DOI:
10.1039/D3MO00141E
(Research Article)
Mol. Omics, 2024,
20, 19-26
Unraveling the role of intra-cellular metabolites in the lactic acid production by novel Bacillus amyloliquefaciens using sugarcane molasses as a substratum†
Received
19th July 2023
, Accepted 30th August 2023
First published on 31st August 2023
Abstract
Lactic acid is a versatile, multi-functional organic monomer in various industries, creating worldwide demand. High titer lactic acid production was achieved by novel Bacillus amyloliquefaciens J2V2AA through sugarcane molasses fermentation up to 178 mg mL−1. A metabolomics approach such as combined GC-MS and LC-MS was applied to elucidate the involvement of key metabolites in lactic acid production. The results revealed the participation of 58 known intra-cellular metabolites at various pathways in lactic acid production. Twenty-eight highly up-regulated and down-regulated metabolites were analyzed, and a schematic diagram of a possible lactic acid production pathway was proposed. The produced lactic acid was analyzed through FTIR, UV-Spectrum, and HPLC analysis.
Introduction
Lactic acid is an essential, multi-functional organic acid precursor for producing biodegradable biopolymers or bio-plastics in polylactic acid.1 Besides being a monomer for polymer build-ups, lactic acid has also been used as a preservative, acidulate, food flavour, and base material in the textile and pharmaceutical industries.2 Due to its versatile nature, lactic acid has been employed for various industrial purposes, raising its demand in the market. It has been cumbersome for the scientific community to produce optically pure lactic acid, which could only be obtained by microbial fermentation. The latest research focuses on lactic acid production using lignocellulosic substrates.3–8 Sugarcane molasses is a substratum used widely to increase lactic acid production through fermentation. Globally, 6.4 million tons of molasses (4%) are generated from 160 million tons of sugar produced.9,10 Molasses contains 75–85% of total dissolved solids, 30–35% sucrose sugars, 10–17% (fructose and glucose), and 10–16% ash and other contents.11 Though lactic acid production has significantly increased, it cannot meet the current market demand. Understanding the metabolomics of pathways involved in lactic acid production through microbial fermentation is paramount to meeting the current and future market demand.12–14 Understanding the metabolomics pathway is the proper way to meet market demand.15–17 Metabolomics is a powerful tool that props up the scientific community to design target-oriented upstream processes such as TILLING (Targeting Induced Local Lesions in Genomes), mutagenesis library construction, genome mining, and over-expression18–21 or knock-out approaches through CRISPR – Cas 9 and downstream processes such as the utilization of agro-industrial waste, food, and beverages wastes would be possible.22–24 Therefore, the present study is designed to unravel the role of metabolites in lactic acid production through microbial fermentation of sugarcane molasses using combined GC-MS and LC-MS analysis.
Materials and methods
Cultivation of the lactic acid-producing bacteria
Bacillus amyloliquefaciens J2V2AA used in this experiment were screened, identified, and characterized for high lactic acid production by our previous work.25 The seed culture was grown in 50 mL MRS medium in a 100 mL conical flask at 37 °C, 150 rpm for 24 hours.
Optimization of lactic acid production using sugarcane molasses as a carbon source
The sugarcane molasses collected from District Co-Operation Sugarcane Industry, Madurai, Tamil Nadu, was directly mixed with MRS medium in different concentrations and autoclaved at 121 °C for 15 minutes. Different concentrations of sugarcane molasses (5% to 30% v/v) were employed to identify the best combination for higher lactic acid production using the best lactic acid-producing bacteria.25 Sugarcane molasses of 30% showed higher lactic acid production. Thus, cultures grown in control MRS, and MRS media supplemented with 30% sugarcane molasses (100 mL) inoculated with 10% inoculum incubated for 24 hours at 150 rpm and 37 °C were taken to analyze the metabolites. All the experiments were performed in triplicate.
Sampling and quenching of the intra-cellular metabolites
A 20 mL bacterial broth triplicate sample was collected from the fermentation broth (normal MRS media and MRS media supplemented with 30% sugarcane molasses) and centrifuged at 8500 rpm for 15 minutes at −4 °C and the collected pellets containing bacterial cells were washed twice with 10 mL of 1% saline solution to remove the debris and the filtrate was immediately transferred to a 50 mL centrifuge tube along with 20 mL of 60% methanol pre-chilled at −80 °C. Quenching of the metabolism was done in 10–20 seconds. Then, the mixture was centrifuged at 8500 rpm for 5 minutes at −4 °C. The pellet was stored at −80 °C until further analysis.18,26,27
Extraction of the intra-cellular metabolites
The cell pellet was retrieved from the −80 °C freezer after 48 hours, suspended with 2.5 mL of ice-cold methanol–water solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and thawed in an ice bathtub for 5 minutes then rigorously vortexed for a couple of minutes and frozen at −80 °C for 30 minutes. This cycle was repeated thrice and centrifuged at 10
1), and thawed in an ice bathtub for 5 minutes then rigorously vortexed for a couple of minutes and frozen at −80 °C for 30 minutes. This cycle was repeated thrice and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 minutes at −20 °C. An additional 2.5 mL of ice-cold methanol–water solution was added to the cell pellets and combined with the former. This supernatant was lyophilized at −88 °C condensation with a 1.000 Pa vacuum for 24 hours and used for further LC-MS and GC-MS analysis.18,26,27
000 rpm for 20 minutes at −20 °C. An additional 2.5 mL of ice-cold methanol–water solution was added to the cell pellets and combined with the former. This supernatant was lyophilized at −88 °C condensation with a 1.000 Pa vacuum for 24 hours and used for further LC-MS and GC-MS analysis.18,26,27
Derivatization of intra-cellular metabolites
The samples were subjected to a two-step derivatization procedure. Firstly, 200 μL of methoxyamine pyridine hydrochloride was mixed with the sample for 30 seconds and incubated at 37 °C for 90 minutes without disturbance. Secondly, 200 μL of N,O-bistrifluoroacetamide was added to the mixture, incubated at 70 °C for 30 minutes, and incubated at room temperature for 30 minutes. The derivatized samples were transferred to a GC-MS vial for analysis.27,28
GC-MS analysis
The GC-MS coupled Shimadzu Q.P. 2020 system was used in this study. As the carrier gas, helium was used, with a flow rate of 1.20 mL min−1 at the pressure of 68.1 kPa with a total flow of 16.2 mL min−1. 1 μL of derivatized samples were injected in GC-MS at a split ratio of 10.0. The initial capillary column temperature was 50 °C for 20 minutes, then rose with a linear velocity to 280 °C in 40.33 minutes and was held at 280 °C for 2 minutes. Mass spectra were recorded from 50 m/z to 500 m/z at an acquisition rate of 0.30 seconds per spectra. The MS spectra were matched with standard libraries NIST14.LIB and WILEY8.LIB.
LC-MS analysis
The LC-MS coupled Agilent LC/Q-TOF 6530 system was used in this study. A mass analyzer with an electronic spray of E.S.I. source methanol was used as a mobile phase, with a constant flow rate of 0.2 mL min−1. 5 μL of the sample was injected in negative ionization mode, 175.0 V fragmentation, 0.3–0.7 min retention time, scanned between 100–3000 m/z, the pressure was 50 psi, and the nebulizer flow rate was 10 mL min−1. A 300 °C nebulizer gas temperature was used for the nitrogen carrier. Functional metabolite abundance differences were assessed using the KEGG Database and the heat map generated using HemI 2.0.18,29
UV-visible and FT-IR spectrum analysis of the produced lactic acid
The FTIR spectrophotometer (Bruker, Alpha II Model Advanced, Berlin, Germany) measured the spectrum ranges from 500 to 4000 nm for 100 μL of the separated liquid LA sample. According to the instructions in the instrument manual, the standard smoothening, baseline correction, atmospheric compensation, and peak picking were carried out. To identify functional groups, the FTIR spectrum is used. The 200–800 nm range was scanned for the separated LA in a UV-vis spectrophotometer. Additionally, the analysis of the fermentation broth's lactic acid determination followed previously reported30 instructions.
Statistical analysis
The statistical analysis was performed using Origin 8.5, and principal component analysis (PCA) was performed using OriginPro 2022b.
Results and discussion
From the author's previous study, the comparison between control MRS media and MRS media enriched with 30% sugarcane molasses showed significantly improved lactic acid production up to 178 mg mL−1.25 In this study, the intra-cellular metabolites comparison between control MRS media and MRS media enriched with 30% sugarcane molasses was analyzed using LC-MS and GC-MS analysis. A total of 58 intra-cellular metabolites were detected using LC-MS and GC-MS analysis. Twenty-three highly up-and down-regulated intra-cellular metabolites were identified through LC-MS analysis, followed by 35 intra-cellular metabolites detected by GC-MS analysis from 24 hours control and MRS media enriched with 30% sugarcane molasses fermentation samples. From the LC-MS analysis, 11 metabolites are up-regulated in the treated sample compared to the control sample, and 12 are down-regulated. Hierarchical cluster analysis and PCA analysis of the metabolites detected through LC-MS analysis (Fig. 1, 3 and Fig. S4, ESI†) revealed the presence of relatively abundant ribose (Fig. 3, metabolite 1) in the control than in the treated (Fig. S1 and S2, ESI†). Ribose is the “molecular currency of the cell” due to its NAD, FAD, and NADP energy transfers as they act as electron acceptors in the fermentation metabolic pentose phosphate pathway.31,32
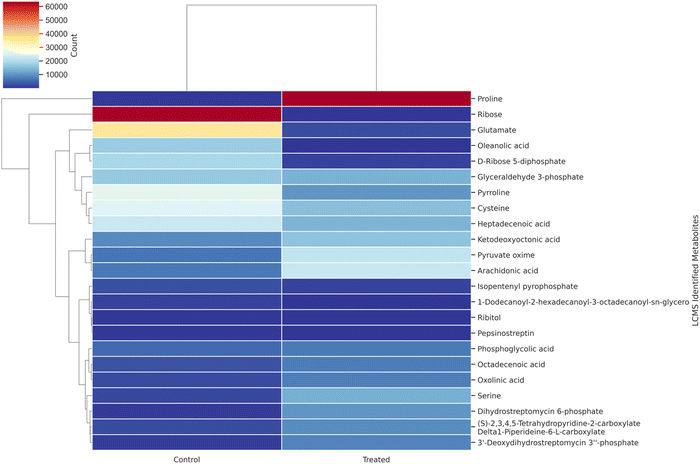 |
| | Fig. 1 Hierarchical cluster analysis of LC-MS metabolite correlation in control (MRS medium) and treated samples (MRS supplemented with 30% sugarcane molasses). | |
Also, ribose acts on nucleic acid synthesis, thus leading to vigorous biomass production. The results are comparable to the previous study.33 The higher glucose utilization in sugarcane molasses produces more ribose, promoting more biomass and lactic acid production. A similar trend was also obtained in the pyruvate (Fig. 3, metabolite 16), which was abundant in the treated rather than the control. The same trend continued at the serine level; a 6-fold increase was found in the treated versus control sets and the more abundant serine could be converted directly into pyruvate through serine deaminase,28 thus leading to the production of lactic acid. Also, 13 fold increases were found in proline (Fig. 3, metabolite 23) in the treated samples compared with the control. Increased proline levels in the intra-cellular metabolites contribute to the osmoregulation of the bacterial cells, thus protecting bacterial cells from managing stress.33,34
Interestingly, pyrroline (Fig. 3, metabolite 8), an intermediate compound in the heterofermentative lactic acid pathway,35 was detected in both the control and treated samples, indicating that this strain, Bacillus amyloliquefaciens, is a heterofermentative bacteria. Meanwhile, abundant other vital metabolites such as phosphoglycolic acid and glyceraldehyde-3-phosphate in the Embden–Meyerhof–Parnas Pathway (EMP pathway) were detected. The EMP pathway provides lactic acid-producing precursors.33 High flux in the precursor compounds shows the maximized production of lactic acid due to flux.33 Simultaneously, heptadecenic acid and octadecenic acid were found to involve fatty acid synthesis36 and promote increased biomass production, resulting in higher lactic acid levels.33
Similar results were observed in the hierarchical cluster analysis and PCA analysis of intra-cellular metabolites detected through GC-MS analysis (Fig. 2 and Fig. S3, ESI†). Hexadecanoic acid, octadecanic acid, and decanoic acid were detected. Unsaturated fatty acids play a substantial role in stress tolerance and the perpetuation of the microbial community.26 Hexadecanoic and octadecanoic acids are oxygen and NADH-dependent and become a precursor for oleic and palmitelaidic acids. These two polyunsaturated fatty acids are responsible for overcoming the inhibitory effects of other chemical compounds in the lignocellulosic substratum and cell viability.26 GC-MS detected metabolites besides the EMP pathway and the Pentose Phosphate Pathway (PPP pathway). PCA analysis was performed and analyzed (Fig. 2, 4 and Tables S1, S2, ESI†).
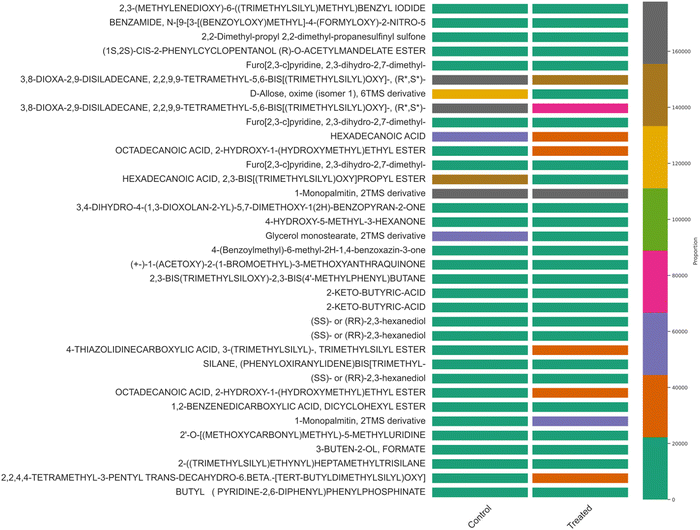 |
| | Fig. 2 Hierarchical cluster analysis of GC-MS metabolite correlation in control (MRS medium) and treated samples (MRS supplemented with 30% sugarcane molasses). | |
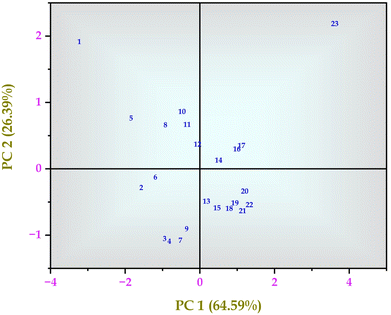 |
| | Fig. 3 Principal component analysis (PCA) of intra-cellular metabolites of Bacillus amyloliquefaciens J2V2AA identified by LC-MS spectrum. | |
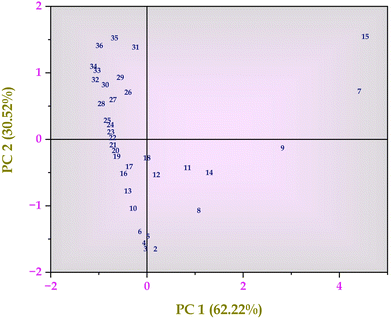 |
| | Fig. 4 Principal component analysis (PCA) of intra-cellular metabolites of Bacillus amyloliquefaciens J2V2AA identified by GC-MS spectrum. | |
Based on the GC-MS and LC-MS metabolomic profiling of Bacillus amyloliquefaciens J2V2AA lactic acid production through sugarcane molasses fermentation, a possible schematic metabolic pathway has been elucidated (Fig. 5). Differences in the intra-cellular metabolites and the path are represented below, indicating a green upside arrow for the up-regulation and a red downside arrow for the down-regulation while comparing the control. The produced lactic acid has been identified by UV-visible and FTIR spectra (Fig. 6 and 7). One carboxylic acid with a hydroxyl group makes up the bi-functional molecule called lactic. The microbially produced lactic acid was removed from the MRS media through downstream processing. The LA was examined through FT-IR analysis and comparison with standard lactic acid. According to Fig. 6, the stretching of the –OH bond was seen at 3396 cm−1 in LA. The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O lengths are represented by bands at 1700 cm−1 and 1200 cm−1, respectively.37 A UV-visible spectrophotometer was used to characterize the microbially fermented lactic acid further. Lactic acid can be detected in the sample as a peak between 200 and 300 nm.25 Understanding of the metabolic pathway will open up the arena to other omics approaches to play a role to enhance the quantity and quality of the lactic acid production. The modern revolutionary genomic tool CRISPR-cas9 genome editing tool has been used to metabolically engineer the lactic acid production from glucose and cellobiose as well as xylose.22,24 Understanding the role of metabolites in each step will lead to the addition or deletion/knock-out of the appropriate gene from the bacteria to enhance lactic acid production.20,23
O and C–O lengths are represented by bands at 1700 cm−1 and 1200 cm−1, respectively.37 A UV-visible spectrophotometer was used to characterize the microbially fermented lactic acid further. Lactic acid can be detected in the sample as a peak between 200 and 300 nm.25 Understanding of the metabolic pathway will open up the arena to other omics approaches to play a role to enhance the quantity and quality of the lactic acid production. The modern revolutionary genomic tool CRISPR-cas9 genome editing tool has been used to metabolically engineer the lactic acid production from glucose and cellobiose as well as xylose.22,24 Understanding the role of metabolites in each step will lead to the addition or deletion/knock-out of the appropriate gene from the bacteria to enhance lactic acid production.20,23
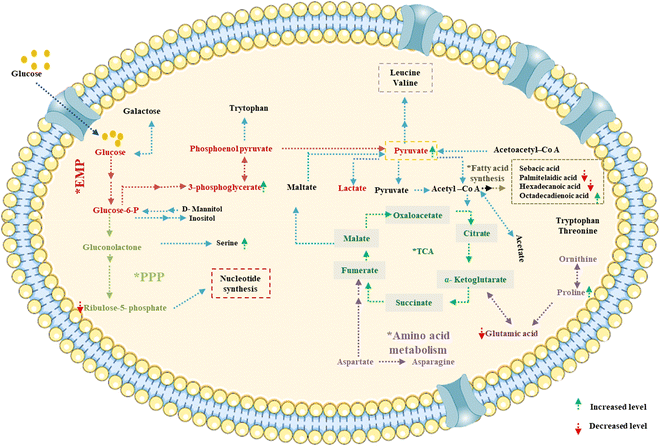 |
| | Fig. 5 Predicted metabolic pathway of lactic acid production by Bacillus amyloliquefaciens J2V2AA utilizing sugarcane molasses. | |
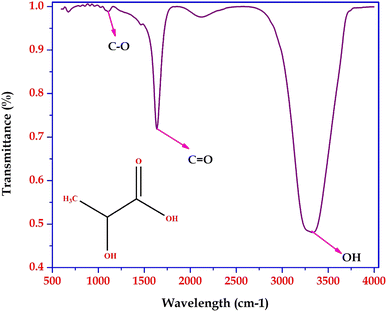 |
| | Fig. 6 FTIR spectrum of lactic acid produced by Bacillus amyloliquefaciens J2V2AA. | |
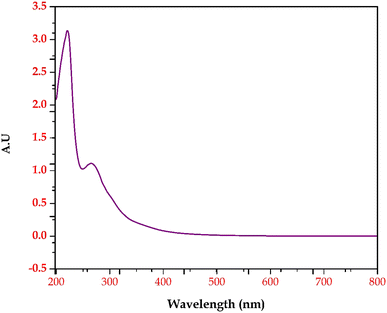 |
| | Fig. 7 UV-visible spectrum of lactic acid produced by Bacillus amyloliquefaciens J2V2AA. | |
Conclusion
A metabolomics profiling of microbial fermentation done by Bacillus amyloliquefaciens showed insightful knowledge of the metabolomics approach to improve the desired level of lactic acid production using sugarcane molasses. Other than the reported and discussed metabolite profiling, about 20 unknown metabolites were also found. Furthermore, to unravel the mechanism of action of the novel metabolite lactic acid production, extensive research has to be done with isobaric tags for relative and absolute quantitation (iTRAQ). Also, it equips the data with combined transcriptomics and metabolomics profiling during the entire fermentation period, which is paramount.
Data availability
Supplementary data for this work can be found online alongside the paper.
Author contributions
B. V. K.: conceptualization; data curation; formal analysis; methodology; writing – original draft; writing – review and editing B. M.: formal analysis; methodology; M. K.: formal analysis; methodology; J. K. J. P. K.: formal analysis; methodology; M. J. B.: conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; supervision; validation; visualization; writing – review and editing. All authors have read and agreed to the published version of the manuscript.
Abbreviation
| LA | Lactic acid |
| SM | Sugarcane molasses |
| BCP | Bromocresol purple |
| MRS | de Man, rogosa & sharpe |
| FM | Fermentation media |
| LAB | Lactic acid-producing bacteria |
| NAD | Nicotinamide adenine dinucleotide |
| FAD | Flavin adenine dinucleotide |
| NADP | Nicotinamide adenine dinucleotide phosphate |
| HPLC | High-performance liquid chromatography |
| FTIR | Fourier-transform infrared spectroscopy |
| GC-MS | Gas chromatography-mass spectrometry |
| LC-MS | Liquid chromatography-mass spectrometry |
| Q-TOF | Quadrupole time-of-flight |
| E. M. P | Embden, Meyerhof, and Parnas pathway |
| P. P. P | Pentose phosphate pathway |
| KEGG | Kyoto encyclopedia of genes and genomes |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
All the authors thank DST-PURSE, Department of Botany (Phase II—3rd Installment), and RUSA 2.0 (letter no. F. 24-51/2014-U, Policy (TN-Multi-Gen), Dept. of Edn. Govt. of India, Dt. 09.10.2018), Alagappa University for the financial support of this research work, and also Research Instrumentation facilities of Department of Botany, Department of Animal Health and Management, Department of Microbiology, Alagappa University, LC-MS Facility, and SEM facility, The Gandhigram Rural Institute – Deemed to be University, GC-MS Facility, Heber analytical instrumentation facility (HAIF), Bishop Heber College are significantly acknowledged. Special thanks go to B. Vaseeharan, NM. Prabhu, K. Pannerselvam, M. Vimalraj, M. Kannan, J. Jeyavani, R. Rajagopal, S. Sivaranjani, M. Jeyaprabha, S. Sudhabose, M. Manikandakrishnan and D. Subashri for their academic support. This research received no specific grant from public, commercial, or not-for-profit funding agencies.
References
- L. F. Coelho, C. J. B. De Lima, M. P. Bernardo and J. Contiero,
D(−)-Lactic Acid Production by Leuconostoc Mesenteroides B512 Using Different Carbon and Nitrogen Sources, Appl. Biochem. Biotechnol., 2011, 164(7), 1160–1171, DOI:10.1007/s12010-011-9202-6.
- A. Dumbrepatil, M. Adsul, S. Chaudhari, J. Khire and D. Gokhale, Utilization of Molasses Sugar for Lactic Acid Production by Lactobacillus Delbrueckii Subsp. Delbrueckii Mutant Uc-3 in Batch Fermentation, Appl. Environ. Microbiol., 2008, 74(1), 333–335, DOI:10.1128/AEM.01595-07.
- M. A. Abdel-Rahman, Y. Tashiro and K. Sonomoto, Lactic Acid Production from Lignocellulose-Derived Sugars Using Lactic Acid Bacteria: Overview and Limits, J. Biotechnol., 2011, 156(4), 286–301, DOI:10.1016/j.jbiotec.2011.06.017.
- M. A. Abdel-Rahman, S. E. D. Hassan, A. Fouda, A. A. Radwan, M. G. Barghoth and S. G. Desouky, Evaluating the Effect of Lignocellulose-Derived Microbial Inhibitors on the Growth and Lactic Acid Production by Bacillus Coagulans Azu-10, Fermentation, 2021, 7(1), 17, DOI:10.3390/fermentation7010017.
- H. Azaizeh, H. N. Abu Tayeh, R. Schneider, A. Klongklaew and J. Venus, Production of Lactic Acid from Carob, Banana and Sugarcane Lignocellulose Biomass, Molecules, 2020, 25(13), 2956, DOI:10.3390/molecules25132956.
- Y. Li, S. S. Bhagwat, Y. R. Cortés-Penã, D. Ki, C. V. Rao, Y. S. Jin and J. S. Guest, Sustainable Lactic Acid Production from Lignocellulosic Biomass, ACS Sustainable Chem. Eng., 2021, 9(3), 1341–1351, DOI:10.1021/acssuschemeng.0c08055.
-
E. O. Ajala; Y. O. Olonade; M. A. Ajala and G. S. Akinpelu, Lactic Acid Production from Lignocellulose – A Review of Major Challenges and Selected Solutions. ChemBioEng Reviews, Wiley-Blackwell, 2020. DOI:10.1002/cben.201900018.
- E. Cubas-Cano, C. González-Fernández, M. Ballesteros and E. Tomás-Pejó, Biotechnological Advances in Lactic Acid Production by Lactic Acid Bacteria: Lignocellulose as Novel Substrate, Biofuels, Bioprod. Biorefin., 2018, 12(2), 290–303, DOI:10.1002/bbb.1852.
- A. Vidra, A. J. Tóth and Á. Németh, Lactic Acid Production from Cane Molasses, Waste Treat. Recovery, 2017, 2(1), 13–16, DOI:10.1515/lwr-2017-0003.
-
G. Karthiga Devi; K. Vignesh and S. Chozhavendhan, Effective Utilization of Sugarcane Trash for Energy Production, Elsevier Inc., 2019. DOI:10.1016/B978-0-12-818996-2.00012-0.
- K. Xu and P. Xu, Efficient Production of L-Lactic Acid Using Co-Feeding Strategy Based on Cane Molasses/Glucose Carbon Sources, Bioresour. Technol., 2014, 153, 23–29, DOI:10.1016/j.biortech.2013.11.057.
- A. M. Ferreira and A. Mendes-Faia, The Role of Yeasts and Lactic Acid Bacteria on the Metabolism of Organic Acids during Winemaking, Foods, 2019, 9(9), 1–19, DOI:10.3390/foods9091231.
- P. Filannino, R. Di Cagno and M. Gobbetti, Metabolic and Functional Paths of Lactic Acid Bacteria in Plant Foods: Get out of the Labyrinth, Curr. Opin. Biotechnol, 2018, 49, 64–72, DOI:10.1016/j.copbio.2017.07.016.
- S. J. Lee, H. S. Jeon, J. Y. Yoo and J. H. Kim, Some Important Metabolites Produced by Lactic Acid Bacteria Originated from Kimchi, Foods, 2021, 10(9), 2148, DOI:10.3390/foods10092148.
- P. Huang, L. Yu, F. Tian, J. Zhao, H. Zhang, W. Chen and Q. Zhai, Untargeted Metabolomics Revealed the Key Metabolites in Milk Fermented with Starter Cultures Containing Lactobacillus Plantarum CCFM8610, LWT, 2022, 165, 113768, DOI:10.1016/j.lwt.2022.113768.
- F. Mozzi, M. E. Ortiz, J. Bleckwedel, L. De Vuyst and M. Pescuma, Metabolomics as a Tool for the Comprehensive Understanding of Fermented and Functional Foods with Lactic Acid Bacteria, Food Res. Int., 2013, 54(1), 1152–1161, DOI:10.1016/j.foodres.2012.11.010.
- Y. Wang, J. Wu, M. Lv, Z. Shao, M. Hungwe, J. Wang, X. Bai, J. Xie, Y. Wang and W. Geng, Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry, Front. Bioeng. Biotechnol., 2021, 9, 1–19, DOI:10.3389/fbioe.2021.612285.
- S. Liang, D. Gao, H. Liu, C. Wang and J. Wen, Metabolomic and Proteomic Analysis of D-Lactate-Producing Lactobacillus Delbrueckii under Various Fermentation Conditions, J. Ind. Microbiol. Biotechnol., 2018, 45(8), 681–696, DOI:10.1007/s10295-018-2048-y.
- F. Mozzi, M. E. Ortiz, J. Bleckwedel, L. De Vuyst and M. Pescuma, Metabolomics as a Tool for the Comprehensive Understanding of Fermented and Functional Foods with Lactic Acid Bacteria, Food Res. Int., 2013, 1152–1161, DOI:10.1016/j.foodres.2012.11.010.
- T. Pan, Z. Pei, Z. Fang, H. Wang, J. Zhu, H. Zhang, J. Zhao, W. Chen and W. Lu, Uncovering the Specificity and Predictability of Tryptophan Metabolism in Lactic Acid Bacteria with Genomics and Metabolomics, Front. Cell. Infect. Microbiol., 2023, 13, DOI:10.3389/fcimb.2023.1154346.
- W. Fu, Ó. Guomundsson, G. Paglia, G. Herjólfsson, Ó. S. Andrésson, B. O. Palsson and S. Brynjólfsson, Enhancement of Carotenoid Biosynthesis in the Green Microalga Dunaliella Salina with Light-Emitting Diodes and Adaptive Laboratory Evolution, Appl. Microbiol. Biotechnol., 2013, 97(6), 2395–2403, DOI:10.1007/s00253-012-4502-5.
- S. Huang, Y. Xue, C. Zhou and Y. Ma, An Efficient CRISPR/Cas9-Based Genome Editing System for Alkaliphilic Bacillus Sp. N16-5 and Application in Engineering Xylose Utilization for D-Lactic Acid Production, Microb. Biotechnol., 2022, 15(11), 2730–2743, DOI:10.1111/1751-7915.14131.
- Z. Q. Xiong, Y. Y. Wei, L. H. Kong, X. Song, H. X. Yi and L. Z. Ai, Short Communication: An Inducible CRISPR/DCas9 Gene Repression System in Lactococcus Lactis, J. Dairy Sci., 2020, 103(1), 161–165, DOI:10.3168/jds.2019-17346.
- A. Ozaki, R. Konishi, C. Otomo, M. Kishida, S. Takayama, T. Matsumoto, T. Tanaka and A. Kondo, Metabolic Engineering of Schizosaccharomyces Pombe via CRISPR-Cas9 Genome Editing for Lactic Acid Production from Glucose and Cellobiose, Metab. Eng. Commun., 2017, 5, 60–67, DOI:10.1016/j.meteno.2017.08.002.
- B. Vignesh Kumar, B. Muthumari, M. Kavitha, J. K. J. P. Kumar, S. Thavamurugan, A. Arun and M. J. Basu, Studies on Optimization of Sustainable Lactic Acid Production by Bacillus Amyloliquefaciens from Sugarcane Molasses through Microbial Fermentation, Sustainability, 2022, 14(12), 7400, DOI:10.3390/SU14127400.
- H. Li, M. L. Ma, S. Luo, R. M. Zhang, P. Han and W. Hu, Metabolic Responses to Ethanol in Saccharomyces Cerevisiae Using a Gas Chromatography Tandem Mass Spectrometry-Based Metabolomics Approach, Int. J. Biochem. Cell Biol., 2012, 44(7), 1087–1096, DOI:10.1016/j.biocel.2012.03.017.
- M. Xia, D. Huang, S. Li, J. Wen, X. Jia and Y. Chen, Enhanced FK506 Production in Streptomyces Tsukubaensis by Rational Feeding Strategies Based on Comparative Metabolic Profiling Analysis, Biotechnol. Bioeng., 2013, 110(10), 2717–2730, DOI:10.1002/bit.24941.
- T. Ming, J. Han, Y. Li, C. Lu, D. Qiu, Y. Li, J. Zhou and X. Su, A Metabolomics and Proteomics Study of the Lactobacillus Plantarum in the Grass Carp Fermentation, BMC Microbiol., 2018, 18(1), 1–13, DOI:10.1186/s12866-018-1354-x.
- J. Wang, J. Huang, H. Laffend, S. Jiang, J. Zhang, Y. Ning, M. Fang and S. Liu, Optimization of Immobilized Lactobacillus Pentosus Cell Fermentation for Lactic Acid Production, Bioresour. Bioprocess., 2020, 7, 15, DOI:10.1186/s40643-020-00305-x.
- L. N. Borshchevskaya, T. L. Gordeeva, A. N. Kalinina and S. P. Sineokii, Spectrophotometric Determination of Lactic Acid, J. Anal. Chem. (Transl. of Zh. Anal. Khim.), 2016, 71(8), 755–758, DOI:10.1134/S1061934816080037.
- P. Bork, C. Sander and A. Valencia, Convergent Evolution of Similar Enzymatic Function on Different Protein Folds: The Hexokinase, Ribokinase, and Galactokinase Families of Sugar Kinases, Protein Sci., 1993, 2(1), 31–40, DOI:10.1002/pro.5560020104.
- J. Park and R. S. Gupta, Adenosine Kinase and Ribokinase – The RK Family of Proteins, Cell. Mol. Life Sci., 2008, 65(18), 2875–2896, DOI:10.1007/s00018-008-8123-1.
- S. Liang, W. Jiang, Y. Song and S. F. Zhou, Improvement and Metabolomics-Based Analysis of D-Lactic Acid Production from Agro-Industrial Wastes by Lactobacillus Delbrueckii Submitted to Adaptive Laboratory Evolution, J. Agric. Food Chem., 2020, 68(29), 7660–7669, DOI:10.1021/acs.jafc.0c00259.
- M. Van de Guchte, P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich and E. Maguin, Stress Responses in Lactic Acid Bacteria, Antonie van Leeuwenhoek, 2002, 82(1–4), 187–216, DOI:10.1023/A:1020631532202.
- P. J. Costello and P. A. Henschke, Mousy Off-Flavor of Wine: Precursors and Biosynthesis of the Causative N-Heterocycles 2-Ethyltetrahydropyridine, 2-Acetyltetrahydropyridine, and 2-Acetyl-1-Pyrroline by Lactobacillus Hilgardii DSM 20176, J. Agric. Food Chem., 2002, 50(24), 7079–7087, DOI:10.1021/jf020341r.
- M. Spaggiari, A. Ricci, L. Calani, L. Bresciani, E. Neviani, C. Dall’Asta, C. Lazzi and G. Galaverna, Solid State Lactic Acid Fermentation: A Strategy to Improve Wheat Bran Functionality, LWT, 2020, 118, 108668, DOI:10.1016/j.lwt.2019.108668.
- S. Kumar, N. Yadav, L. Nain and S. K. Khare, A Simple Downstream Processing Protocol for the Recovery of Lactic Acid from the Fermentation Broth, Bioresour. Technol., 2020, 318, 124260, DOI:10.1016/j.biortech.2020.124260.
|
| This journal is © The Royal Society of Chemistry 2024 |
Click here to see how this site uses Cookies. View our privacy policy here.  *b
*b
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and thawed in an ice bathtub for 5 minutes then rigorously vortexed for a couple of minutes and frozen at −80 °C for 30 minutes. This cycle was repeated thrice and centrifuged at 10
1), and thawed in an ice bathtub for 5 minutes then rigorously vortexed for a couple of minutes and frozen at −80 °C for 30 minutes. This cycle was repeated thrice and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 minutes at −20 °C. An additional 2.5 mL of ice-cold methanol–water solution was added to the cell pellets and combined with the former. This supernatant was lyophilized at −88 °C condensation with a 1.000 Pa vacuum for 24 hours and used for further LC-MS and GC-MS analysis.18,26,27
000 rpm for 20 minutes at −20 °C. An additional 2.5 mL of ice-cold methanol–water solution was added to the cell pellets and combined with the former. This supernatant was lyophilized at −88 °C condensation with a 1.000 Pa vacuum for 24 hours and used for further LC-MS and GC-MS analysis.18,26,27




![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O lengths are represented by bands at 1700 cm−1 and 1200 cm−1, respectively.37 A UV-visible spectrophotometer was used to characterize the microbially fermented lactic acid further. Lactic acid can be detected in the sample as a peak between 200 and 300 nm.25 Understanding of the metabolic pathway will open up the arena to other omics approaches to play a role to enhance the quantity and quality of the lactic acid production. The modern revolutionary genomic tool CRISPR-cas9 genome editing tool has been used to metabolically engineer the lactic acid production from glucose and cellobiose as well as xylose.22,24 Understanding the role of metabolites in each step will lead to the addition or deletion/knock-out of the appropriate gene from the bacteria to enhance lactic acid production.20,23
O and C–O lengths are represented by bands at 1700 cm−1 and 1200 cm−1, respectively.37 A UV-visible spectrophotometer was used to characterize the microbially fermented lactic acid further. Lactic acid can be detected in the sample as a peak between 200 and 300 nm.25 Understanding of the metabolic pathway will open up the arena to other omics approaches to play a role to enhance the quantity and quality of the lactic acid production. The modern revolutionary genomic tool CRISPR-cas9 genome editing tool has been used to metabolically engineer the lactic acid production from glucose and cellobiose as well as xylose.22,24 Understanding the role of metabolites in each step will lead to the addition or deletion/knock-out of the appropriate gene from the bacteria to enhance lactic acid production.20,23


