are colored by a staining component; after being suitably colored, different types of cells, structures of cells, and other foreign substances are clearly noticeable under high resolution.48,49 Because all structures appear the same dark grey when the tissue is examined under a microscope, it is impossible to distinguish the structural details, and hence must be colored.47 Following this, pathologists examine the stained tissue slide under a microscope or with the help of high-resolution images taken by the camera. To find tumors, a histopathology test is required. It is a time-and-tested method for predicting the presence of invasive cancer cells in tissues that are stained with H&E. In H&E staining, the chromatin in the nucleus and the nucleic acid in the cytoplasm turn pink, while the components in the cytoplasm and extracellular matrix turn purple-blue. The benefit of free scaling in histopathological images is that they may accurately display the minute features in tissue images and increase the precision of detection. Pathologists directly examine, evaluate, and grade these tissues under a microscope as part of conventional cancer testing, which is a well-established practice for identifying cancer cells. But while working under a microscope, pathologists are prone to feeling dizzy and neck strain. It takes time for the pathologist to make notes and write out their ultimate judgment in the interim.48 It takes a lot of time and effort for pathologists to manually analyze many sections because of the continually rising number of cancer patients. A lack of traceability between observers, internal alterations, and inconsistent results can also result from manual diagnosis. Cancer diagnosis accuracy varies by 20% between pathologists with expertise and those with less experience. Individual diagnoses and expert diagnoses differed by 75.3%, according to recent research.47,48 The objective is to determine if a tumor is benign or malignant since cancerous tumors need to be treated right away to lessen and avoid subsequent consequences. Numerous problems with this approach include intra-observer variability, the potential for recurrence in tissues and cancer cells, and the identification difficulty caused by the fact that numerous additional structures in cells have the same hyperchromatic properties. Due to the fact that the surgery only impacts a small amount of tissue, the site must be carefully considered. It is best to choose a region around the borders of the tumor.
ia), is the study of changes in any tissue, whether it be human, animal, or plant, connected with a disease or disorder. In a medical setting, sampling occurs either during surgery or during an autopsy, and the tissue is then processed for microscopic study.45 The examination of a slide for research and diagnostic purpose is known as histopathology. It is a field of study that only relies on microscopic interpretation and analysis. For an accurate diagnosis to be made, processing procedures, suitable fixation and suitable biopsy technique, staining, and sufficient sectioning are crucial. It is crucial to distinguish the structural and morphological characteristics of tissue components for a complete diagnosis.46 The majority of tissue-based diagnostics require staining. Haematoxylin and eosin (H&E) staining is a staining technique that is often used in histology labs.47 In this procedure, different tissue components are colored by a staining component; after being suitably colored, different types of cells, structures of cells, and other foreign substances are clearly noticeable under high resolution.48,49 Because all structures appear the same dark grey when the tissue is examined under a microscope, it is impossible to distinguish the structural details, and hence must be colored.47 Following this, pathologists examine the stained tissue slide under a microscope or with the help of high-resolution images taken by the camera. To find tumors, a histopathology test is required. It is a time-and-tested method for predicting the presence of invasive cancer cells in tissues that are stained with H&E. In H&E staining, the chromatin in the nucleus and the nucleic acid in the cytoplasm turn pink, while the components in the cytoplasm and extracellular matrix turn purple-blue. The benefit of free scaling in histopathological images is that they may accurately display the minute features in tissue images and increase the precision of detection. Pathologists directly examine, evaluate, and grade these tissues under a microscope as part of conventional cancer testing, which is a well-established practice for identifying cancer cells. But while working under a microscope, pathologists are prone to feeling dizzy and neck strain. It takes time for the pathologist to make notes and write out their ultimate judgment in the interim.48 It takes a lot of time and effort for pathologists to manually analyze many sections because of the continually rising number of cancer patients. A lack of traceability between observers, internal alterations, and inconsistent results can also result from manual diagnosis. Cancer diagnosis accuracy varies by 20% between pathologists with expertise and those with less experience. Individual diagnoses and expert diagnoses differed by 75.3%, according to recent research.47,48 The objective is to determine if a tumor is benign or malignant since cancerous tumors need to be treated right away to lessen and avoid subsequent consequences. Numerous problems with this approach include intra-observer variability, the potential for recurrence in tissues and cancer cells, and the identification difficulty caused by the fact that numerous additional structures in cells have the same hyperchromatic properties. Due to the fact that the surgery only impacts a small amount of tissue, the site must be carefully considered. It is best to choose a region around the borders of the tumor.
Consequently, a detection strategy based on machine vision is necessary. It is able to carry out accurate, reliable, and efficient quantitative analysis. So, to improve the consistency of cancer detection and accuracy, therapy machine vision technology eliminates differences between internal and external observers.
3.2 Bioimaging
Bioimaging plays a crucial role in documenting the information details about biological substances for diagnosis, staging, and treatment using a variety of imaging devices to treat various diseases including cancer. Bioimaging also helps in the evaluation of the cellular process (ion or metabolite level) in cell biology. These imaging methods can be employed for non-invasive recognition or for the purpose of recording and extracting information from biological samples externally, without any physical intervention. Bioimaging is a powerful tool which is also used in cancer for visualizing the abnormal state at the target site.50 This technique helps healthcare professionals to visualize internal structures and identify abnormalities such as tumor, by providing a more detailed image to help in early detection and plan a course of action.51 The rapid growth and significant advancement in bioimaging technology have led to wide application based on different principles and instruments with various types of biological techniques developed such as X-ray, X-ray computed tomography (CT), hyperspectral imaging, magnetic resonance imaging (MRI), optical imaging and thermal imaging which required a large setup of instruments.52 The basic components of an imaging system are an illumination system, camera, frame grabber and image processing software and hardware.53 Camera is used for capturing the biological image of the object but for the evaluation of the external morphology including size, color, shape and surface structure there is the need of a sensor such as X-ray, ultrasound, MRI, X-ray CT and charge-coupled device (CCD) camera. Mostly a charge-coupled device (CCD) camera is used for this purpose. After that, the analog video signal is converted into digital signal, and the process of conversion is known as digitization. A digitizer (also known as a frame grabber) is utilized to transform visual images into a digital format. Processing of the digital image is done by hardware and software to analyze the obtained images. Image processing involves a number of phases that must be completed, including image capturing, partitioning, initial image processing, augmentation, depiction, and detailing. X-rays, magnetic resonance imaging (MRI), computed tomography (CT), positron emission tomography (PET) and single photon emission CT (SPECT) are among the conventional bioimaging techniques but their practical use is constrained by complicated processes, expensive instruments, time-consuming steps and low resolution of the image. Therefore, alternative bioimaging techniques are required that are less expensive, and more precise technologies are needed to get rapidly fast-growing domain of super-resolution; three new techniques such as two-photon fluorescence excitation microscopy, fluorescence recovery/redistribution after photobleaching (FRAP), and fluorescence resonance energy transfer (FRET) are recently developed. In vitro imaging can give important details of the distribution, cytotoxicity, and imaging ability of probes in cells. For clinical imaging applications, there is the requirement of good biocompatibility and minimal cytotoxicity of probes. Conventional quantum dots and similar core–shell nanoparticles were previously employed for optical bioimaging both in vitro and in vivo. But those containing toxic heavy metals cause problem to health and the environment. Other nanomaterials such as gold and silver nanoparticles are a safe alternative but they are costly and lack photostability. On the other hand, there are interesting alternatives of QDs which contain toxic heavy metals but in the biological system they are exposed to oxidative degradation which makes them slightly toxic. Therefore, there is the need of excellent nanoparticles which have good photoluminescence properties and which are biocompatible, highly photostable and of low toxicity for bioimaging.54 Nanotechnology can also be used in bioimaging techniques. Nanoparticles are regarded as promising candidates for bioimaging due to their applicability in fluorescence imaging within the second near-infrared spectral region (NIR-II, 1000–1700 nm). Additionally, they can offer the benefits of exceptional spatial resolution and significant penetration depth, facilitated by reduced light scattering.55 It has been demonstrated that NIR-II quantum dots (QDs) have the potential to be utilized in biomedical and clinical applications for deep tissue imaging application because of their remarkable biocompatibility, photostability, and luminosity.56 Because of their innovative fluorescent carbon nanomaterial, robust resistance to photobleaching, absence of blinking, biocompatibility, minimal toxicity, and environmentally friendly attributes, carbon dots (CDs) have attracted a lot of interest for use in bioimaging, catalysis, and sensing techniques.57–61 Small sized carbon dots with strong biocompatibility can easily penetrate cells. Nanoparticles that exhibit multi-imaging characteristics have drawn a lot of interest and can be employed in composite systems. Nanocarriers with cell-targeting drug storage, detection, cancer cell separation and controlled drug release capabilities62,63 have been designed to enhance the bioimaging capability. Currently, a diverse range of inorganic nanoparticles (NPs) such as biomolecules, CDs, gold and silica nanoparticles, and carbon nanotubes are being utilized for the purpose of drug and gene delivery.64,65 Due to their outstanding chemical inertness, biocompatibility, and precise sizing for mesopore entry, CDs have emerged as a fitting option for a novel bioimaging agent.66 Utilization of carbon dot delivery techniques can reduce cytotoxicity, improve clinical results, and offer promising nano-carriers for the advancement of multifunctional nanomedicine in the future,67 when compared to carbon nanotubes and gold and silica nanoparticles.68 CDs have been utilized for bioimaging across various normal and cancer cells, as illustrated in Table 2.| Nanoparticles | Synthesis method | Technique | Target | Limit of detection range | Limit of detection | Quantum yield (%) | Size (nm) | Ref. |
|---|---|---|---|---|---|---|---|---|
| N,S-CDs | — | Cell imaging | A549 cells | — | — | 80.3 | 3 | 69 |
| CD incorporated polymeric hydrogels | — | Fluorescence imaging | A549 cells | — | — | — | — | 70 |
| CQDs | Hydro thermal | Bioimaging | A549 cells | — | — | — | 1.8 | 71 |
| AA-CDs | Hydro thermal | Bio-imaging (cancer cells) | HeLa, SMMC-7721 and A549 cells | — | — | 56.5 | 1.6 | 72 |
| Amino-functionalized fluorescent carbon nanoparticles | — | Bio-imaging (cancer cells) | A549 cells | — | — | 7.8 | 4–7 | 73 |
| N-P-doped CDs | Hydro thermal | Bio-imaging | HeLa cells, A549 cells | — | — | — | 3.8 | 74 |
| m-Phenylenediamine and L-cysteine | Hydro thermal | Nucleolus imaging | HeLa, A549, HepG2, L02, AT II, RAW264.7 | — | — | — | 3.8 ± 0.5 | 75 |
| Spinach | One-step solvo-thermal | Bio-imaging | A549 cells | — | — | 15.34 | 3–11 | 76 |
| Citric acid (CA) and glutathione (GSH) | Hydro thermal | Cell imaging | A549 cells | — | — | 80.3 | 3 | 69 |
| Glycerol and dimethyl sulfoxide (DMSO) | Solvo-thermal | Imaging of cancerous cells | A549 cells | 0.1–100 μM | 16 nM | — | 6.1 ± 1.4 | 77 |
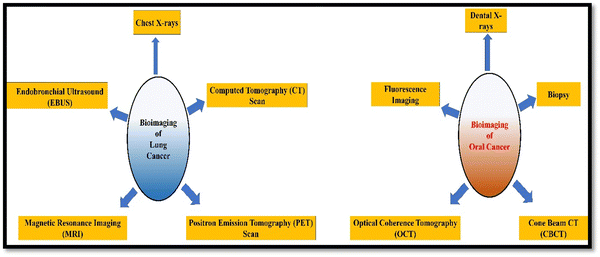
Bioimaging applications in cancer disease are diverse and crucial to improve diagnosis, treatment, and monitoring of cancer patients. Fig. 3 illustrates the different types of bioimaging techniques used for oral and lung cancer detection and their applications are depicted in a pictorial form in Fig. 4. These imaging techniques provide valuable information about the tumor's characteristics and location. They provide informational data to healthcare professionals and help in making informed decisions related to the treatment of cancer.
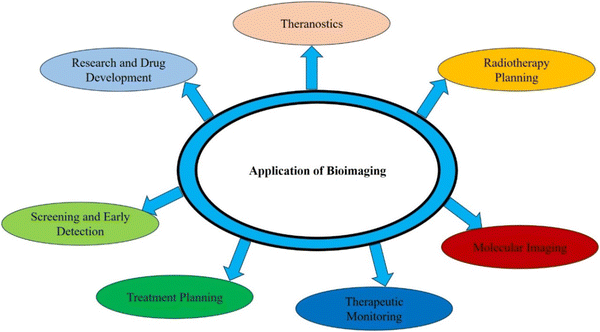 | ||
| Fig. 4 Application of bioimaging in various fields of diagnosis to improve the quality of human health. | ||
3.3 Enzyme linked immunoassay (ELISA)
Among the conventional diagnostic methodologies, enzyme-linked immunoassay (ELISA) stands out as a predominant technique employed to validate and quantify biomarkers associated with the proliferation of several cancer types, including lung and oral cancer. Recognized as a cornerstone in analytical chemistry, ELISA has been refined into multiple variants to suit varied analytical needs. Mainly four primary types of ELISA are identified: direct ELISA, indirect ELISA, sandwich ELISA, and competitive ELISA.78 Of these, competitive ELISA, direct ELISA, and sandwich ELISA are the most frequently utilized immunosorbent assays in the context of cancer biomarker detection.In a study by Danae et al., the direct ELISA technique was harnessed to ascertain elevated levels of ProTα. The study employed antibodies targeted against the C-end peptide sequence 101–109 of ProTα. They compared its presence in breast tumor tissue samples with that in normal breast tissue controls.79 Furthermore, in another significant study, Neal et al. devised a direct ELISA-based assay specifically for detecting bone sialoprotein, a vital biomarker. The presence of bone sialoprotein is especially crucial in the context of several cancers, notably breast, prostate, colon, and lung cancers. The development of such assays is instrumental in the early diagnosis and monitoring of these malignancies, underscoring the critical role of direct ELISA in oncological research and clinical applications.80
• The capture antibody is first added to the plate wells, which are then sealed and incubated overnight at 4 °C.
• After the incubation, the plate is rinsed with phosphate-buffered saline (PBS) to remove any unbound capture antibody.
• The wells are then buffered or blocked using bovine serum albumin (BSA) to prevent non-specific binding. This buffering step is executed at room temperature and spans roughly 1 to 2 hours.
• Another PBS wash follows.
• The antigen sample, containing the molecule of interest, is added. It binds specifically to the capture antibody. This binding phase is maintained for 90 minutes at 37 °C.
• After this, the plate undergoes another wash.
• The primary detection antibody, which recognizes a different epitope or region of the antigen than the capture antibody, is introduced and allowed to bind to the antigen. This incubation lasts for another 1 to 2 hours at ambient temperature, followed by a subsequent wash.
• Next, a secondary antibody, conjugated to an enzyme, is added. This incubation also extends for about 1 to 2 hours.
• A final wash is performed, post which a substrate solution is introduced, which, upon interaction with the enzyme, yields a colorimetric change.
One of the significant merits of sandwich ELISA is its superior sensitivity, arguably the highest among all ELISA variations. However, this method does have its disadvantages, most notably the extended time required, increased costs, and the need for “matched pair” antibodies (those that can simultaneously bind different sites on a divalent or multivalent antigen) along with a suitable secondary antibody.
Here's a breakdown of the process:
• Two specific antibodies are utilized: one that is enzyme-conjugated and another that may be present in the test serum (if the serum is positive for that particular antibody).
• When both antibodies are introduced into the wells, they compete for binding sites on the antigen.
• If a color change manifests, the test result is negative. This indicates that the enzyme-conjugated antibody has effectively bound to the antigen, signifying the absence of the test serum's specific antibodies.
• Conversely, a lack of color change signifies a positive test, indicative of the presence of antibodies in the test serum.
While competitive ELISA offers various advantages, such as the reduced need of the sample, the ability to assess a vast spectrum of antigens to be detected in a sample, suitability for smaller antigens, and minimized variability, it also has its share of drawbacks. Its specificity is relatively low, and it is less effective in analysing dilute samples. To emphasize the utility of competitive ELISA in oncology, numerous studies have showcased its effectiveness in detecting biomarkers pertinent to cancer diagnostics and research. Two notable studies highlight the utility of ELISA in detecting specific biomarkers relevant to lung cancer. In the first study, carried out by Yagihashi, Atshuhito, et al., the researchers reported a significant presence of anti-livin antibodies in lung cancer patients using a specific ELISA. They employed recombinant proteins since both survivin and livin are predominantly expressed in cancer and transformed cells but are scarcely present in normal, differentiated tissues.81 The outcomes of their research revealed the following:
• 19 out of 37 lung cancer patients (51.3%) tested positive for anti-livin antibodies.
• Of 31 samples drawn from these lung cancer patients, 18 (58.1%) exhibited the presence of anti-survivin antibodies.
• When examining sera from 31 lung cancer patients for both anti-survivin and anti-livin antibodies, a striking 71% tested positive for either survivin or livin, or both.
• However, the study also found that the intensity of anti-livin antibody responses did not have any correlation with the intensity of anti-survivin responses.
The second investigation, performed by Yamashita et al., aimed to determine the potential diagnostic value of detecting epidermal growth factor receptor expression on exosomal membranes.82 The researchers attempted to identify lung cancer by using a targeted ELISA that used an anti-CD81 antibody as the capture antibody. Together, these studies underscore the versatility and importance of ELISA as a diagnostic tool in oncology, especially for detecting and understanding lung cancer. Ruay-Sheng et al. designed a study to investigate whether CYFRA2I-1 is a sensitive and specific tumor marker for non-small cell lung cancer (NSCLC) or not. CYFRA 2I-1, a newly developed sandwich ELISA, was used to measure the soluble cytokeratin 19 fragment in serum that is expressed in simple epithelium and its malignant counterpart.83 As an oncogene and crucial component in the diagnosis of lung cancer, Wang et al. described a double-antibody sandwich ELISA kit for the detection of MUC1 in serum from lung cancer patients. Through this work a double-antibody sandwich ELISA kit was successfully constructed and the sensitivity was up to 0.5 μg L−1.84
Two distinct studies focused on the applicability and efficacy of ELISA in diagnosing lung cancer through different biomarkers:
CYFRA 21-1 as a biomarker for NSCLC. In one study, the researchers sought to determine the diagnostic efficacy of CYFRA 21-1 as a marker for NSCLC. For this purpose, they employed the CYFRA 21-1 sandwich ELISA technique to measure the levels of the soluble cytokeratin 19 fragment in the serum. Cytokeratin 19 is expressed in simple epithelia and their malignant counterparts. By quantifying this fragment, the researchers aimed to discern its utility as a sensitive and specific tumor marker for NSCLC.
MUC1 as a biomarker for lung cancer. In another study the central focus was the MUC1 oncogene and its significance in diagnosing lung cancer. MUC1 plays a pivotal role in several cancers, including lung cancer. In this investigation, a double-antibody sandwich ELISA kit was devised to detect serum levels of MUC1 in lung cancer patients. This constructed ELISA kit exhibited a high degree of sensitivity, being able to detect MUC1 concentrations as low as 0.5 μg L−1.84
Together, these investigations illustrate the power and versatility of the ELISA technique, with different adaptations, in the realm of oncological diagnostics. They highlight the potential of specific biomarkers, when quantified and analyzed accurately, in aiding early detection and understanding of lung cancer.
The study by the cited authors indicates that chemokines play a significant role in inflammation and carcinogenesis. Techniques like enzyme-linked immunosorbent assays (ELISA) have provided reliable methods for their detection in saliva. Based on this, there's a proposition that individuals with oral cancer or precancerous oral lesions might exhibit increased concentrations of specific chemokines in their oral fluids. Such elevations could potentially serve as indicators for early malignant detection or for monitoring progression towards malignancy. In a related study, the utilization of ELISA revealed notably higher levels of CXCL8, CXCL10, and CCL14 in the oral fluids of individuals diagnosed with cancer.85
4. Fabrication of electrochemical sensors for cancer detection
From the last decade, progressive exploration has been achieved for the fabrication of sensors for cancer detection using nanobiotechnology and nanomaterial science. The overall performance of the sensor could be improved by the use of nanomaterials such as carbon-based materials, metal oxides, transition metal dichalcogenides, nanoparticles, quantum dots, etc. These nanomaterials possess tremendous potential related to optical, electrical and catalytic properties such as wide band gap, excellent conductivity, biocompatibility, improved surface to volume ratio, good permeability, electronic conductivity, etc. Therefore, the use of nanomaterials in the fabrication of sensors helps to promote the progress of electro-analytical sensors in various aspects such as enhanced detection range, sensitivity, selectivity, specificity, stability and limit of detection.Nanomaterials are low dimension materials compared to building units of sub-micron size, and at least one of their dimensions is in the nanometer range. These materials can be nanotubes (carbon nanotubes), nanosheets (graphene oxide, reduced graphene oxide, molybdenum disulfide, tungsten disulphide), nanoparticles (zinc sulphide, gold, silver, platinum), functionalized materials (acid, base, or capping agents/ligands) or their composites which enhance the intriguing properties of the materials, by providing a large surface to volume ratio, biocompatibility, fast electrochemical signal propagation, amplification of the sensing response, etc.
The electrochemical sensing technique has been extensively explored for the diagnosis purpose and for monitoring clinical as well as non-clinical samples. Electrochemical transduction between the recognition element and the analyte occurs at the electrode surface which can be measured as an electrochemical signal in terms of potential, current, or impedance. Various types of electrochemical sensors have been fabricated such as nucleic acid sensors, aptasensors, enzyme sensors, immunosensors or MIP based sensors for oral or lung cancer biomarker detection in human body fluids. These sensors have been broadly discussed further based on the recent literature articles and analytical sensing responses. The sensitivity, selectivity, specificity, stability, detection range and limit of detection of the fabricated sensor have been discussed for cancer biomarker detection. For the construction of the sensor, the target analyte as a cancer biomarker is used for the specific and selective detection of particular cancer. The sensor matrix surface can be modified by the use of nanomaterials to enhance the surface immobilization of the recognition element, biocompatibility, stability, conductivity and the sensing signal. Therefore, a biorecognition element such as antibodies, enzymes, DNA, and artificial recognition sites as cavities for MIPs is used for the construction of sensors. A few of the oral and lung cancer biomarkers for cancer detection are described in Table 3.
| Cancer type | Biomarker | Ref. | |
|---|---|---|---|
| Lung | RNA | miR-106a-5p, miR-10b-5p, miR-141-3p | 86,87 |
| Protein | ALK, CEA, KRAS, EGFR, ROS, KRAS, MET, NTRK1, FGFRA, HER2, BRAF, PIK3CA, RET, DDR2, PTEN | ||
| Oral | Protein | IL-8, IL-1b, TNF-α, defensin-1, P53, Cyfra 21-1, tissue polypeptide-specific antigen | 88 |
| DNA | Human papillomavirus type 16 and 18 | 89 | |
4.1. Types of sensors
Nucleic acid based sensor for oral cancer detection. Advanced invasive and metastatic oral cancer can now be diagnosed using cancer cell lines or biopsy samples. The most efficient way to decrease oral cancer-related deaths is by early, non-invasive detection. In current times, there has been an evident rise in the number of cancer-related nucleic acids discovered in oral fluid, making it an ideal diagnostic fluid. Oral fluid offers several clear advantages over blood.90 Yet, there are still a number of significant difficulties with the diagnostic method used with saliva, including low sensitivity and poor specificity. Therefore, it is crucial to create procedures that are very sensitive and focused for using saliva to diagnose oral cancer.91
The ability of catalytic nucleic acid (DNAzyme)-based techniques to catalyse a variety of chemical processes, including DNA self-modification and metal ion-dependent DNA/RNA breakage, has garnered growing interest.92,93 One of the most intriguing DNAzymes is the G-quadruplex-hemin DNAzyme, which is made up of hemin and a G-rich nucleic acid and can efficiently catalyse the H2O2-mediated oxidation. For the creation of diverse DNA biosensors, it has therefore demonstrated tremendous promise as a novel type of catalytic label.
For quick, sensitive, dependable, and affordable DNA detection in clinical diagnostics, DNA electrochemical biosensors have shown considerable promise. The sensitivity and selectivity of electrochemical DNA biosensors have been improved via the development of several sensing techniques.94,95 Conventional biosensors are not able to detect a minute amount present of analyte therefore, nucleic acid based sensor helps to quantify the biomarkers present in saliva as they present in a very low concentration.
Chen et al. in 2011 described the fabrication of an electrochemical sensor for the detection of DNA associated with the oral cancer overexpressed 1 gene (ORAOV1) in saliva.96 Through S–Au bonding, the single probe (Sp) is covalently joined to a gold electrode (GE). Then, 2-mercaptoethanol (MCH) is used to inhibit the Sp-modified GE, resulting in the formation of a mixed monolayer. When a target is precisely matched, Sp attaches with the DNA and forms a double-helical structure with a complete N.BstNB I recognition site. Sp gets detached from the DNA when the affinity between the two decreases. When the target DNA is released, it can combine with a fresh, unnicked Sp to start a new cycle of probe transformation. This enables a single target DNA to permanently change a number of Sps into G-rich nucleic acid sequences.97,98 After the strand-scission cycle is over, washing is a simple way to get rid of the target recognised Sp fragment. As a result, only G-rich pieces are left on the sensor surface. Therefore, following hemin addition, the G-rich sequences can interact with hemin to produce the G-quadruplex-hemin DNAzyme. 3,3′,5,5′-Tetramethylbenzidine (TMB) is subsequently oxidised by H2O2 with the help of this DNAzyme, which results in a modified color in the reaction solution and an increase in the current signal. The nicking endonuclease, however, is unable to cleave the Sp in the absence of the target DNA. Consequently, the steric-hindrance effect prevents Sp from binding hemin to create the G-quadruplex-hemin DNAzyme.
In order to confirm the catalytic nature of the DNAzyme, it was further compared with the CV signals of the TMB substrate and oxidation as well as reduction peaks were observed in the bare electrode when CV was performed and after the DNAzyme was added indicating that the DNAzyme effectively reduced hydrogen peroxide with the aid of TMB (an electroactive co-substrate) and amplified electrochemical current signals. During this period, the hue of the solution that could be seen with the naked eye changed progressively from colorless to blue.99
Nucleic acid based sensor for lung cancer detection. Lung cancer is the most common cause of death globally. Flexible bronchoscopy (FB), imaging diagnosis and sputum cytology are the three primary traditional methods for diagnosing lung cancer. Sputum cytology is an invasive procedure that has limited sensitivity and relies on exact specimen collection and preservation procedures to provide accurate diagnoses.100 Similarly, FB can be helpful in the evaluation of a patient who is suspected of having lung cancer, but its sensitivity is constrained by the location and size of the lesion.101 As the most popular approach for identifying lung cancer, computed tomography (CT) has proven to be far more effective than chest radiography.102 CT screening, however, has not been regarded as a cost-effective method because there is still room for improvement in terms of performance.102,103
Small, non-coding RNAs called microRNAs serve as regulators. Although tiny, these microRNAs may be crucial regulators of a variety of human tumors. Therefore, it is thought that a possible method for early cancer diagnostics that would result in a better treatment for patients is quick and sensitive detection of cancer-related microRNAs. However, because of the short sequence and low abundance of microRNAs, it is incredibly difficult to create a quick, low-cost, and straightforward microRNA detection platform. Numerous studies have been conducted in recent years to develop quick and accurate microRNA detection methods, such as Northern blotting and polymerase chain reaction (PCR). The limitations of Northern blotting are its insensitivity and time-consuming process.104 For PCR, small primers are used in the microRNA due to which it gives false positive results. Widespread fluidic management, separation, and other engineering detection situations have been combined and hyphenated with electrochemical sensors. Electrochemical biosensors have many benefits including relatively low cost, quick response times, ease of use for continuous on-site analysis, high selectivity and sensitivity, minimal sample preparation, and reliable reproducibility. These advantages make electrochemical biosensors a promising technology for biomolecule detection. The development of electrochemical genosensors as point-of-care diagnostic tools and multiplexing platforms for quick, easy, and affordable nucleic acid detection has shown considerable promise.105
Lung cancer-related microRNAs (miRNAs) are evolving. Let-7, miR-141, and miR-21 are frequently used as diagnostic targets. MiR-25, miR-145, and miR-126 are a few recent lung cancer miRNAs that have drawn more and more attention. Although there are several methods for analysing lung cancer miRNAs, electrochemistry has gained popularity for its great sensitivity, low cost, and quick response. Electrochemistry has been praised for its high sensitivity, cheap cost, and quick response despite the fact that there are several methods available for the investigation of lung cancer miRNAs. An electrochemical genosensor's specificity and sensitivity depend heavily on the design of the probes. The typical properties of microRNAs include high sequence similarity, low melting points, and short sequences.101 Because of this, high sensitivity detection of microRNAs using conventional sandwich structure DNA probes and single strand DNA probes is difficult. On the other side, single-base mismatch nucleic acid detection has been effectively accomplished using DNA probes with a stem-loop structure. A stem-loop DNA probe-based electrochemical sensor has also been proposed in order to detect the complementary sequence and significantly boost the sensitivity of nucleic acid detection.106 But because it was a signal-off sensor—meaning that its detecting signals were bigger than its baseline—it needed a consistent, steady background level in order to prevent being readily influenced by false positive signals.107
Consequently, there is a need for a promising method for early diagnosis of lung cancer which is very sensitive for the detection of particular microRNAs. In this regard, a 3D DNA origami structure was developed that allows for the electrochemical detection of microRNAs associated with lung cancer. The 3D DNA origami structure is composed of a thiolated tetrahedron DNA nanostructure at the bottom and a stem-loop DNA structure with ferrocene tags at the top. The top section hybridised with the microRNA linked to lung cancer, whereas the bottom piece self-assembled on a gold electrode surface that had been treated with gold nanoparticles (Au NPs) and blocked with mercaptoethanol (MCH). Cyclic voltammetry (CV), scanning electron microscopy (SEM), differential pulse voltammetry (DPV) and atomic force microscopy (AFM) were used to characterize the fabrication procedure and the performance of the proposed electrochemical genosensor. The proposed genosensor demonstrated tremendous promise for very sensitive clinical cancer diagnostic application for detecting microRNA at concentrations ranging from 100 pM to 1 M, with a detection limit of 10 pM with outstanding linearity.104
Aptamers have gained significant attention in various fields, including sensor fabrication due to their unique properties and potential applications. In sensor fabrication, aptamers can be used as unique recognition elements to develop aptamer-based sensors. These sensors utilize the binding affinity of aptamers to detect and quantify target molecules.
Aptamers are first selected or synthesized to specifically bind to the target molecule of interest. This involves an in vitro selection process, where a library of random sequences is iteratively enriched for sequences that bind to the target. The SELEX procedure involves two alternating steps. During the initial stage, the original oligonucleotides are subjected to amplification through a polymerase chain reaction until the desired concentration is achieved. In the subsequent phase, the amplified pool is exposed to specific targets, and the oligonucleotides that interact with these targets are utilized for the initial step of the subsequent SELEX round. Using this method, it is possible to isolate aptamers that exhibit elevated specificity and affinity toward their target molecules from larger collections of diverse oligonucleotides through a profoundly iterative process. Within this scenario, the oligonucleotides that form a bond with a target molecule are extracted and subjected to amplification through a polymerase chain reaction, while the non-bound oligonucleotides are effectively removed through a washing process.
Aptamers are immobilized onto a sensor surface, which could be a solid substrate such as a glass slide, a metal electrode, nanoparticles or carbon nanotubes and polymers.111–113 Aptamers have the capability to be directly attached to a sensor's surface or indirectly linked through chemical coupling methods. Direct immobilization of aptamers primarily arises from electrostatic interactions, hydrogen bonding, or van der Waals forces. The range of feasible approaches here implies the convenient adaptability of aptamers for modification.114 The selection of an immobilization method is contingent upon various considerations, including the characteristics of the biological recognition component and the transducer surface, the attributes of the target molecule, and the operational circumstances of the aptasensor.115 Immobilization methods can include physical adsorption, covalent attachment, or hybridization with complementary sequences on the surface such as biotin/avidin affinity, covalent bonds, complementary nucleic acid chain connections and gold–sulfur self-assembled monolayers. Upon binding of the target molecule to the immobilized aptamer, a conformational alteration occurs within the aptamer structure. This modification can be converted into a quantifiable signal through diverse techniques, including electrochemical, thermal, optical, and piezoelectric methods, or mass-sensitive approaches.116 The alteration in the signal is directly correlated with the concentration of the target molecule present in the sample. The generated signal is detected and quantified using appropriate instrumentation by correlating the measured signal with known concentrations of the analyte. Calibration curves are constructed to establish a quantitative relationship between signal intensity and analyte concentration. The sensitivity and specificity of aptamer-based sensors can rival or even surpass traditional antibody-based sensors. They have the capability to detect an extensive array of targets, encompassing small molecules as well as large proteins.
Nanomaterials play a crucial role in enhancing the performance and capabilities of aptasensors, which are biosensors that utilize aptamers as the recognition element for target molecule detection. It is important to note that the selection of nanomaterials depends on the specific requirements of the aptasensor and the target molecule to be detected. The choice of nanomaterial, surface functionalization, and sensor architecture can greatly influence the sensor's sensitivity, selectivity, and overall performance. Ongoing research in nanomaterial synthesis, modification, and integration continues to expand the capabilities of aptasensors and their potential applications in various fields. The integration of nanomaterials such as nanoparticles (e.g., gold nanoparticles, quantum dots) and nanowires with aptasensors offers several advantages, such as a high surface area-to-volume ratio, allowing for increased immobilization of aptamers and target binding sites. This leads to improved sensitivity as more target molecules can interact with the aptamer molecules. Nanomaterials can be used to amplify the signal generated upon target binding. For example, gold nanoparticles can serve as labels to enhance the electrochemical or optical signal. When a target molecule binds to the immobilized aptamer–nanoparticle complex, the resulting signal change is magnified due to the presence of multiple nanoparticles. Many nanomaterials exhibit good biocompatibility, making them suitable for bioanalytical applications and potential use in medical diagnostics. Specific nanomaterials, like carbon nanotubes and graphene, exhibit remarkable electrical conductivity. They can be used as transducing elements in electrochemical aptasensors, where the binding event between the aptamer and the target molecule leads to a change in electrical conductivity, which can be easily measured. Nanomaterials can provide stability to the aptamer molecules, preventing their degradation and maintaining their functionality over time. Nanomaterials can be functionalized with multiple aptamers or other biomolecules, allowing for the simultaneous detection of multiple target molecules or the integration of additional functionalities, such as controlled release of captured targets. Nanomaterials facilitate the creation of compact and portable aptasensor devices, well-suited for point-of-care and field-based applications. Tiwari et al. in 2016 reported Cys-La(OH)3 nanoparticles electrophoretically deposited onto an ITO glass substrate to immobilize anti-Cyfra-21-1 for the electrochemical detection of Cyfra-21-1 (oral cancer biomarker). This immunosensor demonstrates a wide detection span ranging from 0.001 to 10.2 ng mL−1 using artificial saliva, a limit of detection (LOD) of 0.001 ng mL−1, and notable sensitivity at 12.044 μA (ng per mL cm−2)−1, accompanied by a response time of 5 minutes.117 The practical utility of the developed sensor for Cyfra-21-1 was tested in artificial saliva samples and the results indicate good correlation with the relative standard deviation which was within the acceptable range.
Khaksari et al. in 2023 reported a novel microfluidic electrochemical aptasensor to identify A549 cancer cells. These cells act as a representation of integrin α6β4-containing cells and stand as a circulating tumor cell (CTC) model for non-small cell lung cancer (NSCLC). This aptasensor relies on a target-triggered structural change in an IDA aptamer's conformation, which has a specific affinity for A549 cells. By observing this conformational change in the aptamer structure, the presence of A549 cancer cells can be accurately and selectively determined. The microfluidic biosensor is composed of a microchip that includes a screen-printed gold electrode. The electrode is modified with SH-IDA aptamers through covalent chemical bonding. The biosensor provides an extensive linear detection span ranging from 50 × 105 cells mL−1, achieving a notable level of detection sensitivity with a limit of detection (LOD) as low as 14 cells mL−1 for A549 cancer cells. This microfluidic biosensor has demonstrated successful performance in detecting A549 cells within complex samples like human serum.118 In another report, Chen et al. in 2023 developed an innovative and highly sensitive aptasensor that utilizes surface-modified graphene to detect the lung cancer biomarker CA125. This aptasensor takes advantage of the combined effects of incorporating graphene surfaces and electrodeposited gold nanoparticles (AuNPs), resulting in an exceptional level of sensitivity and specificity for accurate biomarker detection. The introduction of AuNPs through deposition significantly reduces electron transfer resistance, thereby greatly improving the electrochemical performance of the sensor. The findings validate a direct correlation between the sensor response and the concentrations of the lung cancer biomarker CA125, encompassing a range from 0.2 to 15.0 ng mL−1. The results also reveal a limit of detection (LOD) of 0.085 ng mL−1.119
Electrochemical aptasensors are based on the detection of electrochemical signals generated by the binding of a target biomarker to an immobilized aptamer. The change in the electrochemical signal can be detected using various techniques, such as cyclic voltammetry, impedance spectroscopy, and amperometry. For example, an aptamer-based electrochemical biosensor was developed for the detection of TNF-α in three serum samples collected from healthy individuals.120 The aptamer was immobilized onto Ag@Pt core–shell nanoparticles supported on reduced graphene oxide, and showed changes in the electrochemical signal, which were quantified using differential pulse voltammetry and square wave voltammetry. The biosensor showed high sensitivity and specificity, and the results indicate its broad application in bioassay and protein diagnostics.
Conventional techniques such as ELISA, western blot, and PCR can show cross-reactivity with other proteins or molecules in a sample, leading to false-positive results, which can lead to unnecessary treatment and anxiety for patients. Specificity is another crucial factor in cancer biomarker detection with aptasensors. Aptamers can bind to specific biomarkers, allowing for the detection of a particular cancer biomarker without cross-reactivity with other proteins or molecules in a sample. Aptasensors have shown higher specificity than conventional techniques for cancer biomarker detection. An electrochemical aptasensor was developed for the detection of human epidermal growth factor receptor 2 (HER2) in human serum samples with albumin depletion for the diagnosis of early stage breast cancer.121 Also, clinical application of the developed sensor was tested in patient's blood samples with HER2 positive breast cancer and the results showed good correlation with standard samples. The aptasensor showed high selectivity for HER2, with minimal cross-reactivity with other proteins. Another study developed an aptamer-based biosensor for the detection of carcinoembryonic antigen (CEA) in human serum samples suffering from lung disease and the results showed accurate correlation with standard chemiluminescence immunoassay.122 The performance of hairpin oligonucleotide aptamer-functionalized gold nanorod labels and the graphene-streptavidin nanomatrix was quantified using DPV. The biosensor showed high selectivity and sensitivity for CEA with a wide dynamic range of 5 pg mL−1 and 50 ng mL−1 toward CEA standards with a low detection limit of 1.5 pg mL−1. Moreover, a few of the recently published research articles related to aptasensors for cancer biomarker detection using the electrochemical technique and their findings of biosensing parameters are summarized in Table 4.
| S. no | Biomarker | Cancer type | Nanomaterials | Techniques | Linear range | LOD | Year | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1. | α-Enolase | Lung | SPE/AuNP | Voltammetry | 10−8–10−12 g mL−1 | 2.38 pg mL−1 | 2010 | 123 |
| 2. | CEA | Multiple | Graphene films | Voltammetry | 5–60 ng mL−1 | 5 ng mL−1 | 2011 | 124 |
| 3. | α-Fetoprotein | Multiple | SPCE/nanogold-enriched carbon nanohorn | Voltammetry | 0.1 pg mL−1 ng mL−1 | 0.07 pg mL−1 | 2013 | 125 |
| 4. | miRNA-24 | Multiple | GCE/SWCNT-polyamidoamine dendrimer hybrid | Voltammetry | 10 fM–1 nM | 0.5 fM | 2015 | 126 |
| 5. | CEA | Multiple | GCE/AuNP-thionine-reduced GO nanocomposite | Voltammetry | 10–500 pg mL−1 | 4 pg mL−1 | 2011 | 127 |
| 6. | Platelet-derived growth factor | Multiple | GCE/SWCNT/platinum NP/AuNP | Voltammetry | 0.01–35 nM | 8 pM | 2012 | 128 |
| 7. | L-lactate | Multiple | SPCE/AuNP anchored-reduced GO | Amperometry | 5 mM–10 mM | 0.13 mM | 2015 | 129 |
| 8. | CEA | Multiple | CPE/TiO2 and Au nanocomposite + chitosan | Amperometric | 0.01–1 ng mL−1 | 0.01 ng mL−1 | 2014 | 130 |
| 9. | Let-7b | Multiple | ITO/Ru(PD)2Cl2 + oxidation of hydrazine | Amperometric | 0.50–400 pM | 0.20 pM | 2007 | 131 |
| 10. | Osteopontin | Multiple | AuE/3D scaffold of mercapto-peptides + AB | Impedimetric | 2.27–20.43 nM | 0.17 nM | 2014 | 132 |
| 11. | Interleukin-6 | Multiple | SiO2/Si-wafer/aligned SWCNT + AuNP | Impedimetric | 0.01–100 fg mL−1 | 0.01 fg mL−1 | 2013 | 133 |
| 12. | Lysozyme | Multiple | AuE/AuNP | Impedimetric | 0.1 pM–500 pM | 0.01 pM | 2011 | 134 |
Immunosensors are made to be simple for untrained staff to operate, have quick test times, and have sensitivity similar to ELISA techniques. Antibody–antigen interactions have been used in surface plasmon resonance, resonant mirror, continuous-flow immunosensors, fiber-optic waveguides, and planar waveguides for the detection and measurement of therapeutically relevant analytes143
Because antibodies are sensitive and specific, immunoassay tools have become essential in clinical diagnosis. Immunosensors, which are compact, portable devices for the measurement of analytes in complicated fluids, find useful application in the quick and accurate diagnostic examination of clinical samples.143
The biosensor class known as electrochemical immunosensors are widely known for their high selectivity, ease of use, point of care, and quickness in providing data.144 The analyte of choice, the biological recognition component, and an appropriate immobilization matrix are all important in the fabrication of immunosensors. The manufacture of biosensors has utilized a variety of immobilization matrix types, including polymers and nanomaterials, and immobilization techniques.145,146 The construction of biosensors frequently makes use of electrochemical transducers using platinum, carbon, and gold bases.
As 70% of oral cancers are discovered at advanced stages, it might be difficult to detect OC at an early stage.142 As a result, OC patients often have a bad prognosis because the overall 5-year survival rate for OCs is less than 40%.147 In an effort to improve prognosis, a wide variety of OC screening techniques have been developed including visual, toluidine blue (TB) staining, auto-fluorescence spectroscopy,148 exfoliative cytology, and biopsy/histopathology.
However, these methods have drawbacks such as limited sensitivity, lack of specificity for malignant tissue, time consumption, high cost, and tedious work-up. Therefore, there is a need for the creation of a quick, inexpensive, and highly sensitive technology that may be employed for early diagnosis of lung cancer. Recent research has concentrated on methods using biosensors for a quicker, less expensive, and early cancer diagnosis.149,150
Deepa et al. in 2016 fabricated a label-free impedimetric immunosensor that can identify CD 59, an essential and early-stage OC biomarker, and be used to diagnose OC. The self-assembled molecular layer of L-cysteine (Cys) on a gold electrode is used to immobilize CD 59 antibodies (anti-CD 59) in order to create the immunosensor probe. In order to get the highest level of sensitivity, the experimental settings were optimized for antibody concentration, temperature, pH, and incubation duration. The CD 59 is subjectively recognized by EIS, which has a detection limit of 0.38 (0.03) fg mL−1 and a linear range of detection from 1 fg mL−1 to 1000 fg mL−1. The developed immunosensor successfully detects CD 59 in clinically relevant quantities.151 Moreover, real application of the oral cancer biosensor was investigated in human saliva samples and the detection limit was found to be 0.84 fg mL−1. This study demonstrates that the developed sensor is highly efficient towards the detection of CD59 in saliva samples, and hence the developed sensor shows precise clinical application.
Verma et al. in 2017 found that salivary interleukin-8 (IL8), an oral cancer biomarker, can be used to detect cancer without using labels or other intrusive methods. They fabricated an electrochemical immunosensor using a transducer matrix made of an extremely stable composite material of reduced graphene oxide and gold nanoparticles (AuNPs-rGO). Due to the increased electron transfer abelled of the composite made possible by the synergy between rGO and AuNPs, the immunosensor could now respond quickly and with great sensitivity. The immunosensor had an experimental linear range of detection from 500 fg mL−1 to 4 ng mL−1 and a LOD of 72.73 ± 0.18 pg mL−1. It also exhibited great sensitivity and enabled extremely quick detection of IL8 (9 min). The created immunosensor had outstanding specificity for detecting IL8 in samples of human saliva. Additionally, the immunosensor showed reusability and stability for up to three months revealing the commercial potential of this nanoplatform for the detection of other clinically relevant biomarkers.152
Malhotra et al. in 2010 found another biomarker for oral cancer detection based on the observation that interleukin-6 (IL-6)-mediated angiogenic, immunological, and inflammatory responses are linked to head and neck squamous cell carcinoma (HNSCC). In this the authors fabricated an electrochemical immunosensor for human IL-6 along with proof-of-concept tests on HNSCC cells to demonstrate IL-6 detection. In an electrochemical sandwich immunoassay approach employing enzyme abelled horseradish peroxidase (HRP), single wall carbon nanotube (SWNT) forests with connected capture antibodies (Ab1) for IL-6 were utilized to quantify extremely low (≤30 pg mL−1) and increased amounts of IL-6. A representative panel of HNSCC cells was employed to quantify a wide concentration range of IL-6 using two levels of multienzyme labelling. The best detection limit (DL) for IL-6 in 10 L of calf serum was 0.5 pg mL−1 (25 fM) and the maximum sensitivity was 19.3 nA mL (pg IL-6)−1 cm2. This was achieved using secondary antibodies (Ab2) linked to carboxylated multiwall carbon nanotubes with 106 HRP labels per 100 nm. In order to give 14–16 labels for each antigen, biotinylated Ab2 coupled to streptavidin-HRP was used. The excellent correlations between this immunosensor's measurements of secreted IL-6 in a variety of HNSCC cells and the results of standard enzyme-linked immunosorbent assay (ELISA) suggest that SWNT immunosensors in combination with multilabel detection hold great promise for detecting IL-6 in both research and human serum clinical applications.153
Also, Pachauri et al. fabricated a noninvasive electrochemical immunosensor using silver molybdate nanoparticles for the detection of another oral cancer biomarker interleukin-8 (IL-8). This sensor had a linear range of detection of 1 fg mL−1 to 40 ng mL−1 and showed an LOD of 90 pg mL−1 and a sensitivity of 7.03 μA ng−1 mL cm−2.154 Also, experiments were conducted for practical utility of the developed immunosensor in human saliva samples by spiking of various concentrations ranging from 400 fg mL−1 to 40 ng mL−1 and current values were recorded using the DPV technique. It was found that the current values increased with the increasing concentration of the IL-8 antigen revealing the successful binding of the antigen of IL-8 to the antibody of IL-8. This study shows that the developed immunosensor is suitable for real sample sensing applications.
Lung cancer is screened for at an early stage of the disease using a variety of techniques, including a chest radiograph, X-ray, CT scan, tissue biopsy, and sputum cytology. However, due to several restrictions, these techniques are not applicable on all patients. Other therapeutic approaches have also been employed, including chemotherapy, radiation therapy, and surgical procedures.155,156 The present diagnostic techniques do, however, have several limitations, such as cost, long-term therapy, pricey instruments, and limited sensitivity.
Cytokeratin fragment antigen 21-1 (CYFRA21-1) is a sensitive diagnostic marker for detecting non-small cell lung cancer (NSCLC). CYFRA21-1 is recognized as a protein marker for sensitive and specific diagnosis of NSCLC since lung SCC patients express it more than cancer patients with other types of disease.157,158 Enzyme-linked immunosorbent assay (ELISA), fluorometric immunoassay,159,160 and inductively coupled plasma source mass spectrometer (ICP-MS) detection are only a few of the methodologies currently being used for the detection and profiling of CYFRA21-1.161,162 These techniques can be utilized to detect CYFRA21-1; however they still have limitations, such as low sensitivity, high analytical costs, or high equipment costs.163 Due to its simplicity, sensitivity, quick reaction, and low cost, the electrochemical biosensor has emerged as a new research hotspot in the detection of protein markers.160 Over the years, several types of high-performing materials have been created to build more sensitive electrochemical sensors, including graphene, molybdenum disulfide (MoS2), and others. AuNPs@MoS2@Ti3C2Tx composite materials labelled with secondary antibody (Ab2) and toluidine blue (TB) as the signal probe were integrated with Nafion-AuNPs as the electrode material to create a sensor for the first time that can detect CYFRA21-1 quantitatively. Nafion-AuNPs may offer a high conductivity interface, and its large surface area can offer a variety of places for the loading of Ab1. In the meanwhile, the Ab2-TB-AuNPs@MoS2@Ti3C2Tx composite materials were employed as signal amplification labels; they have the all-inclusive benefits of Ti3C2Tx's large specific surface area, MoS2's strong catalytic performance, and AuNPs’ biocompatibility. The sensor therefore demonstrated excellent electrochemical performance with respect to CYFRA21-1 in real patient serum samples, a strong electrochemical signal, a low detection limit (LOD) of 0.03 pg mL−1 (S/N = 3), good selectivity, and sensitivity, indicating that it has a potential application prospect in the early diagnosis of NSCLC.164
Along with this approach for detecting CYFRA21-1, an electrochemical immunosensor was fabricated based on a silicon nitride (Si3N4)–molybdenum disulfide (MoS2) composite on multi-walled carbon nanotubes (Si3N4/MoS2–MWCNTs) as an electrochemical sensor platform. First, the capture antibody (Ab1) was immobilized on Si3N4/MoS2-MWCNT modified on a glassy carbon electrode (Si3N4/MoS2-MWCNTs/GCE) through stable electrostatic/ionic contacts between the –NH2 groups of the capture antibody and the polar groups of Si3N4/MoS2. A unique voltammetric CYFRA21-1 immunosensor was then fabricated through specific antibody–antigen interactions between the electrochemical sensor platform and the signal amplifier. For analytical applications, a linearity range of 0.01–1.0 pg mL−1 and a low detection limit (LOD) of 2.00 fg mL−1 were also attained.165 Moreover, a validation study of the fabricated immunosensor for CYFRA21-1 detection was performed in four different plasma samples by the voltammetric technique and the percentage recoveries were found to be within the acceptable range (from 99.34 to 99.43%).
Pachauri et al. also constructed an electrochemical immunosensor for CYFRA21-1 detection using cubic CeO2 implanted rGO [Fig. 5].166 A highly sensitive and selective non-invasive biosensor for oral cancer detection was fabricated using the electrochemical technique. In situ reduction of GO to rGO in the presence of CeO2 materials was performed using hydrazine hydrate as the reducing agent. The synthesized nanocomposite of CeO2-rGO was decorated onto an ITO substrate and further anti-CYFRA21-1 antibodies were immobilized using EDC:NHS coupling chemistry. This fabricated immunosensor was used to detect CYFRA21-1 in the concentration range from 0.625 pg mL−1 to 15 ng mL−1 with a LOD of 0.625 pg mL−1 and the sensitivity of the sensor was 14.5 μA ng−1 mL cm−2 and the sensor was highly selective towards the CYFRA21-1 antigen. To check the practical utility of the developed sensor, the authors conducted a spiked human saliva sample study using the DPV technique.% of relative standard deviation was found in the range of 0.18 to 2.10% which was within the acceptable range.
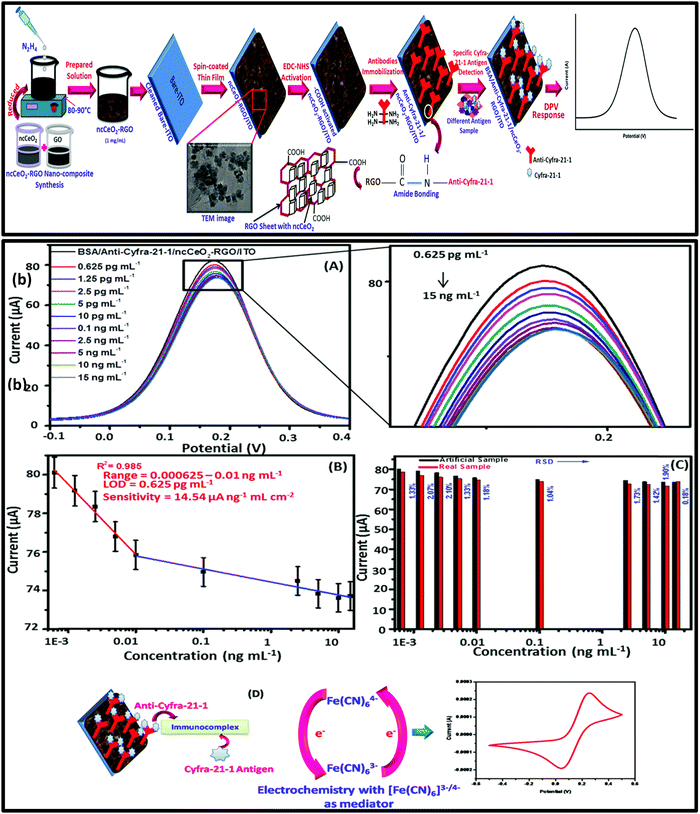 | ||
| Fig. 5 (a) Fabrication of an electrochemical sensor for CYFRA21-1 detection, (b) the electrochemical sensing results using the immunosensor for CYFRA21-1 detection166 | ||
Similarly, other biomarkers of lung cancer are also detected such as enolase (ENO). In addition, an elevated level of ENO2 can be used as an indicative, useful tumor marker for small cell lung cancer (SCLC). Recent research has additionally dealt with the potential for linking ENO1, a cancer marker that is highly expressed, with non-small cell lung cancer (NSCLC).167,168 Additionally, SCLC tissue frequently contains overexpressed ENO1, and the ratio of ENO2 to ENO1 [ENO1 + ENO2] has been thought to be a useful indicator for SCLC identification.169 In recent years, it has been shown that the amount of ENO1 strongly influences the migration of cancer cells.170 Consequently, it is crucial to measure ENO1 in biological samples, which has turned into a significant difficulty. Although several methods for the detection of ENO1 have been described based on two-dimensional differential in-gel electrophoresis, mass spectrometry and immunobioluminescence assays, these methods frequently require the tedious and time-consuming pretreatment of complex samples.169–172 In order to detect ENO1 at traces of pg mL−1, the author has devised a simple yet creative electrochemical immunosensor that utilizes gold nanoparticle (AuNP) congregates as bioprobes. The altering properties of AuNPs make them flexible materials for the creation of electrochemical sensors when they are dropcast on disposable screen-printed carbon electrodes (SPCE). Their sensors are fabricated by physically adsorbing polyethylene glycol (PEG) onto SPCE. In order to construct 151 nm AuNP congregates sensitized with secondary anti-ENO1 antibody molecules, polyclonal secondary anti-ENO1 and 33.0 nm diameter AuNPs were combined simultaneously to form the signal amplification probes. Following further incubation with the sample ENO1, the secondary anti-ENO1-tagged AuNP congregate bioprobes produced immunocomplexes on the primary monoclonal anti-ENO1-modified SPCE surface. A later research study employing square wave voltammetry revealed that this electrochemical immunosensor was capable of detecting ENO1 levels in the linear dynamic range from 10−12 to 10−8 g mL−1 with a low detection limit of 11.9 fg mL−1.123
Choudhary et.al. found CD59 is a biomarker with therapeutic value for lung cancer and fabricated an immunosensor. To fabricate the functional immunosensor assembly, anti-CD59 antibodies were immobilized on a pencil-shaped graphite electrode covered with graphene oxide nanoparticles (GrONPs). In order to get the greatest degree of activity, the experimental parameters were changed for pH, incubation time, temperature, and antigen concentration. When the current was measured, CD59 could be found, and this revealed a range of 1 fg mL−1 to 10 ng mL−1. The sensor's minimal limit of detection (LOD) was 1 fg mL−1.173 The presence of CD59 was also validated in human urine samples of lung cancer patients and the results were compared with the healthy adult's samples. It was found that the levels of the CD59 protein range from 3 ± 0.02 to 9 ± 0.04 ng mL−1 (n = 7) in apparently healthy persons whereas the levels of CD59 were found to be 0.001 ± 0.02 to 0.35 ± 0.01 ng mL−1 (n = 7) in lung cancer patients. This developed sensor was highly specific, selective, accurate and ultrasensitive towards lung cancer biomarker detection.
Prominent researchers such as Mosbach, Wolf, Shea, Takeuchi, and others who have made significant contributions in this field have continually refined the process of molecular imprinting over time. Their advancements provide potential solutions to challenges associated with natural biorecognition elements in sensors. Introduced first in 1972, the molecular imprinting technique has since witnessed substantial growth.174 Here the key to its efficacy is the intricate chemical interactions that take place during the polymerization phase during which the shape and size of the target molecule get imprinted in the polymer matrix. The interactions within the pre-polymerized mixture can follow either of the molecular imprinting techniques, namely, “covalent imprinting” and “non-covalent imprinting”, based on these interactions.
In the covalent imprinting approach, the template molecule and functional monomers initially form covalent bonds. Subsequently, to remove the template, these bonds are chemically cleaved. The covalent imprinting tends to offer superior host selectivity because the monomer forms a robust bond during polymerization, ensuring precise complementarity. However, a drawback is that the template removal can occasionally be problematic due to the strength of these covalent interactions. In contrast, the non-covalent imprinting method involves the interaction between the template and the functional monomer through non-covalent interactions. Fig. 6 presents the synthesis of an MIP for the targeted analyte. These interactions are readily reversible, allowing for the template's removal simply by washing the polymer in an appropriate solvent. In addition, non-covalent imprinting has the advantage of simplicity. This flexibility means imprinting can be executed across various systems without significant constraints on material selection. The formation of an MIP can be validated using different techniques such as atomic force microscopy (AFM), Fourier transform infrared spectroscopy (FTIR), scanning electron microscopy (SEM), electrochemical techniques, etc.175,176 Some of the relevant studies for the detection of cancer biomarkers are highlighted in further sections.
Moussa et al. reported an rGO-AgNPs–Au electrode-MIP based electrochemical sensor for the detection of lactic acid, an important biomarker for cancer cell determination. The confirmation of rGO-AgNPs was performed using X-ray diffraction, SEM, and FTIR. Here [Fe(CN)6]3−/4− was used as an active redox couple for the cyclic voltammetry study. The fabricated sensor showed a low limit of detection of 0.726 μM. This lactic acid sensor demonstrated superior consistency, selectivity, and durability. Its efficacy remained consistent in varied pH environments. Furthermore, its applicability in real-world scenarios was confirmed through tests on human serum samples, emphasizing its potential for cancer detection and monitoring.177 Also, Oliveira et.al. in 2023 reported a MIP based sensor for the detection of the CA15-3 antigen on a paper based substrate using o-phenylenediamine (oPD) as the polymer matrix to synthesis an MIP. The electrodes were printed using carbon-based ink consisting of silver nanoparticles and were further modified using gold nanoparticles. Here, the MIP was synthesized using the electro-polymerization technique and was further validated using AFM and FTIR. Electro-polymerization was conducted at 50 mV s−1 from −0.2 V to 1.0 V for 20 cycles in the presence of the analyte and the polymer. Oxalic acid was used as the elution solution to remove the molecules from the polymer matrix and left behind CA15-3 specific cavities in the poly-oPD matrix. The fabricated sensor was further validated by using it for the detection of CA15-3 in serum samples, and it gave satisfactory results.178
Moreover, a state-of-the-art electrochemical sensor using Fe3O4@graphene oxide (GO)-MIP was fabricated for the quantification of interleukin-8 (IL-8) in saliva samples. It is crucial to highlight that IL-8, having a molecular weight of 8.5 kDa, plays a pivotal role in oral cancer diagnostics. The imprinting surface used here was GO and the implication of super paramagnetic Fe3O4 nanoparticles helped in the assembly of Fe3O4@(GO)-MIP on a gold film electrode using magnetic forces. The core–shell architecture of these nanoparticles was deciphered through the combined use of SEM and energy dispersive X-ray spectroscopy. Also, cyclic voltammetry was employed to investigate and fine-tune the electrochemical properties of the Fe3O4@(GO)-MIP composite. The outcomes revealed that the sensor could selectively bind with IL-8, leading to a measurable decline in the resultant electrical current, a phenomenon that became more pronounced with increasing IL-8 concentrations. The sensor efficacy was marked by its exceptional specificity, a noteworthy detection limit of 0.04 pM, and an extensive linear detection window ranging from 0.1 to 10 pM for IL-8. Moreover, it demonstrated proficiency in quantifying IL-8 levels within saliva samples. Given these attributes, the Fe3O4@(GO)-MIP nanoparticle-infused sensor is positioned as a front-runner for the rapid, precise, efficient, and economically viable evaluation of IL-8 and related oral cancer markers.179
5. Recent advancement in cancer biomarker detection: miniaturization of the electrochemical diagnostic device and multiplex sensing platform
Over the past few years, researchers have been focused on the development of miniaturized point-of-care analytical devices to resolve the diagnosis problem and provide the easiest, easily accessible, portable, ultra-precise and onset detection. The microfluidic based analytical technique is a highly advanced detecting tool which requires minimum sample volume and provides a compact and portable platform for on-site detection. Microfluidic devices are micro total analysis systems having 10 to 200 μm microchannels that allow a small amount of fluid to flow in a laminar motion.180 Microfluidic device-based sensors for cancer biomarker detection have undergone a substantial improvement in the past few years and they can easily detect multiple analytes in a single chip unit system.The microfluidic system integrated electrochemical detection method is increasingly utilized for the development of sensors and designing of paper based analytical devices. Low fabrication cost, less power consumption and ability to miniaturize the device and portability are the key advantages of paper based electrochemical sensors. Also, paper substrates used in the construction of electrochemical sensors eliminate the disadvantages of non-portable devices. The high availability with a variety of paper as well as excellent mechanical properties such as lightness, specific stiffness, ease of use and stability of the paper, biodegradability, and easy disposal are the uniqueness of paper substrates utilized for the development of electrochemical sensors. Some of the intrinsic properties of fibrous or porous cellulose fiber paper based substrates are high absorption ability for effective storage of aptamers, enzymes, and antibodies, increased sensitivity, faster response time and accuracy of time-dependent measurement, elimination of the air bubble problem, large specific area to improve biomolecule immobilization, reduced need for a pump for transportation of fluid due to their capillary action, better electrochemical detecting ability by the use of conductive ink that can easily be deposited on paper and detection using a low volume of samples.
A microfluidic multiplex immunoarray sensor was developed by Tang et al. using an electrochemical analytical device for the multiplex cancer biomarker protein detection such as prostate specific antigen (PSA), prostate specific membrane antigen, interleukin-6, and platelet factor-4 in serum using eight 32-sensor immunoarrays which were connected to an 8-port manifold that allowed 256 measurements in <1 h.181 Clinically, the relevant detection limit was in the range of 0.05 to 2 pg mL−1 and the 5-decade dynamic range was from sub pg mL−1 to above ng mL−1. Also, validation of the sensor was conducted by using seven serum samples from prostate cancer patients and one sample from a cancer-free patient and the relative standard deviation in the serum sample study was found to be ±3% to ±10%.
Another biosensor for multiple cancer biomarker detection was developed by Wang et al. in 2019. A label-free microfluidic paper based electrochemical aptasensor was constructed for multiplex detection of cancer biomarkers.182 A highly sensitive and selective, and low-cost disposable simultaneous detecting platform was developed using the paper based microfluidic platform for CEA and neuron-specific enolase (NSE) cancer biomarkers in clinical samples. For the construction of the aptasensor, wax printing and screen-printing techniques were used, which enabled the functions of sample filtration and sample auto-injection [Fig. 7]. Surface functionality of the sensor surface for aptamer immobilization was improved by the use of amino functional graphene thionine-gold nanoparticles and Prussian blue-poly(3,4-ethylenedioxythiophene) gold nanocomposite. This nanocomposite was used as a signal transduction element to improve stability as well as intensity of redox signals. The electrochemical sensing results showed that the constructed multiplex aptasensor worked in the linear range from 0.01 to 500 ng mL−1 with a LOD of 2 pg mL−1 for the CEA cancer biomarker whereas the sensor showed ability to detect NSE cancer biomarkers linearly in the detection range of 0.05 to 500 ng mL−1 with a LOD of 10 pg mL−1. In addition to the good linear detection range, the developed aptasensor also worked for the clinical serum samples and the percentage of relative error was found to be 7.81% for CEA and 22.43% for NSE, indicating good accuracy and high sensitivity for the simultaneous detection of CEA and NSE in clinical serum samples.
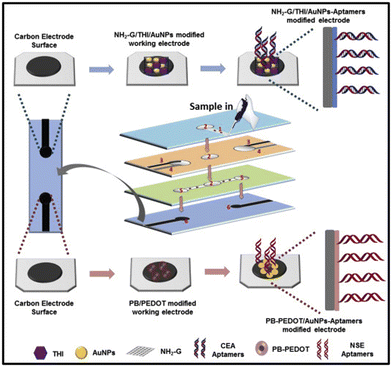 | ||
| Fig. 7 Schematic illustration of the fabrication of an electrochemical paper based aptasensor for multiplex cancer biomarker detection.182 | ||
Recently, Wang et al. in 2021 developed a label-free electrochemical impedimetric aptasensor for lung cancer detection based on the CEA cancer biomarker using a Au NP decorated graphene material.183 For the working electrode surface modification, the graphene electrode was used as the base electrode and gold nanoparticles were decorated using the electrodeposition cyclic voltammetry technique. An optimized concentration of the aptamer was immobilized over the nanoparticle decorated graphene electrode and electrochemical sensing response was evaluated using the EIS technique. A change in the impedance signal was observed upon binding of CEA with the carcinoembryonic antigen aptamer. This developed sensor worked in the range of 0.2 to 15.0 ng mL−1 with a LOD of 0.085 ng mL−1. The developed sensor was highly sensitive and selective but the authors did not perform the real sample study for validation of the sensor.
Yang et al. in 2021 worked on the electrochemical based simultaneous detection of cancer biomarkers CEA and CYFRA-21 using gold doped poly-thionine and poly-m-cresol156 since the use of a single tumor biomarker is not sufficient in terms of sensitivity and specificity for accurate detection of markers in the body fluid. Single antigen detection is prone to false negative or false positive results. Therefore, simultaneous detection of biomarkers instead of a single biomarker is precise to diagnose the accurate level in body fluids. This multiple detection of an analyte is a highly encouraging analytical method for more than one analyte detection of biomarkers. In this work, gold nanoparticles are decorated over the three-dimensional graphene material, poly-thionine, and poly-m-cresol which provide a large number of binding sites that helps to amplify the electrochemical sensing signal for better sensitivity performance. This multianalyte sensor worked in the range of 0.5 to 200 ng mL−1 with a limit of detection of 0.18 ng mL−1 and 0.31 ng mL−1 for CYFRA-21 and CEA antigen, respectively. Also, the authors conducted the real spiked serum sample study with different concentrations of CYFRA-21 and CEA. The average recoveries were found to be 92.1–108.4% for CYFRA21-1 and 94.3–107.0% for CEA with RSD less than 4.0% (n = 3). Therefore, the developed sensor showed great potential towards the simultaneous detection of both cancer biomarkers CYFRA-21 and CEA.
Recently, MIP based sensors have gained more attention in the area of biomedical point-of-care device development and diagnosis owing to their low cost, specificity, sensitivity and stability to detect various chemical and biological responses. It has been reported that MIP based sensors are very good alternatives of aptamer- or antibody-based sensors, due to the generation of artificial recognition sites as site-specific cavities for the non-covalent adsorption of the target analyte. Many researchers have worked on the development of electrochemical sensors using MIPs for single analyte detection, but recently, the approach has been focused on the development of sensors for multianalyte detection. Therefore, developing dual template-based MIP sensors for detection of more than one target analyte offers the advantages of low cost, less analysis time, less time needed for the results and less laborious. For the preparation of a MIP, a conductive polymer film is used for the generation of a stable, conductive, sensitive sensor. Polymers such as polypyrrole, polydopamine, poly-methylacrylate, etc. are used for imprinting and template preparation.
Taheri et al. developed imprinting of dual molecules into a single polymer as a dual template based MIP for the two biomarker detection of CEA and alpha-fetoprotein (AFP) as lung cancer biomarkers.184 Artificial recognition sites were created in the electropolymerized polypyrrole on the fluorine doped tin oxide electrode surface. Various characterization techniques were used for the analysis of formation and characterization of the sensing layer such as Raman spectroscopy and scanning electron microscopy, and for electrochemical measurements cyclic voltammetry and electrochemical impedance spectroscopy were used. To improve the conductivity of the synthesized polymer of polypyrrole, methyl orange was used as a doping agent and rectangular shaped nanotubes were synthesized. The impedimetric technique was used for the evaluation of the sensing performance of AFP and CEA target analytes. The linear dynamic range was found to vary from 5 to 104 pg mL−1 with a LOD of 1.6 pg mL−1 for the CEA biomarker. Also, the linear detection range was observed for the AFP biomarker, ranging from 10 to 104 pg mL−1 with a LOD of 3.3 pg mL−1. The dual template-based MIP sensor for AFP and CEA detection was also validated in real serum samples with a satisfactory result. The high sensitivity, selectivity and stability of the dual template-based MIP sensor proved to be promising for AFP and CEA detection in serum samples.
Liu et al. in 2022 demonstrated the development of an electrochemical immunosensor for simultaneous detection of dual lung cancer biomarkers using an electro-active probe system.185 A multiplex electrochemical immunosensor was constructed for CEA and CA-125 detection using electroactive probes of toluidine blue (TB) and Prussian blue (PB) indicators. Copper-tetrakis(4-carboxyphenyl)porphyrin (Cu-TCPP) with a laminar structure was synthesized using a hydrothermal process and was attached to TB and PB indicators to improve electrochemical activity and biocompatibility. The Cu-TCPP-TB complex was labeled with the antibody of CEA and the Cu-TCPP-PB complex was labeled with the antibody of CA-125 for the multiplex sensor. This sensor worked for both analytes of CEA and CA-125. By changing the CEA concentration a linear change in the electrochemical signal was observed in the range of 0.1 to 160 ng mL−1 with a LOD of 0.03 ng mL−1, whereas for CA-125, a linear response in the signal was observed from 0.5 to 200 U mL−1 with a LOD of 0.05 U mL−1. Furthermore, the fabricated multiplex immunosensor was tested in real human blood serum samples and the obtained results were comparable with those of the ELISA kit. Hence, the constructed electrochemical immunosensor showed simultaneous detection of two lung cancer markers CEA and CA-125 on the same substrate matrix.
Jiang et al. in 2023 fabricated an ultrasensitive sandwich type electrochemical aptasensor for dual lung cancer biomarker detection of NSE and epidermal growth factor receptor (EGFR) using the black phosphorus–gold nanocomposite (BP–AuNC).186 BP–AuNC was synthesized using an in situ reduction process without using other reducing agents and the synthesized nanocomposite was decorated onto the working area of an electrode of a cloth substrate matrix and a three electrode system was fabricated by screen printing. This BP–AuNC provides good electrical conductivity and a large number of binding sites to capture the aptamer. Differential pulse voltammetry response was obtained for detection of NSE and EGFR lung cancer biomarkers. Upon changing the concentration of NSE and EGFR analytes, a change in the DPV signal was observed in the linear range of 1 to 400 ng mL−1 for NSE and 1 to 600 ng mL−1 for the EGFR analyte. The limit of detection for the NSE analyte was found to be 0.1 ng mL−1 and for EGFR the LOD was 0.6 ng mL−1. This constructed multiplex aptasensor displayed good selectivity, stability, accuracy and low-cost detection for lung cancer biomarkers but the authors did not perform the real sample study of the developed sensor for further validation.
6. Conclusions and future perspectives
In summary, this review article discusses literature search on cancer disease which is a leading cause of death worldwide. Cancer is estimated to cause nearly 10 million deaths worldwide due to cancer-causing agents including physical (ultraviolet rays or ionizing radiation), chemical (tobacco, smoke, food or drinking water contaminants) or biological carcinogens (infections, bacteria, viruses, parasites). Moreover, information about the cancer cases of oral or lung cancer is discussed thoroughly. Biology of cancer development and definition of the problem related to cancer, both hallmarks and significant challenges in oncology, and six characteristic features commonly exhibited by cancer cells are described in detail. Also, a brief discussion of conventional techniques such as histopathology, bioimaging, and ELISA for cancer detection and their advantages as well as disadvantages is provided. Further advancements in sensing techniques used for cancer detection that are based on significant cancer biomarkers related to oral or lung cancer using nanomaterials are also summarized.A detailed overview of the electrochemical sensors mainly focused on nucleic acids, aptamers, immunosensors, enzyme-based sensors or MIP based sensors using nanomaterials for cancer biomarker detection. Various types of nanomaterials like graphene-based materials, metal oxides, polymers, conducting nanoparticles, etc. are used to improve the electrochemical or optical properties by enhancing the surface to volume ratio, biocompatibility, electrical conductivity, electron transfer process, and conjugating ability. The integration of nanomaterials into the construction of sensors enhances sensitivity, selectivity, specificity, reproducibility, stability as well as detection range or limit of detection of the device. Therefore, nanomaterials owing to their potential physiochemical properties can provide effective ways to design ultrasensitive biosensors. Therefore, recent research articles mainly on lung and oral cancer detection using the electrochemical sensor have been summarized. Electrochemical sensors which provide a low-cost and convenient solution for variable analyte detection are widely used in biomedical applications. The main key advantages of electrochemical sensors are the ability to detect the analyte over a wide range of concentrations, better sensitivity, good stability, reproducibility, low detection limit, affordability and robustness which have led to their high demand compared to other detecting technologies. Apart from the detection of a single cancer biomarker on the sensing surface, recent research has focused on multiple cancer biomarker detection. The detection of two or more cancer biomarkers on a single sensing platform has various advantages compared to the single detection such as low cost, less analysis time requirement, less time needed for the results and less laborious, less sample volume requirement, less average test time, more informative results and more statically reliable analysis results. Sometimes, it becomes difficult to identify cancer by one cancer biomarker; therefore, multiple biomarker detection gives more clear confirmation about the development of cancer in a particular site, i.e., primary cancer as well as metastasis stage of cancer. Simultaneous detection of biomarkers instead of a single biomarker is precise to diagnose the accurate level in the human body. Additionally, a critical overview of integration of the microfluidic-paper based electrochemical sensor is also discussed for cancer biomarker detection. Low fabrication cost, less power consumption and ability to miniaturize the device and portability are the key advantages of the paper based electrochemical sensor. Also, a paper-based substrate is useful in the construction of an electrochemical sensor that is immensely required for portable devices. Although most electrochemical biosensors perform well under ambient working conditions, changes in the temperature conditions particularly in the case of antigen–antibody based biosensors result in drastic changes in the sensing performance. Also, shorter or limited shelf-life, i.e. from 6 months to one year, and sensitivity to humidity conditions are certain limitations of electrochemical sensors. During the past few years, researchers have particularly focused on overcoming these pre-existing limitations for the development of electrochemical sensors using the advanced nanomaterials for particular sensor surface modifications.
In the near future, electrochemical portable designing and construction of sensors for cancer detection will be commercially available for clinical samples’ monitoring with early diagnosis of level of biomarkers in the body fluids. Simple, sensitive, and selective detection by electrochemical sensors is an effective alternative approach for early detection of level of cancer biomarkers in real cancer patient samples than the existing conventional techniques. In the far future of nano biosensor integration with artificial intelligence, machine learning could achieve impressive advances in the construction and fabrication of biosensors and impact the commercialization of devices.
Availability of data and material
Available data and material in this work are properly cited.Author contributions
HB: conceptualization, investigation, writing original draft, review, editing. Archana: conceptualization, writing original draft, review, editing. AN: writing original draft, editing, review. JKH: writing original draft, editing, review. SC: writing original draft, editing, review. PRS: conceptualization, editing, review.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
HB is thankful to DBT for the Research Associate fellowship [DBT-RA/2022/July/N/2383]. PRS is thankful to ICMR for the project [34/13/2019-TF/Nano/BMS]. Archana is thankful to DBT for the JRF fellowship [DBTHRDPMU/JRF/BET-21/I/2021-22/364]. AN is thankful to DST for the project. JKH is thankful to UGC for the non-NET fellowship.References
- S. D. Hohl, K. A. Shoenbill and K. L. Taylor, et al., The Impact of the COVID-19 Pandemic on Tobacco Treatment Program Implementation at National Cancer Institute-Designated Cancer Centers, Nicotine Tob. Res., 2023, 25, 345–349 CrossRef PubMed.
- N. Y. Spencer and R. C. Stanton, The Warburg effect, lactate, and nearly a century of trying to cure cancer, Seminars in nephrology, Elsevier, 2019, pp. 380–393 Search PubMed.
- J. Boutry, S. Tissot and B. Ujvari, et al., The evolution and ecology of benign tumors. Biochim Biophys Acta (BBA)-Reviews, Cancer, 2022, 1877, 188643 CAS.
- R. A. Weinberg, How cancer arises, Sci. Am., 1996, 275, 62–70 CrossRef CAS PubMed.
- K. P. Gunasekaran, Leveraging object detection for the identification of lung cancer, arXiv, arXiv230515813, 2023, Prepr.
- P.-T. Chen, T. Wu and P. Wang, et al., Pancreatic cancer detection on CT scans with deep learning: a nationwide population-based study, Radiology, 2023, 306, 172–182 CrossRef PubMed.
- M. B. Lawson, S. C. Partridge and D. S. Hippe, et al., Comparative Performance of Contrast-enhanced Mammography, Abbreviated Breast MRI, and Standard Breast MRI for Breast Cancer Screening, Radiology, 2023, 308, e230576 CrossRef PubMed.
- J. M. Rafalko, K. M. Kruglyak and A. L. McCleary-Wheeler, et al., Age at cancer diagnosis by breed, weight, sex, and cancer type in a cohort of more than 3,000 dogs: Determining the optimal age to initiate cancer screening in canine patients, PLoS One, 2023, 18, e0280795 CrossRef CAS PubMed.
- S. Connal, J. M. Cameron and A. Sala, et al., Liquid biopsies: the future of cancer early detection, J. Transl. Med., 2023, 21, 118 CrossRef PubMed.
- F. Ye, S. Dewanjee and Y. Li, et al., Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer, Mol. Cancer, 2023, 22, 1–40 CrossRef PubMed.
- M. D. Melisa Hunis and M. D. Adrian Hunis, MAR Gynecol. Urol., 2023, 5, 4 Search PubMed.
- T. Muka, J. J. X. Li, S. J. Farahani and J. P. A. Ioannidis, An umbrella review of systematic reviews on the impact of the COVID-19 pandemic on cancer prevention and management, and patient needs, eLife, 2023, 12 DOI:10.7554/eLife.85679.
- L. A. Hall, S. C. McKay and J. Halle-Smith, et al., The impact of the COVID-19 pandemic upon pancreatic cancer treatment (CONTACT Study): a UK national observational cohort study, Br. J. Cancer, 2023, 128, 1922–1932, DOI:10.1038/s41416-023-02220-2.
- T. Li, B. Nickel and P. Ngo, et al., A systematic review of the impact of the COVID-19 pandemic on breast cancer screening and diagnosis, Breast, 2023, 67, 77–88, DOI:10.1016/j.breast.2023.01.001.
- I. S. Jabbal, S. Sabbagh and B. Dominguez, et al., Impact of COVID-19 on cancer-related care in the United States: An overview, Curr. Oncol., 2023, 30, 681–687 CrossRef PubMed.
- Y.-L. Zhang, W.-S. Feng and X.-K. Zheng, et al., Three new ursane-type triterpenes from the leaves of Rehmannia glutinosa, Fitoterapia, 2013, 89, 15–19, DOI:10.1016/j.fitote.2013.05.013.
- P. Wang, X. Hu and Y. Li, et al., Automatic cell nuclei segmentation and classification of breast cancer histopathology images, Signal Process., 2016, 122, 1–13, DOI:10.1016/j.sigpro.2015.11.011.
- M. de Bruijne, Machine learning approaches in medical image analysis: From detection to diagnosis, Med. Image Anal., 2016, 33, 94–97, DOI:10.1016/j.media.2016.06.032.
- X. Huang, P. H. G. Duijf and S. Sriram, et al., Circulating tumour DNA alterations: emerging biomarker in head and neck squamous cell carcinoma, J. Biomed. Sci., 2023, 30, 1–19 CrossRef PubMed.
- J. M. García-Martín, P. Varela-Centelles and M. González, et al., Epidemiology of oral cancer, Oral cancer detection: novel strategies and clinical impact, Springer, Cham, 2019, pp. 81–93 DOI:10.1007/978-3-319-61255-3_3.
- K. S. Ravi, N. B. Pushpa and S. Kishore, et al., Taxation of micronuclei frequency as a prognostic marker in oral and oropharyngeal carcinoma: A cytogenetic study, Natl. J. Clin. Anat., 2021, 10, 57–60 CrossRef.
- R. Sankaranarayanan, K. Ramadas and H. Amarasinghe, et al., Oral cancer: prevention, early detection, and treatment, Cancer Dis Control priorities, 3rd edn, vol. 3, 2015 Search PubMed.
- E. E. W. Cohen, J. Baru and D. Huo, et al., Efficacy and safety of treating T4 oral cavity tumors with primary chemoradiotherapy, Head Neck J. Sci. Spec., 2009, 31, 1013–1021 CrossRef PubMed.
- J. Balakittnen, C. E. Weeramange and D. F. Wallace, et al., Noncoding RNAs in oral cancer, Wiley Interdiscip. Rev.: RNA, 2023, 14, e1754 CrossRef CAS PubMed.
- Q. Huang, H. Ding and N. Razmjooy, Optimal deep learning neural network using ISSA for diagnosing the oral cancer, Biomed. Signal Process. Control, 2023, 84, 104749 CrossRef.
- N. Al-Rawi, A. Sultan and B. Rajai, et al., The effectiveness of artificial intelligence in detection of oral cancer, Int. Dent. J., 2022, 72, 436–447 CrossRef PubMed.
- A. Leiter, R. R. Veluswamy and J. P. Wisnivesky, The global burden of lung cancer: current status and future trends, Nat. Rev. Clin. Oncol., 2023, 1–16 Search PubMed.
- A. Lahiri, A. Maji and P. D. Potdar, et al., Lung cancer immunotherapy: progress, pitfalls, and promises, Mol. Cancer, 2023, 22, 1–37 CrossRef PubMed.
- B. Wang, Y. Han and Y. Zhang, et al., Overcoming acquired resistance to cancer immune checkpoint therapy: potential strategies based on molecular mechanisms, Cell Biosci., 2023, 13, 1–23 CrossRef PubMed.
- A. Nath, K. Sathishkumar and P. Das, et al., A clinicoepidemiological profile of lung cancers in India–Results from the National Cancer Registry Programme, Indian J. Med. Res., 2022, 155, 264 CrossRef PubMed.
- J. T. Yan, Y. Jin and E. Lo, et al., Real-World Biomarker Test Utilization and Subsequent Treatment in Patients with Early-Stage Non-small Cell Lung Cancer in the United States, 2011–2021, Oncol. Ther., 2023, 1–18 Search PubMed.
- K. Sathishkumar, M. Chaturvedi and P. Das, et al., Cancer incidence estimates for 2022 & projection for 2025: Result from national cancer Registry Programme, India, Indian J. Med. Res., 2022, 156, 598 Search PubMed.
- C. L. Chaffer and R. A. Weinberg, A Perspective on Cancer Cell Metastasis, Science, 2011, 331, 1559–1564, DOI:10.1126/science.1203543.
- A. W. Lambert, D. R. Pattabiraman and R. A. Weinberg, Emerging Biological Principles of Metastasis, Cell, 2017, 168, 670–691, DOI:10.1016/j.cell.2016.11.037.
- X. Zheng, J. L. Carstens and J. Kim, et al., Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer, Nature, 2015, 527, 525–530, DOI:10.1038/nature16064.
- G. Evan and T. Littlewood, A matter of life and cell death, Science, 1998, 281, 1317–1322 CrossRef CAS PubMed.
- D. Hanahan and R. A. Weinberg, Hallmarks of cancer: the next generation, Cell, 2011, 144, 646–674 CrossRef CAS PubMed.
- P. Vineis and C. P. Wild, Global cancer patterns: causes and prevention, Lancet, 2014, 383, 549–557, DOI:10.1016/S0140-6736(13)62224-2.
- M. Iranifam, Analytical applications of chemiluminescence methods for cancer detection and therapy, TrAC, Trends Anal. Chem., 2014, 59, 156–183, DOI:10.1016/j.trac.2014.03.010.
- V. A. Katzke, R. Kaaks and T. Kühn, Lifestyle and Cancer Risk, Cancer J., 2015, 21, 104–110, DOI:10.1097/PPO.0000000000000101.
- K. Gilham, A. Gadermann, T. Dummer and R. A. Murphy, Mental health, cancer risk, and the mediating role of lifestyle factors in the CARTaGENE cohort study, PLoS One, 2023, 18, e0281588 CrossRef CAS PubMed.
- M. Narwaria, Does explainable machine learning uncover the black box in vision applications?, Image Vis. Comput., 2022, 118, 104353, DOI:10.1016/j.imavis.2021.104353.
- N. Sharma, R. Sharma and N. Jindal, Machine Learning and Deep Learning Applications-A Vision, Glob. Transitions Proc., 2021, 2, 24–28, DOI:10.1016/j.gltp.2021.01.004.
- M. L. Smith, L. N. Smith and M. F. Hansen, The quiet revolution in machine vision - a state-of-the-art survey paper, including historical review, perspectives, and future directions, Comput. Ind., 2021, 130, 103472, DOI:10.1016/j.compind.2021.103472.
- D. Komura and S. Ishikawa, Machine learning methods for histopathological image analysis, Comput. Struct. Biotechnol. J., 2018, 16, 34–42 CrossRef CAS PubMed.
- A. Korzynska, L. Roszkowiak, J. Zak and K. Siemion, A review of current systems for annotation of cell and tissue images in digital pathology, Probl. Biocybern. Biomed. Eng., 2021, 41, 1436–1453, DOI:10.1016/j.bbe.2021.04.012.
- O. Kott, D. Linsley and A. Amin, et al., Development of a Deep Learning Algorithm for the Histopathologic Diagnosis and Gleason Grading of Prostate Cancer Biopsies: A Pilot Study, Eur. Urol. Focus, 2021, 7, 347–351, DOI:10.1016/j.euf.2019.11.003.
- Y. Zeng and J. Zhang, A machine learning model for detecting invasive ductal carcinoma with Google Cloud AutoML Vision, Comput. Biol. Med., 2020, 122, 103861, DOI:10.1016/j.compbiomed.2020.103861.
- D. G. Vinsard, Y. Mori and M. Misawa, et al., Quality assurance of computer-aided detection and diagnosis in colonoscopy, Gastrointest. Endosc., 2019, 90, 55–63, DOI:10.1016/j.gie.2019.03.019.
- S. Shaikh, F. U. Rehman and T. Du, et al., Real-Time Multimodal Bioimaging of Cancer Cells and Exosomes through Biosynthesized Iridium and Iron Nanoclusters, ACS Appl. Mater. Interfaces, 2018, 10, 26056–26063, DOI:10.1021/acsami.8b08975.
- M. S. Ghamsari, in Introductory Chapter: Nano-bioimaging—Past, Present, and Future, ed. M. S. Ghamsari, IntechOpen, Rijeka, ch. 1, 2018 Search PubMed.
- Biomedical Signals, Imaging, and Informatics, ed. J. D. Bronzino and D. R. Peterson, 1st edn, 2014 Search PubMed.
- R. Vadivambal and D. S. Jayas, Bio-Imaging: Principles, Techniques, and Applications, CRC Press, 1st edn, 2015 Search PubMed.
- J. Wen, Y. Xu and H. Li, et al., Recent applications of carbon nanomaterials in fluorescence biosensing and bioimaging, Chem. Commun., 2015, 51, 11346–11358, 10.1039/C5CC02887F.
- S. Goel, C. G. England, F. Chen and W. Cai, Positron emission tomography and nanotechnology: A dynamic duo for cancer theranostics, Adv. Drug Delivery Rev., 2017, 113, 157–176, DOI:10.1016/j.addr.2016.08.001.
- A. Zebibula, N. Alifu and L. Xia, et al., Ultrastable and Biocompatible NIR-II Quantum Dots for Functional Bioimaging, Adv. Funct. Mater., 2018, 28, 1–13, DOI:10.1002/adfm.201703451.
- R. Liu, D. Wu and S. Liu, et al., An aqueous route to multicolor photoluminescent carbon dots using silica spheres as carriers, Angew. Chem., Int. Ed., 2009, 48, 4598–4601, DOI:10.1002/anie.200900652.
- S. N. Frank and A. J. Bard, Heterogeneous photocatalytic oxidation of cyanide and sulfite in aqueous solutions at semiconductor powders, J. Phys. Chem., 1977, 81, 1484–1488 CrossRef CAS.
- X. Cui, L. Zhu and J. Wu, et al., A fluorescent biosensor based on carbon dots-labeled oligodeoxyribonucleotide and graphene oxide for mercury(II) detection, Biosens. Bioelectron., 2015, 63, 506–512, DOI:10.1016/j.bios.2014.07.085.
- Q. Xu, P. Pu and J. Zhao, et al., Preparation of highly photoluminescent sulfur-doped carbon dots for Fe(III) detection, J. Mater. Chem. A, 2015, 3, 542–546, 10.1039/C4TA05483K.
- S.-T. Yang, X. Wang and H. Wang, et al., Carbon Dots as Nontoxic and High-Performance Fluorescence Imaging Agents, J. Phys. Chem. C Nanomater Interfaces, 2009, 113, 18110–18114, DOI:10.1021/jp9085969.
- Q. Wang, X. Huang and Y. Long, et al., Hollow luminescent carbon dots for drug delivery, Carbon N Y, 2013, 59, 192–199, DOI:10.1016/j.carbon.2013.03.009.
- J. Tang, B. Kong and H. Wu, et al., Carbon nanodots featuring efficient FRET for real-time monitoring of drug delivery and two-photon imaging, Adv. Mater., 2013, 25, 6569–6574, DOI:10.1002/adma.201303124.
- T. Atabaev, G. Urmanova and N. Hong, Highly Mesoporous Silica Nanoparticles for Potential Drug Delivery Applications, Nano LIFE, 2014, 04, 1441003, DOI:10.1142/S1793984414410037.
- M. Karimi, H. Mirshekari and M. Aliakbari, et al., Smart mesoporous silica nanoparticles for controlled-release drug delivery, Nanotechnol. Rev., 2016, 5, DOI:10.1515/ntrev-2015-0057.
- Z. Wang, H. Liao and H. Wu, et al., Fluorescent carbon dots from beer for breast cancer cell imaging and drug delivery, Anal. Methods, 2015, 7, 8911–8917, 10.1039/C5AY01978H.
- H. Lee, C. Kim and E. Park, et al., [ACS AMI 2014] Photoluminescent green carbon nanodots from food-waste-derived sources Large-scale synthesis Properties and Biomedical applications, ACS Appl. Mater. Interfaces, 2014, 6, 3365–3370, DOI:10.1021/am500159p.
- J.-H. Park, L. Gu and G. von Maltzahn, et al., Biodegradable luminescent porous silicon nanoparticles for in vivo applications, Nat. Mater., 2009, 8, 331–336, DOI:10.1038/nmat2398.
- Y. Zhuo, H. Miao and D. Zhong, et al., One-step synthesis of high quantum-yield and excitation-independent emission carbon dots for cell imaging, Mater. Lett., 2015, 139, 197–200, DOI:10.1016/j.matlet.2014.10.048.
- A. Sachdev, I. Matai and P. Gopinath, Carbon dots incorporated polymeric hydrogels as multifunctional platform for imaging and induction of apoptosis in lung cancer cells, Colloids Surf., B, 2016, 141, 242–252, DOI:10.1016/j.colsurfb.2016.01.043.
- F. Du, M. Zhang and X. Li, et al., Economical and green synthesis of bagasse-derived fluorescent carbon dots for biomedical applications, Nanotechnology, 2014, 25, 315702, DOI:10.1088/0957-4484/25/31/315702.
- J. Qian, F. Quan and F. Zhao, et al., Aconitic acid derived carbon dots: Conjugated interaction for the detection of folic acid and fluorescence targeted imaging of folate receptor overexpressed cancer cells, Sens. Actuators, B, 2018, 262, 444–451, DOI:10.1016/j.snb.2018.01.227.
- Y. Yang, J. Cui and M. Zheng, et al., One-step synthesis of amino-functionalized fluorescent carbon nanoparticles by hydrothermal carbonization of chitosan, Chem. Commun., 2012, 48, 380–382, 10.1039/c1cc15678k.
- M. Zhang, C. Chi and P. Yuan, et al., A hydrothermal route to multicolor luminescent carbon dots from adenosine disodium triphosphate for bioimaging, Mater. Sci. Eng., C, 2017, 76, 1146–1153, DOI:10.1016/j.msec.2017.03.144.
- X.-W. Hua, Y.-W. Bao and F.-G. Wu, Fluorescent Carbon Quantum Dots with Intrinsic Nucleolus-Targeting Capability for Nucleolus Imaging and Enhanced Cytosolic and Nuclear Drug Delivery, ACS Appl. Mater. Interfaces, 2018, 10, 10664–10677, DOI:10.1021/acsami.7b19549.
- L. Li, R. Zhang and C. Lu, et al., In situ synthesis of NIR-light emitting carbon dots derived from spinach for bio-imaging applications, J. Mater. Chem. B, 2017, 5, 7328–7334, 10.1039/C7TB00634A.
- G. Gao, Y.-W. Jiang and H.-R. Jia, et al., On-off-on fluorescent nanosensor for Fe3+ detection and cancer/normal cell differentiation via silicon-doped carbon quantum dots, Carbon N Y, 2018, 134, 232–243, DOI:10.1016/j.carbon.2018.02.063.
- M. Alhajj and A. Farhana, Enzyme linked immunosorbent assay, StatPearls, StatPearls Publishing, 2023 Search PubMed.
- D. Costopoulou, L. Leondiadis and J. Czarnecki, et al., Direct ELISA method for the specific determination of prothymosin alpha in human specimens, J. Immunoassay Immunochem., 1998, 19, 295–316 CrossRef CAS PubMed.
- N. S. Fedarko, A. Jain and A. Karadag, et al., Elevated serum bone sialoprotein and osteopontin in colon, breast, prostate, and lung cancer, Clin. Cancer Res., 2001, 7, 4060–4066 CAS.
- A. Yagihashi, K. Asanuma and D. Kobayashi, et al., Detection of autoantibodies to livin and survivin in Sera from lung cancer patients, Lung Cancer, 2005, 48, 217–221 CrossRef PubMed.
- T. Yamashita, H. Kamada and S. Kanasaki, et al., Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis, Int. J. Pharm. Sci., 2013, 68, 969–973 CAS.
- R.-S. Lai, H.-K. Hsu and J.-Y. Lu, et al., CYFRA 21–1 enzyme-linked immunosorbent assay: evaluation as a tumor marker in non-small cell lung cancer, Chest, 1996, 109, 995–1000 CrossRef CAS PubMed.
- J. Wang, S. Zhang and W. Ni, et al., Development and application of a double-antibody sandwich ELISA kit for the detection of serum MUC1 in lung cancer patients, Cancer Biomarkers, 2016, 17, 369–376 CAS.
- G. Prasad and M. McCullough, Chemokines and cytokines as salivary biomarkers for the early diagnosis of oral cancer, Int. J. Dent., 2013, 2013, 1–7, DOI:10.1155/2013/813756.
- S. N. Topkaya, M. Azimzadeh and M. Ozsoz, Electrochemical Biosensors for Cancer Biomarkers Detection: Recent Advances and Challenges, Electroanalysis, 2016, 28, 1402–1419, DOI:10.1002/elan.201501174.
- P. Villalobos and I. I. Wistuba, Lung Cancer Biomarkers, Hematol. Oncol. Clin. North Am., 2017, 31, 13–29, DOI:10.1016/j.hoc.2016.08.006.
- Z. Khurshid, M. S. Zafar and R. S. Khan, et al., Role of Salivary Biomarkers in Oral Cancer Detection, Adv. Clin. Chem., 2018, 86, 23–70, DOI:10.1016/bs.acc.2018.05.002.
- S. Yete, W. D’Souza and D. Saranath, High-risk human papillomavirus in oral cancer: clinical implications, Oncology, 2018, 94, 133–141 CrossRef PubMed.
- G. Corso, A. Villa, A. Tarsitano and A. Ghoel, Current trends in oral cancer: a review, Cancer cell Microenviron., 2016, 3, DOI:10.14800/ccm.1332.
- S. Balakrishnan, S. Manoharan, L. M. Alias and M. R. Nirmal, Effect of curcumin and ferulic acid on modulation of expression pattern of p53 and bcl-2 proteins in 7, 12-dimethylbenz [] anthracene-induced hamster buccal pouch carcinogenesis, Indian J. Biochem. Biophys., 2010, 47(1), 7–12 CAS.
- J. Rittscher, R. Machiraju and S. T. C. Wong, Microscopic image analysis for life science applications, Artech House, 2008 Search PubMed.
- M. N. Gurcan, L. E. Boucheron and A. Can, et al., Histopathological image analysis: A review, IEEE Rev. Biomed. Eng., 2009, 2, 147–171 Search PubMed.
- C. D. M. Fletcher, Diagnostic histopathology of tumors: 2-volume set with CD-ROMs. Elsevier Health Sciences, 2007 Search PubMed.
- S. Di Cataldo, E. Ficarra and E. Macii, Computer-aided techniques for chromogenic immunohistochemistry: status and directions, Comput. Biol. Med., 2012, 42, 1012–1025 CrossRef CAS PubMed.
- L. Jiang, X. Zeng and H. Yang, et al., Oral cancer overexpressed 1 (ORAOV1): a regulator for the cell growth and tumor angiogenesis in oral squamous cell carcinoma, Int. J. Cancer, 2008, 123, 1779–1786 CrossRef CAS PubMed.
- K. Hsieh, Y. Xiao and H. Tom Soh, Electrochemical DNA detection via exonuclease and target-catalyzed transformation of surface-bound probes, Langmuir, 2010, 26, 10392–10396, DOI:10.1021/la100227s.
- F. Xia, X. Zuo and R. Yang, et al., Label-Free, Dual-Analyte Electrochemical Biosensors: A New Class of Molecular-Electronic Logic Gates, J. Am. Chem. Soc., 2010, 132, 8557–8559, DOI:10.1021/ja101379k.
- S. Bi, J. Zhang and S. Zhang, Ultrasensitive and selective DNA detection based on nicking endonuclease assisted signal amplification and its application in cancer cell detection, Chem. Commun., 2010, 46, 5509–5511 RSC.
- M. P. Rivera and A. C. Mehta, Initial Diagnosis of Lung Cancer: ACCP Evidence-Based Clinical Practice Guidelines, 2nd edn, Chest, 2007, 132, 131S–148S, DOI:10.1378/chest.07-1357.
- K. Han, H. Liu and J. Cui, et al., Recent strategies for electrochemical sensing detection of miRNAs in lung cancer, Anal. Biochem., 2023, 661, 114986, DOI:10.1016/j.ab.2022.114986.
- C. I. Henschke, D. Shaham, D. F. Yankelevitz and N. K. Altorki, CT Screening for Lung Cancer: Past and Ongoing Studies, Semin. Thorac. Cardiovasc. Surg., 2005, 17, 99–106, DOI:10.1053/j.semtcvs.2005.05.002.
- U. Pastorino, M. Bellomi and C. Landoni, et al., Early lung-cancer detection with spiral CT and positron emission tomography in heavy smokers: 2-year results, Lancet, 2003, 362, 593–597, DOI:10.1016/S0140-6736(03)14188-8.
- S. Liu, W. Su, Z. Li and X. Ding, Electrochemical detection of lung cancer specific microRNAs using 3D DNA origami nanostructures, Biosens. Bioelectron., 2015, 71, 57–61, DOI:10.1016/j.bios.2015.04.006.
- G. Bonnet, S. Tyagi, A. Libchaber and F. R. Kramer, Thermodynamic basis of the enhanced specificity of structured DNA probes, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 6171–6176, DOI:10.1073/pnas.96.11.6171.
- L. V. Sequist, S. Nagrath and M. Toner, et al., The CTC-Chip: An Exciting New Tool to Detect Circulating Tumor Cells in Lung Cancer Patients, J. Thorac. Oncol., 2009, 4, 281–283, DOI:10.1097/JTO.0b013e3181989565.
- T. Xiao, W. Ying and L. Li, et al., An Approach to Studying Lung Cancer-related Proteins in Human Blood, Mol. Cell. Proteomics, 2005, 4, 1480–1486, DOI:10.1074/mcp.M500055-MCP200.
- D. S. Wilson and J. W. Szostak, In vitro selection of functional nucleic acids, Annu. Rev. Biochem., 1999, 68, 611–647, DOI:10.1146/annurev.biochem.68.1.611.
- S. M. Shamah, J. M. Healy and S. T. Cload, Complex target SELEX, Acc. Chem. Res., 2008, 41, 130–138, DOI:10.1021/ar700142z.
- C. K. O’Sullivan, Aptasensors--the future of biosensing?, Anal. Bioanal. Chem., 2002, 372, 44–48, DOI:10.1007/s00216-001-1189-3.
- C. Zhu, D. Liu and Z. Chen, et al., An ultra-sensitive aptasensor based on carbon nanohorns/gold nanoparticles composites for impedimetric detection of carbendazim at picogram levels, J. Colloid Interface Sci., 2019, 546, 92–100, DOI:10.1016/j.jcis.2019.03.035.
- M. Adeel, M. M. Rahman and J.-J. Lee, Label-free aptasensor for the detection of cardiac biomarker myoglobin based on gold nanoparticles decorated boron nitride nanosheets, Biosens. Bioelectron., 2019, 126, 143–150, DOI:10.1016/j.bios.2018.10.060.
- X. Wang, F. Gao and Y. Gong, et al., Electrochemical aptasensor based on conductive supramolecular polymer hydrogels for thrombin detection with high selectivity, Talanta, 2019, 205, 120140, DOI:10.1016/j.talanta.2019.120140.
- S. C. B. Gopinath, K. Awazu and M. Fujimaki, Waveguide-mode sensors as aptasensors, Sensors, 2012, 12, 2136–2151, DOI:10.3390/s120202136.
- S. G. Meirinho, L. G. Dias, A. M. Peres and L. R. Rodrigues, Voltammetric aptasensors for protein disease biomarkers detection: A review, Biotechnol. Adv., 2016, 34, 941–953, DOI:10.1016/j.biotechadv.2016.05.006.
- I. E. Tothill, Biosensors for cancer markers diagnosis, Semin. Cell Dev. Biol., 2009, 20, 55–62, DOI:10.1016/j.semcdb.2009.01.015.
- S. Tiwari, P. K. Gupta and Y. Bagbi, et al., L-cysteine capped lanthanum hydroxide nanostructures for non-invasive detection of oral cancer biomarker, Biosens. Bioelectron., 2017, 89, 1042–1052, DOI:10.1016/j.bios.2016.10.020.
- S. Khaksari, A. R. Ameri and S. M. Taghdisi, et al., A microfluidic electrochemical aptasensor for highly sensitive and selective detection of A549 cells as integrin α6β4-containing cell model via IDA aptamers, Talanta, 2023, 252, 123781, DOI:10.1016/j.talanta.2022.123781.
- J. Chen, L. Yu and W. Xu, et al., A graphene-based highly sensitive aptasensor for the detection of lung cancer marker CA125, Carbon Lett., 2023, 33, 1811–1817, DOI:10.1007/s42823-023-00560-w.
- M. Mazloum-Ardakani, L. Hosseinzadeh and Z. Taleat, Synthesis and electrocatalytic effect of Ag@Pt core–shell nanoparticles supported on reduced graphene oxide for sensitive and simple label-free electrochemical aptasensor, Biosens. Bioelectron., 2015, 74, 30–36, DOI:10.1016/j.bios.2015.05.072.
- G. Bezerra, C. Córdula and D. Campos, et al., Electrochemical aptasensor for the detection of HER2 in human serum to assist in the diagnosis of early stage breast cancer, Anal. Bioanal. Chem., 2019, 411, 6667–6676, DOI:10.1007/s00216-019-02040-5.
- W. Wen, J.-Y. Huang and T. Bao, et al., Increased electrocatalyzed performance through hairpin oligonucleotide aptamer-functionalized gold nanorods labels and graphene-streptavidin nanomatrix: Highly selective and sensitive electrochemical biosensor of carcinoembryonic antigen, Biosens. Bioelectron., 2016, 83, 142–148, DOI:10.1016/j.bios.2016.04.039.
- J. A. Ho, H.-C. Chang and N.-Y. Shih, et al., Diagnostic detection of human lung cancer-associated antigen using a gold nanoparticle-based electrochemical immunosensor, Anal. Chem., 2010, 82, 5944–5950 CrossRef CAS PubMed.
- S. Gamagedara and Y. Ma, Biomarker analysis for prostate cancer diagnosis using LC–MS and CE–MS, Bioanalysis, 2011, 3, 2129–2142, DOI:10.4155/bio.11.203.
- C. Zhao, D. Lin and J. Wu, et al., Nanogold-Enriched Carbon Nanohorn Label for Sensitive Electrochemical Detection of Biomarker on a Disposable Immunosensor, Electroanalysis, 2013, 25, 1044–1049, DOI:10.1002/elan.201200423.
- F. Li, J. Peng and Q. Zheng, et al., Carbon Nanotube-Polyamidoamine Dendrimer Hybrid-Modified Electrodes for Highly Sensitive Electrochemical Detection of MicroRNA24, Anal. Chem., 2015, 87, 4806–4813, DOI:10.1021/acs.analchem.5b00093.
- F.-Y. Kong, M.-T. Xu, J.-J. Xu and H.-Y. Chen, A novel lable-free electrochemical immunosensor for carcinoembryonic antigen based on gold nanoparticles–thionine–reduced graphene oxide nanocomposite film modified glassy carbon electrode, Talanta, 2011, 85, 2620–2625, DOI:10.1016/j.talanta.2011.08.028.
- L. Bai, R. Yuan and Y. Chai, et al., Simultaneous electrochemical detection of multiple analytes based on dual signal amplification of single-walled carbon nanotubes and multi-labeled graphene sheets, Biomaterials, 2012, 33, 1090–1096, DOI:10.1016/j.biomaterials.2011.10.012.
- S. Azzouzi, L. Rotariu and A. M. Benito, et al., A novel amperometric biosensor based on gold nanoparticles anchored on reduced graphene oxide for sensitive detection of L-lactate tumor biomarker, Biosens. Bioelectron., 2015, 69, 280–286, DOI:10.1016/j.bios.2015.03.012.
- S. Aslan and Ü. Anik, Development of TiO2 and Au Nanocomposite Electrode as CEA Immunosensor Transducer, Electroanalysis, 2014, 26, 1373–1381, DOI:10.1002/elan.201400086.
- Z. Gao and Y. H. Yu, Direct labeling microRNA with an electrocatalytic moiety and its application in ultrasensitive microRNA assays, Biosens. Bioelectron., 2007, 22, 933–940, DOI:10.1016/j.bios.2006.04.020.
- H. Chen, Q. Mei and S. Jia, et al., High specific detection of osteopontin using a three-dimensional copolymer layer support based on electrochemical impedance spectroscopy, Analyst, 2014, 139, 4476–4481, 10.1039/C4AN00576G.
- T. Yang, S. Wang and H. Jin, et al., An electrochemical impedance sensor for the label-free ultrasensitive detection of interleukin-6 antigen, Sens. Actuators, B, 2013, 178, 310–315, DOI:10.1016/j.snb.2012.12.107.
- Z. Chen, L. Li and H. Zhao, et al., Electrochemical impedance spectroscopy detection of lysozyme based on electrodeposited gold nanoparticles, Talanta, 2011, 83, 1501–1506, DOI:10.1016/j.talanta.2010.11.042.
- J. Sheng, Y. Wu and H. Ding, et al., Multienzyme-like Nanozymes: Regulation, Rational Design, and Application, Adv. Mater., 2023, 2211210, DOI:10.1002/adma.202211210.
- R. Miglani, N. Parveen and A. Kumar, et al., Degradation of xenobiotic pollutants: an environmentally sustainable approach, Metabolites, 2022, 12, 818 CrossRef CAS PubMed.
- G. K. Arora, L. Palamiuc and B. M. Emerling, Expanding role of PI5P4Ks in cancer: a promising druggable target, FEBS Lett., 2022, 596, 3–16 CrossRef CAS PubMed.
- W. Huang, S. Huang, G. Chen and G. Ouyang, Biocatalytic Metal-Organic Frameworks: Promising Materials for Biosensing, ChemBioChem, 2022, 23, e202100567 CrossRef CAS PubMed.
- B. Das, J. Lou Franco and N. Logan, et al., Nanozymes in point-of-care diagnosis: an emerging futuristic approach for biosensing, Nano-Micro Lett., 2021, 13, 1–51 CrossRef PubMed.
- H. Keshavarz Alikhani, M. Pourhamzeh and H. Seydi, et al., Regulatory non-coding RNAs in familial hypercholesterolemia, theranostic applications, Front. Cell Dev. Biol., 2022, 10, 894800 CrossRef PubMed.
- X. Meng, I. Zare, X. Yan and K. Fan, Protein-protected metal nanoclusters: An emerging ultra-small nanozyme. Wiley Interdiscip Rev Nanomedicine, Nanobiotechnology, 2020, 12, e1602 Search PubMed.
- R. Sankaranarayanan, R. Swaminathan and H. Brenner, et al., Cancer survival in Africa, Asia, and Central America: a population-based study, Lancet Oncol., 2009, 11, 165–173, DOI:10.1016/S1470-2045(09)70335-3.
- C. A. Rowe, S. B. Scruggs and M. J. Feldstein, et al., An Array Immunosensor for Simultaneous Detection of Clinical Analytes, Anal. Chem., 1999, 71, 433–439, DOI:10.1021/ac980798t.
- C. A. Rowe, J. S. Bolitho and A. Jane, et al., Rapid detection of D-dimer using a fiber optic biosensor, Thromb. Haemostasis, 1998, 59, 94–98 CrossRef.
- C. A. Rowe, L. M. Tender and M. J. Feldstein, et al., Array Biosensor for Simultaneous Identification of Bacterial, Viral, and Protein Analytes, Anal. Chem., 1999, 71, 3846–3852, DOI:10.1021/ac981425v.
- Z. Liron, L. M. Tender, J. P. Golden and F. S. Ligler, Voltage-induced inhibition of antigen-antibody binding at conducting optical waveguides, Biosens. Bioelectron., 2002, 17, 489–494 CrossRef CAS PubMed.
- M. W. Lingen, J. R. Kalmar, T. Karrison and P. M. Speight, Critical evaluation of diagnostic aids for the detection of oral cancer, Oral Oncol., 2008, 44, 10–22, DOI:10.1016/j.oraloncology.2007.06.011.
- R. Kortum and E. Sevick, Quantitative Optical Spectroscopy for Tissue Diagnosis, Annu. Rev. Phys. Chem., 1996, 47, 555–606, DOI:10.1146/annurev.physchem.47.1.555.
- D. C. Christian, Computer-assisted analysis of oral brush biopsies at an oral cancer screening program, J. Am. Dent. Assoc., 2002, 133, 357–362, DOI:10.14219/jada.archive.2002.0175.
- A. R. Kerr, D. A. Sirois and J. B. Epstein, Clinical evaluation of chemiluminescent lighting: an adjunct for oral mucosal examinations, J. Clin. Dent., 2006, 17, 59–63 CAS.
- Deepa, B. Nohwal and C. S. Pundir, An electrochemical CD59 targeted noninvasive immunosensor based on graphene oxide nanoparticles embodied pencil graphite for detection of lung cancer, Microchem. J., 2020, 156, 104957, DOI:10.1016/j.microc.2020.104957.
- S. Verma, A. Singh and A. Shukla, et al., Anti-IL8/AuNPs-rGO/ITO as an Immunosensing Platform for Noninvasive Electrochemical Detection of Oral Cancer, ACS Appl. Mater. Interfaces, 2017, 9, 27462–27474, DOI:10.1021/acsami.7b06839.
- R. Malhotra, V. Patel and J. P. Vaqué, et al., Ultrasensitive Electrochemical Immunosensor for Oral Cancer Biomarker IL-6 Using Carbon Nanotube Forest Electrodes and Multilabel Amplification, Anal. Chem., 2010, 82, 3118–3123, DOI:10.1021/ac902802b.
- N. Pachauri, G. B. V. S. Lakshmi and S. Sri, et al., Silver molybdate nanoparticles based immunosensor for the non-invasive detection of Interleukin-8 biomarker, Mater. Sci. Eng., C, 2020, 113, 110911, DOI:10.1016/j.msec.2020.110911.
- N. Wang, J. Wang and X. Zhao, et al., Highly sensitive electrochemical immunosensor for the simultaneous detection of multiple tumor markers for signal amplification, Talanta, 2021, 226, 122133, DOI:10.1016/j.talanta.2021.122133.
- H. Yang, J. Bao and D. Huo, et al., Au doped poly-thionine and poly-m-Cresol purple: Synthesis and their application in simultaneously electrochemical detection of two lung cancer markers CEA and CYFRA21-1, Talanta, 2021, 224, 121816 CrossRef CAS PubMed.
- G. Yang, Z. Xiao and C. Tang, et al., Recent advances in biosensor for detection of lung cancer biomarkers, Biosens. Bioelectron., 2019, 141, 111416, DOI:10.1016/j.bios.2019.111416.
- J. Chen, H.-M. Meng and Y. An, et al., Structure-switching aptamer triggering hybridization displacement reaction for label-free detection of exosomes, Talanta, 2020, 209, 120510, DOI:10.1016/j.talanta.2019.120510.
- L. Kashefi-Kheyrabadi, A. Koyappayil and T. Kim, et al., A MoS2@Ti3C2Tx MXene hybrid-based electrochemical aptasensor (MEA) for sensitive and rapid detection of Thyroxine, Bioelectrochemistry, 2021, 137, 107674, DOI:10.1016/j.bioelechem.2020.107674.
- F. Hou, X.-B. Hu and S.-H. Ma, et al., Construction of electrochemiluminescence sensing platform with in situ generated coreactant strategy for sensitive detection of prostate specific antigen, J. Electroanal. Chem., 2020, 858, 113817, DOI:10.1016/j.jelechem.2019.113817.
- B. Feyzi-barnaji, R. Dinarvand and H. Salehzadeh, et al., Construction of a ternary nano-architecture based graphene oxide sheets, toward electrocatalytic determination of tumor-associated anti-p53 autoantibodies in human serum, Talanta, 2021, 230, 122276, DOI:10.1016/j.talanta.2021.122276.
- K. Hu, T. Pang and Y. Shi, et al., Magnetic borate-modified Mxene: A highly affinity material for the extraction of catecholamines, Anal. Chim. Acta, 2021, 1176, 338769, DOI:10.1016/j.aca.2021.338769.
- R. A. Soomro, S. Jawaid and Q. Zhu, et al., A mini-review on MXenes as versatile substrate for advanced sensors, Chin. Chem. Lett., 2020, 31, 922–930, DOI:10.1016/j.cclet.2019.12.005.
- K. Hu, J. Cheng and K. Wang, et al., Sensitive electrochemical immunosensor for CYFRA21-1 detection based on AuNPs@MoS2@Ti3C2Tx composites, Talanta, 2022, 238, 122987, DOI:10.1016/j.talanta.2021.122987.
- M. L. Yola, N. Atar and N. Özcan, A novel electrochemical lung cancer biomarker cytokeratin 19 fragment antigen 21-1 immunosensor based on Si3N4/MoS2 incorporated MWCNTs and core–shell type magnetic nanoparticles, Nanoscale, 2021, 13, 4660–4669 RSC.
- N. Pachauri, K. Dave, A. Dinda and P. R. Solanki, Cubic CeO2 implanted reduced graphene oxide-based highly sensitive biosensor for non-invasive oral cancer biomarker detection, J. Mater. Chem. B, 2018, 6, 3000–3012 RSC.
- P. He, T. Naka and S. Serada, et al., Proteomics-based identification of α-enolase as a tumor antigen in non-small lung cancer, Cancer Sci., 2007, 98, 1234–1240 CrossRef CAS PubMed.
- K.-J. Liu and N.-Y. Shih, The role of enolase in tissue invasion and metastasis of pathogens and tumor cells, J. Cancer Mol., 2007, 3, 45–48 CAS.
- J. L. Viallard, D. Caillaud and B. Kantelip, et al., Enzymatic determination of serum neuron-specific enolase in small cell lung cancers: utility of the serum neuron-specific enolase/serum nonneuronal enolase ratio, Chest, 1988, 93, 1225–1233 CrossRef CAS PubMed.
- R. A. Wevers, A. A. C. Jacobs and O. R. Hommes, A bioluminescent assay for enolase (EC 4.2. 1.11) activity in human serum and cerebrospinal fluid, Clin. Chim. Acta, 1983, 135, 159–168 CrossRef CAS PubMed.
- M. Katayama, H. Nakano and A. Ishiuchi, et al., Protein pattern difference in the colon cancer cell lines examined by two-dimensional differential in-gel electrophoresis and mass spectrometry, Surg. Today, 2006, 36, 1085–1093 CrossRef CAS PubMed.
- D. B. Wall, M. T. Kachman and S. Gong, et al., Isoelectric focusing nonporous RP HPLC: a two-dimensional liquid-phase separation method for mapping of cellular proteins with identification using MALDI-TOF mass spectrometry, Anal. Chem., 2000, 72, 1099–1111 CrossRef CAS PubMed.
- M. Choudhary, P. Yadav and A. Singh, et al., CD 59 Targeted Ultrasensitive Electrochemical Immunosensor for Fast and Noninvasive Diagnosis of Oral Cancer, Electroanalysis, 2016, 28, 2565–2574, DOI:10.1002/elan.201600238.
- J. J. BelBruno, Molecularly Imprinted Polymers, Chem. Rev., 2019, 119, 94–119, DOI:10.1021/acs.chemrev.8b00171.
- S. Bhakta and P. Mishra, Molecularly imprinted polymer-based sensors for cancer biomarker detection, Sens. Actuators Rep., 2021, 3, 100061, DOI:10.1016/j.snr.2021.100061.
- Archana, D. Verma and S. Z. H. Hashmi, et al., Electrochemical Sensor based on Polydopamine-Molecularly Imprinted Polymer for Detection of 4-Ethylphenyl Sulfate “a Novel Gut Metabolite”: Fabrication, Characterization, and Performance Evaluation in Human Urine, Microchem. J., 2023, 193, 108964 CrossRef CAS.
- F. B. Moussa, F. Achi and H. Meskher, et al., Green one-step reduction approach to prepare rGO@ AgNPs coupled with molecularly imprinted polymer for selective electrochemical detection of lactic acid as a cancer biomarker, Mater. Chem. Phys., 2022, 289, 126456, DOI:10.1016/j.matchemphys.2022.126456.
- A. E. F. Oliveira, A. C. Pereira and L. F. Ferreira, Disposable electropolymerized molecularly imprinted electrochemical sensor for determination of breast cancer biomarker CA 15-3 in human serum samples, Talanta, 2023, 252, 123819 CrossRef CAS PubMed.
- P. Tang, H. Zhang, J. Huo and X. Lin, An electrochemical sensor based on iron (II, III)@ graphene oxide@ molecularly imprinted polymer nanoparticles for interleukin-8 detection in saliva, Anal. Methods, 2015, 7, 7784–7791 RSC.
- K. Ren, J. Zhou and H. Wu, Materials for microfluidic chip fabrication, Acc. Chem. Res., 2013, 46, 2396–2406 CrossRef CAS PubMed.
- C. K. Tang, A. Vaze, M. Shen and J. F. Rusling, High-throughput electrochemical microfluidic immunoarray for multiplexed detection of cancer biomarker proteins, ACS Sens., 2016, 1, 1036–1043 CrossRef CAS PubMed.
- Y. Wang, J. Luo and J. Liu, et al., Label-free microfluidic paper-based electrochemical aptasensor for ultrasensitive and simultaneous multiplexed detection of cancer biomarkers, Biosens. Bioelectron., 2019, 136, 84–90 CrossRef CAS PubMed.
- Y. Wang, L. Chen and T. Xuan, et al., Label-free electrochemical impedance spectroscopy aptasensor for ultrasensitive detection of lung cancer biomarker carcinoembryonic antigen, Front. Chem., 2021, 9, 721008 CrossRef CAS PubMed.
- N. Taheri, H. Khoshsafar and M. Ghanei, et al., Dual-template rectangular nanotube molecularly imprinted polypyrrole for label-free impedimetric sensing of AFP and CEA as lung cancer biomarkers, Talanta, 2022, 239, 123146 CrossRef CAS PubMed.
- J. Liu, J. Liu and Y. Shang, et al., An electrochemical immunosensor for simultaneous detection of two lung cancer markers based on electroactive probes, J. Electroanal. Chem., 2022, 919, 116559 CrossRef CAS.
- H. Jiang, H. Zhang and M. Qin, et al., Ultrasensitive Sandwich-type Electrochemical Aptasensor for Dual Detection of Lung Cancer Biomarkers, IEEE Sens. J., 2023, 23(17), 18977–18985, DOI:10.1109/JSEN.2023.3294886.
| This journal is © The Royal Society of Chemistry 2024 |