Open Access Article
Open Access ArticleDesign, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection†
Xavier
Jalencas‡
 a,
Hannes
Berg‡
a,
Hannes
Berg‡
 bc,
Ludvik Olai
Espeland‡
bc,
Ludvik Olai
Espeland‡
 de,
Sridhar
Sreeramulu
de,
Sridhar
Sreeramulu
 bc,
Franziska
Kinnen
bc,
Christian
Richter
bc,
Franziska
Kinnen
bc,
Christian
Richter
 bc,
Charis
Georgiou
bc,
Charis
Georgiou
 d,
Vladyslav
Yadrykhinsky
d,
Vladyslav
Yadrykhinsky
 d,
Edgar
Specker
f,
Kristaps
Jaudzems
d,
Edgar
Specker
f,
Kristaps
Jaudzems
 g,
Tanja
Miletić
g,
Tanja
Miletić
 f,
Robert
Harmel
f,
Robert
Harmel
 f,
Phil
Gribbon
f,
Phil
Gribbon
 hi,
Harald
Schwalbe
hi,
Harald
Schwalbe
 *bcj,
Ruth
Brenk
*bcj,
Ruth
Brenk
 *dk,
Aigars
Jirgensons
*dk,
Aigars
Jirgensons
 *g,
Andrea
Zaliani
*g,
Andrea
Zaliani
 *hi and
Jordi
Mestres
*hi and
Jordi
Mestres
 *al
*al
aResearch Group on Systems Pharmacology, Research Program on Biomedical Informatics (GRIB), IMIM Hospital del Mar Medical Research Institute, Parc de Recerca Biomèdica (PRBB), Doctor Aiguader 88, 08003 Barcelona, Spain. E-mail: jmestres@imim.es
bCenter for Biomolecular Magnetic Resonance (BMRZ), Institute for Organic Chemistry, Max-von-Laue-Str. 7, 60438 Frankfurt/M, Germany. E-mail: schwalbe@nmr.uni-frankfurt.de
cChemical Biology, Goethe University, Max-von-Laue-Str. 7, 60438 Frankfurt/M, Germany
dDepartment of Biomedicine, University of Bergen, Jonas Lies Vei 91, 5020 Bergen, Norway
eDepartment of Chemistry, University of Bergen, Allégaten 41, 5007 Bergen, Norway
fEU-OPENSCREEN ERIC, Robert-Rössle Straße 10, 13125 Berlin, Germany
gLatvian Institute of Organic Synthesis, Aizkraules 21, Riga LV-1006, Latvia. E-mail: aigars@osi.lv
hFraunhofer Institute for Translational Medicine and Pharmacology (ITMP), Schnackenburgallee 114, 22525 Hamburg, Germany. E-mail: andrea.zaliani@itmp.fraunhofer.de
iFraunhofer Cluster of Excellence for Immune-Mediated Diseases (CIMD), Theodor Stern Kai 7, 60590 Frankfurt, Germany
jInstruct-ERIC, Oxford House, Parkway Court, John Smith Drive, Oxford OX4 2JY, UK. E-mail: harald.schwalbe@instruct-eric.org
kComputational Biology Unit, University of Bergen, Thormøhlensgate 55, 5008 Bergen, Norway. E-mail: ruth.brenk@uib.no
lInstitut de Quimica Computacional i Catalisi, Facultat de Ciencies, Universitat de Girona, Maria Aurelia Capmany 69, 17003 Girona, Catalonia, Spain. E-mail: jordi.mestres@udg.edu
First published on 12th February 2024
Abstract
The EU-OPENSCREEN (EU-OS) European Research Infrastructure Consortium (ERIC) is a multinational, not-for-profit initiative that integrates high-capacity screening platforms and chemistry groups across Europe to facilitate research in chemical biology and early drug discovery. Over the years, the EU-OS has assembled a high-throughput screening compound collection, the European Chemical Biology Library (ECBL), that contains approximately 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 commercially available small molecules and a growing number of thousands of academic compounds crowdsourced through our network of European and non-European chemists. As an extension of the ECBL, here we describe the computational design, quality control and use case screenings of the European Fragment Screening Library (EFSL) composed of 1056 mini and small chemical fragments selected from a substructure analysis of the ECBL. Access to the EFSL is open to researchers from both academia and industry. Using EFSL, eight fragment screening campaigns using different structural and biophysical methods have successfully identified fragment hits in the last two years. As one of the highlighted projects for antibiotics, we describe the screening by Bio-Layer Interferometry (BLI) of the EFSL, the identification of a 35 μM fragment hit targeting the beta-ketoacyl-ACP synthase 2 (FabF), its binding confirmation to the protein by X-ray crystallography (PDB 8PJ0), its subsequent rapid exploration of its surrounding chemical space through hit-picking of ECBL compounds that contain the fragment hit as a core substructure, and the final binding confirmation of two follow-up hits by X-ray crystallography (PDB 8R0I and 8R1V).
000 commercially available small molecules and a growing number of thousands of academic compounds crowdsourced through our network of European and non-European chemists. As an extension of the ECBL, here we describe the computational design, quality control and use case screenings of the European Fragment Screening Library (EFSL) composed of 1056 mini and small chemical fragments selected from a substructure analysis of the ECBL. Access to the EFSL is open to researchers from both academia and industry. Using EFSL, eight fragment screening campaigns using different structural and biophysical methods have successfully identified fragment hits in the last two years. As one of the highlighted projects for antibiotics, we describe the screening by Bio-Layer Interferometry (BLI) of the EFSL, the identification of a 35 μM fragment hit targeting the beta-ketoacyl-ACP synthase 2 (FabF), its binding confirmation to the protein by X-ray crystallography (PDB 8PJ0), its subsequent rapid exploration of its surrounding chemical space through hit-picking of ECBL compounds that contain the fragment hit as a core substructure, and the final binding confirmation of two follow-up hits by X-ray crystallography (PDB 8R0I and 8R1V).
Introduction
Over the last twenty-five years, fragment-based screening has become a well-established drug discovery strategy for hit generation.1 In contrast to conventional high-throughput screening (HTS) technologies, fragment-based drug discovery (FBDD) relies on screening low numbers of less complex low molecular weight molecules to find low affinity chemical fragments that bind with high efficiency to disease-relevant targets.2 In the event that fragment hits are identified, grow and link strategies are applied to further optimize fragments into potent small molecule leads.3,4 The use of FBDD has proven to be a successful approach to generate leads for a wide diversity of proteins covering all target classes,5 leading to multiple clinical candidates, some of which ultimately resulting in marketed drugs.6Fragment hits are often efficient protein binders7 but their small size results usually in weak binding affinities.8 Therefore, the techniques used to detect fragment binding have to be highly sensitive at the low μM to mM scale. A range of biophysical methods such as Nuclear Magnetic Resonance (NMR), Surface Plasmon Resonance (SPR), Grating-Coupled Interferometry (GCI), Thermal Shift (TS), Microscale Thermophoresis (MTS), Biolayer Interferometry (BLI), Isothermal Titration Calorimetry (ITC), X-ray crystallography, and biochemical assays at high concentration are used to screen purposely designed fragment libraries and to identify low-affinity binders.9–14 These fragment libraries are usually provided by chemical vendors or assembled in-house,15 and they contain small molecules that comply with pre-defined fragment-like properties, such as the rule-of-three (Ro3),16 but are also chemically diverse and have high aqueous solubility.17
In this work we introduce a new fragment library developed by EU-OPENSCREEN (EU-OS), the European Research Infrastructure Consortium (ERIC) for drug discovery and chemical biology (https://www.eu-openscreen.eu/services/compound-collection/fragment-library-fbld.html). EU-OS is an international, distributed non-for-profit initiative that provides high-throughput screening and medicinal chemistry support to academia and industry. Beyond its academic network of partners, EU-OS offers access to the European Chemical Biology Library (ECBL), a thoroughly designed diversity set consisting of approximately 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 small molecules selected from commercial sources.18 Under the framework of the EC-funded project EU-OS DRIVE, the European Fragment Screening Library (EFSL) poised to ECBL compounds was designed to facilitate rapid fragment-to-lead progression. The EFSL is the result of collaborative efforts between EU-OS ERIC researchers (https://www.eu-openscreen.eu/) with structural biology experts from Instruct ERIC (https://instruct-eric.org/) and the iNEXT-Discovery (https://inext-discovery.eu/) consortia. They joined forces to offer a complete pipeline of services and resources from fragment hit identification and confirmation to follow-up support on fragment growth and optimization, including access to biochemical assays and chemoinformatics tools to accelerate chemical probe discovery and FBDD. By making EFSL available to the worldwide research community,19 both from academia and industry, we have been able to collect intrigued and valuable information on library performance in a variety of projects on multiple diverse targets over the last two years that is helping us to refine the composition of EFSL.
000 small molecules selected from commercial sources.18 Under the framework of the EC-funded project EU-OS DRIVE, the European Fragment Screening Library (EFSL) poised to ECBL compounds was designed to facilitate rapid fragment-to-lead progression. The EFSL is the result of collaborative efforts between EU-OS ERIC researchers (https://www.eu-openscreen.eu/) with structural biology experts from Instruct ERIC (https://instruct-eric.org/) and the iNEXT-Discovery (https://inext-discovery.eu/) consortia. They joined forces to offer a complete pipeline of services and resources from fragment hit identification and confirmation to follow-up support on fragment growth and optimization, including access to biochemical assays and chemoinformatics tools to accelerate chemical probe discovery and FBDD. By making EFSL available to the worldwide research community,19 both from academia and industry, we have been able to collect intrigued and valuable information on library performance in a variety of projects on multiple diverse targets over the last two years that is helping us to refine the composition of EFSL.
Results and discussion
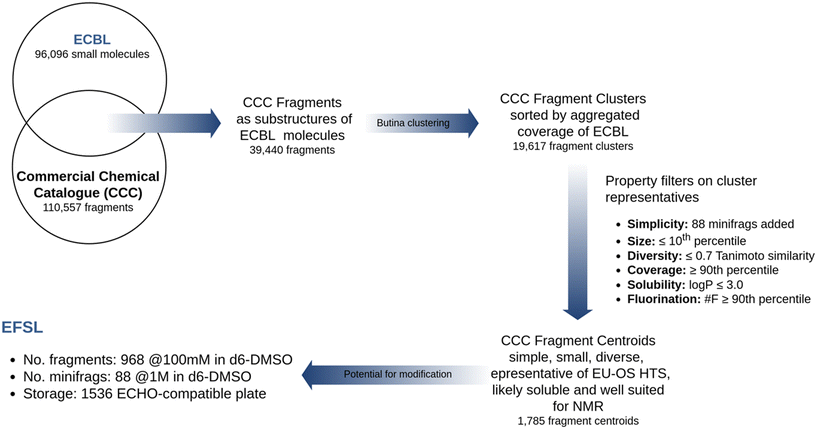
Design criteria of EFSL
To build a library of chemical fragments that has a good representativity of ECBL and that it is also easily accessible, the Enamine Fragment Collection (https://enamine.net/compound-collections/fragment-collection) was taken into account as the reference fragment set. The 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 557 chemical fragments of this collection were used to perform a substructural search on the 96
557 chemical fragments of this collection were used to perform a substructural search on the 96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 compounds in ECBL. The search was done using the RDKit library (https://www.rdkit.org) and resulted in a preliminary list of 39
096 compounds in ECBL. The search was done using the RDKit library (https://www.rdkit.org) and resulted in a preliminary list of 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440 candidate fragments.
440 candidate fragments.
A workgroup comprising members of the EU-OS, Instruct ERIC and iNext-Discovery consortia agreed on a series of priority criteria to select the minimum number of candidate fragments that would represent the widest coverage of ECBL. Additionally, it was decided to include as many of the 80 minifrags as possible described by O'Reilly et al.20 Up to 14 of them were found among the candidate fragments, while another 33 were found to be similar to a selected fragment (MACCS keys with a Tanimoto similarity ≥ 0.8). These 47 fragments were directly included in the final list. The remaining candidate fragments were grouped using a Butina clustering,21 as implemented in RDKit and using a cutoff of 0.1 with MACCS keys as fingerprints, resulting in 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 617 fragment clusters. All fragments in each cluster were then sorted according to their coverage of EU-OS compounds (i.e. the number of compounds in the ECBL that have the fragment as a substructure) and the following priority criteria were applied to select a fragment within each cluster: i) diversity: Tanimoto similarity of MACCS keys ≤ 0.7 to any already selected fragment; ii) coverage: above the 90th percentile of the ECBL coverage distribution for all fragments; iii) size: molecular weight (MW) under the 10th percentile of the MW distribution within the cluster; iv) solubility: log
617 fragment clusters. All fragments in each cluster were then sorted according to their coverage of EU-OS compounds (i.e. the number of compounds in the ECBL that have the fragment as a substructure) and the following priority criteria were applied to select a fragment within each cluster: i) diversity: Tanimoto similarity of MACCS keys ≤ 0.7 to any already selected fragment; ii) coverage: above the 90th percentile of the ECBL coverage distribution for all fragments; iii) size: molecular weight (MW) under the 10th percentile of the MW distribution within the cluster; iv) solubility: log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ≤ 3; v) fluorination: number of fluorine atoms above the 90th percentile of the distribution of fluorine presence in structures within the cluster. The application of these criteria led to a selection of 1785 fragment centroids that covered a substructural portion of 87.7% of the compounds in ECBL.
P ≤ 3; v) fluorination: number of fluorine atoms above the 90th percentile of the distribution of fluorine presence in structures within the cluster. The application of these criteria led to a selection of 1785 fragment centroids that covered a substructural portion of 87.7% of the compounds in ECBL.
Finally, to enhance the probability that a chosen fragment had to be a substructure of multiple ECBL compounds, and thus assuring multiple follow-up compounds to the chosen fragment, we finally ranked fragments by descending number of modification vectors. We defined modification vectors as the number of hydrogens and aromatic halogen atoms contained in the fragment which could be substituted by a heavy atom with a single bond. Naturally, not all hydrogens and halogens can be easily modified by common synthetic routes to replace them with a heavy atom, so we practically rewarded those fragments possessing hydrogens attached to heteroatoms [O,N], aromatic chlorine or bromine (also useful for MS-driven screening22) and number of fluorine atoms (for NMR-driven screening23). Implicitly, this definition also smoothed the obvious tendency to reward larger fragments against smaller ones. In the end, the application of all criteria led to a total of 1056 fragments (968 frags and 88 minifrags) that composed the final EFSL. The entire library design protocol is summarized in Fig. 1.
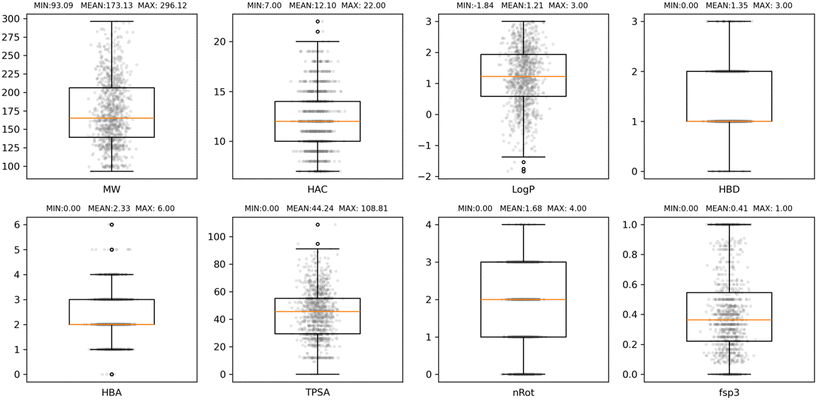
The property profile of the final EFSL composition was calculated with RDKit and included eight descriptors, namely, molecular weight (MW), heavy atom count (HAC), Wildman–Crippen log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value (Log
P value (Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P), number of hydrogen-bond donors (HBD) and acceptors (HBA), topological polar surface area (TPSA), number of rotational bonds (nRot), and the fraction of sp3 carbon atoms (fsp3). The boxplot distributions of these eight properties are collected in Fig. 2. As it can be observed, the EFSL is Ro3 compliant and fit well with the property profiles reported for some of the most recent fragment libraries developed.24–26 A closer look at the comparative property profiles of EFSL with the DSi-poised library from XChem in Oxford24 and F2X library from the Berlin Synchrotron (BESSY)25 reveals that fragments in EFSL are slightly smaller than fragments in F2X and DSi-poised (Fig. S1†).
P), number of hydrogen-bond donors (HBD) and acceptors (HBA), topological polar surface area (TPSA), number of rotational bonds (nRot), and the fraction of sp3 carbon atoms (fsp3). The boxplot distributions of these eight properties are collected in Fig. 2. As it can be observed, the EFSL is Ro3 compliant and fit well with the property profiles reported for some of the most recent fragment libraries developed.24–26 A closer look at the comparative property profiles of EFSL with the DSi-poised library from XChem in Oxford24 and F2X library from the Berlin Synchrotron (BESSY)25 reveals that fragments in EFSL are slightly smaller than fragments in F2X and DSi-poised (Fig. S1†).
Finally, the presence in the structures of EFSL fragments of the most common functional groups and linkers in bioactive molecules27–29 was also analyzed (Table S1†). It was found that each of the top-10 most common functional groups was present in at least 4.2% of the EFSL collection (that is, 44 fragments), except for the carboxyl group that was found to be particularly under-represented (0.3% of EFSL).
Quality control of the EFSL
Screening campaigns can be performed with commercial or user-defined fragment libraries. However, this requires additional structure compliance and quality control of fragments, solubility checks in buffer-of-choice for the target, verification of fragment concentration, and design of fragment mixtures. Instead, for standardization within EU-OS and beyond, we propose to use the defined and verified EFSL that consists of 1056 fragments. For this purpose, we applied NMR methods and protocols for quality assessment which were previously utilized with success and benchmarked for fragment libraries.30The fragments in EFSL were delivered as 100 mM (or 1000 mM for the minifrags) stock solutions in 100% d6-DMSO. A working stock of 50 mM (or 500 mM for the minifrags) was prepared using 90% d6-DMSO and 10% D2O. In order to confirm the integrity and also assess the solubility of the fragments, 1H-NMR spectra of the individual fragments with a final concentration of 1 mM (or 10 mM for the mini fragments) in 25 mM sodium phosphate buffer, 150 mM NaCl, 5% d6-DMSO, and 10 μM sodium trimethyl silyl propane sulfonate (DSS) at pH 7.5 were acquired. DSS was used both as an internal chemical reference and quantification standard. Using our previously established semi-automatic method of analysis,30 we categorized the fragments as either “consistent” (QC passed) or “inconsistent” (QC failed).
Out of the initial 1056 compounds, 913 (86.5%) were deemed “QC passed” and 143 (13.5%) “QC failed” (Fig. 3). The most common reasons for inconsistency included solubility issues and additional or missing signals in the spectra. In general, by NMR the concentration of the substance is determined relative to the known concentration of a standard. To achieve this, a fragment with very good solubility in the buffer was defined as an Eretic Reference.30 The concentration of this reference was set to its nominal concentration of 1 mM that was pipetted and using the automated tool Complete Molecular Confidence-assist (CMCa), the concentration of all other fragments were determined relative to this Eretic Reference. Accordingly, for the minifrags a second Eretic Reference was defined, and its nominal concentration set to 10 mM and then used to determine the relative concentration of the minifrags. All fragments were then clustered into one of three categories according to their solubility of <0.2 mM, 0.2–0.5 mM, and >0.5 mM, the minifrags stayed as their own category. It resulted in a population of the three categories of 15 fragments for <0.2 mM, 48 fragments for 0.2–0.5 mM, and 850 fragments for >0.5 mM. As a result, the identified poorly soluble fragments (<0.2 mM) and the “QC failed” fragments will be replaced by more stable and soluble fragments in a future version of the EFSL.
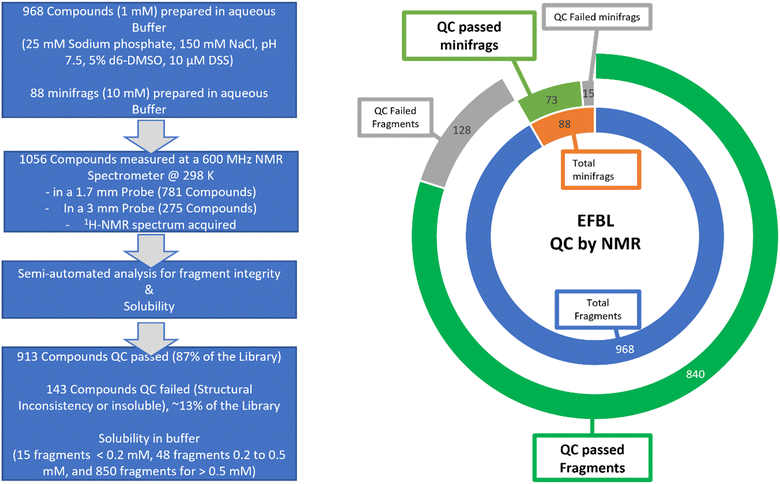 | ||
| Fig. 3 NMR-based quality control of EFSL. (Left) schematic representation of the workflow and results. (Right) doughnut graph summarizing the QC results of EFSL. | ||
Summary of screening campaigns using the EFSL
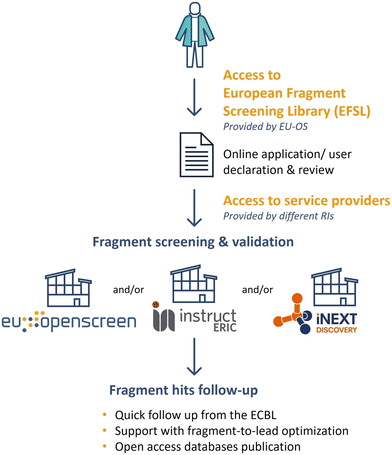
The EFSL is available to researchers for fragment screening projects at several EU-OS and iNEXT-Discovery and/or Instruct ERIC partners. The open access to EFSL is offered by the EU-OS ERIC, which acts as a single point of contact for interested users as outlined in Fig. 4. Depending on the research infrastructure (e.g. EU-OS ERIC or Instruct ERIC), users are invited to submit a full application indicating the project idea and main scientific goals. Proposals are subjected to a peer-review evaluation by independent reviewers to ensure the scientific quality of research. Additionally, users are requested to submit a user declaration form to EU-OS ERIC via the online application platform ARIA (https://apply.eu-openscreen.eu/submit-call/eu-openscreen-fragment-library--). Specific to the details of the proposals, a project can take place at one or multiple EU-OS and/or Instruct-ERIC/iNEXT-Discovery sites. After completion of the fragment screening, every researcher will be able to quickly follow up fragment hits by searching the ECBL for small molecules containing the fragment substructure of one or more fragments. Moreover, users can benefit from the access to EU-OS chemistry partners to start, in a timely and cost-effective manner, their fragment-to-lead optimisation projects. Fragment screening data will be made available to researchers using open access databases such as ECBD (https://ecbd.eu/).One of the intrinsic added assets of EFSL lies in its close link to small molecules available in ECBL, a screening collection of almost 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 diverse commercially available compounds. By consultation of the EU-OS chemistry sites, suitable small molecules from ECBL containing the fragment structures can be quickly tested in the relevant assays. Over the last three years, several research groups have benefited from access to EFSL in their screening campaigns and the statistics on the results obtained are collected in Table 1.
000 diverse commercially available compounds. By consultation of the EU-OS chemistry sites, suitable small molecules from ECBL containing the fragment structures can be quickly tested in the relevant assays. Over the last three years, several research groups have benefited from access to EFSL in their screening campaigns and the statistics on the results obtained are collected in Table 1.
| Target and/or disease area | No. of fragments screened | Primary screening technology | Hit criteria | No. of hitse | Hit rate (%) | Validation technique |
|---|---|---|---|---|---|---|
| a EFSL without the 88 minifrags. b Minimum representative selection of EFSL without the 88 minifrags. c Thirty-seven compounds were found insoluble and thus, not tested. d Nine compounds were not supplied. e Number of hits according to the hit criteria for each screening campaign. | ||||||
| Nsp3 macrodomain of SARS-CoV-2 (ref. 19) | 968a | X-ray crystallography | Crystal-bound fragment at 100 mM | 24 | 2.5% | X-ray crystallography |
| Endothiapepsin | 96b | X-ray crystallography | Crystal-bound fragment at 100 mM as determined by PanDDA maps | 30 | 31.3% | X-ray crystallography |
| Antibiotics E. coli enzyme | 1056 | Small-angle X-ray scattering | 30% of inhibited conformation | 19 | 1.8% | Enzyme kinetics: NADH formation monitored by UV absorbance |
| Nsp10/16 complex/infectious diseases | 1017c | NMR (wLOGSY, T1ρ) | WaterLOGSY – signal sign change T1ρ – at least 20% reduction in signal intensity | 132/3 (catalytic site) | 13.0%/0.3% (catalytic site) | Binders competitive with sinefungin |
| Threonyl t-RNA synthetase/antibiotics | 1056 | Thermal shift assay | ΔTm > 1.5 °C | 46 | 4.4% | N/A |
| FabF/infectious diseases | 1047d | Bio-Layer Interferometry | K D < 2 mM | 17 | 1.6% | X-ray crystallography |
| FabB/infectious diseases | 1047d | Bio-Layer Interferometry | K D < 2 mM | 20 | 1.9% | X-ray crystallography |
| PanK/infectious diseases | 1047d | Bio-Layer Interferometry | K D < 2 mM | 1 | 0.1% | N/A |
This track record analysis on the use of EFSL highlights the diversity of applicable screening technologies used and the variety of hit selection criteria of the projects. Up to five different techniques were employed in this set of eight screening projects: two opted for X-ray crystallography as primary screening technology, another three used BLI, and the remaining three applied SAXS, NMR, and TS. On average, a hit rate of 7.1% ± 10.6% was obtained considering all eight screening campaigns (2.4% ± 1.1% if the three screening campaigns with significantly lower, 0.1%, and higher, 13.0% and 31.3%, hit rates are considered as outliers).
One of the first applications of EFSL, albeit without the 88 minifrags, was in the fragment-based crystallographic screening of the Nsp3 macrodomain of SARS-CoV-2.19 Of the 968 compounds screened, crystal-bound fragments were obtained for 24 at a concentration of 100 mM. Using this criterion, a hit rate of 2.5% was obtained. The other screening project using X-ray crystallography as primary screening technology aimed at identifying fragment hits for endothiapepsin and detected 30 hits from a focused selection of 96 fragments screened (31.25% hit rate). The SAXS project used the full version of the EFSL and it identified 19 compounds with over 30% of inhibited confirmation, resulting in a hit rate of 1.8%. The only screening performed using an NMR methodology, water-ligand observed gradient spectroscopy (wLOGSY), screened a subset of EFSL (1017 compounds) and returned 132 hits for binding to Nsp10/16 complex with at least 20% reduction in signal intensity. This gives a hit rate of 13.0%, a value that is much higher than the average hit rate observed over all campaigns, but it is close to the hit rate obtained by a recent work using the same technique on other fragment libraries.26 The hit rate for fragments binding to a catalytic site of the Nsp10/16 complex was nonetheless considerably less (0.3%). The antibiotics project using a thermal shift assay identified 4.4% of the full contents of EFSL to have ΔTm > 1.5 °C. Finally, for the three screens using BLI, hit rates ranged between 1.5–2.0% for two of them when considering fragments binding with a dissociation constant of less than 2 mM. However, one of them returned a hit rate of 0.1%, much lower than the average hit rate observed over all campaigns. This low hit rate obtained for PanK could be due to the low presence in EFSL of fragments containing carboxylic acids (3/1056), which is precisely the functional group present in the substrate that is deeply buried in the pocket of the PanK binding cavity. This is an illustrative example that the use of EFSL in screening campaigns is providing valuable information on the current limitations and directions on further improvements of both EFSL and ECBL. Accordingly, future enhancements of the EFSL contemplate expanding the current library with fragments containing carboxyl groups and increasing the presence of fragments with some of the most common functional groups and linkers present in bioactive molecules of currently targeted proteins.27–29
Some of the fragment-based screening campaigns summarized in Table 1 took advantage of the poised feature of EFSL and ordered some fragment-containing compounds present in ECBL for fast hit follow-up focused activities and exploration of structure–activity relationships. The results are compiled in Table 2.
| Project/target | Technique | ECBL compounds requested | ECBL binders identified | Hit rate (%) |
|---|---|---|---|---|
| Nsp3 macrodomain of SARS-CoV-2 | X-ray crystallography | 20 | 0 crystal-bound compounds at 1 mM | 0.0% |
| Endothiapepsin | X-ray crystallography | 20 | 2 (as determined by POLDER maps) | 10.0% |
| Antibiotics E. coli enzyme | Small-angle X-ray scattering | 51 | 0 | 0.0% |
| FabF/infectious diseases | Bio-layer interferometry | 147 | 7 with KD < 100 μM | 4.8% |
Based on the 24 fragment hits that were identified from the X-ray crystallography screening of EFSL on the Nsp3 macrodomain of SARS-CoV-2, 20 compounds from ECBL were selected and delivered to Diamond Light Source (XChem group) for X-ray crystallography. Since the objective was to identify larger molecules with more potent affinities, attempts to obtain crystal-bound compounds were made using a concentration of 1 mM (100 times less than the original fragment hits). Unfortunately, this resulted in no hits for this project, an outcome that is not uncommon in real-case scenarios of unconventional protein targets and that deserve to be reported. The same outcome was obtained for the 51 compounds that were ordered from ECBL for the antibiotics E. coli enzyme. In contrast, 20 follow-up compounds from ECBL were also ordered for the endothiapepsin project and in this case 2 hits were found to have higher occupancy of the ligand in the crystal (using POLDER maps) compared to the fragment (using PanDDA maps), which is often qualitatively associated with having higher affinity. Finally, the FabF project discussed above selected 147 compounds from ECBL and this action identified 7 molecules with KD values below 100 μM, four of them with slightly more potent affinities than the original fragment hit (vide infra).
Fragment hit identification and follow-up against PaFabF
There is an urgent need for new antibiotics, in particular against P. aerugionosa and other Gram-negative bacteria.31 A promising target is beta-ketoacyl-ACP synthase 2 (FabF) which is part of the fatty acid synthesis pathway.32–34 In an effort to fuel drug discovery, we screened the EFSL against FabF from P. aeruginosa (PaFabF) using BLI. BLI is, like SPR, a label-free biosensing technology. Compared to SPR, BLI has found little use in the literature for small molecule research, but some screening campaigns have been reported.35–36 Further, we have shown that BLI is suitable for affinity determination of FabF ligands.37 Potent natural product inhibitors of FabF, such as platensimycin, bind to the acyl-bound intermediate state of the enzyme. Thus, our screening efforts included the use of the intermediate state mimicking C164A mutant described earlier in addition to the w.t. enzyme.37,38Fragments in EFSL were screened at a concentration of 500 μM using 96-well plates in a single point assay format. Both FabF variants were biotinylated and loaded to super streptavidin sensors (SSA). Biocytin blocked SSA sensors served as reference sensors. To validate the assay performance, platencin was used as control after each screening plate. In the subsequent data analysis, the mean binding response of all compounds to the reference sensors was determined and those having a response outside one standard deviation (1 STD) of the mean were rejected, leaving 617 compounds for further processing. As hit threshold, a response of at least the mean + 1 STD against FabF loaded double referenced sensor signals was chosen.
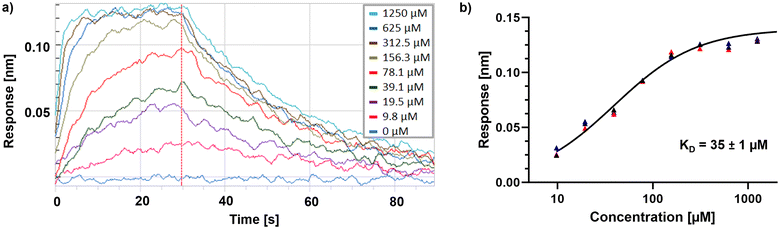
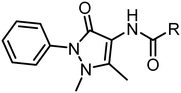
The hit lists from FabF w.t. and FabF C164A were joined and after visual inspection of the raw data, 74 compounds with values above the hit threshold (hit rate of 7.1%) were identified and progressed to dose–response experiment. Of those, 17 compounds (Table S2†) gave a KD lower than 2 mM (confirmed hit rate of 1.6%) against either the C164A or w.t. loaded sensors (Fig. S2†). One of them, fragment hit 1 (EOS102727), exhibited good affinity (KD = 9.8 μM) and selectivity to the mutant enzyme over the w.t., indicating binding close to the modified catalytic triad (Fig. S2†), and it was subsequently repurchased for retesting in triplicate and further validation using X-ray crystallography. The repurchased material of this compound gave a KD = 35 μM for FabF C164A while no binding to the w.t. up to a concentration of 2.5 mM was detectable (Fig. 5).
The cocrystal structure of 1 with FabF C164A was determined at 1.5 Å resolution (Fig. 6a and b, Table S4†). A clear electron density was obtained that allowed to place the ligand unambigously. Based on the binding mode, several favourable interactions with the FabF binding site. The oxygen of the pyrazolone carbonyl was placed in the oxyanion hole formed between the Phe400 and Ala164 backbone amide nitrogen atoms also known to harbour the alcohol of the covalently bound inhibitor cerulenin39 as well as the thioester oxygen of the natural acyl substrates40 (Fig. 6c and d). The aliphatic amide oxygen of 1 formed a hydrogen bond with the side chains of the catalytic His304 and His341, comparable to the interactions of the carboxylate group of platensimycin,37 while the amide nitrogen participated in a water network anchored to the Thr271 backbone via HOH235. The extended network was centred around HOH264, which formed a direct interaction with the Gly399 backbone, and water-bridged interactions to the Asp266 and Asn404 side chains and the His169 backbone (Fig. 6c). The phenyl group formed two T-shaped stacking interactions to the adjacent Phe203 and Phe400.
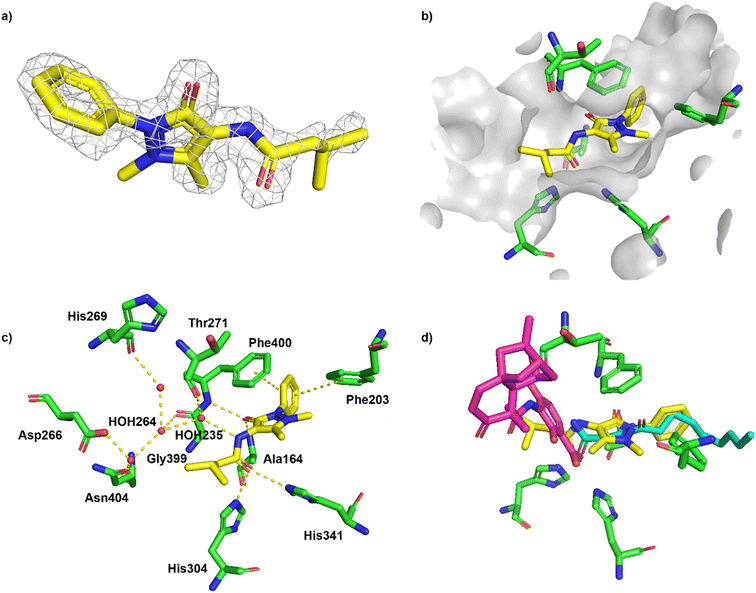 | ||
| Fig. 6 Binding of fragment hit 1 (EOS102727, yellow) to PaFabF C164A (PDB ID 8PJ0). a) |Fo − Fc| omit map of 1 binding to PaFabF C164A, contoured at 3.0 sigma. b) Solvent accessible surface using residues 4 Å from ligand 1. c) Interactions between 1 (yellow) and PaFabF C164A (green). Putative hydrogen bonds and π–π interactions are displayed as yellow dotted lines. d) Binding modes of 1 overlayed with the binding modes of platensimycin (magenta), PDB ID 7OC1, and cerulenin (teal), PDB ID 4LS8. | ||
While compound 1 makes several key interactions found in known potent inhibitors (Fig. 6d), it occupies the same space as the acyl substrate and cerulenin and it is neither binding the w.t. or the acyl intermediate (data not shown). Covalent binding of the catalytic cysteine to the acyl substrate induces a rotation of Phe400, resulting in a more accessible binding site, which is mimicked by C164A. To accommodate a ligand like 1 in the w.t. receptor, we hypothesise that a covalent attack would need to take place between Cys164 and an electrophilic analogue of 1. Efforts are ongoing to synthesize such a compound.
Subsequent hit expansion was rapidly undertaken by virtually screening the ECBL collection for analogues. This revealed 147 compounds containing the same dimethyl-phenyl-pyrazolone motif as found in the hit with variation in the pyrazolone 4-position. All 147 compounds were tested at 100 μM using the BLI assay (five times less than the concentration used in the fragment screening). Twelve compounds were progressed to a dose–response experiment, from which seven compounds had KD values below 100 μM and showed selective binding to the C164A mutant (Table S3 and Fig. S3†). Four of them were found to have slightly more potent affinities than fragment hit 1. Repurchased material for the two compounds that showed clean binding curves, hits 2 (EOS69423) and 3 (EOS21030), gave KD values of 19 and 23 μM, respectively (Table 3 and Fig. S4†). Ligand efficiencies (in kcal mol−1 per heavy atom) for hits 2 and 3 are 0.27 and 0.23, respectively, slightly below the ligand efficiency for the original fragment hit 1 (0.29).
The cocrystal structures of compound 2 and 3 with FabF C164A were determined at 1.5 Å and 2.1 Å resolution, respectively (Fig. 7a). The binding modes of the phenyl–pyrazolone moieties of 2 and 3 were identical to the mode determined for hit 1 (Fig. 7b). The aniline group of 2 was placed in the same position and adopted the same interactions as a water molecule found in the structure of 1 (HOH264, Fig. 6c), namely a direct hydrogen bond to the carbonyl group of Gly399 and water-mediated hydrogen bonds to Asp266, His269 and Asn404 (Fig. 7c). The water molecule found to bridge the amide nitrogen of 1 to Thr271 (HOH325, Fig. 6c) was not present in the complex with 2. Thus, the amide group of 2 lacked a hydrogen-bonding partner, which might explain the marginal affinity gain observed for 2 compared to 1, despite 2 forming more extend hydrogen bonds with the receptor. In contrast, 3 did not form any additional hydrogen bonds with FabF C164A, but instead the anisolyl group extended deeper into the pocket and displaced three structural water molecules that were found in this area in the complexes with 1 and 2 (Fig. 7d).
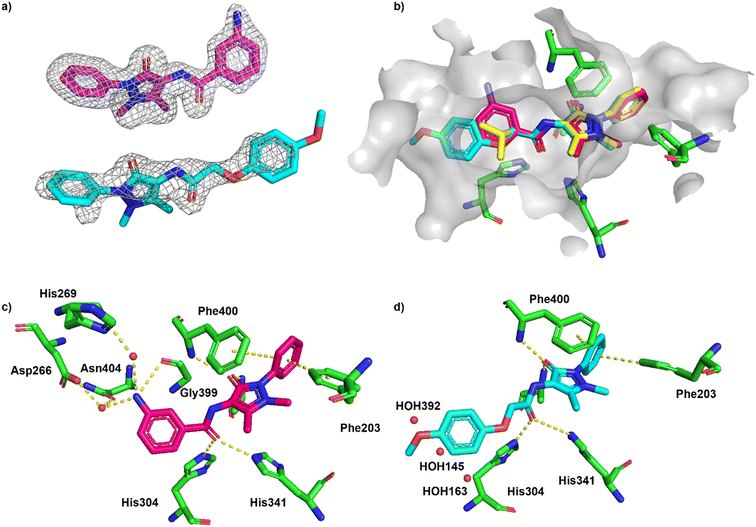 | ||
| Fig. 7 Binding of ECBL compounds 2 and 3 to PaFabF C164A (PDB ID 8R0I and 8R1V, respectively). a) |Fo − Fc| omit map of 2 (top) and 3 (bottom) binding to PaFabF C164A contoured at 3.0 sigma. b) Alignment of binding modes of 1 (yellow), 2 (magenta) and 3 (cyan) together with solvent accessible surface of the binding site using residues 6 Å from ligand 3. For clarity, only selected interacting residues from the complex PaFabF C164A-1 are shown (green). c) Interactions between 2 (magenta) and PaFabF C164A (green). Putative hydrogen bonds and π–π interactions are displayed as yellow dotted lines. d) Binding mode of 3 (cyan) together with three water molecules that were found in the complexes with 1 and 2 but displaced by 3 (HOH392, 145 and 163, numbering from PaFabF C164A-1). | ||
Conclusions
We have introduced a new library of 1056 chemical fragments (EFSL) that was purposely designed to facilitate follow-up fragment hit optimization activities through a direct substructural connection with a high-throughput screening library (ECBL) of almost 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds from commercial vendors and academic partners of the EU-OS ERIC initiative. Both compound collections are openly accessible to the scientific community and examples of their use in screening campaigns, with a range of diverse screening technologies, have demonstrated the successful application in the identification of fragment hits for unconventional protein targets as well as in the fragment progressibility and generation of structure–activity relationships in fast follow-up activities. Besides performance assessments, the general use of EFSL in those screening campaigns provided valuable information on the current strengths, limitations and future improvements of EFSL. In addition, these insights will help us to gain a better understanding of the capacities of ECBL. We will strive to continue supporting worldwide efforts on chemical biology and drug discovery by aligning the expansion of both collections with the feedback received from an increasing number of users.
000 compounds from commercial vendors and academic partners of the EU-OS ERIC initiative. Both compound collections are openly accessible to the scientific community and examples of their use in screening campaigns, with a range of diverse screening technologies, have demonstrated the successful application in the identification of fragment hits for unconventional protein targets as well as in the fragment progressibility and generation of structure–activity relationships in fast follow-up activities. Besides performance assessments, the general use of EFSL in those screening campaigns provided valuable information on the current strengths, limitations and future improvements of EFSL. In addition, these insights will help us to gain a better understanding of the capacities of ECBL. We will strive to continue supporting worldwide efforts on chemical biology and drug discovery by aligning the expansion of both collections with the feedback received from an increasing number of users.
Materials and methods
NMR-based quality control of the EFSL
BLI screening
The DNA sequence coding for PaFabF C164A was synthesized and cloned into pET-28a-TEV vectors by Genscript (New Jersey). The protein was purified and expressed using the same protocol as described for PaAviFabF C164A.37 BLI measurements were performed using an Octet RED96 instrument (FortéBio) at a constant temperature of 298 K. Wells contained a total volume of 200 μL liquid. The biotinylated proteins at a concentration of 500 μg mL−1 in assay buffer [1× PBS, 0.0001% (v/v) TritonX100, 5 mM DTT, 5% (v/v) DMSO] were immobilised onto super streptavidin (SSA) biosensors (FortéBio) as described earlier.36 Fragment DMSO stocks were transferred to polypropylene 96-well F-Bottom microplates (Greiner Bio-One) using a Mosquito liquid handler and buffer was added to a final concentration of 500 μM fragment in 1× PBS, pH = 7.4, 0.0001% (v/v) TritonX100, 5 mM DTT, 5% (v/v) DMSO in all wells besides the outer ones (rows A, H and columns 1, 12) which only contained buffer. Binding of fragments to PaAviFabF C164A, PaAviFabF and w.t. and biocytin blocked reference sensors was measured one at the time by a series of sequential steps. The screening of each plate was initiated by a 300 s washing step of the sensors in buffer, followed by an alternating sequence of measuring 30 s baseline (buffer), 30 s association (fragment containing well) and 30 s dissociation (buffer) for all compound-containing wells. 12 plates containing a total of 1047 fragments were screened in this fashion, while a plate containing platencin at a concentration of 5 μM was run after every fragment plate, serving as a positive control against the FabF C164A and negative control against the w.t. sensors. The average Z′ factor calculated per plate was 0.41. The BLI responses were single-well referenced and imported into KNIME41 (v. 4.3.1). To remove binders with strong response to the biocytin loaded reference sensors the mean single well referenced response of these single well referenced sensors was calculated, and the cut off was set to mean ± 1 STD. Fragments with a higher or lower response than this range were excluded from further analysis. Double referenced responses of the remaining fragments were obtained by subtracting the response of biocytin loaded sensors from protein loaded sensors. These were averaged over all fragments per receptor (FabF w.t. or FabF C164A) and the hit threshold was set to mean + 1 STD. The hit lists for FabF C164A and FabF w.t. were joined and after visual inspection of the raw data, 74 compounds were progressed to 5 concentration dose–response follow-up ranging from 500 to 31.25 μM using 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution and an alternating sequence of 30 s baseline, 30 s association and 30 s dissociation steps. Double referencing against biocytin sensors were applied and the last 5 seconds of the association step was used to determine responses using the FortéBio software. 17 compounds gave a KD lower than 2 mM against either FabF variant (Table S2 and Fig. S2†).
1 dilution and an alternating sequence of 30 s baseline, 30 s association and 30 s dissociation steps. Double referencing against biocytin sensors were applied and the last 5 seconds of the association step was used to determine responses using the FortéBio software. 17 compounds gave a KD lower than 2 mM against either FabF variant (Table S2 and Fig. S2†).
Screening of ECBL compounds
Tanimoto similarities between fragment hit 1 and all compounds in the ECBL collection were calculated using radial fingerprints in Canvas (https://www.schrodinger.com/training/canvas). A threshold of 0.24 was chosen resulting in 147 compounds where all structures retained the phenyl-pyrazolone core scaffold. The set was screened as described above for fragments but a fragment compound concentration of 100 μM and DMSO concentration of 1% was used. 12 compounds were advanced to a 10-concentration dose–response experiment ranging from 125 to 0.24 μM in 1.25% DMSO.K D determination for compounds 1–3
K D values were determined as the average of three experiments using the last 5 seconds of the association step of FabF C164A responses double referenced against biocytin responses. Steady state plots were obtained using GraphPad (Fig. S2 and S3†). Reported errors are standard errors. The repurchased hits were measured as 8 concentration 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilutions ranging from 1250 to 9.77 μM for hit 1, and 500 to 1.96 μM for hit 2 and 3. Ligand efficiencies were calculated using LE = −RT
1 dilutions ranging from 1250 to 9.77 μM for hit 1, and 500 to 1.96 μM for hit 2 and 3. Ligand efficiencies were calculated using LE = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln(KD)/N, where T is 298.15 K, R is the gas constant 1.9872 cal K−1 mol−1 and N is the number of non-H atoms.7
ln(KD)/N, where T is 298.15 K, R is the gas constant 1.9872 cal K−1 mol−1 and N is the number of non-H atoms.7
Chemical compounds
Fragment 1 was purchased from Crea-Chim UAB, whereas compounds 2 and 3 were purchased from Enamine Ltd and ChemBridge Corporation, respectively.X-ray crystallography of the fragment–protein complex
For ligand 1 purified PaFabF C164A (9.2 mg mL−1) was mixed with well buffer (35% PEG 3350, 0.24 M ammonium formate) at 0.6:0.3 μL drop ratio. For ligands 2 and 3PaFabF C164A (10 mg mL−1) was mixed with well buffer at 0.4:0.2 μL drop ratio. Drops were let to equilibrate at 25 °C using the vapor diffusion, sitting drop technique. Crystals were observed within 12 hours and crystals growth continued for up to 2 days. For soaking of compound 1, 100 nL of ligand solution (50 mM in DMSO) was added in the crystal drop and let to equilibrate for a maximum of 30 minutes. For compound 2, 300 nL of ligand solution (10 mM in DMSO) was added to the drop and let to equilibrate for 30 minutes while for compound 3, crystals were dipped into the ligand solution (10 mM in DMSO) for 30 seconds. Soaked crystals were then fished and frozen in liquid nitrogen. X-ray data were collected at 100 K at the P11 beamline, DESY synchrotron radiation facility in Hamburg, Germany for ligand 1, and at the ESRF beamline ID30B for compounds 2 and 3. X-ray diffraction images were processed using the auto process pipelines provided by the beamline. Dimple42 from the CCP4i2 suite43 was used for molecular replacement using the PDB 4JB6 as input model. The modelled structures were manually inspected and corrected using Coot,44 while ref. 45 was used for the refinement of the crystal structures (Table S4†). Atomic coordinates have been deposited in the PDB (8PJ0, 8R0I and 8R1V) and they will be released upon publication of the article.Accession codes
Coordinates and structure factures have been deposited at the PDB with the corresponding structure identification code: 8PJ0 (fragment hit 1), 8R0I (hit 2) and 8R1V (hit 3). The authors will release the atomic coordinates upon article publication.Abbreviations
| ECBD | European Chemical Biology Database |
| ECBL | European Chemical Biology Library |
| EFSL | European Fragment Screening Library |
| ERIC | European Research Infrastructure Consortium |
| EU-OS | EU-OPENSCREEN |
| FBDD | Fragment-based drug discovery |
| HTS | High-throughput screening |
Author contributions
X. J., A. Z. and J. M. performed the chemoinformatics analyses for library design. H. B., S. S., F. K., C. R., and H. S. obtained and analysed the NMR spectra for quality control. L. O. E. and V. Y. carried out the fragment screening against PaFabF and PaFabB. L. O. E. selected compounds for follow-up studies, performed binding assays, and drafted the section about the PaFabF use case with input from C. G. and V. Y. C. G. and V. Y. carried out X-ray crystallography with PaFabF, with input from L. O. E. E. S. formatted and provided the compounds of the EFSL and ECBL to all screening sites. T. M. and R. H. managed access from users. K. J. and A. J. performed the screening campaign against Nsp10/16 complex. P. G., H. S., R. B., A. J., A. Z., and J. M. conceived the project, guided the research, analysed all data and wrote the paper. X. J., H. B., and L. O. E. contributed equally to this work. All authors reviewed and approved the final version of the manuscript.Conflicts of interest
X. J. and J. M. are currently employees of the company Chemotargets, of which J. M. is co-founder and co-owner.Acknowledgements
The work was supported by the Research Council of Norway (RCN, grant number 273588). L. O. E. obtained a PhD studentship through the Global Challenges initiative at the University of Bergen and further support from the Meltzer Research Fund. L. O. E. made use of the Facility for Biophysics, Structural Biology and Screening at the University of Bergen (BiSS), which received funding from the RCN through the NORCRYST (grant number 245828) and NOR-OPENSCREEN (grant number 245922) consortia. V. Y. is funded by the Western Norway Regional Health Authority (Helse Vest; program number F-12602). Work at BMRZ (HS) is supported by the EU project Fragment-Screen, grant agreement ID: 101094131 and by the state of Hesse. A. J. thanks the European Regional Development Fund (Agreement Nr. 1.1.1.1/19/A/019) for financial support. J. M. acknowledges the financial support of the Spanish Ministerio de Ciencia, Innovacion y Universidades (program number PID2020-112539RB-I00). The fragment library and the implementation of the fragment-screen workflow at EU-OPENSCREEN was funded by the EC through the H2020 project EU-OPENSCREEN-DRIVE (grant number 8238939). The collaboration between EU-OPENSCREEN and fragment screening facilities was supported by the EU project iNEXT-Discovery (grant number 871037). We thank Khan Kim Dao for the excellent support with protein purification and Samuel David Sutton for the PanK screening. We also thank all members of the EU-OPENSCREEN ERIC for their contribution to the EU-OS screening platform, chemistry libraries and informatics infrastructure. We thank members of the EU-OPENSCREEN-DRIVE, Instruct ERIC and iNEXT-Discovery consortia for their consultation on the design of the fragment library (Mads H. Clausen, Josan Marquez, Anastassis Perrakis, Manfred S. Weiss and Frank von Delft). We thank EU-OS chemistry sites for providing consultation to users on the compounds follow-up: Camen Gil and Ana Martinez from the Center for Biological Research Margarita Salas (CSIC). We also thank users of the EU-OS fragment library and fragment screening sites for providing summary information of screening campaigns: Manfred Weiss and Jan Wollenhaupt from the Helmholtz-Zentrum Berlin, Germany; Rubén Martínez Buey from University of Salamanca, Spain; Emilio Parisini from the Latvian Institute of Organic Synthesis, Latvia; Ivan Ahel, University of Oxford, UK; Frank von Delf, Daren Fearon and Nathan Cowieson from the Diamond Light Source Ltd, UK. We thank Martin Neuenschwander for his support during the acquisition of the fragments contained in EFSL.References
- J. Quancard, A. Vulpetti, A. Bach, B. Cox, S. M. Guéret, I. V. Hartung, H. F. Koolman, S. Laufer, J. Messinger, G. Sbardella and R. Craft, ChemMedChem, 2018, 18, e202300002 CrossRef PubMed.
- D. A. Erlanson, S. W. Fesik, R. E. Hubbard, W. Jahnke and H. Jhoti, Nat. Rev. Drug Discovery, 2016, 15, 605–619 CrossRef CAS PubMed.
- A. Bancet, C. Raingeval, T. Lomberget, M. Le Borgne, J.-F. Guichou and I. Krimm, J. Med. Chem., 2020, 63, 11420–11435 CrossRef CAS PubMed.
- N. S. Troelsen and M. H. Clausen, Chem. – Eur. J., 2020, 26, 11391–11403 CrossRef CAS PubMed.
- W. Jahnke, D. A. Erlanson, I. J. P. de Esch, C. N. Johnson, P. N. Mortenson, Y. Ochi and T. Urushima, J. Med. Chem., 2020, 63, 15494–15507 CrossRef CAS PubMed.
- M. Bon, A. Bilsland, J. Bower and K. McAulay, Mol. Oncol., 2022, 16, 3761–3777 CrossRef CAS PubMed.
- A. L. Hopkins, C. R. Groom and A. Alex, Drug Discovery Today, 2004, 9, 430–431 CrossRef PubMed.
- T. Fink, H. Gruggesser and J. L. Reymond, Angew. Chem., Int. Ed., 2005, 44, 1504–1508 CrossRef CAS PubMed.
- B. J. Davis, Methods Mol. Biol., 2021, 2263, 247–270 CrossRef CAS PubMed.
- A. Chavanieu and M. Pugnière, Expert Opin. Drug Discovery, 2016, 11, 489–499 CrossRef CAS PubMed.
- C. A. Wartchow, F. Podlaski, S. Li, K. Rowan, X. Zhang, D. Mark and K.-S. Huang, J. Comput.-Aided Mol. Des., 2011, 25, 669–676 CrossRef CAS PubMed.
- H. L. Silvestre, T. L. Blundell, C. Abell and A. Ciulli, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12984–12989 CrossRef CAS PubMed.
- S. J. Pollack, K. S. Beyer, C. Lock, I. Müller, D. Sheppard, M. Lipkin, D. Hardick, P. Blurton, P. M. Leonard, P. A. Hubbard, D. Todd, C. M. Richardson, T. Ahrens, M. Baader, D. O. Hafenbradl, K. Hilyard and R. W. Bürli, J. Comput.-Aided Mol. Des., 2011, 25, 677–687 CrossRef CAS PubMed.
- M. D. Duong-Thi, M. Bergström, T. Fex, R. Isaksson and S. Ohlson, J. Biomol. Screening, 2013, 18, 160–171 CrossRef PubMed.
- G. M. Keserü, D. A. Erlanson, G. G. Ferenczy, M. M. Hann, C. W. Murray and S. D. Pickett, J. Med. Chem., 2016, 59, 8189–8206 CrossRef PubMed.
- H. Jhoti, G. Williams, D. C. Rees and C. W. Murray, Nat. Rev. Drug Discovery, 2013, 12, 644 CrossRef CAS PubMed.
- S. C. C. Lucas, et al. , RSC Med. Chem., 2022, 13, 1052–1057 RSC.
- D. Horvath, M. Lisurek, B. Rupp, R. Kühne, E. Specker, J. P. von Kries, D. Rognan, C. D. Andersson, F. Almqvist, M. Elofsson, P.-A. Enqvist, A.-L. Gustavsson, N. Remez, J. Mestres, G. Marcou, A. Varnek, M. Hibert, J. Quintana and R. Frank, ChemMedChem, 2014, 9, 2309–2326 CrossRef CAS PubMed.
- M. Schuller, et al. , Sci. Adv., 2021, 7, eabf8711 CrossRef PubMed.
- M. O'Reilly, A. Cleasby, T. G. Davies, R. J. Hall, R. F. Ludlow, C. W. Murray, D. Tisi and H. Jhoti, Drug Discovery Today, 2019, 24, 1081–1086 CrossRef PubMed.
- D. Butina, J. Chem. Inf. Comput. Sci., 1999, 39, 747–750 CrossRef CAS.
- C. Tang, J. Tan, Y. Fan, K. Zheng, Z. Yu and X. Peng, RSC Adv., 2020, 10, 13749 RSC.
- N. S. Troelsen, et al. , Angew. Chem., Int. Ed., 2019, 59, 2204–2210 CrossRef PubMed.
- O. B. Cox, et al. , Chem. Sci., 2016, 7, 2322–2330 RSC.
- J. Wollenhaupt, A. Metz, T. Barthel, G. M. A. Lima, A. Heine, U. Mueller, G. Klebe and M. S. Weiss, Structure, 2020, 28, 694–706 CrossRef CAS PubMed.
- M. Bührmann, et al. , J. Med. Chem., 2023, 66, 6297–6314 CrossRef PubMed.
- N. Sánchez-Cruz, B. A. Pilón-Jiménez and J. L. Medina-Franco, F1000Research, 2019, p. 8, Chem Inf Sci-2071 Search PubMed.
- P. Ertl, E. Altmann and J. M. McKenna, J. Med. Chem., 2020, 63, 8408–8418 CrossRef CAS PubMed.
- P. Ertl, E. Altmann and S. Racine, Bioorg. Med. Chem., 2023, 81, 117194 CrossRef CAS PubMed.
- S. Sreeramulu, et al. , J. Biomol. NMR, 2020, 74, 555–563 CrossRef CAS PubMed.
- E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans, Y. Carmeli, M. Ouellette, K. Outterson, J. Patel, M. Cavaleri, E. M. Cox, C. R. Houchens, M. L. Grayson, P. Hansen, N. Singh, U. Theuretzbacher and N. Magrini, Lancet Infect. Dis., 2018, 18, 318–327 CrossRef PubMed.
- J. Yao and C. O. Rock, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2017, 1862, 1300–1309 CrossRef CAS PubMed.
- L. Bibens, J.-P. Becker, A. Dassonville-Klimpt and P. Sonnet, Pharmaceuticals, 2023, 16, 425 CrossRef CAS PubMed.
- L. O. Espeland, C. Georgiou, R. Klein, H. Bhukya, B. E. Haug, J. Underhaug, P. S. Mainkar and R. Brenk, ChemMedChem, 2021, 16, 2715–2726 CrossRef CAS PubMed.
- S. Bothe, P. Hänzelmann, S. Böhler, J. Kehrein, M. Zehe, C. Wiedemann, U. A. Hellmich, R. Brenk, H. Schindelin and C. Sotriffer, Commun. Chem., 2022, 5, 169 CrossRef CAS PubMed.
- C. A. Wartchow, F. Podlaski, S. Li, K. Rowan, X. Zhang, D. Mark and K.-S. Huang, J. Comput.-Aided Mol. Des., 2011, 25, 669–676 CrossRef CAS PubMed.
- L. O. Espeland, C. Georgiou, R. Klein, H. Bhukya, B. E. Haug, J. Underhaug, P. S. Mainkar and R. Brenk, ChemMedChem, 2021, 16, 2715–2726 CrossRef CAS PubMed.
- S. B. Singh, et al. , Bioorg. Med. Chem. Lett., 2009, 19, 4756–4759 CrossRef CAS PubMed.
- F. Trajtenberg, S. Altabe, N. Larrieux, F. Ficarra, D. De Mendoza, A. Buschiazzo and G. E. Schujman, FEBS J., 2014, 281, 2324–2338 CrossRef CAS PubMed.
- J. Wang, et al. , Nature, 2006, 441, 358–361 CrossRef CAS PubMed.
- M. R. Berthold, N. Cebron, F. Dill, T. R. Gabriel, T. Kötter, T. Meinl, P. Ohl, C. Sieb, K. Thiel and B. Wiswedel, in Studies in Classification, Data Analysis, and Knowledge Organization, Springer GfKL, 2007 Search PubMed.
- M. Wojdyr, R. Keegan, G. Winter and A. Ashton, Acta Crystallogr., Sect. A: Found. Crystallogr., 2013, 69, s299 Search PubMed.
- L. Potterton, J. Agirre, C. Ballard, K. Cowtan, E. Dodson, P. R. Evans, H. T. Jenkins, R. Keegan, E. Krissinel, K. Stevenson, A. Lebedev, S. J. McNicholas, R. A. Nicholls, M. Noble, N. S. Pannu, C. Roth, G. Sheldrick, P. Skubak, J. Turkenburg, V. Uski, F. von Delft, D. Waterman, K. Wilson, M. Winn and M. Wojdyr, Acta Crystallogr., Sect. D: Struct. Biol., 2018, 74, 68–84 CrossRef CAS PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- G. N. Murshudov, P. Skubák, A. A. Lebedev, N. S. Pannu, R. A. Steiner, R. A. Nicholls, M. D. Winn, F. Long and A. A. Vagin, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 355–367 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Most frequent functional groups in EFSL, structures of the 17 fragment hits from the BLI screening of EFSL in the FabF project, structures of the 7 hits poised from the ECBL in the FabF project, X-ray data collection and refinement statistics, distributions of eight calculated descriptors for different fragment libraries, sensorgrams and steady-state plots of the 17 fragment hits from the BLI screening of EFSL in the FabF project, sensorgrams and steady-state plots of the 7 hits poised from the ECBL in the FabF project, steady-state plots in triplicate of repurchased material for the two co-crystallised poised hits. See DOI: https://doi.org/10.1039/d3md00724c |
| ‡ X. J., H. B., and L. O. E. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |