Cancer evaluation in dogs using cerumen as a source for volatile biomarker prospection†
João Marcos G.
Barbosa
 *a,
Engy
Shokry
a,
Lurian
Caetano David
*a,
Engy
Shokry
a,
Lurian
Caetano David
 a,
Naiara Z.
Pereira
a,
Adriana R.
da Silva
b,
Vilma F.
de Oliveira
b,
Maria Clorinda S.
Fioravanti
b,
Paulo H. Jorge
da Cunha
b,
Anselmo E.
de Oliveira
a,
Naiara Z.
Pereira
a,
Adriana R.
da Silva
b,
Vilma F.
de Oliveira
b,
Maria Clorinda S.
Fioravanti
b,
Paulo H. Jorge
da Cunha
b,
Anselmo E.
de Oliveira
 c and
Nelson Roberto
Antoniosi Filho
c and
Nelson Roberto
Antoniosi Filho
 *a
*a
aLaboratório de Métodos de Extração e Separação, Instituto de Química, Universidade Federal de Goiás (UFG), Campus II – Samambaia, 74690-900, Goiânia, GO, Brazil. E-mail: joaomarcosquim.ufg@outlook.com; nelsonroberto@ufg.br
bHospital Veterinário – Escola de Veterinária e Zootecnia da UFG, Rodovia Goiânia – Nova Veneza, km 8 Campus II – Samambaia, 74690-900, Goiânia, GO, Brazil
cLaboratório de Química Teórica e Computacional, Instituto de Química, Universidade Federal de Goiás (UFG), Campus II – Samambaia, 74690-900, Goiânia, GO, Brazil
First published on 8th September 2023
Abstract
Cancer is one of the deadliest diseases in humans and dogs. Nevertheless, most tumor types spread faster in canines, and early cancer detection methods are necessary to enhance animal survival. Here, cerumen (earwax) was tested as a source of potential biomarkers for cancer evaluation in dogs. Earwax samples from dogs were collected from tumor-bearing and clinically healthy dogs, followed by Headspace/Gas Chromatography-Mass Spectrometry (HS/GC-MS) analyses and multivariate statistical workflow. An evolutionary-based multivariate algorithm selected 18 out of 128 volatile metabolites as a potential cancer biomarker panel in dogs. The candidate biomarkers showed a full discrimination pattern between tumor-bearing dogs and cancer-free canines with high accuracy in the test dataset: an accuracy of 95.0% (75.1–99.9), and sensitivity and specificity of 100.0% and 92.9%, respectively. In summary, this work raises a new perspective on cancer diagnosis in dogs, being carried out painlessly and non-invasive, facilitating sample collection and periodic application in a veterinary routine.
Introduction
As in the case of humans, cancer is one of the leading causes of death in dogs.1 It is estimated that 50% of the dogs will develop cancer over ten years, and a quarter of them will eventually die from it, resulting in millions of deaths yearly.2 Most cancer classifications, pathology, and treatment responses are the same for dogs and humans.2,3 Despite parallels between the therapeutic interventions taken in response to cancer in humans and dogs, which suggests that they have similar underlying genetics and biochemical reaction mechanisms, the tumor growth and metastasis rates are usually faster in dogs.1 Hence, early cancer detection methods for dogs are desirable as they improve prognosis and facilitate earlier treatment at the first signs of the disease, thereby enhancing the animal survival rate.Most traditional diagnosis methods are invasive and require cytology or histology testing performed directly on the specific suspected type of tumor. However, these approaches may hinder an accurate identification due to the lack of characteristic clinical signs at the earliest stages of cancer growth. Furthermore, despite the current increase in clinical trials for cancer detection in dogs using advanced imaging techniques (computer tomography, magnetic resonance, and positron emission tomography),4 they often present risks due to the inherent risk of excessive exposure of the animal to radiation, the effect of the application of anesthetics and their associated procedures, and the cost-prohibitive nature of the exams.
New omics platform tests have been explored for veterinary purposes to overcome the drawbacks of using conventional diagnostic tools. A recent clinical validation assay applying a next-generation sequencing of blood-derived DNA (liquid biopsy) for multi-cancer detection was used in over 1000 dogs and presented an effective detection rate higher than 85% for the most aggressive canine cancer types.5 Elevated circulating cell-free DNA (cfDNA) levels in dogs with cancer were found to be a potential marker for tumor diagnosis and disease prognosis.6 Moreover, a previous study demonstrated the usefulness of evaluating nucleosome concentrations as a tool in veterinary oncology.7
In addition to genome-based testing, new bioanalytical methods have been developed by screening volatile organic metabolites (VOMs) derived from biological matrices.8,9 VOMs are promising candidates for metabolome-based diagnostics as they can be performed by safe, non-invasive, and specific tests for the early detection of different cancer types.10–12 One primary origin of the volatile biomarkers for cancer may be linked to the reactive oxygen species (ROS) associated with cancer growth and progression.13,14 These oxidative species promote damage in the DNA of cells, endoplasmic reticulum stress, and the oxidation of lipids and proteins, culminating in small metabolites being released into body fluids.15 Although the potential of VOMs to be used in cancer detection in humans has been explored, there is a lack of volatile compound-based methods for cancer detection in dogs. Therefore, to facilitate the exploration of cancer-related VOMs for diagnostic purposes in dogs, it is critical to assess their chemical attributes and diversity.
Cerumen, or earwax, is a biomatrix that seems to fit these criteria. It refers to secretions of the sebaceous and sweat glands containing polar and non-polar substances, mainly metabolic lipid-derived components.16,17 Recently, cerumen VOMs have been successfully applied in diabetes diagnosis,18,19 forensic applications,20–24 toxicological monitoring purposes,25–29 identification of rare otolaryngological disorder (Ménière's disease),30 and human cancer diagnosis.31 The chemical fingerprint of VOMs for use in disease detection is known as Cerumenogram.
In this sense, this work proposes the use of cerumen as a source for valuable biomarker prospection in cancer identification in dogs. It is essential to emphasize that, as far as the authors are aware, this study spotlights the first application of the canine cerumen volatilome for cancer detection purposes, using a non-invasive biomatrix and a qualitative method performed without sample preparation steps by Headspace/Gas Chromatography-Mass Spectrometry (HS/GC-MS) analysis and multivariate statistical approaches.
Materials and methods
Chemicals
As an internal standard (IS), 3-methylcyclohexanone purchased from Sigma-Aldrich (Santa Louis, MO, USA) was used for chromatography analyses. Other analytical standards, such as heptane, tetradecane, pentadecane, hexadecane, and 2-nonanone purchased from Sigma-Aldrich (Santa Louis, MO, USA), and acetone and acetic acid purchased from Neon (Suzano, SP, Brazil) were used to confirm their presence in the cerumen chemical profile. The purities of all chemicals were higher than 95%.Test population
The present study was approved and conducted following the regulatory standards of animal and human research ethic committees of the Universidade Federal de Goiás (UFG, Brazil), under protocol numbers 027/16 and 57880516.9.0000.5083. All animals were clinically evaluated at the Escola de Veterinária e Zootecnia, UFG, Brazil (EVZ-UFG). After veterinary inspection by biochemical and clinical tests, the presumably cancer-free dogs fitting the clinically healthy criteria (i.e., no other comorbidities reported) were enrolled in the study as the control group. The case group was formed with dogs with spontaneously occurring cancer previously diagnosed by conventional clinical means (imaging techniques and suspect tissue biopsy). The owners of the animals signed an informed consent approving the earwax sample collection of the dog.After clinical confirmation, one hundred earwax samples were obtained from 50 cancer-free dogs (control group) and 50 tumor-bearing dogs (case group). The case group was heterogenous and included 31 samples from female dogs with breast cancer (BC), 7 samples (six male and one female) from dogs with skin and soft tissue cancer (STC), 2 samples from two male dogs with colorectal cancer (CC), 6 samples for six male dogs with testicular and prostate cancer (TPC), and 4 samples from four male dogs with transmissible venereal tumor (TVT). Although the TVT are clonally transmitted rather than caused by mutation, which turns this cancer type generally to be considered a different tumor outcome than the rest of the others included, we kept these samples to observe the similarities of the earwax chemical profile to the other cancer types. Also, among the case group, 23 samples were collected from animals undergoing chemotherapy. Then, all the chemotherapeutics and other medications used during the cancer treatment were analyzed by HS/GC-MS to evaluate and exclude the presence of any potential xenobiotic and its derivative in earwax profiles, and its effect on the cerumen chemical profile was tested before any biomarker selection steps. Detailed demographic characteristics of the canine groups and the specific diagnosis for the cancer group are available in Table S1 in the ESI.†
Sample collection and preparation
The veterinary hospital of the EVZ-UFG, Brazil, was used as the sample collection site. Samples were collected directly from the internal auditory canal by gently swabbing the animal's ears using sterile 6 inch cotton-tipped wooden applicators. The applicators were transferred to sterile tubes, immediately closed, and stored in a deep freezer at −20 °C until analysis (maximum of seven days). Earwax samples were transferred into the inner bottom of 20 mL standard headspace vials, and 0.1 μL of 3-methylcyclohexanone (Sigma-Aldrich, Santa Louis, MO, USA) was added as IS. Every vial was tightly sealed with gas-tight polytetrafluoroethylene (PTFE)-lined rubber septum caps and sent to HS/GC-MS analysis. Headspace vials containing the samples were set in a randomized order in the batch tray. Quality control (QC) blank (sterile HS vials) and QC IS blank (sterile HS vials spiked with IS) were used to monitor background noise (such as the carry-over effect), signal intensity over time, peak quality, and retention time drift during the batch. The samples were randomized in a set containing a QC blank at the beginning and final tray position and a QC IS blank before the first sample vial and after every five sample run.HS/GC-MS parameters
Volatile analyses were carried out using an HS/GC-MS Shimadzu GCMS-QP2010 Ultra system and AOC-5000 headspace analyzer (Shimadzu, Kyoto, Japan) equipped with a VT32-20 tray for 20 mL standard headspace vials and a 2500 μL gas-tight syringe using a preheating module LHS0 Combi PAL (PAL System, Zwingen, Switzerland). The headspace sampler parameters were an injection volume of 2500 μL, a syringe temperature of 150 °C, an incubation temperature of 160 °C, an incubation time of 60 min, and an agitation speed of 500 rpm. The injector was configured in splitless mode. High-purity helium (99.999%–5.0, White Martins, Goiânia, Brazil) was used as carrier gas with a linear velocity of 45.8 cm s−1 (continual flow of 1.36 mL min−1). An NST-100 ms polyethylene glycol-based high-polarity stationary phase was utilized as an analytical capillary column (25 m length × 0.25 mm inner diameter × 0.30 μm film thickness) (NST, São Carlos-SP, Brazil). The oven temperature program started with isothermal heating at 30 °C for 5 min, then heated to 45 °C at 2 °C min−1 (held for 5 min), another temperature increase at the same rate to 50 °C (held for 5 min), and then to 120 °C again at 2 °C min−1, heating up to 200 °C at 6 °C min−1 (held for 5 min), and finally to 250 °C by 5 °C min−1 (held for 10 min). The overall chromatographic run was 98.33 min. MS was performed in the electron ionization (EI) mode at 70 eV. Data acquisition was performed in the full scan mode from 40–500 m/z with a scan time and a scan speed of 0.3 s and 1600 u s−1, respectively. All chromatogram files (.qgd) were processed using LabSolutions GCMSsolution version 4.50 SP1 (Shimadzu, Kyoto, Japan).VOM selection criteria, data processing, and statistical analysis
Initial metabolite annotation was performed by comparing their mass spectra fragmentation patterns with NIST and NIST17s (National Institute of Standards and Technology, Gaithersburg, MD, USA) mass spectral libraries matched by their relative retention time (RRT) to the IS. Only metabolites that fit the criteria of presenting the correct base peak, a minimum of 80% match probability in both NIST17s libraries, and confirmed by their RRT information compared to our internal cerumen retention time database, were putatively marked as VOMs. Under the Metabolomics Standards Initiative (MSI),32 this study reached level 3 of identification confidence (putatively characterized/annotated compounds). Authentic standards run using the same HS/GC-MS conditions were used to identify a few metabolites (level 1 of identification).The molecular features detected were subtracted from the QC blank runs. Those corresponding to a chromatographic peak with an area/height (A/H) ratio higher or equivalent than three were pointed out as putative metabolites. In total, 128 peaks were selected corresponding to VOMs. To perform statistical analysis, the 128 VOMs were transformed into a binary dataset output, where “1” indicates the compound presence (peak area > 0 and A/H ≥ 3) and “0” is its absence (peak area = 0 or A/H < 3). Then, aiming to evaluate which variables/VOMs might discriminate between the control and cancer group, a binary dataset was built containing 100 samples (50 cancer-free dogs + 50 tumor-bearing dogs) by the 128 VOMs.
Multivariate statistical analyses employed for variable selection and visualization were carried out using R version 4.1.2 and the online platform Metaboanalyst 5.0 (https://metaboanalyst.ca/).33 A biological evolutionary-based approach using a genetic algorithm with partial least squares (GA-PLS)34 was applied to optimize the discriminant VOM selection in the canine dataset. The parameters used for GA-PLS analysis were: a population size of 100, a window width of 1, the maximum number of variables in each population of 100, a convergence probability of 50%, a mutation probability of 0.5%, and the maximum number of generations of 35 with contiguous cross-validation. Multiple GA-PLS models were run to select the best discriminant variables/VOM panel and avoid overfitting. Each time, the performance of the chromosome (PLS of the chosen variables/VOMs) was evaluated using k-fold cross-validation – k ∈ {10,10} – as a random resample technique. Then, the optimal tuning parameters of each panel of discriminant variables/VOMs were assessed under 100 different sets of the training set (80% of the samples: n = 80), using receiver operating characteristic (ROC) as an efficacy metric for the model. Next, the test/validation set (20% of the remaining samples: n = 20) was used to fit the model. The best panel of discriminant VOMs was based on the diagnostic measurement results of the test set: accuracy, sensitivity, specificity, control and cancer predictive ability, and kappa value.
Hierarchical cluster analysis (HCA) was run applying Hamming distances and the Ward agglomeration method to visualize the distance measures of the best chromosome of the GA-PLS. Principal coordinate analysis (PCoA) was run using Hamming distance to visualize the samples across the multidimensional space in two principal coordinates (PCO1 and PCO2). Venn diagrams were built using the web-based “InteractiVenn (interactivenn.net)” tool.35 The following R packages were used: e1071 (v1.7-9),36 dendextend (v1.15.2),37 plsVarSel (v0.9.7),38 ade4 (1.7-18),39 caret (v6.0-92),40 and ggplot2 (v3.3.5).41
Results and discussion
Canine cerumenomics
Although there have been significant breakthroughs in metabolomics concerning cancer biomarker discovery, therapeutic approaches, and oncometabolic pathways in humans,14 there is still an enormous gap in studying tumor metabolomics in dogs. This lack of information regarding canine cancer metabolomics is a surprise, primarily due to the high mortality of cancer in dogs and the apparent parity in the disease for dogs and humans.42 So far, urinary-based metabolites have been explored to identify bladder cancer43 and canine mammary gland tumors.44,45 Other studies examined plasma chemical profiles to detect canine lymphoma,46 oral melanoma,47 and 23 types of neoplasm.48In this study, the cerumen analysis of dogs led to the annotation of 128 VOMs, covering a broad spectrum of chemical diversity from polar to non-polar metabolites. The cerumen VOMs are described in Table S2 (ESI†) with their respective elution order, IUPAC name, target peak (m/z), the level of identification, canonical simplified molecular input line entry system (SMILES), CAS number, and the occurrence (%) in each group of samples. Typical total ion chromatogram (TIC) fingerprints of dog cerumen analysis are shown in Fig. S1 (ESI†). In sum, the deconvolution of the TICs led to the identification of 22 alcohols, 19 aldehydes, 18 ester/ethers, 17 ketones, 14 carboxylic acids, 13 amine/amide derivatives, 12 hydrocarbons, 7 furan and lactones, 4 pyran metabolites, and 2 pyrrole compounds.
An abundance of organic families in cerumen composition resembles those also detected in the skin metabolome,8 which may be associated with sebum material in both biomatrices.17,31,49 Sebum is an oily substance produced by the sebaceous glands; when it is broken down upon exposure to ROS, it gives rise to many volatile organic compounds from different organic classes, such as aldehydes, ketones, and hydrocarbons,8 also identified herein with a high frequency. Nonetheless, despite the skin and cerumen chemical advantages regarding the vast range of polarity, it might be preferable to work with cerumen rather than skin secretions due to less liability to external contamination, such as cosmetic products, and due to less exposure to ultraviolet (UV) radiation and air pollution. Moreover, the earwax volatile profile associated with the canine volatilome information from other biofluids may be a valuable resource to elaborate a compendium of VOMs detected in the canine organism, as already performed for humans.50
To look for possible demographic bias in the dog cerumenomic data, cluster analyses were run for factors such as age and sex before applying any variable selection procedure using the complete set of 128 VOMs. As done for humans, a binary data approach was used to reduce the noise of demographic factor impact on the earwax chemical profile.31 Dendrograms in Fig. S2 and S3 (ESI†) present no discrimination pattern for sex and age, respectively.
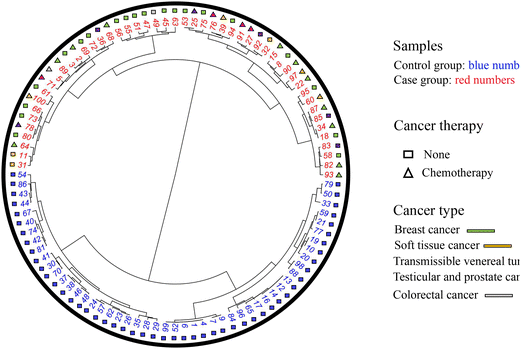
On the other hand, Fig. S4 (ESI†) indicates that cancer samples present, in general, a different VOM pattern (right-hand branch of the dendrogram) when compared to the control samples (left-hand branch). The clusters in Fig. S4 (ESI†) indicate that this discrimination can be improved using a variable selection procedure. Furthermore, as shown in Fig. S5 (ESI†), no pattern of discrimination between dogs under chemotherapy and cancer dogs without therapy was found.
Candidate cancer biomarkers in cerumen of dogs
Cancer cells have a distinct mitochondrial metabolism from healthy cells, resulting in a different fingerprint of small metabolites released in the biofluids/secretion of oncology patients that can be monitored as cancer biomarkers.51–53 Thus, when selecting cancer biomarker candidates in the cerumen secretion of dogs along the 128 VOMs, GA-PLS was applied to optimize and lead to an optimal subset of VOMs that can discriminate between cancer and control groups.Ten GA-PLS models were built to select the best panel of candidate biomarkers. In sum, 65 out of 128 VOMs distributed across ten GA chromosomes were used to build panels of potential cancer biomarkers. In addition, we created two more chromosomes with a cut-off of 0.5 and 0.8 of frequency of variable selection. In summary, the chromosomes from I to X held the variables randomly selected by GA, whilst XI and XII contained the most frequently chosen variables during the ten repeated times (5 out of 10 times and 8 out of 10 times, respectively), as shown in Fig. S6 (ESI†). Table S3 (ESI†) presents a summary of the performance of each candidate model using training and test sets. Fig. S7 (ESI†) presents ROC plots for the optimal tuning parameter results (measured using repeated k-fold cross-validation) for the 12 evaluated chromosomes.
As shown in Table S3 (ESI†), chromosomes I, V, and VII presented similar performances with 95% and 90% in the test dataset regarding the accuracy and kappa value, respectively. Between the three chromosomes, two VOMs are shared (Fig. S8, ESI†). Even though all three chromosomes exhibited similar performances, chromosome I was selected as the best panel of candidate biomarkers for the following reasons: (i) chromosome I presents the most straightforward model, using only 1 PLS component compared to the 3 used by chromosomes V and VII, which makes it a less complex model and less prone to overfitting; (ii) contrary to chromosomes V and VII, the performance of chromosome I in the training set reached the highest values for all metrics used to measure the effectiveness of a diagnostic test (ROC = 100.0%, accuracy = 100.0%, sensitivity and specificity = 100.0%, kappa value = 100.0%, healthy and cancer predictive value = 100.0%); and iii) the candidate biomarkers selected by chromosomes V and VII did not show a full discrimination pattern between samples for cancer and control groups, as shown in Fig. S9 (ESI†). Thus, the 18 VOMs in chromosome I were selected as the best subset of potential cancer biomarkers in dogs. Fig. S10 (ESI†) shows the panel of 18 volatile biomarkers, including their chemical structures and occurrences in cancer and control groups.
Among the 18 VOMs, 3-methylbutanal (VOM 11, Table S2, ESI†), 2-furanmethanol (VOM 23), octanal (VOM 40), 3-decenoic acid (VOM 58), 1,1-dibutoxyhexadecane (VOM 64), 1-dodecanol (VOM 75), hexadecane (VOM 79), octadecane (VOM 88), n-nonadecanoic acid (VOM 108), methyl palmitate (VOM 115), methyl stearate (VOM 121), and heneicosane (VOM 126) presents a higher frequency of occurrence in cancer samples. On the other hand, 2-hydroperoxypentane (VOM 7), 2-methylfuran (VOM 9), 2-dodecanone (VOM 78), 2-tetradecanone (VOM 93), 5-dodecenyl acetate (VOM 94), and butyl stearate (VOM 127) appears more frequently in the control group.
Fig. 1 presents the circular dendrogram run using the 18 VOMs selected as potential cancer biomarkers in the earwax of dogs. This dendrogram shows a clear discrimination pattern between control and cancer samples. On the other hand, over-classification bias driven by the sex, age, or cancer therapy of the dogs was not found since no pattern of discrimination can be noticed in Fig. S11–S15 (ESI†). Moreover, although it is not within the scope of the paper, we monitored if any clusters could be observed due to the cancer type in the cerumenogram model. However, as shown in Fig. S16 (ESI†), no apparent trend was noticed.
Thus, these 18 VOMs selected by GA-PLS arise here as promising potential cancer biomarkers in dogs with the following diagnostic metric figures in the test set of: 95.0% (75.1–99.9, CI = 95%, accuracy), 100.0% (sensitivity), 92.9% (specificity), 90.0% (kappa value), 90.9% (healthy predictive value), and 100.0% (cancer predictive value).
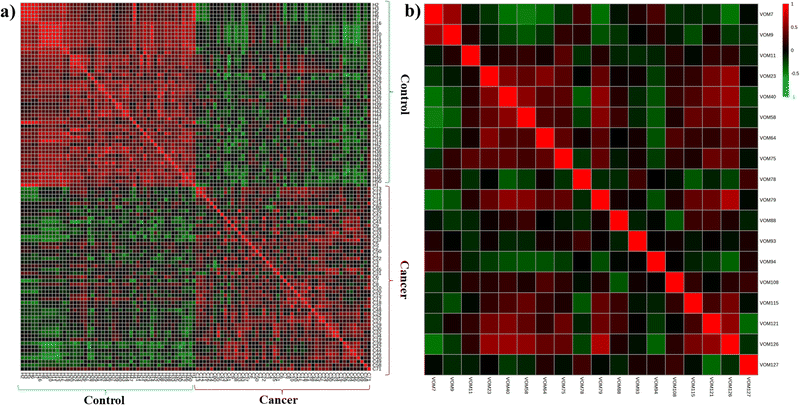
As shown in the Heatmap (Fig. 2a), when observing the matrix correlation for the samples regarding the 18 potential cancer biomarkers, there is a high correlation (>0.5, Spearman rank correlation test) within the samples from the same group (control or cancer), contrasting with a poor correlation of the samples from different groups (control × cancer). Another pattern observed in the Heatmap is the high correlation of VOMs with the same trend of occurrence across the groups. For example, as observed in Fig. 2b, there is a high correlation of VOMs 126, 121, 40, and 79 (heneicosane, methyl stearate, octanal, and hexadecane, respectively), which are related to their higher presence in cancer samples compared to control ones (Fig. S10, ESI†). Similarly, VOMs 7 and 9 (2-hydroperoxypentane and 2-methylfuran, respectively) are highly correlated, appearing more frequently in the control group (Fig. S10, ESI†).
Aiming to test the discrimination power of the selected biomarkers, 18 dogs from the cancer group (n = 9 BC, n = 4 STC, n = 1 TPC, n = 1 CC, and n = 3 TVT) were chosen to obtain earwax samples longitudinally during their routine visits to the veterinary hospital for cancer treatment purposes, totalizing in 33 new earwax samples (Table S4, ESI†). These new samples were added to the healthy/cancer VOM discrimination set without labeling the samples as Y (cancer) or N (cancer-free, control) after tracking the presence/absence of the 18 earwax VOMs. A multivariate ROC (MultiROC) curve-based model function predicted the class for 33 new samples. Table S4 (ESI†) shows that all samples were correctly classified in the Y (cancer) class. Fig. S17 (ESI†) shows the PCoA plot with the longitudinal cancer samples, evidencing their similarities with the cancer group.
Tumor cells, energy production, oxidative stress, and volatile biomarkers in cerumen
Although the biochemical onco-pathway of the volatile biomarkers is still being elucidated, previous studies have proposed that the energy production required to meet the high cancer cell proliferation levels,54,55 and the oxidative stress associated with mitochondrial dysfunction of the tumor cells,51,53 might be responsible for the release of small thermostable metabolites in the organism.8,56,57 Thus, it was anticipated that the 18 VOMs selected as potential cancer biomarkers in dogs may be directly associated with tumoral cell energy production and oxidative stress. For instance, the alcohol marked as a cancer biomarker in the cerumen of dogs, 1-dodecanol (VOM 75), is known as a fatty alcohol metabolite. Long-chain alcohol compounds result from fatty acid reduction by peroxisomal fatty acyl-CoA reductases (FAR1 and FAR2), crucial for plasmalogen production to protect mammalian cells against oxidative stress induced by ROS,58 which are ubiquitous in proliferating cancer cells. Fatty alcohol metabolites in cerumen have already been proposed to indicate hepatic oxidation activity in other mammals, suggesting a link between this marker and cellular oxidative stress.28Moreover, of the aldehydes selected as potential biomarkers for cancer in dogs, 3-methylbutanal (VOM 11) and octanal (VOM 40) have been shown to be side-products of lipid peroxidation.8 Cerumen analysis indicates a slightly higher frequency of the biomarker 3-methylbutanal in samples for cancer dogs compared to cancer-free dogs (≈10%, Table S2, ESI†). Interestingly, a previous study showed the same pattern of occurrence for this metabolite in the human urinary signature of breast cancer patients compared to healthy patients.59 Nevertheless, between the two selected aldehydes as biomarkers, octanal is the one that has been previously described as a cancer biomarker in the blood plasma of dogs.48 In this work, octanal was only detected in earwax samples of dogs from the cancer group. Furthermore, in a previous study, this metabolite was selected as a crucial discriminant metabolite in the hair of canines to identify visceral leishmaniasis,60 which may indicate that this aldehyde is an important marker for metabolic disturbances in dogs.
In total, the evolutionary algorithm indicated six compounds putatively annotated from ester and organic ether classes as candidate markers: 2-hydroperoxypentane (VOM 7), 1,1-dibutoxyhexadecane (VOM 64), 5-dodecenyl acetate (VOM 94), methyl palmitate (VOM 115), methyl stearate (VOM 121), and butyl stearate (VOM 127). Compounds from these organic families are derived from lipid metabolism,61 which is a route that cancer cells use to compensate for the Warburg effect and produce the energy necessary for tumor cell proliferation.62 Among these metabolites, methyl palmitate and methyl stearate were identified as volatile markers of adipogenic differentiation in mesenchymal stromal cells.63 Furthermore, methyl stearate has been detected as a breast cancer biomarker in human blood serum64 and as an important compound in a panel of biomarkers identified in exhaled breath for discriminating between lung cancer patients and those with high-risk factors.65
Fatty acids, also metabolites widely associated with cancer lipid metabolism, play an essential role in the mutation of tumor cells to ensure their growth, proliferation, and survival.62 Found in this work were two compounds that may be potential cancer biomarkers: 3-decenoic acid (VOM 58) and n-nonadecanoic acid (VOM 108), both presenting higher occurrence in cancer samples (Fig. S10, ESI†). Previous studies employing a metabolomic approach for detecting cancer in dogs have revealed higher fatty acid levels in the blood of dogs with oral melanoma47 and lymphoma,46 indicating that compounds in this class may be useful markers for cancer identification in dogs. Also, a previous study has demonstrated that evaluating fatty acid levels in cerumen could be a useful clinical tool for rapidly and accurately detecting Ménière's disease.30
During tumor progression, the higher rate of fatty acid oxidation - associated with negative protein and energy - results in cancer cachexia and is characterized by the involuntary loss of the patient's lean body mass and the release of ketone bodies.66 Ketone bodies have been previously identified as a potential indicator of cancer in dogs.43,48 Here, GA-PLS selected the ketones 2-dodecanone (VOM 78) and 2-tetradecanone (VOM 93) as members of the panel of canine cancer biomarkers in cerumen. Remarkably, these two ketones were previously identified as earwax biomarkers for other metabolic disturbances. For example, increased levels of 2-dodecanone were found in diabetic type 1 patients,18 and higher concentrations of both ketones have been observed in mammals during pre and post-parturition periods.16
The oxidative stress associated with the peroxidation of polyunsaturated fatty acids in cellular and subcellular membrane levels can indicate cancer initiation, and it is the primary mechanism of hydrocarbon release in the body of mammals.66 In this study, three long straight-chain saturated hydrocarbons were designated as probable cancer biomarkers in dogs: hexadecane (VOM 79), octadecane (VOM 88), and heneicosane (VOM 126). Among them, hexadecane has been described as part of the volatile signature for bladder cancer cell lines,67 lung cancer tissue,68 and in the breath samples of ovarian cancer patients where it appears at higher concentrations.69 Also, hexadecane and octadecane increase their levels in exhaled breath of gastric cancer patients.70 Heneicosane is a common metabolite detected in the human body biomatrices, such as skin, saliva, and cerumen.50,71 Here, it is described for the first time as having a higher occurrence in cancer dogs (78.0%) compared to control dogs (2.0%) (Fig. S10 and Table S2, ESI†), being the most prominent variable in the GA-PLS model (Table S3, ESI†).
Endogenous furan derivative formation in mammals may be associated with the natural dehydration of monosaccharides and the oxidation of fatty acids catalyzed by lipoxygenases.72 The two furan compounds detected as biomarkers, 2-methylfuran (VOM 9) and 2-furanmethanol (VOM 23), have already been detected when cancer is present. The metabolite 2-methylfuran has been described as part of the volatilomic footprint of human gastric cancer cell lines73 and in the urine of tumor-bearing mice.74 In addition, both metabolites, 2-methylfuran and 2-furanmethanol, were detected in the urinary profile of human breast cancer patients.59
As noted, lipid metabolism for energy purposes and the oxidative stress associated with the tumor/cancer cell microenvironment are the leading causes of the appearance of these candidate cancer biomarkers in the cerumen of dogs. Nevertheless, many hypotheses explaining the exact origin of the volatile biomarkers have been proposed so far, and further efforts exploring the volatile fingerprint of many cancer cell lines must be conducted to elucidate the cancer volatilome and eliminate the confounding effects associated with clinical samples.66 Some of these hypotheses are: (i) the volatile biomarkers arise due to pathway over-activation in cancer, e.g., glycolysis, fatty acid biosynthesis, mitochondrial β-oxidation of long-chain saturated fatty acids, etc.; (ii) the volatile metabolites are linked to the immune system rather than the tumor environment; (iii) many of the volatile biomarkers arise due to patient exposure to the environment (exposome); and (iv) cancer VOMs emerge in the organism as a consequence of cancer stem cells and their high levels of aldehyde dehydrogenase (ALDH).66 Although these hypotheses are yet to be tested, the origin of the potential biomarkers identified in this study implies that the first hypothesis is the most likely explanation for the metabolic differences noted when cancer is present and which also emphasizes the potential utility of volatile biomarkers in cancer diagnosis.
Limitations and perspectives
The results obtained in this study indicate a promising approach to cancer diagnosis using cerumen VOMs, with an accuracy (in the test dataset) of 95.0% (75.1–99.9, CI = 95%). Although the performance of the cancer discrimination model is highly satisfactory, the limitations of the study should be noted. For instance, some cancer types may be underrepresented due to the small sample size set (n = 100) available for this study. Then, before clinical veterinary implementation, the 18 potential cancer biomarkers in dogs must be validated in double-blind trials in a larger cancer cohort, and samples from new cancer types should also be added. It will also be crucial to evaluate a cohort of cancer-free samples from dogs with infection, inflammation, hyperplasia, metaplasia, dysplasia, benign tumor, and other comorbidities to evaluate the diagnostic merits of the model. In addition, it will be very informative for upcoming studies to assess the metabolome profile differences as a function of the cancer cell subtype (e.g., adenomas, carcinomas, adenocarcinomas) and their influence on cancer discrimination. Last, future efforts exploring the relative concentration of cerumen metabolites rather than their absence/presence may raise interesting new crucial metabolites for cancer evaluation. The exploration of these factors will be beneficial for facilitating clinical diagnosis.In summary, this study sheds light on the perspective that the discriminating cancer-related VOMs in cerumen may be linked to lipid metabolism and oxidative stress, which has the potential to indicate mitochondrial dysfunction during cancer growth and progression. Studies involving other animals, as well as the footprint and fingerprint of a wide variety of cancer cell lines, may be helpful in the search for common biomarkers to elucidate and confirm the oncopathways, which can guide the development of new chemotherapeutic approaches and diagnostic kits focusing on a specific targeted group of molecules.
Conclusion
Here, we present the cerumen volatilome profile as a source of biomarkers for cancer identification in dogs. The experimental pipeline utilizes earwax that can be collected non-invasively and painlessly, with the additional benefit that the analysis can be carried out quickly and efficiently, making it highly suitable for periodic screening in dogs. The highest model prediction accuracy was achieved when using a subset of 18 VOMs, with an accuracy of 95.0% (75.1–99.9, CI = 95%), a sensitivity and specificity of 100.0% and 92.9%, respectively, a kappa value of 90.0%, associated with the performance of a predictive value of 90.9% for cancer-free (control) samples and 100.0% for cancer samples.Declarations
Author contributions
All authors contributed to the study's conception and design. J. M. G. B., E. S., A. E. O., V. F. O., M. C. S. F., P. H. J. C., and N. R. A. F. developed the work conceptualization, visualization, and study design. V. F. O., A. R. S., M. C. S. F., and P. H. J. C. performed the biochemical exams, clinical evaluation of the animals, and earwax sample collection. J. M. G. B., E. S., L. C. D., and N. Z. P. carried out the HS/GC-MS analysis. J. M. G. B., A. E. O., and N. R. A. F. conducted the software analysis and interpretation. J. M. G. B. wrote the original draft. J. M. G. B., A. E. O., and N. R. A. F. performed the formal investigation, data curation, and writing review and editing. N. R. A. F. provided the original idea, funding acquisition, and project coordination. All authors read and approved the final manuscript.Data availability
The code and datasets for R software can be found at https://github.com/Barbosa-JMG/Canine-cancer-dataset.git. Data for this paper, including the GC-MS raw data files (.qgd extension), are available at Mendeley Data Repository at https://www.doi.org/10.17632/9rp53jg3pr.2.Conflicts of interest
All the authors declare no conflict of interest.Acknowledgements
This work was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brazil (CAPES)—Finance Code 001—for fellowship to J.M.G.B. (grant number: # 88887.819724/2023-00); and the management of financial resources by the Agência Nacional do Petróleo, Gás Natural e Biocombustíveis (ANP), Fundação de Apoio à Pesquisa (FUNAPE), Fundação Rádio e TV (RTVE) and Universidade Federal de Goiás.References
- H. L. Gardner, J. M. Fenger and C. A. London, Annu. Rev. Anim. Biosci., 2016, 4, 199–222 CrossRef CAS.
- B. W. Davis and E. A. Ostrander, ILAR J., 2014, 55, 59–68 CrossRef CAS PubMed.
- V. E. Valli, M. S. Myint, A. Barthel, D. Bienzle, J. Caswell, F. Colbatzky, A. Durham, E. J. Ehrhart, Y. Johnson, C. Jones, M. Kiupel, P. Labelle, S. Lester, M. Miller, P. Moore, S. Moroff, P. Roccabianca, J. Ramos-Vara, A. Ross, T. Scase, H. Tvedten and W. Vernau, Vet. Pathol., 2011, 48, 198–211 CrossRef CAS PubMed.
- M. C. Paoloni and C. Khanna, Vet. Clin. North Am. – Small Anim. Pract., 2007, 37, 1023–1032 CrossRef.
- A. Flory, K. M. Kruglyak, J. A. Tynan, L. M. McLennan, J. M. Rafalko, P. C. Fiaux, G. E. Hernandez, F. Marass, P. Nakashe, C. A. Ruiz-Perez, D. M. Fath, T. Jennings, R. Motalli-Pepio, K. Wotrang, A. L. McCleary-Wheeler, S. Lana, B. Phillips, B. K. Flesner, N. F. Leibman, T. LaDue, C. D. Tripp, B. L. Coomber, J. P. Woods, M. Miller, S. W. Aiken, A. Wolf-Ringwall, A. Borgatti, K. Kraska, C. B. Thomson, A. Kosanovich Cahalane, R. L. Murray, W. C. Kisseberth, M. A. Camps-Palau, F. Floch, C. Beaudu-Lange, A. Klajer-Peres, O. Keravel, L.-A. Fribourg-Blanc, P. C. Mazetier, A. Marco, M. B. McLeod, E. Portillo, T. S. Clark, S. Judd, C. K. Feinberg, M. Benitez, C. Runyan, L. Hackett, S. Lafey, D. Richardson, S. Vineyard, M. Tefend Campbell, N. Dharajiya, T. J. Jensen, D. van den Boom, L. A. J. Diaz, D. S. Grosu, A. Polk, K. Marsal, S. C. Hicks, K. M. Lytle, L. Holtvoigt, J. Chibuk, I. Chorny and D. W. Y. Tsui, PLoS One, 2022, 17, e0266623 CrossRef CAS PubMed.
- G. Beffagna, A. Sammarco, C. Bedin, C. Romualdi, M. Mainenti, A. Mollo, L. Cavicchioli, S. Ferro, D. Trez, R. De Maria, D. Nitti, A. Saccani, M. Campanella, M. Agostini and V. Zappulli, PLoS One, 2017, 12, e0169454 CrossRef.
- H. Wilson-Robles, T. Miller, J. Jarvis, J. Terrell, N. Dewsbury, T. Kelly, M. Herzog, T. Bygott, N. Hardat and G. Michel, PLoS One, 2020, 15, 1–16 CrossRef PubMed.
- Y. Y. Broza, P. Mochalski, V. Ruzsanyi, A. Amann and H. Haick, Angew. Chem., Int. Ed., 2015, 54, 11036–11048 CrossRef CAS PubMed.
- S. Giannoukos, A. Agapiou, B. Brkić and S. Taylor, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2019, 1105, 136–147 CrossRef CAS.
- C. Cavaco, J. A. M. Pereira, K. Taunk, R. Taware, S. Rapole, H. Nagarajaram and J. S. Câmara, Anal. Bioanal. Chem., 2018, 410, 4459–4468 CrossRef CAS.
- G. H. Bueno Duarte, A. M. A. de Piloto Fernandes, A. A. R. Silva, H. R. Zamora-Obando, A. G. Amaral, A. de Sousa Mesquita, J. Schmidt-Filho, V. C. Cordeiro de Lima, F. D’Almeida Costa, V. P. Andrade, A. M. Porcari, M. N. Eberlin and A. V. C. Simionato, Anal. Bioanal. Chem., 2020, 412, 7469–7480 CrossRef CAS PubMed.
- C. Wang, C. Ke, X. Wang, C. Chi, L. Guo, S. Luo, Z. Guo, G. Xu, F. Zhang and E. Li, Anal. Bioanal. Chem., 2014, 406, 4757–4763 CrossRef CAS PubMed.
- D. Hanahan, Cancer Discovery, 2022, 12, 31–46 CrossRef CAS PubMed.
- D. R. Schmidt, R. Patel, D. G. Kirsch, C. A. Lewis, M. G. Vander Heiden and J. W. Locasale, CA. Cancer J. Clin., 2021, 71, 333–358 CrossRef PubMed.
- G. Poli, G. Leonarduzzi, F. Biasi and E. Chiarpotto, Curr. Med. Chem., 2012, 11, 1163–1182 CrossRef PubMed.
- E. Shokry, J. Pereira, J. G. Marques Júnior, P. H. J. Da Cunha, A. D. F. Noronha Filho, J. A. Da Silva, M. C. S. Fioravanti, A. E. De Oliveira and N. R. A. Filho, PLoS One, 2017, 12, 1–22 CrossRef PubMed.
- A. M. Coon, A. J. Dane, G. Setzen, R. B. Cody and R. A. Musah, ACS Omega, 2022, 7, 230–239 CrossRef CAS PubMed.
- E. Shokry, A. E. de Oliveira, M. A. G. Avelino, M. M. de Deus and N. R. A. Filho, J. Proteomics, 2017, 159, 92–101 CrossRef CAS PubMed.
- A. Herane-Vives, S. Espinoza, R. Sandoval, L. Ortega, L. Alameda, A. H. Young, D. Arnone, A. Hayes and J. Benöhr, Diagnostics, 2020, 10, 1069 CrossRef CAS PubMed.
- A. G. Nicolaou, I. J. Stavrou, A. P. Louppis, M. S. Constantinou and C. Kapnissi-Christodoulou, J. Chromatogr. A, 2021, 1642, 462035 CrossRef CAS.
- E. Shokry, J. G. Marques, P. C. Ragazzo, N. Z. Pereira and N. R. A. Filho, Forensic Toxicol., 2017, 35, 348–358 CrossRef PubMed.
- E. Shokry, A. E. de Oliveira, M. A. G. Avelino, M. M. de Deus, N. Z. Pereira and N. R. A. Filho, Forensic Toxicol., 2017, 35, 389–398 CrossRef CAS.
- S. I. Meier, S. C. Koelzer, M. Schubert-Zsilavecz and S. W. Toennes, Drug Test. Anal., 2017, 9, 1572–1585 CrossRef CAS.
- S. I. Meier, S. Petzel-Witt, M. Schubert-Zsilavecz, E. B. de Sousa Fernandes Perna, E. L. Theunissen, J. G. Ramaekers and S. W. Toennes, Drug Test. Anal., 2020, 12, 968–974 CrossRef CAS.
- D. R. Gardner, K. D. Welch, K. E. Panter, D. Cook, F. C. dos Santos, B. T. Green, J. A. Pfister, C. A. Stonecipher and S. T. Lee, J. Agric. Food Chem., 2018, 67, 43–49 Search PubMed.
- C. A. Stonecipher, S. T. Lee, B. T. Green, D. Cook, K. D. Welch, J. A. Pfister and D. R. Gardner, Toxicon, 2019, 161, 33–39 CrossRef CAS PubMed.
- S. J. Trumble, S. A. Norman, D. D. Crain, F. Mansouri, Z. C. Winfield, R. Sabin, C. W. Potter, C. M. Gabriele and S. Usenko, Nat. Commun., 2018, 9, 4587 CrossRef.
- J. M. G. Barbosa, M. K. Fernandes Rodrigues, L. C. David, T. C. E. Silva, D. A. Fortuna Lima, N. Z. Pereira, E. B. D’Alessandro, A. E. de Oliveira, P. H. Jorge da Cunha, M. C. S. Fioravanti and N. R. Antoniosi Filho, Biomed. Chromatogr., 2020, 34, e4935 CrossRef CAS.
- J. M. Gonçalves Barbosa, A. F. Machado Botelho, R. H. Santana da Silva, S. S. Ferreira de Almeida, E. R. Ferreira, L. Caetano David, D. Alves Fortuna Lima, T. Cavalcante E Silva, P. H. Jorge da Cunha and N. Roberto Antoniosi Filho, Biomed. Chromatogr., 2021, 35, e5017 CrossRef.
- A. M. Coon, G. Setzen and R. A. Musah, ACS Omega, 2023, 8(30), 27010–27023 CrossRef CAS PubMed.
- J. M. G. Barbosa, N. Z. Pereira, L. C. David, C. G. de Oliveira, M. F. G. Soares, M. A. G. Avelino, A. E. de Oliveira, E. Shokry and N. R. A. Filho, Sci. Rep., 2019, 9, 11722 CrossRef PubMed.
- R. M. Salek, C. Steinbeck, M. R. Viant, R. Goodacre and W. B. Dunn, Gigascience, 2013, 2, 1 CrossRef.
- Z. Pang, J. Chong, G. Zhou, D. A. de Lima Morais, L. Chang, M. Barrette, C. Gauthier, P.-É. Jacques, S. Li and J. Xia, Nucleic Acids Res., 2021, 49, W388–W396 CrossRef CAS.
- R. Leardi and A. L. González, Chemom. Intell. Lab. Syst., 1998, 41, 195–207 CrossRef CAS.
- H. Heberle, G. V. Meirelles, F. R. da Silva, G. P. Telles and R. Minghim, BMC Bioinf., 2015, 16, 169 CrossRef.
- D. Meyer, E. Dimitriadou, K. Hornik, A. Weingessel, F. Leisch, C.-C. Chang and C.-C. Lin, R Packag. version 1.7-9, 2021, pp. 1–66 Search PubMed.
- T. Galili, Y. Benjamini, G. Simpson, G. Jefferis, M. Gallotta, J. Renaudie, K. Hornik, U. Ligges, A.-N. Spiess, S. Horvath, P. Langfelder, M. Van Der Loo, A. de Vries, Z. Gu, Cath, J. Ma, K. G. M. Hummel, C. Clark, L. Graybuck, B. Ho, S. Perreault, C. Hennig, D. Bradley and H. Huang, R Packag. version 1.15.2, 2021, pp. 1–209 Search PubMed.
- K. H. Liland, T. Mehmood and S. Sæbø, R Packag. version 0.9.7, 2022, pp. 1–26 Search PubMed.
- S. Dray, A.-B. Dufour, J. Thioulouse, T. Jombart, S. Pavoine, J. R. Lobry, S. Ollier, D. Borcard, P. Legendre, S. Bougeard and A. Siberchicot, R Packag. version 1.7-18, 2021, pp. 1–404 Search PubMed.
- M. Kuhn, J. Wing, S. Weston, A. Williams, C. Keefer, A. Engelhardt, T. Cooper, Z. Mayer, A. Ziem, L. Scrucca, T. Hunt and M. Kuhn, R Packag. version 6.0-92, 2022, pp. 1–224 Search PubMed.
- H. Wickham, W. Chang, L. Henry, T. L. Pedersen, K. Takahashi, C. Wilke, K. Woo, H. Yutani and D. Dunnington, R Packag. version 3.3.5, 2021 Search PubMed.
- G. Carlos, F. P. dos Santos and P. E. Fröehlich, Metabolomics, 2020, 16, 1–19 CrossRef PubMed.
- J. Zhang, S. Wei, L. Liu, G. A. Nagana Gowda, P. Bonney, J. Stewart, D. W. Knapp and D. Raftery, Biochim. Biophys. Acta, Mol. Basis Dis., 2012, 1822, 1807–1814 CrossRef CAS PubMed.
- M. Valko-Rokytovská, P. Očenáš, A. Salayová, R. Titková and Z. Kostecká, J. Vet. Sci., 2020, 21, 1–10 CrossRef PubMed.
- S. Lee, B. J. Seung, I. S. Yang, J. Lee, T. Ha, H. M. Park, J. H. Cheong, S. Kim, J. H. Sur, G. S. Hwang and H. Nam, Sci. Data, 2022, 9, 1–8 CrossRef CAS.
- R. Tamai, M. Furuya, S. Hatoya, H. Akiyoshi, R. Yamamoto, Y. Komori, S. I. Yokoi, K. Tani, Y. Hirano, M. Komori and S. Takenaka, J. Vet. Med. Sci., 2014, 76, 1513–1518 CrossRef CAS PubMed.
- M. Kawabe, Y. Baba, R. Tamai, R. Yamamoto, M. Komori, T. Mori and S. Takenaka, J. Vet. Med. Sci., 2015, 77, 1025–1028 CrossRef CAS PubMed.
- R. Selyanchyn, T. Nozoe, H. Matsui, T. Kadosawa and S.-W. Lee, Diagnostics, 2013, 3, 68–83 CrossRef CAS PubMed.
- K. A. Prokop-Prigge, E. Thaler, C. J. Wysocki and G. Preti, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2014, 953–954, 48–52 CrossRef CAS PubMed.
- B. De Lacy Costello, A. Amann, H. Al-Kateb, C. Flynn, W. Filipiak, T. Khalid, D. Osborne and N. M. Ratcliffe, J. Breath Res., 2014, 8, 014001 CrossRef CAS PubMed.
- J. S. Bhatti, G. K. Bhatti and P. H. Reddy, Biochim. Biophys. Acta, Mol. Basis Dis., 2017, 1863, 1066–1077 CrossRef CAS PubMed.
- L. Sainero-Alcolado, J. Liaño-Pons, M. V. Ruiz-Pérez and M. Arsenian-Henriksson, Cell Death Differ., 2022, 29, 1304–1317 CrossRef CAS PubMed.
- T. N. Seyfried, R. E. Flores, A. M. Poff and D. P. D’Agostino, Carcinogenesis, 2014, 35, 515–527 CrossRef CAS PubMed.
- M. G. V. Heiden, L. C. Cantley and C. B. Thompson, Science, 2009, 324, 1029–1033 CrossRef PubMed.
- D. Wishart, Metabolites, 2022, 12, 1–33 Search PubMed.
- M. Hakim, Y. Y. Broza, O. Barash, N. Peled, M. Phillips, A. Amann and H. Haick, Chem. Rev., 2012, 112, 5949–5966 CrossRef CAS PubMed.
- A. Amann, B. D. L. Costello, W. Miekisch, J. Schubert, B. Buszewski, J. Pleil, N. Ratcliffe and T. Risby, J. Breath Res., 2014, 8, 034001 CrossRef CAS PubMed.
- M. Dasouki, Peroxisomal disorders: Clinical and biochemical laboratory aspects, Elsevier Inc., 2017, ch. 11, vol. 2 Search PubMed.
- C. L. Silva, M. Passos and J. S. Câmara, Talanta, 2012, 89, 360–368 CrossRef CAS PubMed.
- J. T. Magalhães-Junior, P. R. R. Mesquita, W. F. D. S. Oliveira, F. S. Oliveira, C. R. Franke, F. D. M. Rodrigues, J. B. De Andrade and S. M. Barrouin-Melo, Anal. Bioanal. Chem., 2014, 406, 6691–6700 CrossRef PubMed.
- M. Carbone and G. Melino, Cell Death Differ., 2019, 26, 2516–2519 CrossRef PubMed.
- E. Currie, A. Schulze, R. Zechner, T. C. Walther and R. V. Farese, Cell Metab., 2013, 18, 153–161 CrossRef CAS PubMed.
- D. K. Lee, T. Yi, K. E. Park, H. J. Lee, Y. K. Cho, S. J. Lee, J. Lee, J. H. Park, M. Y. Lee, S. U. Song and S. W. Kwon, Sci. Rep., 2014, 4, 1–8 Search PubMed.
- N. I. Hadi, Q. Jamal, A. Iqbal, F. Shaikh, S. Somroo and S. G. Musharraf, Sci. Rep., 2017, 7, 1–11 CrossRef CAS PubMed.
- A. Peralbo-Molina, M. Calderón-Santiago, F. Priego-Capote, B. Jurado-Gámez and M. D. Luque De Castro, J. Breath Res., 2016, 10, 026002 CrossRef CAS PubMed.
- H. Haick, Y. Y. Broza, P. Mochalski, V. Ruzsanyi and A. Amann, Chem. Soc. Rev., 2014, 43, 1423–1449 RSC.
- D. Rodrigues, J. Pinto, A. M. Araújo, S. Monteiro-Reis, C. Jerónimo, R. Henrique, M. de Lourdes Bastos, P. G. de Pinho and M. Carvalho, Metabolomics, 2018, 14, 1–15 CrossRef CAS PubMed.
- W. Yishan, Y. Hub, D. Wanga, Y. Kai, W. Ling, Z. Yingchang, Z. Cong, X. Zhanga, W. Ping and K. Ying, Cancer Biomarkers, 2012, 11, 129–270 Search PubMed.
- H. Amal, D.-Y. Shi, R. Ionescu, W. Zhang, Q.-L. Hua, Y.-Y. Pan, L. Tao, H. Liu and H. Haick, Int. J. Cancer, 2015, 136, E614–E622 CrossRef CAS PubMed.
- C. M. Durán-Acevedo, A. L. Jaimes-Mogollón, O. E. Gualdrón-Guerrero, T. G. Welearegay, J. D. Martinez-Marín, J. M. Caceres-Tarazona, Z. C. Sánchez- Acevedo, K. de, J. Beleño-Saenz, U. Cindemir, L. Österlund and R. Ionescu, Oncotarget, 2018, 9, 28805–28817 CrossRef PubMed.
- C. N. Burkhart, M. A. Kruge, C. G. Burkhart and C. Black, Otol. Neurotol., 2001, 22, 715–722 CrossRef CAS PubMed.
- A. Jiménez-Pacheco, M. Salinero-Bachiller, M. C. Iribar, A. López-Luque, J. L. Miján-Ortiz and J. M. Peinado, Urol. Oncol. Semin. Orig. Investig., 2018, 36, 243.e21–243.e27 Search PubMed.
- A. Leiherer, D. Ślefarska, M. Leja, C. Heinzle, A. Mündlein, I. Kikuste, L. Mezmale, H. Drexel, C. A. Mayhew and P. Mochalski, Front. Mol. Biosci., 2021, 7, 1–16 Search PubMed.
- Y. Hanai, K. Shimono, H. Oka, Y. Baba, K. Yamazaki and G. K. Beauchamp, Cancer Cell Int., 2012, 12, 1–13 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3mo00147d |
| This journal is © The Royal Society of Chemistry 2024 |