Open Access Article
Open Access ArticleThe impact of macrophage phenotype and heterogeneity on the total internalized gold nanoparticle counts†
Henry
Lee
 a,
Dimitri
Vanhecke
a,
Dimitri
Vanhecke
 a,
Sandor
Balog
a,
Sandor
Balog
 a,
Patricia
Taladriz-Blanco
a,
Patricia
Taladriz-Blanco
 a,
Alke
Petri-Fink
a,
Alke
Petri-Fink
 ab and
Barbara
Rothen-Rutishauser
ab and
Barbara
Rothen-Rutishauser
 *a
*a
aAdolphe Merkle Institute, University of Fribourg, Switzerland, Chemin des Verdiers 4, Fribourg, Switzerland. E-mail: barbara.rothen@unifr.ch; Tel: +41-26-300-9502
bDepartment of Chemistry, University of Fribourg, Chemin du Musée 9, Fribourg, Switzerland
First published on 1st July 2024
Abstract
Macrophages play a pivotal role in the internalization and processing of administered nanoparticles (NPs). Furthermore, the phagocytic capacity and immunological properties of macrophages can vary depending on their microenvironment, exhibiting a spectrum of polarization states ranging from pro-inflammatory M1 to anti-inflammatory M2. However, previous research investigating this phenotype-dependent interaction with NPs has predominantly relied on semi-quantitative techniques or conventional metrics to assess intracellular NPs. Here, we focus on the interaction of human monocyte-derived macrophage phenotypes (M1-like and M2-like) with gold NPs (AuNPs) by combining population-based metrics and single-cell analysis by focused ion beam-scanning electron microscopy (FIB-SEM). The multimodal analysis revealed phenotype-dependent response and uptake behavior differences, becoming more pronounced after 48 hours. The study also highlighted phenotype-dependent cell-to-cell heterogeneity in AuNPs uptake and variability in particle number at the single-cell level, which was particularly evident in M2-like macrophages, which increases with time, indicating enhanced heteroscedasticity. Future efforts to design NPs targeting macrophages should consider the phenotypic variations and the distribution of NPs concentrations within a population, including the influence of cell-to-cell heterogeneity. This comprehensive understanding will be critical in developing safe and effective NPs to target different macrophage phenotypes.
Introduction
Nanoparticles (NPs) have shown tremendous potential in various biomedical applications, including drug delivery and imaging capabilities.1–3 However, the successful development and implementation of NPs-based therapies require a comprehensive and holistic understanding of how these NPs interact with a biological system, particularly with immune cells. As an integral part of our innate immune system, macrophages have emerged as critical players in nanomedicine.4,5 They play an essential role in the uptake and clearance of NPs from the human body after administration, thus contributing significantly to their fate in the human body.6–8 Macrophages exhibit remarkable phenotypic plasticity and can adopt a spectrum of activation states in response to their microenvironment, ranging from the pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype.9 Previous investigations showed that the interplay between macrophages and NPs is modulated by their activation state, resulting in varying levels of NPs uptake.10–14 Furthermore, cell-to-cell heterogeneity, a phenomenon in which individual cells within a population exhibit differences in their properties, functions, and behavior, may influence the uptake and fate of NPs.15,16 Consequently, there is a recognized need to understand the impact of the variability in interactions between individual cells on the collective response of a cell population when exposed to NPs.Commonly used methods to detect and measure NPs uptake encompass fluorescence-based methods, including flow cytometry17,18 and confocal microscopy,17,19 inductively coupled plasma-optical emission spectroscopy20,21 (ICP-OES) for elemental analysis and transmission electron microscopy22,23 for high-resolution imaging. The choice of technique depends on the properties of the NPs material and labeling strategy.24 Given the particular focus on understanding NPs interactions at the single-cell level, quantitative methods can be categorized into population-averaged data and those providing information on the single-cell level. For instance, flow cytometry is a high throughput method that offers valuable information on the presence and distribution of NPs in a cell population by utilizing fluorescent and optical signals, including scattering phenomena from the particles.25,26 However, it is considered a semi-quantitative technique as it does not directly attribute the optical signals to an absolute number of NPs but derives mean intensities. Additionally, it cannot distinguish between internalized and membrane-bound NPs.24 In this respect, flow cytometry is most effectively used in conjunction with other techniques to provide comprehensive information about NP uptake, such as with ICP-OES, a powerful tool with a very sensitive detection range. In particular, ICP-OES is widely used to quantify the cumulative mass of different elements derived from metal-based NPs, including entities such as gold (AuNPs) or silver NPs (AgNPs) within cells or tissues.27 As it requires sample digestion before analysis, this technique does not provide spatial information on intracellular or membrane-associated NPs. To overcome these limitations and to complement the previously mentioned methods, we employed focused ion beam scanning electron microscopy (FIB-SEM) tomography for three-dimensional (3D) imaging. This technique uses a focused gallium (Ga+) ion beam to generate cross-sectional cell slices for SEM imaging, producing a 3D image in silico.28 As metallic NPs have inherent contrast for SEM imaging due to their increased secondary electron yield, no additional particle staining or labeling is required.29 With a lateral resolution of approximately 1 nm and a slice thickness in the tens of nm,30 FIB/SEM can be considered optimal for distinguishing between membrane-associated NPs and internalized NPs and accurately quantifying NPs numbers at the single-cell level.
We investigated the internalization and the downstream effects of AuNPs on the colony-stimulating factor (GM-CSF, i.e., M1-like) and macrophage colony-stimulating factor (M-CSF, i.e., M2-like) differentiated primary human monocyte-derived macrophages (MDM),31–34 which are post-mitotic and do not undergo division, thereby reducing an additional variable that affects the quantification of internalized NPs.35 This investigation involved the integration of the cell sorting capabilities of flow cytometry with ICP-OES analysis, which allowed the determination of the average AuNPs content associated with the previously sorted cell population. The population-averaged observations for AuNPs counts were then examined using FIB-SEM tomography at the single-cell level of entire cells. We showed that the cell-to-cell heterogeneity of variance, also referred to as heteroscedasticity, in the number of internalized particles was phenotype-dependent and more pronounced over time in M-CSF macrophages.
Results
Modulation of surface marker expression and cytokine secretion by AuNPs is phenotype-dependent
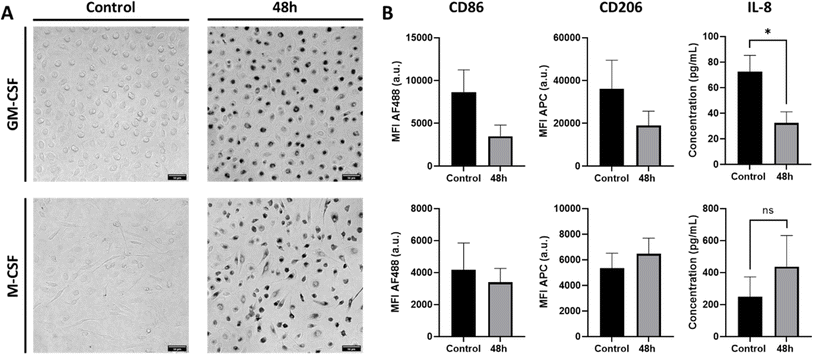
Macrophage polarization and the downstream effects of AuNPs exposure were assessed by examining cell morphology, surface marker expression, and basal cytokine/chemokine production (Fig. 1). The AuNPs used in the study had a diameter of 45 nm as determined by transmission electron microscopy (TEM) counting 203 particles and remained stable in the cell culture medium for 48 h (ESI (Fig. S1 and Table S1†)). Cell viability in macrophages, as determined by WST-1 assays, showed no significant reduction in formazan formation, suggesting that the cells maintain metabolic activity and remain viable despite in presence of NPs (Fig. S2†). Initially, bright-field imaging revealed that the morphology of GM-CSF macrophages differed from that of M-CSF macrophages, and the morphology did not reveal any adverse effect (Fig. 1A). GM-CSF macrophages tended to be round, whereas M-CSF macrophages appeared more heterogeneous with rounded and elongated shapes. After 48 h of exposure to AuNPs, the concentration of AuNPs used here did not result in any visible cell damage, including apoptotic or necrotic events. Furthermore, over time, there was an increased interaction between AuNPs and macrophages, accompanied by variations in the distribution of cell-associated AuNPs, resulting in amplified contrast within the cell population (Fig. 1A). Subsequently, after the exclusion of cell debris, duplets, and DAPI positive events (Fig. S3†), flow cytometry data showed that GM-CSF-treated macrophages exhibited higher expression levels of the mannose receptor CD206 and the costimulatory membrane-associated protein CD86 for T-cell activation than M-CSF macrophages (Fig. S4†), suggesting phenotypic differences. While no significant changes in CD86 and CD206 expression levels were observed after exposure to AuNPs at any time point (Fig. S5†), distinct trends were observed after 48 hours of incubation (Fig. 1B). Specifically, AuNPs caused a decrease in the expression levels of both markers in GM-CSF macrophages. In contrast, M-CSF macrophages responded with a decrease in CD86 and an increase in CD206 expression levels. Finally, M-CSF-treated macrophages showed a higher basal interleukin (IL)-8 production than GM-CSF macrophages (Fig. S4†). At the same time, other cytokines such as IL-1β, IL-6, and TNF-α were not detected in either macrophage phenotype. In addition, different IL-8 release profiles were observed after 48 hours of AuNP exposure, with significantly reduced levels detected in the medium of GM-CSF macrophages and no change observed in M-CSF macrophages.Multimodal analysis of AuNPs interaction with GM-CSF and M-CSF macrophages
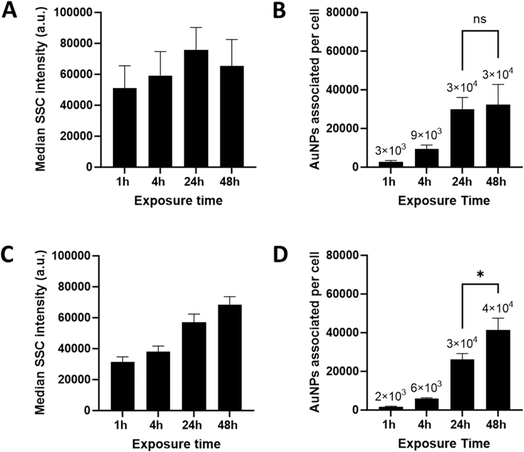
A multimodal strategy was used to investigate the interaction of AuNPs with GM-CSF and M-CSF macrophages in a cell population. First, flow cytometry side scatter intensities were used to assess the interaction of AuNPs with cells. Since the internalization of AuNPs affects intracellular properties, such as granularity, side scattering (SSC) can be used to detect the association of AuNPs with cells.25 However, SSC signals cannot distinguish between AuNPs adhering to the outer cell membrane and intracellular AuNPs. Mean SSC intensity shows a trend toward uptake saturation in GM-CSF macrophages after 24 hours of exposure (Fig. 2A). In contrast, SSC intensity in M-CSF macrophages increases continuously up to 48 hours after AuNP exposure (Fig. 2C). Additionally, ICP-OES was used to quantify the cell-associated number of AuNPs. Both macrophage phenotypes showed similar behavior up to 24 hours of exposure. Slightly higher average particle counts of 3400 NPs associated per cell at 1 hour and 9000 NPs associated per cell at 4 hours were detected in GM-CSF macrophages (Fig. 2B) compared to 1700 and 5800 NPs associated per cell in M-CSF macrophages (Fig. 2D). An average of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 NPs associated per cell were quantified after 24 hours of exposure. Interestingly, only a small increase in NPs associated per cell was observed in GM-CSF-differentiated macrophages (Fig. 2B) after 48 hours (31
000 NPs associated per cell were quantified after 24 hours of exposure. Interestingly, only a small increase in NPs associated per cell was observed in GM-CSF-differentiated macrophages (Fig. 2B) after 48 hours (31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 particles per cell). In contrast, following 48 h of exposure, M-CSF macrophages show a significant increase of up to 41
600 particles per cell). In contrast, following 48 h of exposure, M-CSF macrophages show a significant increase of up to 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 NPs associated per cell (Fig. 2D) compared to 24 h.
400 NPs associated per cell (Fig. 2D) compared to 24 h.
Absolute AuNPs number determined at the single cell level by FIB-SEM
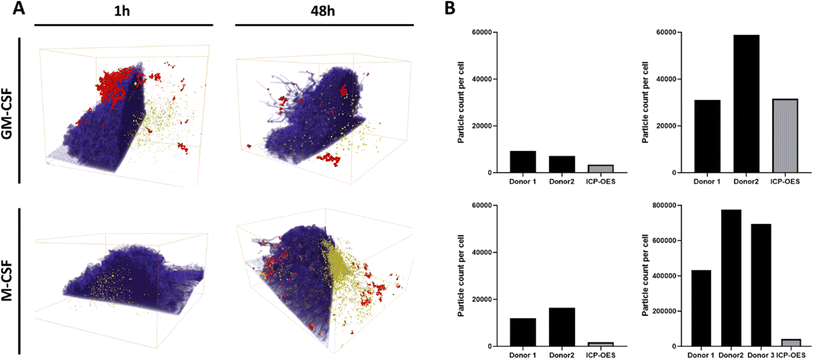
To differentiate between internalized and adherent AuNPs, the total number of internalized AuNPs per cell was counted using FIB-SEM tomography, the slice and view microscopy imaging method, which allows the collection of a volume of up to several tens of μm with a spatial resolution in X, Y, and Z down to a few nm.28,36 FIB-SEM is a demanding low throughput technique;37,38 therefore, only the 1 h and 48 h time points were analyzed. Fig. 3 shows representative cross-sections for each of the above conditions. Intracellular AuNPs were identified in both macrophage phenotypes at both time points examined. Interestingly, larger agglomerates adhering to the outer cell membrane were observed, for instance, after 1 h in GM-CSF macrophages and after 48 h in M-CSF macrophages. FIB-SEM images also show clear morphological differences between GM-CSF and M-CSF macrophages: GM-CSF macrophages have more hemispherical bodies with short and dense filopodia, whereas M-CSF macrophages have flatter cell bodies with longer filopodia's. These differences were also observed by bright-field microscopy, as shown in Fig. 1A. Due to phenotype-derived and cell-to-cell heterogeneity, the cell volume differs, respectively (see Table 1). The 3D reconstruction (Fig. 4A) and the calculation of the absolute number of AuNPs per cell (Fig. 4B) were performed by segmenting the raw data into the cell body, intracellular and extracellular AuNPs (see ESI† for the algorithm). The renderings of the 3D data show one of the major strengths of this technique, as membrane-associated, i.e., adherent, and internalized AuNPs can be clearly distinguished. The volume for the cell, the extracellular and intracellular AuNPs were derived from the number of voxels assigned to each class time (see Table 1). In the case of the AuNPs, the number of NPs within individual cells, referred to as cardinality, was derived by assuming that all AuNPs had an ideal spherical shape with a uniform diameter of 45 nm: each AuNPs thus had a volume of ∼47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 713 nm3. The data at the single-cell level revealed cell-to-cell variability for both macrophage phenotypes after 1 hour of exposure, as the numbers ranged between 7000 and 16
713 nm3. The data at the single-cell level revealed cell-to-cell variability for both macrophage phenotypes after 1 hour of exposure, as the numbers ranged between 7000 and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 AuNPs per cell. The numbers of intracellular AuNPs per cell increased significantly after 48 h of exposure. Approximately 31
000 AuNPs per cell. The numbers of intracellular AuNPs per cell increased significantly after 48 h of exposure. Approximately 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 56
000 and 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 intracellular AuNPs per cell were counted in GM-CSF macrophages, while the number of AuNPs ranged from 433
100 intracellular AuNPs per cell were counted in GM-CSF macrophages, while the number of AuNPs ranged from 433![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 775
000 to 775![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in M-CSF macrophages. The numbers of AuNPs per cell in GM-CSF macrophages measured by FIB-SEM were close to the number of AuNPs determined by ICP-OES after 48 h of exposure. In contrast, we found approximately 10 to 20 times more particles per cell by FIB-SEM at the single-cell level for M-CSF macrophages than ICP-OES.
000 in M-CSF macrophages. The numbers of AuNPs per cell in GM-CSF macrophages measured by FIB-SEM were close to the number of AuNPs determined by ICP-OES after 48 h of exposure. In contrast, we found approximately 10 to 20 times more particles per cell by FIB-SEM at the single-cell level for M-CSF macrophages than ICP-OES.
| 1 h AuNPs | 48 h AuNPs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | GM-CSF | M-CSF | GM-CSF | M-CSF | |||||
| Donor | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 3 |
| Cell volume (μm3) | 1938 | 1361 | 1068 | 2751 | 2267 | 2129 | 1593 | 2960 | 2167 |
| NPs volume per cell (μm3) | 0.44 | 0.34 | 0.57 | 0.78 | 1.48 | 2.68 | 20.65 | 36.96 | 33.10 |
| Cardinal number (NPs per cell) | 9300 | 7147 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 857 857 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 349 349 |
31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 050 050 |
56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 136 136 |
432![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 774 774 |
774![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 553 553 |
693![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 606 606 |
Discussion
Macrophages are functionally versatile immune cells that play a crucial role in the immune system, serving as sentinels to detect and eliminate pathogens, regulate tissue homeostasis, and repair processes.39 It is well-known that their phenotypic heterogeneity is influenced by the microenvironment and various factors, resulting in different activation states along the M1/M2 polarization spectrum.40 The interaction between NPs and macrophages has received considerable attention, and it is widely recognized that the polarization status of macrophages strongly influences NPs uptake and cellular responses.5,13,41 Nevertheless, previous investigations of this phenotype-dependent interaction with NPs have primarily relied on semi-quantitative techniques or other standard metrics, overlooking the existing cellular variability within the population and the possibility of distinguishing between internalized and adsorbed particles.In our study, we differentiated and polarized macrophages using the hematopoietic growth factors GM-CSF and M-CSF, commonly used to mimic the pro-inflammatory M1 and anti-inflammatory M2 activation states, respectively.33,42 We found that prolonged exposure to AuNPs, up to 48 h, induced phenotype-dependent changes in macrophage polarization and cytokine release status. These results highlight the dynamic nature of macrophage responses to NPs stimuli. While the effects of AuNPs on the immune status of macrophages vary due to different sources of macrophages and the physicochemical properties of the AuNPs,43,44 it is evident that NPs can re-educate macrophages.45,46
Depending on the polarization profile, macrophages show time-dependent differences in AuNPs internalization, which could be attributed to differences in cell surface receptor profiles and expressions. Several studies have provided evidence for CD206 as a potential mediator of particle internalization in macrophages.13,41,47,48 We observed time-resolved differences in AuNPs uptake by GM-CSF and M-CSF macrophages that correlated positively with the trends in CD206 expression. GM-CSF macrophages showed higher particle numbers than M-CSF macrophages within the first 4 hours of exposure. Around 24 h after exposure, GM-CSF macrophages reached its plateau regarding uptake, with parallel trends in CD206 expression levels decreasing over time. In contrast, M-CSF macrophages internalize AuNPs continuously up to 48 h of exposure, where a trend of a slight increase in the expression level of CD206 was observed. However, it is important to note that the observed modulation in the expression levels of CD206 did not reach statistical significance. Furthermore, the interaction of macrophages with NPs involves different receptors and internalization mechanisms. For instance, phagocytosis can be mediated by different receptors, such as scavenger and complement receptors.49,50 In the case of M-CSF macrophages, they express higher levels of scavenger and lectin receptors, which may contribute to increased NPs clearance.41
Notably, the analysis of AuNPs–macrophage interaction at the single-cell level by FIB-SEM tomography highlights the importance of cell-to-cell heterogeneity in AuNPs uptake and its progression over time. This detailed information is not accessible through population-averaging techniques like ICP-OES or flow cytometry analysis. Cell-to-cell heterogeneity refers to the inherent variability within a population, and its impact on the interaction with NPs has been emphasized by previous studies. For instance, Åberg et al.51 demonstrated that uptake is influenced by cell-derived sources of variability, such as cell size and differences in the gene expression profile. Another study demonstrated the importance of population heterogeneity in terms of uptake, where they found amounts of Ag ranging from 17 to 121 fg per cell in human T-lymphocytes by mass cytometry.52 Finally, regarding the number of NPs per cell, various studies have reported 103 to 107 NPs per cell for different cell types using various techniques.20,37,38,53–55 For instance, Chou et al.53 estimated, using fluorescence, that there were 8000 nanoparticles per cell after 4 h of exposure at a concentration of 3.0 × 1011 per mL in RAW 264.7 macrophages. Another study found a total of 3138 ± 722 AgNPs in a single THP-1-derived macrophage after 24 h of exposure at a concentration of 10 μg mL−1, using FIB-SEM tomography.38 Our findings on the number of NPs per cell, acquired through population-averaging and single-cell methodologies, align with the previously reported data. Intriguingly, we observed a consistent pattern of uptake behavior dependent on the cell phenotype in both techniques. Specifically, macrophages stimulated with GM-CSF and M-CSF showed a similar rate of internalizing AuNPs up to 24 hours of exposure, as evidenced by the average count of NPs per cell measured via ICP-OES. This similarity was also evident at the individual cell level, with closely corresponding NPs counts. However, after 48 hours, we noticed a divergence in the internalization behavior of AuNPs between these two phenotypes. GM-CSF macrophages seemed to reach a saturation point in uptake, as the average number of NPs per cell did not significantly differ from the number after 24 hours. Conversely, M-CSF macrophages exhibited continuous internalization behavior, evidenced by the average number of NPs per cell measured through ICP-OES and further supported by higher counts at the single-cell level obtained using FIB-SEM. While ICP-OES solely provides information on the average NPs concentration and cannot delineate the distribution within a cell population, our single-cell sampling through FIB-SEM also allowed us to address the variance and heterogeneity in NPs count at the individual cell level to some extent. We observed that M-CSF macrophages had a considerably higher capacity for internalizing NPs, as revealed by FIB-SEM tomography, with uptake levels ranging from 10 to 20 times more than the average of 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 NPs per cell measured by ICP-OES. Conversely, in GM-CSF macrophages, we did not observe the same deviation in particle numbers at the single-cell level after 48 h of exposure, as the numbers were closer to the measurements obtained from the population-based analysis using ICP-OES. Additional flow cytometry analysis (Fig. S6†), focusing on the standard deviation of SSC signals, showed a significant increase in SSC signal variability after 24 hours in M-CSF macrophages, whereas such variability was not observed in GM-CSF macrophages. These findings may suggest an additional layer of complexity in understanding the interaction between macrophages and NPs, which is not only derived from the polarization status of macrophages but also from the variation in cell-to-cell heterogeneity, which may differ among phenotype populations and becomes more apparent over time through NPs internalization behavior. This means that GM-CSF macrophages exhibit a more homogeneous population. In contrast, M-CSF macrophages exhibit inherent variability and extremes in functionalities and signatures, which leads to heteroscedasticity in the number of NPs taken up by each cell. Moreover, the number of NPs revealed at the single-cell level using FIB-SEM tomography highlights the need to re-evaluate the efficacy and safety of engineered nanomaterials for therapeutic applications. The observed variance in delivered NPs doses between cells may lead to different cell effects. This underscores the significance of considering cell-to-cell variability when assessing the impact of nanomaterials on biological systems, as it can have substantial implications for their safety and effectiveness across diverse applications. In particle counting by FIB-SEM, several factors significantly impact accuracy and quality. Key elements include the data acquisition setup, where high x, y, and z resolutions capture finer particle details, enhancing segmentation accuracy despite increasing acquisition time and dataset size. Using 16 bit grayscale levels improves contrast and detail, aiding in precise particle identification. Beam current and dwell time must be optimized to balance signal intensity, signal-to-noise ratio, and sample integrity. The segmentation process also plays a crucial role. State-of-the-art segmentation algorithms, such as in Ilastik, classify data into predefined categories, such as cells and AuNPs, reducing experimenter bias and improving accuracy and speed over manual methods. Conversely, AuNPs aggregation does not affect particle counting. The emission intensity of electromagnetic radiation is governed by the Coulomb law, and is thus dependent solely on the atomic number of the material in question, regardless of the presence of neighboring structures. Consequently, the high atomic number of Au (Z = 79) ensures efficient electron scattering, resulting in a strong backscattered electron signal in a SEM,56 which enables clear visualization regardless of the aggregation state. This ensures consistent imaging quality, making AuNPs reliable markers in FIB-SEM analysis. Detecting non-metallic NPs using FIB-SEM is challenging due to their low electron density. However, meticulous sample preparation, including fixation, staining with heavy metals, and embedding, can enhance detection. Adjusting imaging parameters, such as acceleration voltage and dwell time, and using energy-dispersive X-ray spectroscopy (EDS) for elemental analysis further improve visibility and identification. Advanced algorithms, like Ilastik, assess features such as texture and edge distance for successful segmentation. Thus, FIB-SEM, combined with these techniques, enables accurate quantification of both metallic and non-metallic NPs within cells. Additionally, each technique employed has its limitations. Population-based techniques provide statistically significant data, but retrieving quantitative data at the single-cell level regarding absolute particle numbers is challenging. Conversely, while FIB-SEM tomography allows for the determination of absolute numbers, it is, however, a low-throughput method, and achieving statistical significance requires analyzing a larger number of cells, which can be time-consuming. By integrating various techniques, each providing valuable quantitative data, we were able to highlight the role of cellular heterogeneity and phenotype dependence in NP uptake. Further elucidating this heterogeneity and the underlying genes or mechanisms driving variance in NPs uptake requires additional experiments. Isolating populations with low and high uptake and expanding studies to include different NPs concentrations, sizes, and macrophage phenotypes will result in a more in-depth understanding of NPs-macrophage interactions at the quantitative level.
000 NPs per cell measured by ICP-OES. Conversely, in GM-CSF macrophages, we did not observe the same deviation in particle numbers at the single-cell level after 48 h of exposure, as the numbers were closer to the measurements obtained from the population-based analysis using ICP-OES. Additional flow cytometry analysis (Fig. S6†), focusing on the standard deviation of SSC signals, showed a significant increase in SSC signal variability after 24 hours in M-CSF macrophages, whereas such variability was not observed in GM-CSF macrophages. These findings may suggest an additional layer of complexity in understanding the interaction between macrophages and NPs, which is not only derived from the polarization status of macrophages but also from the variation in cell-to-cell heterogeneity, which may differ among phenotype populations and becomes more apparent over time through NPs internalization behavior. This means that GM-CSF macrophages exhibit a more homogeneous population. In contrast, M-CSF macrophages exhibit inherent variability and extremes in functionalities and signatures, which leads to heteroscedasticity in the number of NPs taken up by each cell. Moreover, the number of NPs revealed at the single-cell level using FIB-SEM tomography highlights the need to re-evaluate the efficacy and safety of engineered nanomaterials for therapeutic applications. The observed variance in delivered NPs doses between cells may lead to different cell effects. This underscores the significance of considering cell-to-cell variability when assessing the impact of nanomaterials on biological systems, as it can have substantial implications for their safety and effectiveness across diverse applications. In particle counting by FIB-SEM, several factors significantly impact accuracy and quality. Key elements include the data acquisition setup, where high x, y, and z resolutions capture finer particle details, enhancing segmentation accuracy despite increasing acquisition time and dataset size. Using 16 bit grayscale levels improves contrast and detail, aiding in precise particle identification. Beam current and dwell time must be optimized to balance signal intensity, signal-to-noise ratio, and sample integrity. The segmentation process also plays a crucial role. State-of-the-art segmentation algorithms, such as in Ilastik, classify data into predefined categories, such as cells and AuNPs, reducing experimenter bias and improving accuracy and speed over manual methods. Conversely, AuNPs aggregation does not affect particle counting. The emission intensity of electromagnetic radiation is governed by the Coulomb law, and is thus dependent solely on the atomic number of the material in question, regardless of the presence of neighboring structures. Consequently, the high atomic number of Au (Z = 79) ensures efficient electron scattering, resulting in a strong backscattered electron signal in a SEM,56 which enables clear visualization regardless of the aggregation state. This ensures consistent imaging quality, making AuNPs reliable markers in FIB-SEM analysis. Detecting non-metallic NPs using FIB-SEM is challenging due to their low electron density. However, meticulous sample preparation, including fixation, staining with heavy metals, and embedding, can enhance detection. Adjusting imaging parameters, such as acceleration voltage and dwell time, and using energy-dispersive X-ray spectroscopy (EDS) for elemental analysis further improve visibility and identification. Advanced algorithms, like Ilastik, assess features such as texture and edge distance for successful segmentation. Thus, FIB-SEM, combined with these techniques, enables accurate quantification of both metallic and non-metallic NPs within cells. Additionally, each technique employed has its limitations. Population-based techniques provide statistically significant data, but retrieving quantitative data at the single-cell level regarding absolute particle numbers is challenging. Conversely, while FIB-SEM tomography allows for the determination of absolute numbers, it is, however, a low-throughput method, and achieving statistical significance requires analyzing a larger number of cells, which can be time-consuming. By integrating various techniques, each providing valuable quantitative data, we were able to highlight the role of cellular heterogeneity and phenotype dependence in NP uptake. Further elucidating this heterogeneity and the underlying genes or mechanisms driving variance in NPs uptake requires additional experiments. Isolating populations with low and high uptake and expanding studies to include different NPs concentrations, sizes, and macrophage phenotypes will result in a more in-depth understanding of NPs-macrophage interactions at the quantitative level.
Conclusion
Taken together, the data obtained indicate several relevant aspects for understanding the interaction of NPs with different macrophage phenotypes. First, macrophage polarization plays an important role in determining the uptake behavior of NPs. This is evident from the differences in NPs numbers observed through different methods after 48 h of exposure. These differences highlight the importance of considering the macrophage phenotype when studying the interaction with NPs and the potential translation of a promising particle for therapeutic purposes. Second, further exploration of the mechanisms underlying cell-to-cell heterogeneity and their impact on NPs uptake at the single-cell level will be essential for the development of more precise and effective nanomedicine approaches. Finally, we have demonstrated that the FIB-SEM is the only technique capable of highly precise and accurately revealing the total number of high-contrast intracellular NPs at the single-cell level. Its unique capabilities to visualize and count NPs within cellular structures have significant implications for future research in nanomedicine, the assessment of NPs, and the development of more precise and effective NPs-based therapies, which will advance our understanding of NPs-cell interactions from a different perspective and contribute to the safe and effective implementation of nanotechnology in various biomedical fields.Methods
Synthesis of gold nanoparticles
Briefly, 15 nm AuNPs and further used as seeds were prepared by boiling 0.5 mM of gold salt (tetrachloroauric acid, HAuCl4·3H2O, 99%, Sigma-Aldrich, Switzerland) in sodium citrate (1.5 mM, C6H5Na3O7·H2O, 98%, Sigma-Aldrich, Switzerland). The dispersion was cooled down to room temperature and stored in the fridge until further use. AuNPs with a diameter of ∼45 nm were prepared by seeded growth following the Brown method et al.57 1.34 mL of hydroxylamine hydrochloride (0.22 M, NH2OH·HCl, ACS Reagent ≥ 99%, Sigma-Aldrich, Switzerland) was added under magnetic stirring to a solution containing gold salt (0.25 mM), as-prepared gold seeds of 15 nm by the Turkevich method (0.0125 mM), and sodium citrate (0.5 mM).58 Particles were washed by centrifugation for 20 min at 3500 rpm and concentrated in a 1 mM sodium citrate solution.Physicochemical characterization
The AuNPs were imaged using a Tecnai Spirit transmission electron microscope (TEM) operating at 120 kV (TEM, FEI, Columbia, MD, USA). The core diameter and size distribution were calculated using an open-source image processing program (ImageJ, Wayne Rasband National Institute of Health, Bethesda, MD, USA59). The UV-Vis extinction spectrum of AuNPs was measured at 25 °C using Suprasil-grade quartz cuvettes with a 10 mm optical path length in a Jasco V-670 spectrophotometer (Hellma Analytics in Plainview, NY, USA). The phase-amplitude light scattering technique (ZetaPALS, Brookhaven Instruments Corp., USA) was used to determine the zeta potential of particles suspended in water. The hydrodynamic diameter was measured via polarized dynamic light scattering (3D LS Spectrometer, LS Instruments AG, Fribourg, Switzerland) in Milli-Q H2O at 25 °C and in complete cell culture media at 0 and 48 h at 37 °C.Endotoxin content
The endotoxin concentration of the AuNPs suspensions was measured using the Pierce™ LAL Chromogenic Endotoxin Quantitation Kit (Thermo Fisher Scientific, Basel, Switzerland), according to the manufacturer's instructions. The AuNPs suspension was less than 0.5 EU mL−1.Cell culture
The study on primary human MDM was approved by the Federal Office for Public Health Switzerland committee (reference number: 611-1, Meldung A110635/2). Macrophages were prepared from a buffy coat (donation service by Interregionale Blutspende SRK AG, Bern, Switzerland) according to previously developed protocol.60 Peripheral blood mononuclear cells were obtained by fractionation of human blood using the density gradient medium (Lymphoprep, Grogg Chemie, Stettlen, Switzerland), and monocytes were further isolated and purified with magnetic CD14+ beads (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) according to the manufacturer's protocol. Monocytes were cultured in Roswell Park Memorial Institute (RPMI-1640) medium supplemented with 10% (v/v) fetal bovine serum, 2 mM L-glutamine, and penicillin/streptomycin (100 units per mL and 100 mg mL−1, respectively) at a density of 106 cells per mL with either 10 ng per mL GM-CSF or M-CSF for 6 days (Miltenyi Biotec, Bergisch Gladbach, Germany). The cells were seeded and cultured in 6-well plates (Corning, USA).Cytotoxicity assay
The WST-1 assay (Roche Diagnostics, Mannheim, Germany) was used for the spectrometric assessment of cell viability. After removal of the cell culture medium, macrophages were incubated for 30 min with 100 μL of freshly prepared WST-1 solution diluted in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 cRPMI. Absorbance was measured at a wavelength of 450 nm. Cells lysed in Triton X-100 were used as a positive control. All assays were performed in triplicates.
10 cRPMI. Absorbance was measured at a wavelength of 450 nm. Cells lysed in Triton X-100 were used as a positive control. All assays were performed in triplicates.
Phase contrast imaging
Cells were seeded in 8-well μ-slides and exposed to AuNPs for 1 h, 4 h, 24 h, and 48 h. After rinsing three times with PBS, cells were fixed with 4% paraformaldehyde (PFA, in PBS, w/v) for 15 min at room temperature. After each well was washed three times with PBS, 300 μL was added to each well before image acquisition with Leica Stellaris 5 confocal microscope.Cell sorting
6-Well cell culture plates were used to grow the cells for flow cytometry. The supernatant was collected and stored at −80 °C for the ELISA experiment. The cells were gently washed once with PBS before incubation with 1 mL of Accutase™ (cat. no. 00-4555-56, Thermo Fisher Scientific) for 15 min at 37 °C. The macrophages were then scraped off with a cell scraper in 1 mL of flow cytometry buffer and collected in a flow cytometry tube (5 mL polystyrene round-bottom tube, Corning® Falcon, Reinach, Switzerland). The supernatant was discarded after the centrifugation step for 5 min at 300 × g. The cell pellet was resuspended in 100 μL Fc-Block (eBioscience, Thermo Fisher) diluted in cold flow cytometry buffer 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 for 10 min on ice. Cells were stained with an antibody cocktail containing Alexa Fluor 488 conjugated anti-human CD86 (B7-2) monoclonal antibody (clone IT2.2) and APC conjugated anti-human CD206 (MMR) monoclonal antibody (clone 19.2) at a 1
10 for 10 min on ice. Cells were stained with an antibody cocktail containing Alexa Fluor 488 conjugated anti-human CD86 (B7-2) monoclonal antibody (clone IT2.2) and APC conjugated anti-human CD206 (MMR) monoclonal antibody (clone 19.2) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution each for 20 min on ice in the dark. After washing, cells were incubated for 5 min on ice in a flow cytometry buffer containing 2 μM 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, Switzerland) to exclude dead cells. Finally, the cells were washed with 3 mL flow cytometry buffer and centrifuged at 500 RCF for 5 min at 4 °C. The cell pellet was resuspended with cold flow cytometry buffer and stored on ice until data acquisition and sorting using CD FACSARIA Fusion (BD Biosciences). Analysis was performed using FlowJo software (version 10.8.1, Treestar, United States).
100 dilution each for 20 min on ice in the dark. After washing, cells were incubated for 5 min on ice in a flow cytometry buffer containing 2 μM 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, Switzerland) to exclude dead cells. Finally, the cells were washed with 3 mL flow cytometry buffer and centrifuged at 500 RCF for 5 min at 4 °C. The cell pellet was resuspended with cold flow cytometry buffer and stored on ice until data acquisition and sorting using CD FACSARIA Fusion (BD Biosciences). Analysis was performed using FlowJo software (version 10.8.1, Treestar, United States).
Inductively coupled plasma-optical emission spectra
After cell sorting, a total of 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 300
000 to 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 collected cells per sample were then resuspended in 300 μL of a mixture of H2O2
000 collected cells per sample were then resuspended in 300 μL of a mixture of H2O2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HNO3 (2
HNO3 (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Sigma-Aldrich, USA) overnight at room temperature in a flow cytometry tube. The next day, 400 μL of HCl (37%, VWR, USA) was added to each well and incubated overnight. The samples were transferred to 15 mL Falcon tubes. Each flow cytometer tube was washed with Milli-Q H2O, and the washes were added to the Falcon tubes. Every Falcon tube was then diluted to a final volume of 3 mL with Milli-Q H2O. The samples were further analyzed by ICP-OES (Optima 7000 DV, PerkinElmer, USA, axial-viewing, λ = 267.6 nm) using a radio frequency power of 1500 W, gas flow rates of 8 L min−1 (Ar, plasma), 0.2 L min−1 (N2, auxiliary), pump 0.7 L min−1 (nebulizer), and 1 L min−1 sample flow rate (equilibration delay of 15 s) and compared to a standard in the range of 0.0 to 7 mg L−1 prepared gravimetrically using a gold standard (Sigma-Aldrich, USA). To determine the number of AuNPs associated with each cell, the total mass of Au per sample was first calculated by extrapolating the mass of gold using the calibration curve multiplied by the dilution factor after blank correction. The total Au mass per sample was then divided by the mass of a single gold nanoparticle. The average number of AuNPs per cell was finally obtained by dividing the total number of AuNPs by the number of sorted cells for each sample.
1, Sigma-Aldrich, USA) overnight at room temperature in a flow cytometry tube. The next day, 400 μL of HCl (37%, VWR, USA) was added to each well and incubated overnight. The samples were transferred to 15 mL Falcon tubes. Each flow cytometer tube was washed with Milli-Q H2O, and the washes were added to the Falcon tubes. Every Falcon tube was then diluted to a final volume of 3 mL with Milli-Q H2O. The samples were further analyzed by ICP-OES (Optima 7000 DV, PerkinElmer, USA, axial-viewing, λ = 267.6 nm) using a radio frequency power of 1500 W, gas flow rates of 8 L min−1 (Ar, plasma), 0.2 L min−1 (N2, auxiliary), pump 0.7 L min−1 (nebulizer), and 1 L min−1 sample flow rate (equilibration delay of 15 s) and compared to a standard in the range of 0.0 to 7 mg L−1 prepared gravimetrically using a gold standard (Sigma-Aldrich, USA). To determine the number of AuNPs associated with each cell, the total mass of Au per sample was first calculated by extrapolating the mass of gold using the calibration curve multiplied by the dilution factor after blank correction. The total Au mass per sample was then divided by the mass of a single gold nanoparticle. The average number of AuNPs per cell was finally obtained by dividing the total number of AuNPs by the number of sorted cells for each sample.
Focused ion beam-scanning electron microscopy (FIB-SEM) sample preparation
Macrophages were seeded in a 35 mm glass-bottom dish (MatTek Inc., Ashland, MA, USA), exposed to AuNPs for 1 h and 48 h, and then fixed with 2 vol% PFA and 2.5 vol% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) in PBS for 2 h on ice. After two washes with 0.2 M cacodylate buffer (Electron Microscopy Sciences, Hatfield, PA, USA), the samples were immersed in a solution containing 3 w/v% potassium hexacyanoferrate(II) trihydrate (≥99.95%, Sigma-Aldrich, Buchs, Switzerland) and 0.25 vol% osmium tetroxide (1% in Milli-Q H2O, 4% Electron Microscopy Sciences, Hatfield, PA, USA) in 0.2 M cacodylate buffer for 1 h on ice. The solution was then replaced by 1 w/v% thiocarbohydrazide (98%, Sigma-Aldrich, Buchs, Switzerland) in Milli-Q H2O for 20 min at room temperature, previously warmed to 60 °C for 1 hour with gentle shaking. The samples were incubated with 0.25% osmium tetroxide diluted in mQ H2O for 30 min. Dehydration was performed using a graded series of anhydrous ethanol and 100% anhydrous acetone. Finally, Epon resin (Epoxy Embedding Kit, Sigma-Aldrich, Buchs, Switzerland) was infiltrated in 3 steps of 2 h, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, 1
4, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 4
1, and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 resin
1 resin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetone. Finally, the samples were left in fresh epoxy overnight on a shaker and then embedded in fresh epoxy for 48 h at 60 °C to polymerize. The Epon (Epoxy Embedding Kit, Sigma-Aldrich, Buchs, Switzerland) block was detached from the dish by thermal shock in a liquid nitrogen bath and then attached to an aluminum stub with carbon tape and silver paste. A layer of ∼3 nm Au was sputtered prior to the experiment.
acetone. Finally, the samples were left in fresh epoxy overnight on a shaker and then embedded in fresh epoxy for 48 h at 60 °C to polymerize. The Epon (Epoxy Embedding Kit, Sigma-Aldrich, Buchs, Switzerland) block was detached from the dish by thermal shock in a liquid nitrogen bath and then attached to an aluminum stub with carbon tape and silver paste. A layer of ∼3 nm Au was sputtered prior to the experiment.
FIB-SEM tomography
All experiments were performed using a Thermo Scientific Scios 2 Dual Beam microscope (Thermo Fisher Scientific, Waltham, MA, USA). A dice provided randomization in selecting the cell of interest, where we previously numbered 6 cells in a restricted region. The selected cell was protected with a platinum (Pt) layer of 1 μm thickness (30 kV and current of 1 nA). Then, a trench of 5 μm in depth was milled on the front face and each side of the cell of interest using the ion beam at 7 nA current. Finally, the front face of the volume of interest was polished with an ion beam current of 1 nA until the beginning of the cell was visible to optimize the focus and contrast. FIB-SEM tomography of the whole cell was performed using the FEI Slice and View software (Thermo Fisher Scientific, Waltham, MA, USA, version 4.1). The electron beam acceleration voltage was set to 5 kV, the current to 0.4 nA, the resolution to 1536 × 1024 pixels, and the dwell time to 30 μs. Images were acquired in immersion mode with the backscattered electron detector, yielding clear signals from AuNPs due to the detector's sensitivity to backscattered electrons, which correlates with the atomic number of Au (Z = 79).56 The ion beam operating with a current of 1 nA current at 30 kV was used to slice through the cell of interest at an interval of 18 nm and a depth of 5 μm. This study imaged a total of nine whole cells by FIB-SEM tomography: two cells per condition for both 1 h and 48 h AuNPs exposure in GM-CSF macrophages, two cells for 1 h exposure in M-CSF macrophages, and three cells for 48 h exposure in M-CSF macrophages.Image processing and particle counting
The acquired SEM image stacks were aligned and cropped using Avizo software (Thermo Fisher Scientific, USA). Segmentation of the cell, NPs, and background was performed using the pixel classification workflow of Ilastik61 (https://www.ilastik.org/publications.html). Before applying the model to the entire image stack, a minimum of five random images were used for training. The segmented AuNPs were separated into extracellular and intracellular objects using the segmentation result of the cell as a mask and executed in FIJI (ImageJ, National Institutes of Health, Bethesda, MD, USA).59 The intracellular gold volume per cell was calculated by multiplying the number of voxels segmented as Au in an entire cell with the known volume of one voxel. To obtain the number of AuNPs per cell, the calculated gold volume of the entire cell was divided by the volume of a sphere measuring 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 713 nm3, corresponding to a single spherical AuNPs with a diameter of 45 nm. 3D rendering of the cell, the intracellular and extracellular AuNPs was performed with the Avizo 3D software (Thermo Fisher Scientific, Waltham, MA, USA, version 2021.1).
713 nm3, corresponding to a single spherical AuNPs with a diameter of 45 nm. 3D rendering of the cell, the intracellular and extracellular AuNPs was performed with the Avizo 3D software (Thermo Fisher Scientific, Waltham, MA, USA, version 2021.1).
As part of our commitment to scientific transparency and collaboration, we have decided to upload our single-cell level data sets to an open repository. In doing so, we aim to make these datasets accessible to the wider scientific community, allowing other researchers to explore and analyze them for further insights.
Enzyme-linked immunosorbent assay
The amount of interleukin 8 (IL-8) released by both macrophage phenotypes after 48 h of exposure to AuNPs was determined using the Human IL-8 DuoSet ELISA Kit (R&D Systems, Zug, Switzerland). The supernatant was collected and stored at −80 °C and assayed in triplicate for the presence of IL-8. Exposure to LPS at a concentration of 10 ng mL−1 was used as a positive control.Statistical analyses
Particle size and zeta potential were expressed as mean ± standard deviation. Determination of the AuNPs number per cell, surface marker expression levels, and cytokine secretion was performed with four biologically independent replicates from different donors. Statistical significance between conditions was determined using a one-way ANOVA followed by Dunnett's and Tukey's post hoc tests for multiple comparisons with a significance level of p < 0.05. Analyses were performed using Prism 8 (GraphPad Software).Data availability
As part of our commitment to scientific transparency and collaboration, we have decided to upload our single-cell level data sets to an open repository. In doing so, we aim to make these datasets accessible to the wider scientific community, allowing other researchers to explore and analyze them for further insights. The data is reposited on zenodo https://doi.org/10.5281/zenodo.11186312.Author contributions
H. L. performed the experiments, analyzed the results, and wrote the manuscript; D. V. supervised the FIB-SEM experiments and analysis and reviewed the manuscript; S. B. performed DLS data analysis; P. T.-B. revised the manuscript and provided scientific suggestions; A. P.-F. supervised the project; B. R.-R. thoroughly reviewed the manuscript, supervised, and designed the project. All authors contributed to the scientific discussions and manuscript revisions. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The authors thank Liliane Ackermann Hirschi for nanoparticle synthesis and Shui Ling Chu for help with the cell cultures. The authors acknowledge funding from the Swiss National Science Foundation (SNSF) (project 310030_159847/1) and the Adolphe Merkle Foundation. Additionally, this work benefitted from support from the Swiss National Science Foundation through the National Center of Competence in Research Bio-Inspired Materials.References
- K. McNamara and S. A. M. Tofail, Nanoparticles in biomedical applications, Adv. Phys.: X, 2017, 2, 54–88 CAS.
- R. Weissleder, M. Nahrendorf and M. J. Pittet, Imaging macrophages with nanoparticles, Nat. Mater., 2014, 13, 125–138 CrossRef CAS PubMed.
- L. Boselli, V. Castagnola, A. Armirotti, F. Benfenati and P. P. Pompa, Biomolecular Corona of Gold Nanoparticles: The Urgent Need for Strong Roots to Grow Strong Branches, Small, 2023, 20, 2306474 CrossRef PubMed.
- M. Medrano-Bosch, A. Moreno-Lanceta and P. Melgar-Lesmes, Nanoparticles to Target and Treat Macrophages: The Ockham's Concept?, Pharmaceutics, 2021, 13, 1340 CrossRef CAS PubMed.
- G. Hu, et al., Nanoparticles Targeting Macrophages as Potential Clinical Therapeutic Agents Against Cancer and Inflammation, Front. Immunol., 2019, 10, 1998 CrossRef CAS PubMed.
- Y.-N. Zhang, W. Poon, A. J. Tavares, I. D. McGilvray and W. C. W. Chan, Nanoparticle-liver interactions: cellular uptake and hepatobiliary elimination, J. Controlled Release, 2016, 240, 332–348 CrossRef CAS PubMed.
- D. E. Owens and N. A. Peppas, Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles, Int. J. Pharm., 2006, 307, 93–102 CrossRef CAS PubMed.
- S.-D. Li and L. Huang, Pharmacokinetics and Biodistribution of Nanoparticles, Mol. Pharm., 2008, 5, 496–504 CrossRef CAS.
- C. D. Mills, K. Kincaid, J. M. Alt, M. J. Heilman and A. M. Hill, M-1/M-2 macrophages and the Th1/Th2 paradigm, J. Immunol., 2000, 164, 6166–6173 CrossRef CAS PubMed.
- K. A. Binnemars-Postma, H. W. Ten Hoopen, G. Storm and J. Prakash, Differential uptake of nanoparticles by human M1 and M2 polarized macrophages: protein corona as a critical determinant, Nanomedicine, 2016, 11, 2889–2902 CrossRef CAS.
- H. L. Herd, K. T. Bartlett, J. A. Gustafson, L. D. McGill and H. Ghandehari, Macrophage silica nanoparticle response is phenotypically dependent, Biomaterials, 2015, 53, 574–582 CrossRef CAS PubMed.
- J. Hoppstädter, et al., M2 polarization enhances silica nanoparticle uptake by macrophages, Front. Pharmacol, 2015, 6, 55 Search PubMed.
- S. A. MacParland, et al., Phenotype Determines Nanoparticle Uptake by Human Macrophages from Liver and Blood, ACS Nano, 2017, 11, 2428–2443 CrossRef CAS.
- P. A. S. Kinaret, G. Scala, A. Federico, J. Sund and D. Greco, Carbon Nanomaterials Promote M1/M2 Macrophage Activation, Small, 2020, 16, 1907609 CrossRef CAS PubMed.
- M. J. Ware, et al., Analysis of the Influence of Cell Heterogeneity on Nanoparticle Dose Response, ACS Nano, 2014, 8, 6693–6700 CrossRef CAS PubMed.
- P. Rees, J. W. Wills, M. R. Brown, C. M. Barnes and H. D. Summers, The origin of heterogeneous nanoparticle uptake by cells, Nat. Commun., 2019, 10, 2341 CrossRef.
- A. Salvati, et al., Quantitative measurement of nanoparticle uptake by flow cytometry illustrated by an interlaboratory comparison of the uptake of labelled polystyrene nanoparticles, NanoImpact, 2018, 9, 42–50 CrossRef.
- Y. Ibuki and T. Toyooka, Nanoparticle uptake measured by flow cytometry, Methods Mol. Biol., 2012, 926, 157–166 CrossRef CAS PubMed.
- C. Gottstein, G. Wu, B. J. Wong and J. A. Zasadzinski, Precise quantification of nanoparticle internalization, ACS Nano, 2013, 7, 4933–4945 CrossRef CAS.
- L. Rodriguez-Lorenzo, et al., Fluorescence-Encoded Gold Nanoparticles: Library Design and Modulation of Cellular Uptake into Dendritic Cells, Small, 2014, 10, 1341–1350 CrossRef CAS.
- B. Drasler, D. Vanhecke, L. Rodriguez-Lorenzo, A. Petri-Fink and B. Rothen-Rutishauser, Quantifying nanoparticle cellular uptake: which method is best?, Nanomedicine, 2017, 12, 1095–1099 CrossRef CAS.
- M. Malatesta, Transmission Electron Microscopy as a Powerful Tool to Investigate the Interaction of Nanoparticles with Subcellular Structures, Int. J. Mol. Sci., 2021, 22, 12789 CrossRef CAS PubMed.
- M. Reifarth, S. Hoeppener and U. S. Schubert, Uptake and Intracellular Fate of Engineered Nanoparticles in Mammalian Cells: Capabilities and Limitations of Transmission Electron Microscopy—Polymer-Based Nanoparticles, Adv. Mater., 2018, 30, 1703704 CrossRef.
- D. Vanhecke, et al., Quantification of nanoparticles at the single-cell level: an overview about state-of-the-art techniques and their limitations, Nanomedicine, 2014, 9, 1885–1900 CrossRef CAS PubMed.
- Y. Wu, M. R. K. Ali, K. Dansby and M. A. El-Sayed, Improving the Flow Cytometry-based Detection of the Cellular Uptake of Gold Nanoparticles, Anal. Chem., 2019, 91, 14261–14267 CrossRef CAS PubMed.
- K. M. McKinnon, Flow Cytometry: An Overview, Curr. Protoc. Immunol., 2018, 120, 5.1.1–5.1.11 Search PubMed.
- A. Albanese, K. M. Tsoi and W. C. W. Chan, Simultaneous quantification of cells and nanomaterials by inductive-coupled plasma techniques, J. Lab. Autom., 2013, 18, 99–104 CrossRef CAS PubMed.
- C. Kizilyaprak, Y.-D. Stierhof and B. M. Humbel, Volume microscopy in biology: FIB-SEM tomography, Tissue Cell, 2019, 57, 123–128 CrossRef PubMed.
- A. Goldstein, Y. Soroka, M. Frušić-Zlotkin, I. Popov and R. Kohen, High resolution SEM imaging of gold nanoparticles in cells and tissues, J. Microsc., 2014, 256, 237–247 CrossRef CAS PubMed.
- C. S. Xu, et al., Enhanced FIB-SEM systems for large-volume 3D imaging, eLife, 2017, 6, e25916 CrossRef PubMed.
- F. A. W. Verreck, et al., Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 4560–4565 CrossRef CAS.
- S. Hashimoto, T. Suzuki, H.-Y. Dong, N. Yamazaki and K. Matsushima, Serial Analysis of Gene Expression in Human Monocytes and Macrophages, Blood, 1999, 94, 837–844 CrossRef CAS.
- D. C. Lacey, et al., Defining GM-CSF- and Macrophage-CSF-Dependent Macrophage Responses by In Vitro Models, J. Immunol., 2012, 188, 5752–5765 CrossRef CAS.
- G. Peng, et al., Biodegradation of graphdiyne oxide in classically activated (M1) macrophages modulates cytokine production, Nanoscale, 2021, 13, 13072–13084 RSC.
- J. Bourquin, et al., Reduction of Nanoparticle Load in Cells by Mitosis but Not Exocytosis, ACS Nano, 2019, 13, 7759–7770 CrossRef CAS.
- C. Kizilyaprak, A. G. Bittermann, J. Daraspe and B. M. Humbel, FIB-SEM tomography in biology, Methods Mol. Biol., 2014, 1117, 541–558 CrossRef CAS.
- A. W. Sanders, K. M. Jeerage, C. L. Schwartz, A. E. Curtin and A. N. Chiaramonti, Gold Nanoparticle Quantitation by Whole Cell Tomography, ACS Nano, 2015, 9, 11792–11799 CrossRef CAS.
- E. Guehrs, et al., Quantification of silver nanoparticle uptake and distribution within individual human macrophages by FIB/SEM slice and view, J. Nanobiotechnol., 2017, 15, 21 CrossRef.
- Y. Oishi and I. Manabe, Macrophages in inflammation, repair and regeneration, Int. Immunol., 2018, 30, 511–528 CrossRef CAS PubMed.
- D. M. Mosser and J. P. Edwards, Exploring the full spectrum of macrophage activation, Nat. Rev. Immunol., 2008, 8, 958–969 CrossRef CAS PubMed.
- A. Gallud, et al., Macrophage activation status determines the internalization of mesoporous silica particles of different sizes: exploring the role of different pattern recognition receptors, Biomaterials, 2017, 121, 28–40 CrossRef CAS PubMed.
- M. Jaguin, N. Houlbert, O. Fardel and V. Lecureur, Polarization profiles of human M-CSF-generated macrophages and comparison of M1-markers in classically activated macrophages from GM-CSF and M-CSF origin, Cell. Immunol., 2013, 281, 51–61 CrossRef CAS PubMed.
- A. K. Dey, et al., Impact of Gold Nanoparticles on the Functions of Macrophages and Dendritic Cells, Cells, 2021, 10, 96 CrossRef CAS.
- D. Reichel, M. Tripathi and J. M. Perez, Biological Effects of Nanoparticles on Macrophage Polarization in the Tumor Microenvironment, Nanotheranostics, 2019, 3, 66–88 CrossRef.
- M. Cao, et al., Ginseng-derived nanoparticles alter macrophage polarization to inhibit melanoma growth, J. ImmunoTher. Cancer, 2019, 7, 326 CrossRef.
- W. Chen, F. Zhang, Y. Ju, J. Hong and Y. Ding, Gold Nanomaterial Engineering for Macrophage-Mediated Inflammation and Tumor Treatment, Adv. Healthcare Mater., 2021, 10, 2000818 CrossRef CAS.
- H. R. Ali, S. A. Selim and D. Aili, Effects of macrophage polarization on gold nanoparticle-assisted plasmonic photothermal therapy, RSC Adv., 2021, 11, 25047–25056 RSC.
- P. Scodeller, et al., Precision Targeting of Tumor Macrophages with a CD206 Binding Peptide, Sci. Rep., 2017, 7, 14655 CrossRef.
- D. Acharya, X. R. L. Li, R. E.-S. Heineman and R. E. Harrison, Complement Receptor-Mediated Phagocytosis Induces Proinflammatory Cytokine Production in Murine Macrophages, Front. Immunol., 2020, 10, 3049 CrossRef.
- S. A. Freeman and S. Grinstein, Phagocytosis: receptors, signal integration, and the cytoskeleton, Immunol. Rev., 2014, 262, 193–215 CrossRef CAS PubMed.
- C. Åberg, V. Piattelli, D. Montizaan and A. Salvati, Sources of variability in nanoparticle uptake by cells, Nanoscale, 2021, 13, 17530–17546 RSC.
- A. Ivask, et al., Single Cell Level Quantification of Nanoparticle–Cell Interactions Using Mass Cytometry, Anal. Chem., 2017, 89, 8228–8232 CrossRef CAS.
- H.-C. Chou, S.-J. Chiu and T.-M. Hu, Quantitative Analysis of Macrophage Uptake and Retention of Fluorescent Organosilica Nanoparticles: Implications for Nanoparticle Delivery and Therapeutics, ACS Appl. Nano Mater., 2024, 7, 3656–3667 CrossRef CAS.
- H. D. Summers, et al., Quantification of Nanoparticle Dose and Vesicular Inheritance in Proliferating Cells, ACS Nano, 2013, 7, 6129–6137 CrossRef CAS PubMed.
- A. Roth, et al., Quantification of Polystyrene Uptake by Different Cell Lines Using Fluorescence Microscopy and Label-Free Visualization of Intracellular Polystyrene Particles by Raman Microspectroscopic Imaging, Cells, 2024, 13, 454 CrossRef CAS.
- R. F. Egerton, Physical Principles of Electron Microscopy, Springer US, Boston, MA, 2005, DOI:10.1007/b136495.
- K. R. Brown and M. J. Natan, Hydroxylamine Seeding of Colloidal Au Nanoparticles in Solution and on Surfaces, Langmuir, 1998, 14, 726–728 CrossRef CAS.
- J. Turkevich, G. Garton and P. C. Stevenson, The color of colloidal gold, J. Colloid Sci., 1954, 9, 26–35 CrossRef.
- J. Schindelin, et al., Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- H. Barosova, B. Drasler, A. Petri-Fink and B. Rothen-Rutishauser, Multicellular Human Alveolar Model Composed of Epithelial Cells and Primary Immune Cells for Hazard Assessment, J. Visualized Exp., 2020, e61090, DOI:10.3791/61090.
- S. Berg, D. Kutra, T. Kroeger, C. N. Straehle, B. X. Kausler, C. Haubold, M. Schiegg, J. Ales, T. Beier, M. Rudy, K. Eren, J. I. Cervantes, B. Xu, F. Beuttenmueller, A. Wolny, C. Zhang, U. Koethe, F. A. Hamprecht and A. Kreshuk, ilastik: interactive machine learning for (bio)image analysis, Nat. Methods, 2019, 16(12), 1226–1232 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00104d |
| This journal is © The Royal Society of Chemistry 2024 |