Open Access Article
Open Access ArticleHybrid bio-nanoporous peptide loaded-polymer platforms with anticancer and antibacterial activities†
Madalina
Icriverzi‡
 a,
Paula Ecaterina
Florian‡
a,
Paula Ecaterina
Florian‡
 a,
Anca
Bonciu
a,
Anca
Bonciu
 b,
Luminita Nicoleta
Dumitrescu
b,
Antoniu
Moldovan
b,
Diana
Pelinescu
c,
Robertina
Ionescu
c,
Ionela
Avram
b,
Luminita Nicoleta
Dumitrescu
b,
Antoniu
Moldovan
b,
Diana
Pelinescu
c,
Robertina
Ionescu
c,
Ionela
Avram
 c,
Cristian V. A.
Munteanu
c,
Cristian V. A.
Munteanu
 a,
Livia Elena
Sima
a,
Valentina
Dinca
a,
Livia Elena
Sima
a,
Valentina
Dinca
 b,
Laurentiu
Rusen
b,
Laurentiu
Rusen
 *b and
Anca
Roseanu
*b and
Anca
Roseanu
 *a
*a
aInstitute of Biochemistry of the Romanian Academy, 060031 Bucharest, Romania. E-mail: anca.roseanu@biochim.ro
bNational Institute for Lasers, Plasma, and Radiation Physics, 409 Atomistilor Street, 077125 Magurele, Romania. E-mail: laurentiu.rusen@inflpr.ro
cFaculty of Biology, University of Bucharest, Department of Genetics, Intrarea Portocalelor no. 1-3, Sector 6, Bucharest, Romania
First published on 27th February 2024
Abstract
In this study, hybrid bio-nanoporous peptides loaded onto poly(N-isopropylacrylamide-co-butylacrylate) (pNIPAM-co-BA) coatings were designed and obtained via matrix-assisted pulsed laser evaporation (MAPLE) technique. The incorporation of cationic peptides magainin (MG) and melittin (Mel) and their combination was tailored to target synergistic anticancer and antibacterial activities with low toxicity on normal mammalian cells. Atomic force microscopy, scanning electron microscopy, X-ray photoelectron spectroscopy, Fourier transform infrared spectroscopy as well as contact angle and surface energy measurements revealed the successful and functional incorporation of both the peptides within porous polymeric nanolayers as well as surface modifications (i.e. variation in the pore size diameter, surface roughness, and wettability) after Mel, MG or Mel-MG incorporation compared to pNIPAM-co-BA. In vitro testing revealed the impairment of biofilm formation on all the hybrid coatings while testing with S. aureus, E. coli and P. aeruginosa. Moreover, MG was shown to modulate the effect of Mel in the combined Mel-MG extract formulation released via pNIPAM-platforms, thus significantly reducing cancer cell proliferation through apoptosis/necrosis as revealed by flow cytometry analysis performed in vitro on HEK293T, A375, B16F1 and B16F10 cells. To the best of our knowledge, Mel-MG combination entrapped in the pNIPAM-co-BA copolymer has not yet been reported as a new promising candidate with anticancer and antibacterial properties for improved utility in the biomedical field. Mel-MG incorporation compared to pNIPAM-co-BA in in vitro testing revealed the impairment of biofilm formation in all the hybrid formulations.
1. Introduction
During the past few decades, there is a great deal of interest in the biomedical device technology field owing to the need for the development of new and more efficient multifunctional hybrid platforms for research and clinical application. In this context, much attention is paid to smart systems/platforms based on polymers, which are responsive to single or multiple external stimuli (e.g. pH, temperature, redox potential, ultrasound, electric field, light and enzyme activity).1–6Owing to the advantage of adjusting and controlling their physicochemical properties, smart polymers with tailored characteristics are able to entrap a variety of bioactive compounds, molecules and drugs with active roles in different pathological processes such as cancer, inflammation and skin wound repair.7,8
In response to external stimuli, nanoparticle-based drug delivery platforms undergo changes in their conformation, stability, solubility, precipitation, structure porosity and hydrophilic/hydrophobic balance, finally leading to the release of entrapped active molecules.9,10
Poly(N-isopropylacrylamide) (pNIPAM) containing both hydrophobic and hydrophilic groups is one of the most studied smart polymers. Considering its pH sensitivity, low toxicity, solubility in water, low critical solution temperature (LCST) and thermo-responsive properties, it is a good candidate for applications in cell culture, drug delivery systems, scaffolds for tissue engineering and skin wound repair.11–13
Despite its attractive characteristics, the medical application of pNIPAM is limited by its poor mechanical and biodegradable properties and reduced loading capacity.14 To overcome these drawbacks, different strategies have been used to optimize the properties of pNIPAM in order to meet the requirements of practical applications in the biomedical field. One effective approach is to produce pNIPAM-based copolymers using biocompatible and biodegradable polymeric components such as polysaccharides, polyesters, and polyethylene glycol.15–18 Among them, butyl acrylate (BA) was used to produce pNIPAM-BA, a copolymer that exhibits a lower LCST (24 °C) compared to the homopolymer (32 °C) and better capacity to incorporate/release entrapped compounds owing to its swelling behaviour. Moreover, a hydrophobic character induced by the presence of BA in the pNIPAM copolymer structure enables its interactions with the proteins (i.e. fibronectin) of the extracellular matrix, thus favouring cell attachment.19 Different pNIPAM-BA based formulations have been obtained and proposed as potential optical detection, drug delivery systems or as a promising substrate for cell sheet engineering.20,21 A synthetic biointerface of pNIPAM-BA with moderate hydrophilic character, good biocompatibility and characteristics suitable to be used as a vehicle for biological active compounds' delivery obtained by Matrix-Assisted Pulsed Laser Evaporation (MAPLE) was reported by Dumitrescu et al.22
With the view to obtain a more efficient biomaterial for medical applications, we have used nanoporous pNIPAM-co-BA coatings obtained by MAPLE upon loading with two cationic peptides belonging to the antimicrobial peptides family (AMPs), namely, melittin (Mel) and magainin (MG). Despite the main characteristic of those peptides of exhibiting antimicrobial activities, our purpose was to obtain a hybrid nanoporous bioplatform that can exhibit anticancer effect and a low level of toxicity on normal cells. An advantage on combining the three types of compounds/polymer is related to the ability of pNIPAM-co BA to respond to pH that can be correlated to tumour or bacteria environment.
Mel, a major component of bee venom (∼50%), is a 26 amino acids natural hydrophobic and amphipathic peptide that has been shown to exhibit a variety of activities of special interest for medical applications, such as antimicrobial, anti-inflammatory, anti-cancer and antiviral effects.23,24
As an antimicrobial peptide, Mel interacts with bacterial cell membranes and induces transmembrane pore formation, leading finally to bacterial killing.25,26
Regarding the anti-cancer capacity, Mel was reported to be effective against different types of cancer, i.e., ovarian, prostate, breast, lung by affecting cell cycle, proliferation/growth, motility, migration, through a complex mechanism implying different signal transduction and regulatory pathways.27–30
Despite the beneficial effects, the clinical application of Mel is limited due to its low stability, haemolytic activity and nonspecific cytotoxicity. To overcome these disadvantages, different approaches were used such as chemically modified Mel or its encapsulation in delivery carriers to specifically target tumour cells and thus improving its therapeutic efficacy.24 For instance, a C-terminus truncated Mel peptide conjugated with monoclonal antibodies against prostate cancer cells epitopes was found to inhibit tumour growth and improved mice survival after systemic or intratumoral injection.31 Hyaluronic acid-modified melittin-liposomes and melittin-loaded niosomes were proposed as more suitable formulations for specific melittin delivery to melanoma and breast cancer cells compared to free melittin form.32,33 A redox-sensitive polymer-based nanocomplex containing Mel or magnetic nanoparticles containing Mel-doxorubicin combination showed an improved antitumor efficiency versus the single-agent regimen.34,35 A decrease in cytotoxicity against normal cells and an increase in preventing biofilm formation capacity were obtained using Mel combination with antibiotics such as vancomycin and rifampin.36
Magainins (MGs) isolated from the African frog Xenopus laevis skin are small peptides (23 amino acids) belonging to AMPs family, which exhibit a broad spectrum of antimicrobial activity against pathogens (bacteria and fungi) through transmembrane pore formation, leading to membrane permeabilization and finally to cell death. Possessing cationic and amphiphilic properties, MGs are able also to act as anti-cancer compounds being cytotoxic to malignant cells but not to normal mammalian cells.37 The mechanisms of MGs interaction with cell membrane follows toroidal or, in some cases, carpet model, depending on membrane origin and its lipid composition.38
Although MGs and Mel act via a similar mechanism, there in vitro cytotoxicity is different, Mel being more potent than MG and other AMPs. Thereby, while Mel is haemolytic, MG does not affect red the blood cells' viability, even at a high concentration.39
Since MGs exhibit a low membrane binding affinity, efforts have been done to improve their biological effectiveness in medical field by enhancing the selectivity against bacterial and cancer cells. The envisaged strategy is based on the use of synthetic analogues, conjugation/combination with other cationic peptides or conventional anti-cancer drugs for use in drug delivery systems.40–43
Starting from the above exposed findings, we have proposed in our work to investigate whether the combination of the two cationic peptides, Mel and MG, embedded into pNIPAM-co-BA scaffold resulted in a more efficient anticancer effect compared to each peptide alone. For this purpose, pNIPAM-co-BA porous coatings prepared by MAPLE were subsequently loaded with Mel, MG and Mel-MG combination. Regarding the potential of cationic peptides combination to increase the individual efficiency of each compound, it is worth mentioning that most studies focused on the antimicrobial activity and less on the anticancer capacity.
The morphological and physico-chemical characteristics of the obtained scaffolds have been investigated by Atomic Force Microscopy (AFM), Scanning Electron Microscopy (SEM), Fourier Transform Infrared Spectroscopy (FTIR), contact angle (CA) and X-ray Photoelectron Spectroscopy (XPS). The antimicrobial effect and the capacity to inhibit biofilm formation of the new developed biomaterials were validated against clinically relevant bacteria. As Gram-positive bacteria, Staphylococcus (S.) aureus, a human pathogen responsible for many skin, tissue and post-surgical infections, was chosen.44 The effect on Gram-negative bacteria was investigated against Escherichia (E.) coli and also against highly antibiotic resistant opportunistic pathogen associated with nosocomial infections, Pseudomonas (P.) aeruginosa.45 Further, the capacity of the MG, Mel and Mel-MG combination released by the new obtained scaffolds to inhibit cell proliferation and cell cycle progression as well as to induce an apoptotic effect was assessed in vitro on normal (human HEK239T) and melanoma model cell lines (human A375, murine B16F1 and B16F10 cells).
Our study revealed that the pNIPAM-co-BA platforms obtained by MAPLE using DMSO as the matrix exhibit suitable physico-chemical characteristics for incorporation of the cationic peptides MG, Mel and their combination.
PNIPAM-based scaffolds containing Mel-MG combination impaired bacterial growth and delayed the development and evolution of biofilm formation. Mel-MG extract released by pNIPAM-co-BA scaffolds was found to reduce cancer cell proliferation through apoptosis/necrosis process, an effect enhanced compared to individual Mel peptide action.
To the best of our knowledge, Mel-MG combination included/entrapped into pNIPAM-co-BA copolymeric porous nanoplatform has not been reported until now as a potential candidate with anticancer and antibacterial for improved utility in the biomedical field.
2. Experimental
2.1 Materials and reagents
Poly(N-isopropyl acrylamide)-butylacrilate copolymer (pNIPAM-co-BA) (C6H11NO)m(C7H12O2)n (butyl acrylate, 5 mol%, average Mn 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, LCST of 250 °C, (Sigma-Aldrich)) Magainin (MG Sigma M7152), Melittin, (Mel Sigma M 4171), and dimethyl sulfoxide (DMSO solvent, ACS reagent, ≥99.8) were purchased from Merck (Sigma-Aldrich, Spruce Street, 3050, Saint Louis, MO 63103, USA).
000, LCST of 250 °C, (Sigma-Aldrich)) Magainin (MG Sigma M7152), Melittin, (Mel Sigma M 4171), and dimethyl sulfoxide (DMSO solvent, ACS reagent, ≥99.8) were purchased from Merck (Sigma-Aldrich, Spruce Street, 3050, Saint Louis, MO 63103, USA).
2.2 Preparation of Mel, MG and Mel-MG loaded pNIPAM-co-BA coatings
Preliminary studies on the copolymer coating optimization showed that the surface characteristics are highly dependent on solvent, laser fluence and target concentration.22 Therefore, in this work, in order to obtain copolymeric layers with nanoporous interfaces, fixed conditions were used: 2% pNIPAM-co-BA in DMSO (in weight concentration), laser spot measured at the target level of 1 mm2, fluence 350 mJ cm−2 and 72k pulses. Solutions of 2% Mel and 2% MG in distilled water were filtered through 0.22 μm Millipore membranes were placed on the samples, allowing slow evaporation under the hood at a constant temperature of 20 °C.
2.3 Materials morphological and physical-chemical analysis
The surface free energy of the layers was determined using the Owens, Wendt, Rabel, and Kaelble (OWRK) method by measuring the contact angles of deionized water as a polar liquid and di-iodomethane as a completely dispersive liquid.
2.4 Biological investigations
Prior to biological study, all copolymeric coatings were sterilized by UV-C irradiation (254 nm, Thermo Scientific HeraSafe KS15) at 250 °C (<LCST) for 15 min.Bacterial cell viability was evaluated by counting the number of colonies (UFC per ml) grown for 24 h at 37 °C on solid nutritional broth using decimal dilution assay.49 Non-functionalized glass was used as the first control and Luria–Bertani liquid medium inoculated with bacterial strain as the second control. All experiments were evaluated in a minimum of three independent determinations.
Murine melanoma B16F1 (CRL-6323) and B16F10 (CRL-6475) cells (ECACC, Porton Down, UK) were cultured in RPMI 1640 medium (PAN Biotech, Aidenbach, Germany) containing glutamine, 10% heat inactivated FBS (v/v), and 1% penicillin/streptomycin.
All cell lines were maintained at 37 °C in a humidified 5% CO2 atmosphere and used in a biological study at more than 90% viability as checked by trypan blue dye staining (Invitrogen, Thermo Fischer Scientific, Life Technologies Co, Eugen, Oregon, USA).
All in vitro experiments were performed in triplicate, twice.
All materials were immersed in 500 μL complete culture medium for 24 h at 23 °C (T0 < LCST), and then the eluates were added to the cells previously grown for 24 h in 96 wells (1 × 104 per well). After 24 h of incubation (37 °C, 5% CO2), the medium was removed and a mix of MTS reagent and cell culture medium (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) was added to each well. After 10–15 min of incubation, 100 μL of the culture solution was transferred to a 96-well clear bottom plate (Nunc, Thermo Fisher Scientific, CA, USA), and the optical density at 450 nm was measured by a microplate reader (Mithras LB 940 Berthold Technology, Bad Wildbad, Germany). Cells grown on coverslip (CS) in the presence of culture medium represented the positive control (CTRL+), while cells treated with 70% ethanol for 5 min were the negative control (CTRL−).
5) was added to each well. After 10–15 min of incubation, 100 μL of the culture solution was transferred to a 96-well clear bottom plate (Nunc, Thermo Fisher Scientific, CA, USA), and the optical density at 450 nm was measured by a microplate reader (Mithras LB 940 Berthold Technology, Bad Wildbad, Germany). Cells grown on coverslip (CS) in the presence of culture medium represented the positive control (CTRL+), while cells treated with 70% ethanol for 5 min were the negative control (CTRL−).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, MA5-14520, Thermo Fisher Scientific, CA, USA) for 30 min at RT and then in the absence of light for 1 h with goat anti-rabbit Alexa-Fluor 594 antibodies (R37117, Invitrogen, Thermo Fisher Scientific, CA, USA) (emission into red TxRed channel). The negative control (Ki67-absent) was obtained by treating the cells with 70% ethylic alcohol (3 min) while untreated cells represent the positive control (Ki67+). Actin filaments were labelled with Alexa Fluor 488-conjugated Phalloidin (dil. 1
200, MA5-14520, Thermo Fisher Scientific, CA, USA) for 30 min at RT and then in the absence of light for 1 h with goat anti-rabbit Alexa-Fluor 594 antibodies (R37117, Invitrogen, Thermo Fisher Scientific, CA, USA) (emission into red TxRed channel). The negative control (Ki67-absent) was obtained by treating the cells with 70% ethylic alcohol (3 min) while untreated cells represent the positive control (Ki67+). Actin filaments were labelled with Alexa Fluor 488-conjugated Phalloidin (dil. 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, A-12379 Invitrogen, Thermo Fisher Sci., CA, USA) (emission into green FITC channel), and Hoechst (1 μg mL−1 H-21492 Life Technologies, Molecular Probes, Eugene, OR, USA) for nuclei staining (emission into the blue DAPI channel). ProlongGold Antifade Mountant Reagent (P-36934 Molecular Probes, Life Technologies, Eugene, OR, USA) was applied on all specimens and mounted on Marienfeld Superior™ glass slides. Fluorescence images were acquired using a Zeiss Axiocam ERc5s Apotom 2 microscope with EC Plan-Neofluar 40× objective and finally analysed with the AxioVision Rel. 4.8 software (Zeiss).
50, A-12379 Invitrogen, Thermo Fisher Sci., CA, USA) (emission into green FITC channel), and Hoechst (1 μg mL−1 H-21492 Life Technologies, Molecular Probes, Eugene, OR, USA) for nuclei staining (emission into the blue DAPI channel). ProlongGold Antifade Mountant Reagent (P-36934 Molecular Probes, Life Technologies, Eugene, OR, USA) was applied on all specimens and mounted on Marienfeld Superior™ glass slides. Fluorescence images were acquired using a Zeiss Axiocam ERc5s Apotom 2 microscope with EC Plan-Neofluar 40× objective and finally analysed with the AxioVision Rel. 4.8 software (Zeiss).
2.4.6.1 Apoptosis. Cells were seeded (5 × 104 per well in 24 wells microplates) and left to attach overnight before incubation with the material extracts for 24 h. Next day, to retain any cells lifting from the cultures due to apoptosis, culture supernatants were collected together with the fractions containing the PBS wash, and the cells still attached to the plastic harvested using trypsin-0.05% EDTA solution. The identification of apoptotic and necrotic cells was performed using Annexin V Apoptosis Detection Kit with 7-AAD (BioLegend, San Diego, CA, USA). Briefly, cells were washed twice with cold Cell Staining Buffer, resuspended in Annexin V Binding Buffer before staining for 15 min with Annexin V-FITC reagent for early apoptotic cells' detection. To distinguish between the necrotic and apoptotic cells, the samples were also stained with 7-amino-actinomycin D (7-AAD) solution. Early apoptotic cells exclude 7-AAD, while late stage apoptotic and necrotic cells will stain positive for 7-AAD, which binds to DNA in the nucleus. Cells were analysed using a BD FACSVerse™ Flow Cytometer (BD Biosciences). The signal emitted by 7-AAD labelled cells was detected on the PerCP-Cy5.5 channel of the cytometer, while Annexin V positive cells were detected on the FITC channel.
2.4.6.2 Cell cycle. Cells used for apoptosis investigation were fixed in 70% ethanol solution, a process that preserves the cell cycling state, makes cells permeable and allows for extended period of storage at −20 °C until it was further assayed for cell cycle investigation. Briefly, the suspension cells were washed twice in cold PBS, then stained with 10 μg mL−1 Hoechst at 37 °C for 30 min and finally analysed using a BD FACSAria™ III Cell Sorter (BD Biosciences) in order to quantify the percentage of cells in each cell cycle phase. The fluorescence intensity of Hoechst was detected on a DAPI channel in a linear scale. Single stained controls for FITC, 7AAD and Hoechst were acquired to assess potential spectral overlap between signals and to exclude any interference in the DAPI channel.
All cytometry data were exported and analysed using the web-based Cytobank software: https://community.cytobank.org. The gating strategy included steps to eliminate doublets (FSC-A vs. FSC-H and DAPI-A vs. DAPI-W dot plots, respectively) before analysing the populations of apoptotic and necrotic cells or cell cycle phases. FMO controls were used for carefully setting up the apoptosis assay gates.
2.5 Statistical analysis
All values from cell viability experiments are expressed as mean ± SD. Data were analysed with one-way ANOVA, followed by Tukey's multiple-comparison post-test using GraphPad Prism software for Windows version 5 (GraphPad Software, Inc., version 5; La-Jolla, CA, US). Differences in statistical significance between groups were set at p values <0.05.The results obtained in microbiological investigation were analysed using GraphPad Prism 9.5.1 software, and statistical interpretation was carried out with a 2-way ANOVA test, followed by Dunnett's multiple comparisons test. Differences with statistical significance between groups were set at p values <0.0001.
3. Results and discussion
3.1 Preparation of pNIPAM-co-BA coatings
pNIPAM-co-BA Mel, pNIPAM-co-BA MG and pNIPAM-co-BA Mel-MG were prepared as described in Materials and methods Section 2.2.First, to establish the amount of Mel and MG to be coated on pNIPAM films, we determined the half maximal inhibitory concentrations (IC50). HEK239T, A375, B16F1 and B16F10 cells were seeded at 1 × 104 cells per well in 96 microplates. Next day, the cells were incubated with serial dilutions 0–16 μg mL−1 for Mel and 0–100 μg mL−1 for MG for another 24 h, and the cell viability was determined by the MTS assay.
It is worth noting that the values obtained for Mel depicted in Fig. S2† are in the range reported by others for the same type of cells.32,52–54 Based on the IC50 values for Mel, the 10 μg per sample was chosen for single as well as dual coating. In the case of MG, the peptide was not cytotoxic up to 100 μg mL−1 for all cell lines tested; thus, 100 μg was used for single peptide as well as in Mel-MG combinatorial formulation. The sample used in our further experiments are presented below: Cop (pNIPAM-co-BA), Cop-Mel (pNIPAM-co-BA_Melittin), Cop-MG (pNIPAM-co-BA_Magainin), and Cop-Mel-MG (pNIPAM-co-BA_Melittin_Magainin).
The morphology and thickness of the copolymer layer are key factors to consider in the development of future bio-intelligent cell cultures or drug delivery platforms/carriers. For example, the presence of pores can be beneficial for a variety of applications, and it has been reported that increasing the size of the pores facilitates the diffusion and adsorption of drugs and metabolites, such as peptides or proteins from the extracellular matrix.19,55 The results of the previous work related to the composition of the target, the type of solvent and fluence showed that the use of DMSO solvent and a fluence of 350 mJ cm−2 resulted in a uniform, homogeneous coating with an undamaged chemical profile. Also, the surface morphology analyses revealed the presence and uniform distribution of nanopores on the material surface.22
Within the context of porous polymeric platforms relevant for loading with antitumor and antimicrobial bioactive compounds, in this study, pNIPAM-co-BA coatings (Cop coatings) were obtained by MAPLE using DMSO as a matrix and 72k pulses.
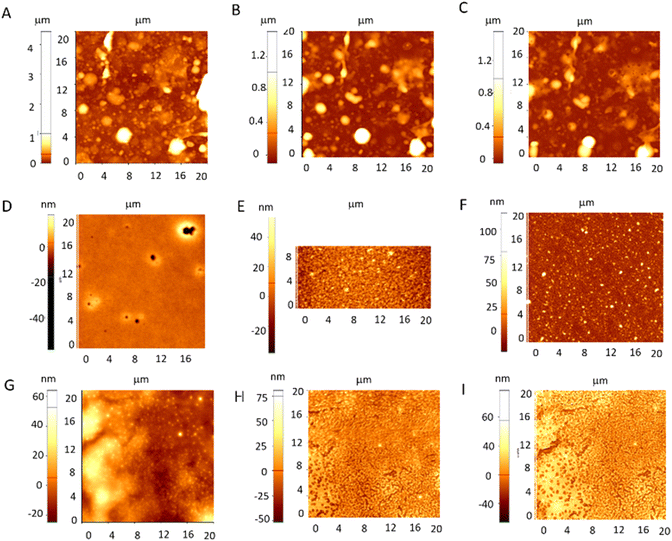
As shown in Fig. S3,† coatings with porous interface, with the dimensions of pores varying between 20 to 100 nm, uniformly distributed and a layer average thickness of 460 nm were obtained. The AFM and SEM image analysis of the dried samples (Fig. 1) revealed major changes in the surface morphology of the platforms, after MG, Mel or Mel-MG incorporation into the matrix. Thus, the pore diameter varied from up to 2 microns to a surface smoothening or decreased below 100 nm or even pore disappearance when both peptides are incorporated. This was especially observed in the case of MG incorporation, where an increase in pore diameter and reorganization occurs as compared to the pNIPAM-co-BA alone (Fig. S3†).
In the case of combinatorial formulation, the lack of pores was reported. As the loading of peptides was done by casting, we could relate the differences observed in pore formation to the thermodynamic stability and biomaterial-solvent affinity, which can be involved in differences in liquid–liquid demixing during phase separation, resulting in surfaces characterized by different pore sizes.
These observations were in concordance also with the measured average value of surface roughness for pNIPAM-co-BA-MG (119 nm) as compared to the significantly decreased values after the incorporation of Mel (up to 55 nm) or both compounds (up to 4 nm) into the copolymer films.
3.2 Physical-chemical characteristics of pNIPAM-co-BA coatings
When a polymer substrate is in contact with cells, its capacity from protein adsorption to cell adhesion, growth and proliferation depend mainly on the surface characteristics (i.e., wettability, energy of the substrate, surface functional groups, and topography). Therefore, understanding both the physical and chemical characteristic of the interface is crucial for correlating to the biological output.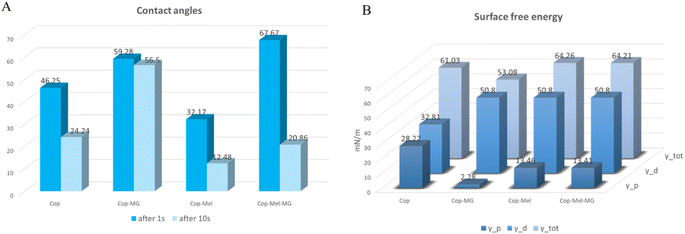 | ||
| Fig. 2 (A) Contact angle values for pNIPAM-co-BA-based coatings (at 1 second and 10 seconds after the droplet was deposited). (B) Surface energy measurements for pNIPAM-co-BA-based coatings. | ||
After 10 seconds, a clear decrease in contact angle values for all samples was observed, except the one containing MG. Exhibiting amphipathic property, MG can interact with both water and lipid-based membranes through the hydrophobic and hydrophilic domains of its structure.38,56 Nonetheless, at 10 seconds, the Cop-Mel-MG sample showed lower contact angle (∼21°), suggesting that the incorporation of both peptides may improve the long-term wettability of the copolymer. The incorporation of active compounds, Mel and MG into the copolymer structure, appears to enhance the wettability of the resulting material, a property which could potentially lead to better cellular adhesion and biocompatibility suitable for biomedical applications. For example, the relationship between cellular behaviour and the surface energy of polymer surface investigations revealed that cells generally prefer to grow on surfaces with higher energy as the polar groups on the polymer surfaces interact with the cell surface groups and form chemical bonds. In the case of nonpolar groups, nonspecific interactions occur over short distances, such as the van der Waals interaction.57
In our case, the results of the surface free energy (SFE) analysis show that the copolymer containing Mel had the highest total SFE value among all the samples, 64.26, indicating a greater affinity towards polar and dispersive liquids. The copolymer loaded with both Mel and MG had the second highest SFE value (64.21), followed by the copolymer alone (61.03) and the copolymer containing MG (53.08). These findings suggest that the incorporation of Mel or Mel-MG combination into the copolymer structure significantly improves its surface properties.
In addition, the polar and dispersive components of surface free energy can provide information on the chemical composition of the surface and the nature of intermolecular forces that are involved in surface interactions. Materials with higher polar components on their surface, such as hydroxyl or carbonyl groups, exhibit stronger hydrogen bonding and dipole–dipole interactions with other materials. The polar component value of surface free energy was highest for the copolymer (28.22), followed by the copolymer incorporated with Mel (13.46), copolymer incorporated with Mel and MG (13.41), and the copolymer with MG (2.28) (Fig. 2B).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of the Cop-BA copolymer.22
O stretching of the Cop-BA copolymer.22
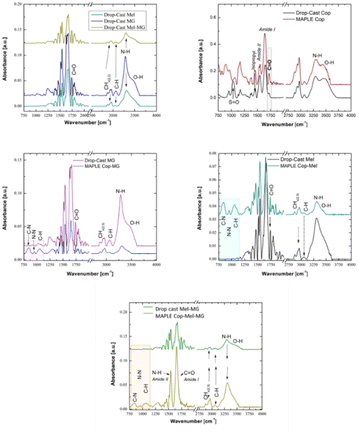 | ||
| Fig. 3 FTIR spectra of the reference AMPs as separate compounds and combination formula. Comparison MAPLE obtained at 350 mJ cm−2 and DMSO as the solvent vs. the control copolymer layers. | ||
From FTIR analyses, we can observe that the bands assigned to C–H deformation modes are observable in the range of 1350–1450 cm−1 (absorption bands at 1388–1368 cm−1), bands that are characteristic to the symmetric deformation of the hydrophobic isopropyl group –C(CH3)2. These observations are in addition to the specific peaks corresponding to the hydrophilic amide bonds that were specified earlier. We also observe a number of absorption bands, unique to the peptide/protein secondary structure,22 in the “fingerprint region” (spectral region between 1700 and 700 cm−1). The peaks observed at 1645 cm−1 and 1537 cm−1, corresponding to the most prominent vibrations in this region, corresponds to the amide I (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching) and amide II bands (N–H bending and stretching vibrations, respectively C–N).
O stretching) and amide II bands (N–H bending and stretching vibrations, respectively C–N).
It is known that the modification of surface chemistry can be performed to decrease or increase protein/surface interactions. Since such a change can occur using the different bioactive compounds, the changes in the surface chemical composition of the Cop containing Mel, MG or their combination were further analysed by XPS. The obtained XPS survey spectra show the typical signals associated with C 1s (carbon), N 1s (nitrogen) and O 1s (oxygen) for the copolymer layers deposited by the MAPLE technique at the laser fluence of 350 mJ cm−2 (Table 1).
| Atomic% | Drop-cast cop. 2% | Drop-cast MG | Drop-cast Mel | Drop-cast Mel-MG | Cop 2 wt% | Cop-MG | Cop-Mel | Cop-Mel-MG |
|---|---|---|---|---|---|---|---|---|
| O 1s | 13.39 | 20.86 | 24.84 | 20.47 | 15.6 | 16.27 | 16.7 | 16.1 |
| N 1s | 13.22 | 17.39 | 7.65 | 18.39 | 13.45 | 11.26 | 16.7 | 14.41 |
| C 1s | 73.39 | 61.75 | 67.51 | 61.14 | 70.95 | 72.47 | 66.6 | 69.49 |
The lack of the Si 2p peak in all other deposition types is an indicator for a homogeneous coverage of the Si substrate with Cop (i.e., comparable C 1s intensity in different areas after MAPLE and peptide incorporation). The Cop before (drop-cast) and after MAPLE deposition showed XPS peaks corresponding to C 1s at 284.8 eV, N 1s at 399 eV and O 1s at 531.5–533 eV, in agreement with the coating elemental composition. XPS spectra analysis shows that all samples presented a peak at 284.8 eV, corresponding to aliphatic carbons. Furthermore, the peak position at 533 eV (i.e., C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) for MAPLE deposition is consistent with the presence of organic matter on the surface of the copolymer layer containing the C
O) for MAPLE deposition is consistent with the presence of organic matter on the surface of the copolymer layer containing the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups of pNIPAM-co-BA.
O groups of pNIPAM-co-BA.
3.3 Morphology and stability of films containing active compounds
Further, morphological changes of the films during immersion in tumour (pH 6.4)-relevant environments were investigated by AFM, while the modifications occurred after immersion for 24 h in different pH solutions simulating normal (pH 7.3), and tumoral pH values were analysed by SEM.Previous studies22 reported that the exposure of pNIPMA-co-BA surfaces to solutions with basic pH value and T = 36 °C was characterized by the hydration phenomenon, resulting in a reduced number of pores. Fig. 4A shows the AFM image of pNIPAM-co-BA films deposited by MAPLE at the fluence of 350 mJ cm−2, 72k pulses, before being immersed in the pH 6.4 solutions, where the surface is characterized by the presence of pores and microglobular structures as the effect of polymer deposition by the MAPLE method. The in-liquid scan revealed an increase in the roughness, from its initial value of 168 nm in air to 221 nm (Fig. 4B). After drying, despite some slight modifications of the morphology, such as a decrease in the diameter of microglobular structures, the value of the roughness was close to the initial one (Fig. 4C, 167 nm).
Nevertheless, as pNIPAM-co-BA has LCST 25 °C, we performed AFM analysis in liquid with both pH of 6.4 and temperature of 25 °C. When compared to the copolymer loaded with peptides coatings in air (Fig. 4A, D and G), those analysed upon immersion in solution of pH 6.4 (Fig. 4B, E and H), the modifications observed for the coatings with incorporated peptides followed similar trend, a decrease of roughness being observed when measured both in liquid, and also after drying. The large pores observed for the MG-loaded samples disappeared (Fig. 4F), and uniformly distributed small nonglobular structures are observed. The coatings with incorporated Mel suffered less changes, with no pores evidenced on the surface after immersion (data not shown), due to the fact that Mel could make considerable hydrogen bonds within copolymeric matrix when hydration occurs. When both peptides are loaded within pNIPAM-co-BA platform and the surface is exposed to acid pH medium, nanopores are predominant (Fig. 4H). When both peptides are loaded within the pNIPAM-co-BA platform and the surface is exposed to acid pH medium, nanopores are predominant (Fig. 4H). Nevertheless, as measured in liquid, the surface roughness decreased below 5 nm for all the samples containing peptides, showing a swollen characteristic, as shown in Fig. S4.† One explanation can also be related to the fact that at pH with different values than pKa, the charge–charge repulsion and an increase in the material's osmotic pressure are responsible for the swelling behaviour. As a result of swelling, the pores are diminished as well and could therefore delay the fast release. The swelling behavior of pNIPAM-co-BA-based coatings, when exposed to acidic fluids, is contingent upon several factors, including the hydrophilicity of its components, the presence of static charges, and the counter ions within. Upon contact with a liquid medium, hydrophilic and polar groups are attracted to water molecules, initiating absorption and diffusion into the coating's pores. Subsequently, as polymer chains relax, a loosening and expansion occurs, akin to the three stages of hydrogel swelling.58 This interaction causes hydrophobic BA segments to engage with water molecules, creating an osmotic driving force that enhances water retention. Furthermore, the swelling of hydrogels containing acidic or basic pendant groups on polymer chains is influenced by the pH of the surrounding medium relative to the respective pKa values of these pendant groups. In the case of a cationic network, the protonation (ionization) of pendant groups leads to an increase in fixed positive charges along polymer chains and mobile negative charges within the solution.59,60
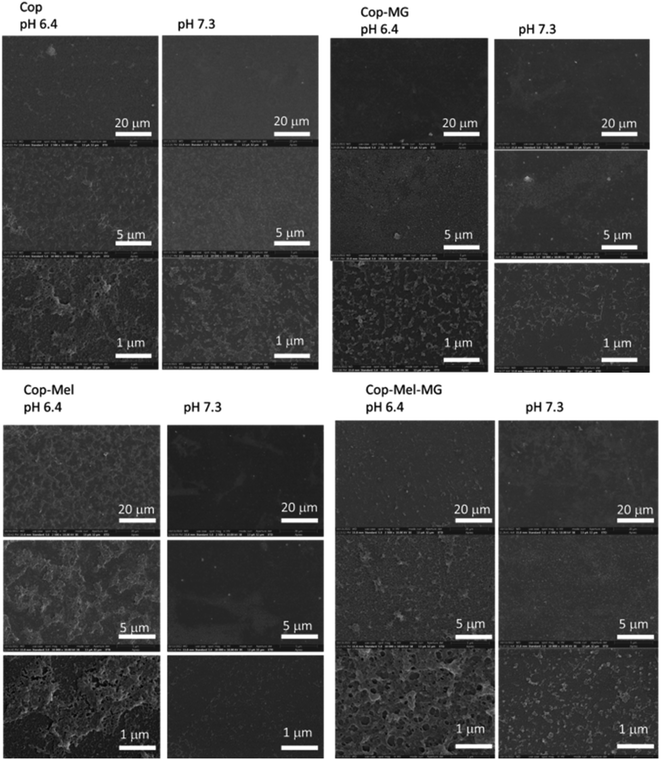
As our targeted application of the designed porous polymeric coatings imply bacteria and cancer cells, the exposure of both type of coatings, polymeric as well as peptides loaded coatings were exposed for 24 h to acidic medium, and then the samples were analysed by SEM for understanding the changes induced by pH. Solutions of 7.3 pH were used as controls. As shown in the sequence of SEM images, there are significant differences when the coatings were subjected to acidic medium as compared to those simulating normal cellular environment. If the modifications observed by AFM after 30 minutes immersion in solution with pH 6.4 revealed a decrease in roughness values; after 24 h, the SEM images revealed an opposite trend, especially for copolymeric coatings containing Mel and Mel-MG peptides, with surface revealing an increased number of pores. One possible explanation for the increased number of pores and their diameters observed in the coatings containing peptides could be related to considering the analogy on how the water-soluble amphipathic peptides, in our case Mel, have the ability to bind to cell membranes and form transmembrane pores (Fig. 5).
3.4 Validation of antimicrobial capacity of pNIPAM-based coatings containing active compounds
One way to prevent bacterial infection and biofilm formation could be the use of antimicrobial peptides (AMPs), which act against a broad-spectrum of microorganisms through multiple complex mechanisms including cell wall- and membrane-targeting, intracellular effects and inhibition of biofilm formation.61–63 Several studies revealed that their combination with conventional antibiotics or a combination between cationic peptides proved to be an efficient strategy for dealing mainly with biofilm-associated bacterial infections.64–68In our study, the antibacterial efficiency of scaffolds embedded with MG, Mel and their combination was validated using Staphylococcus (S.) aureus ATCC 29213, Escherichia (E.) coli ATCC 25922, and Pseudomonas (P.) aeruginosa ATCC 27853. The results presented in Fig. 6A revealed that the cell viability of bacterial strains was not influenced by the presences of glass (used as reference), Cop, Cop-MG and all samples including controls generated growth of around 1010 cells per mL. Cop-Mel and Cop-Mel-MG reduced the number of E. coli cells from 1010 cells per mL to below 10 cells per mL. In the case of S. aureus, the cell viability decreased to 103 cells per mL for Cop-Mel and to 52 cells per mL for Cop-Mel-MG, respectively, for the most resistant bacterial strain P. aeruginosa, the presence of Cop-Mel and Cop-Mel-MG lowered the bacterial cells number to 107 cells per mL and to 106 cells per mL, respectively, compared to the control (1013 cells per mL). The results are in accordance with other reported data regarding antimicrobial activity of Mel against Gram positive and Gram-negative bacteria.69–71
Although in our study MG did not induce an inhibitory effect (probably too low amount), its presence in the combinatory formulation seems to modulate the inhibitory action of Mel and further studies are needed to elucidate these findings. The results presented in Fig. 6B confirmed the data obtained by SEM analyses. The ability of the prepared samples, with or without AMPs, to inhibit the biofilm formation of P. aeruginosa ATCC 27653, S. aureus ATCC 29213 and P. aeruginosa ATCC 27653 strain. S. aureus ATCC 29213 was tested by staining with crystal violet, a dye able to stain both bacterial cells and the extracellular matrix. As shown in Fig. 6B, the coating containing Mel-MG was shown to have the highest ability to inhibit biofilm formation of all the tested strains as compared to cells incubated on glass only. In particular, the percentages related to reduction in biofilm formation obtained were shown to be depending on both the compound/composition as well as on the bacterial species tested. Interestingly, in the case of E. coli, a reduction of more than 75% in biofilm formation was obtained in the copolymeric layer embedding Mel and the copolymeric layer embedding both Mg and Mel, while for S. aureus ATCC 29213, the biofilm was not inhibited for the MG-containing sample (Fig. 6B) but inhibited by more than by 50% polymer alone, Cop-Mel and Cop-Mel-MG. In the case of P. aeruginosa ATCC 27653 strains, the copolymeric layer inhibited the biofilm formation almost 50% in a similar manner to Cop-Mel-MG. These observations suggests that the prevention of the biofilm can be attributed to a classical inhibitory/bactericidal effect of the compounds against planktonic biofilm forming cells but also that the copolymer alone could prevent biofilm formation in the case of the specific strain of P. aeruginosa by interfering with adhesion or with the bacterial communication machinery by hydration/swelling.
Further, the changes in bacterial adherence, cell morphology and biofilm inhibition were assessed by SEM. The images presented in Fig. 6C showed that in the case of Cop, all bacterial cells display normal, unaffected adherence and morphology. On the contrary, both Cop-MG and Cop-Mel exhibited similar effect, altering the membrane integrity and inducing significant changes in the morphology of bacterial strains. Thus, membrane disruption, leading to cytoplasmic leakage, was observed, and the leaked material was found to be around the membrane. According to the above images, the number of vesicles on bacterial membranes (only in the case of Mel for Pseudomonas sp.), number of ghost doughnut shapes and induction of hole formation in S. aureus, extent of disruption in membranes, aggregation of bacterial cells, and cell lysis can be seen (Fig. 6C). A similar effect on roughness, depression and dent formation induced by antimicrobial compound Curcumin I on S. aureus or cell degradation and hole formation action of caerin on Klebsiella sp. was reported by others.72,73 Anti-biofilm activity of Mel alone or in combination with other AMPs or antibiotics against Gram-negative and Gram-positive bacteria was reported.25,72
In the case of Mel-MG combination, an enhanced effect compared to single treatment was observed (Fig. 6B). We appreciate that this is due to the capacity of the both peptides to disrupt the cell membrane through pore formation and thereby increasing the permeabilization of bacterial cells. The difference observed in the level of inhibitory capacity of Mel-MG combination against the tested strains could be explained by differences in membrane fluidity and lipid composition of bacterial membrane (lipopolysaccharide on Gram-negative and lipoteichoic acid on Gram-positive, respectively) involved in the interactions with antimicrobial peptides. Another important factor could be the concentration ratio between the two components. In the study focused on the effect of different AMPs combinations on E. coli MG1655 strain, Yu et al.73 reported that Mel-apidaecin was synergistic in low-concentrations but antagonistic in high concentrations. However, the most common phenomenon in antimicrobial peptides killing action, including MelPex (Pex, MG analog) combination, was synergism. The molecular mechanism of antagonism is not entirely known, but the synergism could be the result of the conjugation of the two peptides, leading to the formation of supermolecules that stabilize the pores.73
3.5 In vitro cancer activity of peptides released from pNIPAM-based coatings
Cationic AMPs exhibit both antimicrobial and anticancer activity through the electrostatic interactions with negatively charged phospholipids on the surfaces of bacterial and cancer cells followed by pore formation and subsequently either membrane disruption or cell penetration, leading to apoptosis/necrosis. The affinity of AMPs for cancer cells is weaker compared to that of the bacteria due to a low density of negative charges present on mammalian cell surface and the rigidifying effect induced by cholesterol on the membrane.74,75 The multifunctional capacity of AMPs raised the interest for their applications as antimicrobial and antitumor drugs. In this respect, several studies focused on the evaluation of combinatory treatment potential on a stronger effect than that of individual cationic peptide, most of the investigations being performed from a microbiological point of view and less on the antitumor activity.24,73Here, the anticancer capacity of Mel, MG and their combination released from a stimuli responsive pNIPAM-based polymer on the cell proliferation and morphology was investigated using mammalian, normal and melanoma cell lines. By applying in vitro approaches, we aimed to assess whether a two-way peptide combination extract from scaffolds exhibits a more efficient anticancer effect compared to single peptide action.
Further, cell viability and proliferation were analysed also by immunofluorescence microscopy using Ki-67 protein as a marker for discriminating proliferating cells within a cell population (Fig. 8B). The results were correlated with observations regarding adhesion and morphology obtained by fluorescence and scanning electron microscopy of normal human HEK293T, human melanoma A375, murine B16F1 and B16F10 melanoma cells incubated for 24 h at 37 °C with the extracts released from the copolymer support at 23 °C. The supernatants obtained from the matrices alone as well as the one containing MG only did not induce morphological or proliferation changes for the normal human cell line HEK293T within cells being Ki-67+ (red), similar to untreated cells and displaying strong actin filaments (green). The images show that cells exhibit normal stellate shape, keep their adhesion capacity and growth in clusters due to good cell–cell contact. Contrary to that, treatment with Mel and Mel-MG combination led to a decrease in the number of adhered cells. However, cells show a normal morphology and are distributed in cell groups. Moreover, adhered cells are proliferating, as indicated by Ki-67+ (Fig. 8B). Like in the case of non-cancerous HEK293T cells, the morphology of A375, B16F1 and B16F10 melanoma cells is less or not affected by the treatment with supernatants released from the Cop and Cop-MG matrices. Cells exhibit morphology similar to the normal phenotype (untreated cells, CTRL) in all cases, and many adherent Ki-67+ cells presenting cell–cell and cell-surface contacts can be observed (Fig. 7B). Previously, cytotoxicity observed by the MTS assay of Mel and Mel-MG eluates released from pNIPAM-co-BA coatings (Fig. 8A) was confirmed by the reduction of adhesion in all cancer cell lines studied compared to untreated controls or cells treated with Cop or Cop-MG supernatants observed in immunofluorescence microscopy investigation. The few remaining attached cells generally present changes in morphology, have a round or spherical shape with cortical actin, shrinkage similar to the apoptotic cells and do not register a signal for the Ki-67 protein (Ki-67−). Also, multiple nuclear debris can be found (no cytoplasmic body).
Cell morphology observations obtained by SEM technique are strongly in accordance with the fluorescence microscopy ones; electronic micrographs (Fig. 8C) present similar effects of the released peptides alone or in combination on normal and tumoral cells. All cells treated with supernatants obtained from Cop matrices alone or loaded with MG lacked any significant impact on cell morphology. Cells displayed normal shape similar with that observed on untreated cells. Mel and Mel-MG extracts are effective against both human and murine melanoma cells, exhibiting significant cytotoxic activity, resulting in dramatically decreased cell size and modified morphology toward a rounded shape similar to that observed in fluorescent microscopy analysis.
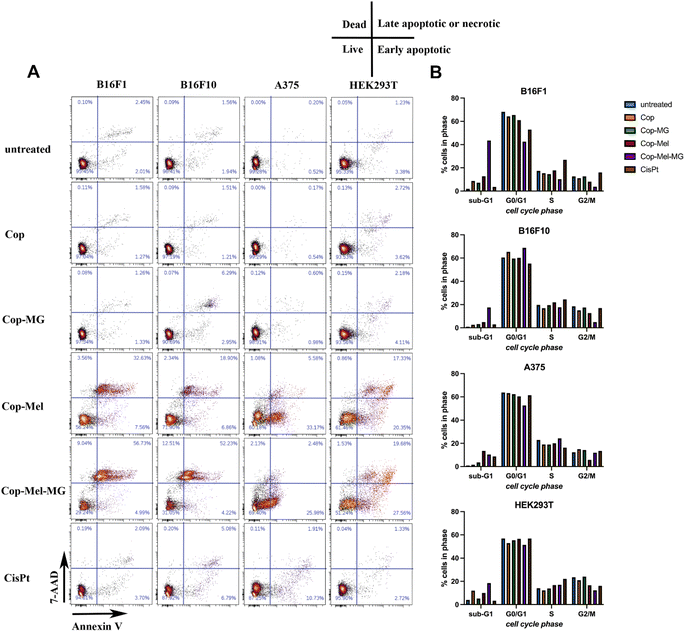
Mechanistically, DNA content analysis shows an opposite effect of Mel-containing formulations to cisplatin (CisPt), a standard cytostatic drug used in chemotherapy.76,77 CisPt-treated cells (at the used concentration of 72 μM) suffered an overall increase in DNA content due to the inhibition of cell division (Fig. S6†) and a blockage in the S to G2/M transition in drug-sensitive cell lines (B16F1, B16F10, Hek293T) (Fig. 9A). These observations are also supported by the FACS morphological parameters analysis (Fig. S7–S9†), which show a progressive decrease in size and granularity of cells treated with Mel and Mel-MG formulations as opposed to cells treated with CisPt that show an increase in median cell size and granularity for CisPt-sensitive cells. The morphological modifications induced by Mel and Mel-MG extracts are in line with those observed by fluorescence and SEM analysis (Fig. 8B and C). Further, we have performed experiments to see whether the cytotoxicity induced by Cop-Mel and Cop-Mel-MG extracts is mediated by apoptosis and/or necrosis. The quantitative analysis of cell death by flow cytometry following annexin V-FITC and 7AAD dual labelling indicated that Cop-Mel and Cop-Mel-MG cytotoxicity were dominantly mediated by late apoptosis/necrosis in murine melanoma cells (Fig. 9B). Thus, after treatment with Cop-Mel extract alone for 24 h, the amount of late apoptotic/necrotic cells increased compared to untreated cells and Cop extract from 2.55% and 1.69%, to 36.19% in B16 F1 cells and from 1.65% and 1.6% to 21.24% in B16F10 cells, respectively. Interestingly, the Cop-MG extract does not affect the cell viability of primary melanoma cells (1.34% late apoptotic/necrotic cells) and only in a limited extend (6.36%) that of metastatic cell line with respect to the controls. The absence of the MG cytotoxic effect is probably due to the amount of peptide released from Cop (Fig. S5†), which is less than the IC50 (≳100 μmol L−1) reported for many cancer cell lines.78 However, the Cop-Mel-MG combination exposure induced a dramatic enhance in the late apoptosis/necrosis process in both primary and metastatic mouse melanoma cells investigated (65.77% and 64.74%, respectively).
In the case of human normal and melanoma cells, the effect of either Cop-Mel or Cop-Mel-MG combination resulted in early apoptosis with only a small percentage of cells in late apoptosis/necrosis (Fig. 9B).
The ability of Mel to trigger early apoptosis in A375 human melanoma was shown by an increased percentage of cells in the sample treated with Cop-Mel extract alone (33.17%) when compared with controls (0.52% for untreated and 0.54% for Cop). In HEK293T cells, the toxicity of Mel was reflected by increased early apoptosis rates (20.35%), after 24 h treatment with the extract as compared to the controls (3.38% for untreated and 3.62% for Cop extract). Although the Cop-MG early apoptotic rate was 4.11%, this is not statistical relevant as compared to the untreated and Cop (3.38% and 3.62%, respectively). Also, the percent of HEK293T cells in late apoptosis/necrosis was significantly higher for the Cop-Mel sample (18.19%) as well as for the combination Cop-Mel-MG (21.21%) as compared to the untreated and Cop controls (1.28% and 2.85%, respectively). This behaviour of HEK293T cells after treatment with extracts containing either the peptide alone or in combination can partially be explained by the enhanced sensitivity of the normal cell line as suggested by the higher percentage (3.38%) of early apoptosis observed in untreated cells compared to all tumour cells investigated (0.51–2.01%) (Fig. 9B). Here, the apoptotic effect induced by Mel and Mel-MG was supported by the observed morphological modifications (Fig. 8B and C) and MTS results (Fig. 8A), such as cell shrinkage, round and spherical cells, decrease in viable cells and presence of multiple nuclear debris.
Altogether the assessment of cell growth, morphology and cell cycle analysis indicated that Mel and Mel-MG released by polymer coatings produces damage to mitochondrial activity and impairs the growth of all tested cells, normal cells are less sensitive compared to cancer cells.
The more damaging cancer cell membrane capacity of Mel compared to that exhibited on normal cells is well documented.79,80 The mechanism of Mel action on cancer cells is through either apoptosis or necrosis depending on the cell line.52,81 Thus, in the experiment performed by Lim et al.53 with human A375S and SKMEL-28 cells and murine B16F10 melanoma cells, Mel was found to induce an increased amount of early and late apoptosis when compared to the basal level in untreated cells. Mel derived from Apis florea was proposed as an apoptosis inducer in cancer cells through its capacity to activate cytochrome-c and to trigger caspase-3 release in A375 cells.82 An increase in the proportion of late apoptotic and necrotic cells was also reported by Tipgomut et al.83 in Mel-treated human bronchogenic carcinoma cells, normal fibroblasts being less sensitive to peptide action. Also, the effect of Mel could be influenced by the capacity of cells to form clumps as revealed in the investigations performed on gastric and colorectal cancer cells.84
In addition, the membrane lipid profile of various cancer cells also differs, a reduced level of certain lipids resulted in increased membrane permeability, while high lipid content account for more rigidity, leading to drug resistance.85 Ganpule et al.86 have demonstrated that the Mel action is influenced by membrane cholesterol content and changes in lipid membrane organization, consequently affecting cell viability. It has been reported that the lower level of cholesterol in some cancer cell membranes compared to normal cells led to an increased fluidity, thus potentiating cationic peptides effect.75
Another important aspect regarding the anticancer action of cationic peptides is the possibility to modulate their action using a combinatorial formula with other molecules, thus reducing the non-specific toxicity and potentiating the biological effect. For example, a synergistic potential of Mel combination with plasma or phospholipase A2 was reported as a promising approach for cancer therapy.54,87 Also, magainin II was conjugated with penetratin in order to enhance its cytotoxic effect in tumour cells.88
All these observations could explain, at least in part, the different response induced by pNIPAM copolymer Mel-MG extracts to the different cell lines, normal and tumoral used in our experiments. Also, some other factors such as the difference between Mel and MG regarding the pore formation rate and stability should be taken into account to understand the mechanism(s) of their action in combinatorial formulations.37
Overall, for the cell lines investigated, the outcome indicated that MG modulates the apoptotic/necrotic effect of Mel when combinatorial formulation is used through a mechanism(s) that requires further studies to be elucidated. Also, it will be important to understand the potential enhanced effect of Mel-MG treatments on primary cancer cells as compared to metastatic ones.
4. Conclusions
Our study revealed that the nanoporous pNIPAM-co-BA coatings obtained by MAPLE were suitable to be used as bio-nanoplatforms for Mel and MG incorporation, allowing the penetration of the peptides within the copolymeric layer and not only at the surface. The presence of pores with dimensions from 20 nm to 100 nm uniformly distributed on the surface of the copolymeric coatings provided an advantage for the incorporation of MG, Mel and Mel-MG. AFM and SEM images analysis of platforms surface morphology revealed a variation in the size of pores diameter—from 2 microns to a surface smoothening—depending on the presence of MG, responsible for large pores, or Mel, responsible for lowering the surface roughness to a minimum of 4 nm when both MG and Mel are incorporated into the copolymer films.The hydrophilic characteristic of the copolymeric coatings was maintained after the incorporation of the two peptides; however, the highest decrease in the contact angle was obtained when Mel was embedded. The FT-IR studies revealed that neither the MAPLE process nor the peptides' incorporation affect the presence and characteristics of the functional groups of copolymer or peptides.
As the nanoporous bioplatforms were designed to be functional in acidic medium, when subjected to environment simulating tumour and normal cell medium, the surfaces revealed an increased surface porosity for those immersed in acid pH as compared to pH 7.3, especially for those containing MG. pNIPAM-based scaffolds embedded with Mel-MG exhibited the capacity to impair bacterial growth and to delay the development and evolution of biofilm formation. The results of in vitro study evidenced the potential of Mel-MG extracts released by pNIPAM-co-BA to significantly reduce cancer cell proliferation through apoptosis/necrosis process induced mainly in murine melanoma cell line B16F1 and B16F10. The presence of MG in two-peptide formulation was shown to modulate the cytotoxic action of Mel depending on the cell lines.
Together, our findings are promising in terms of how a nanoporous polymeric platform embedded with two cationic peptides could be directed towards modified nano-biointerfaces with a beneficial impact for the treatment of key pathological processes, such as cancer and bacterial infection.
Author contributions
MI: investigation, methodology, visualization, formal analysis, writing-original draft; PF: investigation, methodology, visualization, formal analysis, writing-original draft; CM: investigation, methodology and visualization, formal analysis; LS: investigation, methodology, formal analysis, writing-original draft; AM: AFM investigation, methodology and visualization, formal analysis; AB: SEM, CA and SE investigation, methodology and analysis, writing-original draft; ND: XPS investigation and analysis, MAPLE methodology, writing-original draft; DP: antibacterial and antibiofilm investigations; RI: antibacterial and antibiofilm investigations; IA: antibacterial and antibiofilm investigations; VD: conceptualization, methodology, MAPLE methodology, validation, writing – review & editing, methodology, and supervision; LR: conceptualization, methodology, validation, writing – review & editing, methodology, funding and supervision; AR: conceptualization, methodology, validation, writing – review & editing, methodology, and supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by grant of the Romanian Ministry of Education and Research, CCCDI-UEFISCDI, project number PN-III-P2-2.1-PED-2019-2695 and Romanian National Nuclear Program LAPLAS VII—contract no. 30N/2023.Notes and references
- A. Gandhi, A. Paul, S. O. Sen and K. K. Sen, Studies on thermoresponsive polymers: phase behaviour, drug delivery and biomedical applications, Asian J. Pharm. Sci., 2015, 10(2), 99–107, DOI:10.1016/j.ajps.2014.08.010.
- R. M. K. Ramanan, P. Chellamuthu, L. Tang and K. T. Nguyen, Development of a temperature-sensitive composite hydrogel for drug delivery applications, Biotechnol. Prog., 2006, 22(1), 118–125, DOI:10.1021/bp0501367.
- L. Rusen, V. Dinca, B. Mitu, C. Mustaciosu and M. Dinescu, Temperature responsive functional polymeric thin films obtained by matrix assisted pulsed laser evaporation for cells attachment–detachment study, Appl. Surf. Sci., 2014, 302, 134–140, DOI:10.1016/j.apsusc.2013.09.122.
- A. Aghabegi Moghanjoughi, D. Khoshnevis and A. Zarrabi, A concise review on smart polymers for controlled drug release, Drug Delivery Transl. Res., 2016, 6(3), 333–340, DOI:10.1007/s13346-015-0274-7.
- Y. Lu, A. A. Aimetti, R. Langer and Z. Gu, Bioresponsive materials, Nat. Rev. Mater., 2016, 2(1), 1–17, DOI:10.1038/natrevmats.2016.75.
- P. Das and N. R. Jana, Biomedical Applications of Functional Polyaspartamide-Based Materials, ACS Appl. Polym. Mater., 2021, 3(10), 4791–4811, DOI:10.1021/acsapm.1c00785.
- P. Mi, Stimuli-responsive nanocarriers for drug delivery, tumor imaging, therapy and theranostics, Theranostics, 2020, 10(10), 4557, DOI:10.7150/thno.38069.
- M. Zhang, W. Hu, C. Cai, Y. Wu, J. Li and S. Dong, Advanced application of stimuli-responsive drug delivery system for inflammatory arthritis treatment, Mater. Today Bio, 2022, 14, 100223, DOI:10.1016/j.mtbio.2022.100223.
- P. Das, S. Pujals, L. M. A. Ali, M. Gary-Bobo, L. Albertazzi and J. O. Durand, Super-resolution imaging of antibody-conjugated biodegradable periodic mesoporous organosilica nanoparticles for targeted chemotherapy of prostate cancer, Nanoscale, 2023, 15(28), 12008–12024, 10.1039/d3nr01571h.
- X. Wang, H. Lu, B. Liao, G. Li and L. Chen, Facile synthesis of layered double hydroxide nanosheets assembled porous structures for efficient drug delivery, RSC Adv., 2023, 13(18), 12059–12064, 10.1039/d3ra01000g.
- V. Capella, R. E. Rivero and A. C. Liaudat, et al., Cytotoxicity and bioadhesive properties of poly-N-isopropylacrylamide hydrogel, Heliyon, 2019, 5(4), e01474, DOI:10.1016/j.heliyon.2019.e01474.
- X. Fan, S. Gu, L. Wu and L. Yang, Preparation and characterization of thermoresponsive poly(N-isopropylacrylamide) copolymers with enhanced hydrophilicity, E-Polym., 2020, 20(1), 561–570, DOI:10.1515/epoly-2020-0061/machinereadablecitation/ris.
- M. J. Ansari, R. R. Rajendran and S. Mohanto, et al., Poly(N-isopropylacrylamide)-Based Hydrogels for Biomedical Applications: A Review of the State-of-the-Art, Gels, 2022, 8(7), 454, DOI:10.3390/gels8070454.
- M. A. Haq, Y. Su and D. Wang, Mechanical properties of PNIPAM based hydrogels: a review, Mater. Sci. Eng., C, 2017, 70, 842–855, DOI:10.1016/j.msec.2016.09.081.
- L. Yang, X. Fan, J. Zhang and J. Ju, Preparation and Characterization of Thermoresponsive Poly(N-Isopropylacrylamide) for Cell Culture Applications, Polymers, 2020, 12(2), 389, DOI:10.3390/polym12020389.
- X. Xu, Y. Liu and W. Fu, et al., Poly(N-isopropylacrylamide)-Based Thermoresponsive Composite Hydrogels for Biomedical Applications, Polymers, 2020, 12, 580, DOI:10.3390/polym12030580.
- K. Thirupathi, T. T. V. Phan, M. Santhamoorthy, V. Ramkumar and S. C. Kim, pH and Thermoresponsive PNIPAm-co-Polyacrylamide Hydrogel for Dual Stimuli-Responsive Controlled Drug Delivery, Polymers, 2022, 15(1), 167, DOI:10.3390/polym15010167.
- S. Lanzalaco and E. Armelin, Poly(N-isopropylacrylamide) and Copolymers: A Review on Recent Progresses in Biomedical Applications, Gels, 2017, 3(4), 36, DOI:10.3390/gels3040036.
- N. Y. Becerra, B. L. López and L. M. Restrepo, Thermosensitive behavior in cell culture media and cytocompatibility of a novel copolymer: poly(N-isopropylacrylamide-co-butylacrylate), J. Mater. Sci.: Mater. Med., 2013, 24(4), 1043–1052, DOI:10.1007/S10856-013-4861-1.
- A. Baranowska-Korczyc, E. Stelmach, B. Paterczyk, K. Maksymiuk and A. Michalska, Ultrasmall self-assembly poly(N-isopropylacrylamide-butyl acrylate) (polyNIPAM-BA) thermoresponsive nanoparticles, J. Colloid Interface Sci., 2019, 542, 317–324, DOI:10.1016/j.jcis.2019.02.004.
- B. Natalia, A. Henry, L. Betty, R. L. Marina and R. Roberto, Probing poly(N-isopropylacrylamide-co-butylacrylate)/cell interactions by atomic force microscopy, J. Biomed. Mater. Res., Part A, 2015, 103(1), 145–153, DOI:10.1002/jbm.a.35163.
- N. L. Dumitrescu, M. Icriverzi and A. Bonciu, et al., New Poly(N-isopropylacrylamide-butylacrylate) Copolymer Biointerfaces and Their Characteristic Influence on Cell Behavior In Vitro, Int. J. Mol. Sci., 2022, 23(7), 3988, DOI:10.3390/ijms23073988.
- J. Chen, S. M. Guan, W. Sun and H. Fu, Melittin, the Major Pain-Producing Substance of Bee Venom, Neurosci. Bull., 2016, 32(3), 265–272, DOI:10.1007/s12264-016-0024-y.
- S. Guha, R. P. Ferrie and J. Ghimire, et al., Applications and evolution of melittin, the quintessential membrane active peptide, Biochem. Pharmacol., 2021, 193, DOI:10.1016/j.bcp.2021.114769.
- T. Picoli, C. M. Peter and J. L. Zani, et al., Melittin and its potential in the destruction and inhibition of the biofilm formation by Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa isolated from bovine milk, Microb. Pathog., 2017, 112, 57–62, DOI:10.1016/j.micpath.2017.09.046.
- E. Galdiero, A. Siciliano and R. Gesuele, et al., Melittin Inhibition and Eradication Activity for Resistant Polymicrobial Biofilm Isolated from a Dairy Industry after Disinfection, Int. J. Microbiol., 2019, 2019, 125654442, DOI:10.1155/2019/4012394.
- C. Lv, J. Chen, F. Huang, F. Fang and B. Li, Melittin inhibits the proliferation migration and invasion of HCC cells by regulating ADAMTS9-AS2 demethylation, Toxicon, 2023, 222, DOI:10.1016/j.toxicon.2022.106996.
- J. K. Badawi, Bee Venom Components as Therapeutic Tools against Prostate Cancer, Toxins, 2021, 13(5), 337, DOI:10.3390/toxins13050337.
- N. Y. Kwon, S. H. Sung, H. K. Sung and J. K. Park, Anticancer Activity of Bee Venom Components against Breast Cancer, Toxins, 2022, 14(7), 460, DOI:10.3390/toxins14070460.
- J. Yao, Z. Zhang, S. Li, B. Li and X. H. Wang, Melittin inhibits proliferation, migration and invasion of bladder cancer cells by regulating key genes based on bioinformatics and experimental assays, J. Cell. Mol. Med., 2020, 24(1), 655–670, DOI:10.1111/jcmm.14775.
- P. J. Russell, K. T. Ow and P. N. Tam, et al., Immunohistochemical characterisation of the monoclonal antibody BLCA-38 for the detection of prostate cancer, Cancer Immunol., Immunother., 2004, 53(11), 995–1004, DOI:10.1007/S00262-004-0527-7.
- Y. Li, S. Ruan, Z. Wang, N. Feng and Y. Zhang, Hyaluronic Acid Coating Reduces the Leakage of Melittin Encapsulated in Liposomes and Increases Targeted Delivery to Melanoma Cells, Pharmaceutics, 2021, 13(8), 1235, DOI:10.3390/pharmaceutics13081235.
- F. Dabbagh Moghaddam, I. Akbarzadeh and E. Marzbankia, et al., Delivery of melittin-loaded niosomes for breast cancer treatment: an in vitro and in vivo evaluation of anti-cancer effect, Artif. Cells, Nanomed., Biotechnol., 2021, 12(1), 1–35, DOI:10.1186/S12645-021-00085-9/figures/9.
- B. Cheng and P. Xu, Redox-Sensitive Nanocomplex for Targeted Delivery of Melittin, Toxins, 2020, 12(9), 582, DOI:10.3390/toxins12090582.
- M. Hematyar, M. Soleimani, A. Es-haghi and A. Rezaei Mokarram, Synergistic co-delivery of doxorubicin and melittin using functionalized magnetic nanoparticles for cancer treatment: loading and in vitro release study by LC-MS/MS, 2018, 46, 3, S1226–S1235, DOI:10.1080/21691401.2018.1536063.
- R. Mirzaei, M. Y. Alikhani, C. R. Arciola, I. Sedighi, R. Yousefimashouf and K. P. Bagheri, Prevention, inhibition, and degradation effects of melittin alone and in combination with vancomycin and rifampin against strong biofilm producer strains of methicillin-resistant Staphylococcus epidermidis, Biomed. Pharmacother., 2022, 147, DOI:10.1016/j.biopha.2022.112670.
- A. Marquette and B. Bechinger, Biophysical Investigations Elucidating the Mechanisms of Action of Antimicrobial Peptides and Their Synergism, Biomolecules, 2018, 8(2), 18, DOI:10.3390/biom8020018.
- K. A. M. McMillan and M. R. P. Coombs, Review: Examining the Natural Role of Amphibian Antimicrobial Peptide Magainin, Molecules, 2020, 25(22), 5436, DOI:10.3390/molecules25225436.
- S. Maher and S. McClean, Investigation of the cytotoxicity of eukaryotic and prokaryotic antimicrobial peptides in intestinal epithelial cells in vitro, Biochem. Pharmacol., 2006, 71(9), 1289–1298, DOI:10.1016/j.bcp.2006.01.012.
- A. Shin, E. Lee and D. Jeon, et al., Peptoid-Substituted Hybrid Antimicrobial Peptide Derived from Papiliocin and Magainin 2 with Enhanced Bacterial Selectivity and Anti-inflammatory Activity, Biochemistry, 2015, 54(25), 3921–3931, DOI:10.1021/acs.biochem.5b00392/suppl_file/bi5b00392_si_001.
- S. Liu, H. Yang and L. Wan, et al., Enhancement of cytotoxicity of antimicrobial peptide magainin II in tumor cells by bombesin-targeted delivery, Acta Pharmacol. Sin., 2011, 32(1), 79–88, DOI:10.1038/aps.2010.162.
- D. W. Hoskin and A. Ramamoorthy, Studies on anticancer activities of antimicrobial peptides, Biochim. Biophys. Acta, 2008, 1778(2), 357–375, DOI:10.1016/j.bbamem.2007.11.008.
- L. Nyström and M. Malmsten, Membrane interactions and cell selectivity of amphiphilic anticancer peptides, Curr. Opin. Colloid Interface Sci., 2018, 38, 1–17, DOI:10.1016/j.cocis.2018.06.009.
- A. R. Taylor, Methicillin-resistant Staphylococcus aureus infections, Prim Care., 2013, 40(3), 637–654, DOI:10.1016/j.pop.2013.06.002.
- Z. Pang, R. Raudonis, B. R. Glick, T. J. Lin and Z. Cheng, Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies, Biotechnol. Adv., 2019, 37(1), 177–192, DOI:10.1016/j.biotechadv.2018.11.013.
- L. Rusen, S. Brajnicov and P. Neacsu, et al., Novel degradable biointerfacing nanocomposite coatings for modulating the osteoblast response, Surf. Coat. Technol., 2017, 325, 397–409, DOI:10.1016/j.surfcoat.2017.06.045.
- L. N. Dumitrescu, P. Neacsu and M. G. Necula, et al., Induced Hydrophilicity and In Vitro Preliminary Osteoblast Response of Polyvinylidene Fluoride (PVDF) Coatings Obtained via MAPLE Deposition and Subsequent Thermal Treatment, Molecules, 2020, 25, 582, DOI:10.3390/molecules25030582.
- V. Dincă, A. Mocanu and G. Isopencu, et al., Biocompatible pure ZnO nanoparticles-3D bacterial cellulose biointerfaces with antibacterial properties, Arabian J. Chem., 2020, 13(1), 3521–3533, DOI:10.1016/j.arabjc.2018.12.003.
- U. Ben-David, G. Arad and U. Weissbein, et al., Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells, Nat. Commun., 2014, 5, 4825, DOI:10.1038/ncomms5825.
- C. V. A. Munteanu, G. N. Chiritoiu, M. Chiritoiu, S. Ghenea, A. J. Petrescu and S. M. Petrescu, Affinity Proteomics and Deglycoproteomics Uncover Novel EDEM2 Endogenous Substrates and an Integrative ERAD Network, Mol. Cell. Proteomics, 2021, 20, 100125, DOI:10.1016/j.mcpro.2021.100125.
- G. N. Chiritoiu, M. Chiritoiu and C. V. A. Munteanu, Dataset of human EDEM2 melanoma cells proteomics, affinity proteomics and deglycoproteomics, Data Brief, 2021, 39, 107471, DOI:10.1016/j.dib.2021.107471.
- S. H. Rajabnejad, A. Mokhtarzadeh, K. Abnous, S. M. Taghdisi, M. Ramezani and B. M. Razavi, Targeted delivery of melittin to cancer cells by AS1411 anti-nucleolin aptamer, Drug Dev. Ind. Pharm., 2018, 44(6), 982–987, DOI:10.1080/03639045.2018.1427760.
- H. N. Lim, S. B. Baek and H. J. Jung, Bee Venom and Its Peptide Component Melittin Suppress Growth and Migration of Melanoma Cells via Inhibition of PI3K/AKT/mTOR and MAPK Pathways, Molecules, 2019, 24(5), 929, DOI:10.3390/molecules24050929.
- P. Shaw, N. Kumar, D. Hammerschmid, A. Privat-Maldonado, S. Dewilde and A. Bogaerts, Synergistic Effects of Melittin and Plasma Treatment: A Promising Approach for Cancer Therapy, Cancers, 2019, 11(8), 1109, DOI:10.3390/cancers11081109.
- Q. L. Loh and C. Choong, Three-dimensional scaffolds for tissue engineering applications: role of porosity and pore size, Tissue Eng., Part B, 2013, 19(6), 485–502, DOI:10.1089/ten.teb.2012.0437.
- B. Bechinger, Detergent-like properties of magainin antibiotic peptides: a 31P solid-state NMR spectroscopy study, Biochim. Biophys. Acta, Biomembr., 2005, 1712(1), 101–108, DOI:10.1016/j.bbamem.2005.03.003.
- B. Majhy, P. Priyadarshini and A. K. Sen, Effect of surface energy and roughness on cell adhesion and growth – facile surface modification for enhanced cell culture, RSC Adv., 2021, 11(25), 15467–15476, 10.1039/d1ra02402g.
- L. Ferreira, M. M. Vidal and M. H. Gil, Evaluation of poly(2-hydroxyethyl methacrylate) gels as drug delivery systems at different pH values, Int. J. Pharm., 2000, 194(2), 169–180, DOI:10.1016/S0378-5173(99)00375-0.
- D. Buenger, F. Topuz and J. Groll, Hydrogels in sensing applications, Prog. Polym. Sci., 2012, 37(12), 1678–1719, DOI:10.1016/j.progpolymsci.2012.09.001.
- M. Rizwan, R. Yahya and A. Hassan, et al., pH Sensitive Hydrogels in Drug Delivery: Brief History, Properties, Swelling, and Release Mechanism, Material Selection and Applications, Polymers, 2017, 9(4), 137, DOI:10.3390/polym9040137.
- E. Galdiero, L. Lombardi, A. Falanga, G. Libralato, M. Guida and R. Carotenuto, Biofilms: Novel Strategies Based on Antimicrobial Peptides, Pharmaceutics, 2019, 11(7), 322, DOI:10.3390/pharmaceutics11070322.
- J. Lei, L. C. Sun and S. Huang, et al., The antimicrobial peptides and their potential clinical applications, Am. J. Transl. Res., 2019, 11(7), 3919 CAS.
- Y. Luo and Y. Song, Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities, Int. J. Mol. Sci., 2021, 22(21), 11401, DOI:10.3390/ijms222111401.
- Y. Cao, R. Q. Yu and Y. Liu, et al., Design, recombinant expression, and antibacterial activity of the cecropins-melittin hybrid antimicrobial peptides, Curr. Microbiol., 2010, 61(3), 169–175, DOI:10.1007/s00284-010-9592-7.
- A. M. Bardbari, M. R. Arabestani and M. Karami, et al., Highly synergistic activity of melittin with imipenem and colistin in biofilm inhibition against multidrug-resistant strong biofilm producer strains of Acinetobacter baumannii, Eur. J. Clin. Microbiol. Infect. Dis., 2018, 37(3), 443–454, DOI:10.1007/s10096-018-3189-7.
- L. Grassi, G. Maisetta, S. Esin and G. Batoni, Combination Strategies to Enhance the Efficacy of Antimicrobial Peptides against Bacterial Biofilms, Front. Microbiol., 2017, 8, 2409, DOI:10.3389/fmicb.2017.02409.
- R. Akbari, M. Hakemi-Vala, F. Pashaie, P. Bevalian, A. Hashemi and K. P. Bagheri, Highly Synergistic Effects of Melittin with Conventional Antibiotics Against Multidrug-Resistant Isolates of Acinetobacter baumannii and Pseudomonas aeruginosa, Microb. Drug Resist., 2019, 25(2), 193–202, DOI:10.1089/mdr.2018.0016.
- H. B. Koo and J. Seo, Antimicrobial peptides under clinical investigation, Pept. Sci., 2019, 111(5), e24122, DOI:10.1002/pep2.24122.
- J. H. Choi, A. Y. Jang and S. Lin, et al., Melittin, a honeybee venom-derived antimicrobial peptide, may target methicillin-resistant Staphylococcus aureus, Mol. Med. Rep., 2015, 12(5), 6483, DOI:10.3892/mmr.2015.4275.
- I. Haktanir, M. Masoura, F. T. Mantzouridou and K. Gkatzionis, Mechanism of antimicrobial activity of honeybee (Apis mellifera) venom on Gram-negative bacteria: Escherichia coli and Pseudomonas spp, AMB Express, 2021, 11(1), 54, DOI:10.1186/s13568-021-01214-8.
- W. G. Lima, J. C. M. de Brito, V. N. Cardoso and S. O. A. Fernandes, In-depth characterization of antibacterial activity of melittin against Staphylococcus aureus and use in a model of non-surgical MRSA-infected skin wounds, Eur. J. Pharm. Sci., 2021, 156, 105592, DOI:10.1016/J.EJPS.2020.105592.
- S. Dosler, E. Karaaslan and A. Alev Gerceker, Antibacterial and anti-biofilm activities of melittin and colistin, alone and in combination with antibiotics against Gram-negative bacteria, J. Chemother., 2016, 28(2), 95–103, DOI:10.1179/1973947815y.0000000004.
- G. Yu, D. Y. Baeder, R. R. Regoes and J. Rolff, Combination Effects of Antimicrobial Peptides, Antimicrob. Agents Chemother., 2016, 60(3), 1717–1724, DOI:10.1128/AAC.02434-15.
- A. Roy, N. K. Sarangi, S. Ghosh, A. Prabhakaran and T. E. Keyes, Leaflet by Leaflet Synergistic Effects of Antimicrobial Peptides on Bacterial and Mammalian Membrane Models, J. Phys. Chem. Lett., 2023, 14(16), 3920–3928, DOI:10.1021/acs.jpclett.3c00119/asset/images/large/jz3c00119_0005.jpeg.
- B. Deslouches and Y. Peter Di, Antimicrobial peptides with selective antitumor mechanisms: prospect for anticancer applications, Oncotarget, 2017, 8(28), 46635–46651, DOI:10.18632/oncotarget.16743.
- C. Sorenson and A. Eastman, Mechanism of cis-diamminedichloroplatinum(II)-induced cytotoxicity: role of G2 arrest and DNA double-strand breaks, Cancer Res., published online, 1988 Search PubMed.
- A. Krajewski, M. Gagat, A. Żuryń, M. Hałas-Wiśniewska, D. Grzanka and A. Grzanka, Cyclin F is involved in response to cisplatin treatment in melanoma cell lines, Oncol. Rep., 2020, 43(3), 765, DOI:10.3892/or.2020.7465.
- J. Lehmann, M. Retz and S. S. Sidhu, et al., Antitumor activity of the antimicrobial peptide magainin II against bladder cancer cell lines, Eur. Urol., 2006, 50(1), 141–147, DOI:10.1016/j.eururo.2005.12.043.
- P. Erkoc, B. M. von Reumont and T. Lüddecke, et al., The Pharmacological Potential of Novel Melittin Variants from the Honeybee and Solitary Bees against Inflammation and Cancer, Toxins, 2022, 14, 818, DOI:10.3390/toxins14120818.
- C. Lyu, F. Fang and B. Li, Anti-Tumor Effects of Melittin and Its Potential Applications in Clinic, Curr. Protein Pept. Sci., 2019, 20(3), 240–250, DOI:10.2174/1389203719666180612084615.
- G. Gajski and V. Garaj-Vrhovac, Melittin: a lytic peptide with anticancer properties, Environ. Toxicol. Pharmacol., 2013, 36(2), 697–705, DOI:10.1016/j.etap.2013.06.009.
- S. Sangboonruang, K. Kitidee, P. Chantawannakul, K. Tragoolpua and Y. Tragoolpua, Melittin from Apis florea Venom as a Promising Therapeutic Agent for Skin Cancer Treatment, Antibiotics, 2020, 9(8), 1–18, DOI:10.3390/antibiotics9080517.
- C. Tipgomut, A. Wongprommoon, E. Takeo, T. Ittiudomrak, S. Puthong and C. Chanchao, Melittin Induced G1 Cell Cycle Arrest and Apoptosis in Chago-K1 Human Bronchogenic Carcinoma Cells and Inhibited the Differentiation of THP-1 Cells into Tumour-Associated Macrophages, Asian Pac. J. Cancer Prev., 2018, 19(12), 3427, DOI:10.31557/apjcp.2018.19.12.3427.
- C. Soliman, S. Eastwood, V. K. Truong, P. A. Ramsland and A. Elbourne, The membrane effects of melittin on gastric and colorectal cancer, PLoS One, 2019, 14(10), e0224028, DOI:10.1371/journal.pone.0224028.
- W. Szlasa, I. Zendran, A. Zalesińska, M. Tarek and J. Kulbacka, Lipid composition of the cancer cell membrane, J. Bioenerg. Biomembr., 2020, 52(5), 321, DOI:10.1007/S10863-020-09846-4.
- S. Ganpule, A. K. Vijaya, A. Sukova and G. Preta, Membrane Cholesterol Content and Lipid Organization Influence Melittin and Pneumolysin Pore-Forming Activity, Toxins, 2022, 14(5), 346, DOI:10.3390/toxins14050346.
- C. Yaacoub, M. Rifi and D. El-Obeid, et al., The Cytotoxic Effect of Apis mellifera Venom with a Synergistic Potential of Its Two Main Components—Melittin and PLA2—On Colon Cancer HCT116 Cell Lines, Molecules, 2021, 26(8), 2264, DOI:10.3390/molecules26082264.
- S. Liu, H. Yang, L. Wan, J. Cheng and X. Lu, Penetratin-mediated delivery enhances the antitumor activity of the cationic antimicrobial peptide Magainin II, Cancer Biother.Radiopharm., 2013, 28(4), 289–297, DOI:10.1089/cbr.2012.1328.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00947e |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2024 |