Circular RNA oligonucleotides: enzymatic synthesis and scaffolding for nanoconstruction†
Shijie
Li
a,
Yanxin
Chu
a,
Xin
Guo
 b,
Chengde
Mao
b,
Chengde
Mao
 *c and
Shou-Jun
Xiao
*c and
Shou-Jun
Xiao
 *a
*a
aSchool of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, Jiangsu, China
bBruker (Beijing) Scientific Technology Co. Ltd, China
cDepartment of Chemistry, Purdue University, West Lafayette, Indiana 47907, USA. E-mail: mao@purdue.edu; sjxiao@nju.edu.cn
First published on 12th July 2024
Abstract
We report the efficient synthesis of monomeric circular RNAs (circRNAs) in the size range of 16–44 nt with a novel DNA dumbbell splinting plus T4 DNA ligation strategy. Such a DNA dumbbell splinting strategy was developed by one group among ours recently for near-quantitative conversion of short linear DNAs into monomeric circular ones. Furthermore, using the 44 nt circRNA as scaffold strands, we constructed hybrid RNA:DNA and pure RNA:RNA double crossover tiles and their assemblies of nucleic acid nanotubes and flat arrays.
New conceptsSmall-sized noncoding RNAs, including double-stranded small interfering RNAs (siRNAs) and single-stranded 20–24 nt microRNAs (miRNAs), are receiving much more attention because of the current standard-of-care linear messenger RNAs for coronavirus infections, which indicates their great therapeutic and huge market potential. However, the natural, short, and linear RNAs are sensitive to exonuclease degradation, which is a key delivery issue for RNA pharmaceutical applications currently. One potential solution is to convert the linear RNAs into circular RNAs (circRNAs) and/or fold RNAs into nanostructures because these products are more resistant to exonuclease degradation than their cognate linear ones. Herein, we report the efficient synthesis of monomeric circRNAs in the size range of 16–44 nt with a novel DNA dumbbell splinting strategy in the T4 DNA ligation system. Such a DNA dumbbell splinting strategy was developed by the Mao group (one among ours) recently for near-quantitative conversion of short linear DNAs into monomeric circular DNAs via T4 DNA ligation. In the past, in vitro cyclisation of the short linear single-stranded RNAs via linear splinting plus enzymatic ligation often generated byproducts of concatemers rather than the monomeric circRNAs because oligomerisation releases a higher strain which should be resulted by monomerisation. Furthermore, using the synthesized 44 nt circRNA as scaffold strands, we constructed hybrid RNA:DNA and pure RNA:RNA double crossover tiles and their assemblies of nucleic acid nanotubes and flat arrays. We believe that the dumbbell splinting strategy will be highly efficient and user friendly for synthesis of both monomeric DNA and RNA oligonucleotide rings. The convenient and relatively larger scale production of circular DNA and RNA oligonucleotide molecules will expedite their application research in depth and breadth. |
Introduction
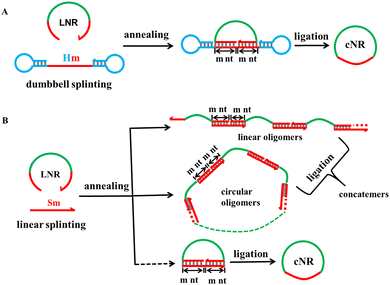
Small-sized noncoding RNAs, including double-stranded (ds) small interfering RNAs (siRNAs) and single-stranded (ss) 20–24 nt microRNAs (miRNAs), are receiving much more attention because of the current standard-of-care linear messenger RNAs for coronavirus infections, which indicates their great therapeutic and huge market potential.1–4 However, the natural, short, and linear RNAs are sensitive to exonuclease degradation, which is a key delivery issue for RNA pharmaceutical applications currently. One potential solution is to convert the linear ssRNAs into circular RNAs (circRNAs) and/or fold RNAs into nanostructures because these products are more resistant to exonuclease degradation than their cognate linear ones.5,6 Herein, we report the preparation of small circRNAs using a novel DNA dumbbell splint plus T4 DNA ligation approach, which was invented by the Mao group, one among ours, recently to convert short linear DNAs (16–40 nt) into monomeric circular DNAs in near-quantitative yields and on a large-scale.7 Furthermore, we adapted three RNA folding approaches to construct hybrid RNA:DNA and pure RNA:RNA nanostructures using the core motif of double crossover (DX) tiles.In vitro cyclisation of short linear ssRNAs (tens of nucleotides long) often applies chemical and enzymatic ligations. In the past, all approaches exhibited drawbacks such as low yields, the higher potential of oligomerisation rather than monomerisation for ligation,8,9 and the byproduct formation of non-physiological linkages such as 2′-5′ phosphodiester bonds especially via chemical ligation.8,10 When the targeting RNA or DNA monomer ring evolves to a much smaller size less than 40 nt, the monomeric looping should generate a higher bending strain, this is why oligomerisation becomes severe for these small-sized DNA and RNA molecules because intermolecular splinting releases a higher bending strain. To overcome the competition of oligomerisation via intermolecular ligation, a novel DNA dumbbell splinting approach has been reported to be successful recently by the Mao group. In this approach, they genuinely designed a dumbbell splint structure and realised near-quantitative synthesis of monomeric circular DNAs from their cognate, short, linear ssDNAs between 16–40 nt in the concentration range of 1–100 μM through the most convenient T4 DNA ligation. They explained that the two terminal hairpin loops should result in a much higher coulombic repulsion force if intermolecular oligomerisation occurred, which is why formation of a much more stable monomer loop rather than linear or circular oligomers is preferred.7
Inspired by this innovation, we applied the dumbbell splinting strategy successfully in synthesis of three short circRNAs (16, 22, and 44 nt) with high efficiency via T4 DNA ligation.
Triggered by DNA nanotechnology,11–15 RNA nanotechnology has also been developed over 30 years, mainly based on construction of rigid motifs plus connection via regular sticky end and specific kissing-loop (KL) base pairings.16–32 For example, via the regular sticky end cohesion, the Mao group constructed hybrid RNA:DNA DAE (double crossovers made of antiparallel duplexes with an even number of half-turns distance), three-point-star, and four-point-star tiles and their corresponding 2D planar O-tiling arrays and curved E-tiling discrete polyhedra (E-tiling and O-tiling mean the inter-tile connection with a distance at an even and an odd number of half turns, respectively).33 The Franco group constructed pure RNA:RNA nanotubes via the DAE-E assembly (-E means E-tiling).34 The Andersen group transcribed long ssRNAs for self-association to form one-strand DX tiles with peripheral kissing-loops for specific base pairing, resulting in larger patches of 2D RNA arrays.35 The Xiao group, one among ours, used small circular DNAs as scaffolds to construct 1–3D DNA nanostructures.36–39 However, as we know until now, using the small circular RNAs as scaffolds to construct a 1–3D hybrid and pure RNA nanostructures has not yet been reported. Therefore, we further used the 44 nt circRNA to construct hybrid RNA:DNA and pure RNA:RNA DX tiles and nanostructures.
Results and discussion
Synthesis of circRNAs
The monomeric cyclisation and oligomerisation of a linear ssRNA via T4 DNA ligation are shown with their corresponding schematic approaches in Fig. 1. The T4 DNA ligation requires a splint strand to pair precisely with both 3′ and 5′ ends each by at least 6 nt long40 because the ligase binds a little longer duplex than one helical turn (≥12 bps) for proper working.41 The dumbbell splinting strategy (A) has much higher monomeric cyclisation efficiency due to the stronger coulombic repulsion force between the terminal hairpin loops when the sticky end cohesion occurs through intermolecular splinting as shown in (B), whereas oligomerisation occurs dominantly in the conventional linear splinting strategy (B) because it generates less free energy than monomeric cyclisation does. It is obvious that the shorter the linear ssRNAs, the stronger the bending strain generated during the monomeric cyclisation process. According to the DNA cyclisation procedure, we examined the following parameters for monomeric cyclisation of RNA: the ssRNA size and concentration, Hm with m at 4, 5, 6 nt for dumbbell splinting and Sm with m at 6 and 10 nt as controls for linear splinting, the hairpin's nT (T represents thymine) loop size in Hm with n at 1, 4, and 8, and combination of them.Similar to the linear DNA size range chosen for the dumbbell splinting strategy, we selected three appropriate, short, linear ssRNAs at lengths of 16, 22, and 44 nt (abbreviated as L16R, L22R, and L44R molecules, respectively) as examples to examine the cyclisation efficiency. We first demonstrate the cyclisation results of miR-16, which is a natural 22 nt miRNA (L22R) and acts as a tumour suppressor in the development of diverse malignancies including breast cancer, lung cancer, cervical cancer and so on.42 The ATP-dependent T4 DNA ligase usually catalyses the joining of nicks located in dsDNA substrates with the help of Mg2+.8,10 Therefore, it is necessary to explore the appropriate concentrations of enzyme (Fig. S1, ESI†), ATP (Fig. S2, ESI†), Mg2+ (Fig. S3, ESI†), and even the reaction temperature and time (Fig. S4, ESI†) for monomeric cyclisation of short linear ssRNAs because the nick locates at the hybrid RNA:DNA substrate. Based on the above orthogonal screening experiments (Section S2 of the ESI†), the standard ligation conditions were determined and used for our RNA cyclisation reactions, unless otherwise noted. (1) The 5′-monophosphated ssRNA concentration is 10 μM and (2) the ligation is carried out with a T4 DNA ligase concentration of 2500 U per nanomolar RNA at 16 °C for 16 h.
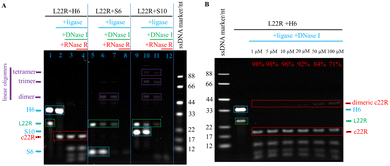
The L22R cyclisation results were analysed through denaturing polyacrylamide gel electrophoresis (dPAGE) (Fig. 2). We tried H6, S6, and S10 splints for ligation, separately. To clearly demonstrate the cyclisation efficiency, we generally analysed four solution samples from a series of consecutive reactions in the same ligation pot, the LNR and DNA splint mixture after mixing for blank control, after T4 DNA ligation, after DNase I digestion, and after RNase R digestion. As shown in the L22R and H6 ligation system of Fig. 2A, a strong H6 band and a medium L22R band as the blank control evolved with carrying on the enzymatic relay reactions in series. After T4 DNA ligation, the L22R band disappeared completely and a new band moving faster than L22R occurred. After consecutive DNase I and RNase R digestions of all linear DNA and RNA residues, respectively, the H6 band disappeared in the first case, and the left band traces exhibited as the same in both cases. Because the new band remained intact, meaning that it corresponds to our target circRNA (c22R), which is resistant to both DNase I and RNase R digestions. Additional fast-moving traces below the c22R band after DNase I and RNase R digestions could be assigned to the broken pieces of DNA hairpin residues or enzyme/DNA hairpin complexes.
To compare the cyclisation results of the dumbbell splint H6 with the linear splints S6 and S10, we provided S6 and S10 mediated splinting and ligation results in Fig. 2A. The L22R band was weakened in both S6 and S10 mediated ligation reactions to some degree, instead of the c22R band, new linear oligomer bands moving more slowly than L22R appeared, which were suggested as the linear dimer, trimer, and tetramer of L22R. More slow-moving oligomer bands appeared in S10 than in S6 mediated ligations demonstrated that the S10 splinting has stronger base pairing strength for oligomerisation. After the RNase R digestion, all RNA oligomer bands disappeared, illustrating that they are linear but not circular oligomers derived from L22R.
Furthermore, we also examined the monomeric cyclisation efficiency by changing the nT loop size in H6 from 4T to 1T and 8T. Both monomeric c22R and dimeric c22R bands appeared. With the loop size at 8T, the ratio of monomeric c22R against dimeric c22R was estimated to be 6 (86/14); however, with the loop size at 1T, the ratio was estimated to be 0.8 (44/56). The above results indicated that the loop sizes at 4–8T are appropriate for highly efficient monomeric cyclisation of short linear RNAs (Fig. S5, ESI†).
To promote monomeric cyclisation and prevent oligomerisation of single-stranded nucleic acids in conventional linear splinting and ligation reactions, much low concentrations of the nucleic acid substrate, such as 0.1 μM, are generally used.8,10 However, such low concentrations decrease the enzymatic ligation efficiency tremendously and the reaction system is not suitable for large-scale production. We previously succeeded in converting the short linear ssDNAs to circular ones with very high yields in quite a wide concentration range from 1 to 100 μM,7 so we also examined the monomeric cyclisation efficiencies of converting L22R to c22R with the L22R concentrations changing from 1, to 5, 10, 20, 50, and 100 μM, accordingly (Fig. 2B). The dPAGE results illustrated that L22R almost changed to c22R quantitatively in the concentration range of 1–10 μM; but with increasing the L22R concentrations to 20, 50, and 100 μM, the c22R yields slowly decreased to 92%, 84% and 71%, respectively; simultaneously, we also observed the increasing evolution of minor oligomer byproducts, especially the dimeric c22R. Overall, the relatively large-scale production of circRNAs from synthetic 5′-monophosphorylated ssRNA oligonucleotides is realisable.
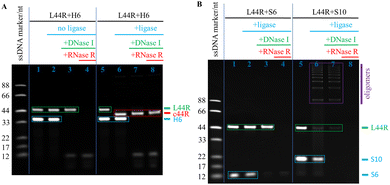
Because the shortest linear DNA for near-quantitative cyclisation examined is 16 nt long,7 we also examined both short linear RNAs of L16R and L14R here. The T4 DNA ligase normally ligates a nick when it braces a 12 bp or an even longer duplex segment, however, monomeric cyclisation of the short nucleic acids such as L16R and L14R should require even shorter effective binding domains. Thereby, except the normal H6, we also tested H5 and H4 as splint strands for cyclisation of L22R, L16R, and L14R, accordingly. The dPAGE results demonstrated that H5 converted L22R partially to c22R with a yield of 56%, but H4 failed (Fig. S6, ESI†). For L16R, H6 converted L16R to both monomeric c16R with a yield of 22% and dimeric c16R with a yield of 78%, H5 converted L16R partially to c16R with a yield of 38%, but H4 failed to convert L16R to c16R or any other oligomers (Fig. S7, ESI†). For L14R, H6 converted L14R only to dimeric c14R with a yield of 82% but not monomeric c14R, neither H5 nor H4 converted L14R to any monomeric c14R or other oligomers (Fig. S8, ESI†). The above circular monomer and dimer yields for each LNR (N = 22, 16, 14 nt) were plotted in a histogram against H6, H5, and H4, along with their LNR-ligation dPAGE photograph together, accordingly (Fig. S6–8). Still, the unpredictable cyclisation results of much short RNA oligonucleotides less than 22 nt need to be explained through further investigations of the dumbbell splinting mechanisms for cyclisation.
The dsRNA displays the A-form structure, so its periodic length is 11 bp per turn. For construction of a DAE tile, a circular ssRNA of c44R is needed to act as the scaffold. That is why we adapted a 44 nt ssRNA sequence from the literature34 and cyclised it with the H6 splinting strategy. As shown in the left half panel of the control dPAGE photograph of Fig. 3A, both L44R and H6 bands appeared clearly without addition of T4 DNA ligase; with sequential additions of DNase I and RNase R, the DNA H6 and RNA L44R bands disappeared accordingly. In the right half panel of Fig. 3A, after addition of T4 DNA ligase, L44R converted to c44R in a near-quantitative yield, and c44R remained intact after 30 min RNase R digestion at 37 °C. Similar to the c22R experiments, we executed additional control ligation experiments with S6 and S10 as splints, separately. As shown in Fig. 3B, L44R remained nearly intact in the S6 group after both T4 DNA ligation and DNase I digestion, but disappeared after RNase R digestion; in the S10 group, most of the L44R changed to linear oligomers, which displayed a series of ladder bands above L44R after T4 DNA ligation and DNase I digestion, but disappeared after RNase R digestion. From both the S6 and S10 mediated ligation results, L22R and L44R cannot be transformed into the target products c22R and c44R, respectively, meaning that the short linear splinting prefers to form linear RNA concatemers rather than monomeric cNR molecules.
Assembly of hybrid RNA:DNA and pure RNA:RNA nanostructures
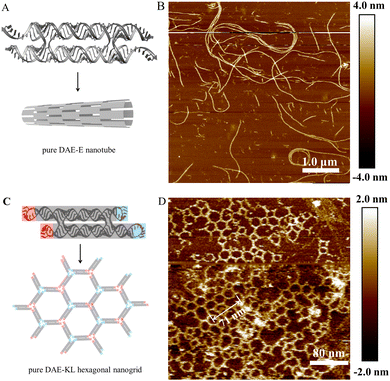
We have reported the use of small circular DNAs serving as scaffold strands to construct 1–3D DNA nanostructures.36–39 Herein, we report the use of the small circular RNA molecule, c44R, serving as scaffold strands to assemble hybrid RNA:DNA and pure RNA:RNA nanostructures via DAE-Ep/q and DAE-Op/q designs, where subscripts p and q represent the numbers of inter-tile base pairs and sticky end nucleotides, respectively. Perturbations of p and q around their optimum values (generally for pure RNA:RNA nanostructures, the optimum p is 22 bp for E-tiling and 27 bp for O-tiling, and q is 5 nt for E-tiling and 6 nt for O-tiling) will screen the optimum inter-tile distance and sticky end cohesion strength for assembling perfect 2D arrays. It is accepted that a tile has both distinctively right- and left-handed faces and generally possesses an intrinsic curvature.38 E-tiling requires that all tiles must orient with the same left- or right-handed faces toward one direction, resulting in accumulation of individual tile curvatures and formation of homogeneous nanotubes; whereas O-tiling alternates the adjacently cohered tiles between left- and right-handed faces, cancels out completely or partly the overall curvature, thus producing planar arrays and/or nonhomogenous nanotubes.43 A DAE tile is normally composed of five short oligonucleotides, an oblang-looped scaffold strand (22 × 2 nt long for A-form nucleic acids), two main helper strands, and two auxilary helper strands (Section S4 of the ESI†). Using a transcribed, linear RNA strand serving as the scaffold, the Mao group first reported the successful assembly of hybrid DAE-Op/q (all helper strands are short linear DNA oligonucleotides, p/q at 29/5) 2D ribbons;33 mimicking the DNA DX assembling strategies, the Franco group reported pure RNA:RNA DAE-Ep/q assemblies, mainly composed of RNA nanotubes (monolayered rectilinear strips with p/q at 22/6, nanotubes with p/q at 23/7 and 24/8, and nanofibers with p/q at 25/9 and 26/10).34 The Andersen group reported the cotranscriptional ssRNA DX-KL origami nanotechnology to assemble pure RNA 2D nanogrids. The intertile KL interactions with a 120° arrangement result in hexagonal nanogrids.35Using the circularised c44R to replace the transcriptional linear RNA molecule and serve as the scaffold strand, we constructed hybrid RNA:DNA and pure RNA:RNA DAE tiles; further applying the simple one-tile assembly strategy, we constructed hybrid DAE-Ep/q nanotubes and DAE-Op/q nanogrids, pure DAE-Ep/q nanotubes via the sticky end cohesion, and DAE-KL honeycomb-like nanogrids via the kissing-loop interaction.
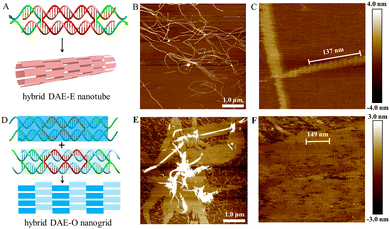
The hybrid DAE-E21/5 and DAE-O27/5 designs and their nanostructures are shown in Fig. 4. Following the general E- and O-tiling rules in DNA nanotechnology, the hybrid DAE-E21/5 provided high-yield mature nanotubes with a diameter of ∼30 nm and lengths ranging between 4 and 9 μm (Fig. 4B and Fig. S9, ESI†). In the zoomed-in AFM image (Fig. 4C), the periodic distance of a single tile length was measured to be 13.7 nm (137 nm/10), which is close to the theoretical value 13.6 nm (3.4 nm/turn × 4 turns). Perturbations of p/q to 20/4 and 22/6 also generated nanotubes, which have the similar tube diameter to that of p/q at 21/5, but in a bit shorter lengths ranging 2–7 μm and with relatively lower yields (Fig. S10 and S11, ESI†). In addition, we also imaged the hybrid DAE-E21/5 nanotubes stored at 4 °C for about a month and found that they kept intact (Fig. S12, ESI†), demonstrating that the hybrid RNA:DNA nanostructures are quite stable under our routine storage and processing conditions. The hybrid DAE-O27/5 provided perfect 2D flat arrays shown in Fig. 4E and F and Fig. S13 (ESI†), where the periodic distance of 14.9 nm (149 nm/10) between stripes corresponds to a DAE-O27/5 unit length (the theoretical value is 3.4 nm/turn × 4.5 turns = 15.3 nm). Perturbations of p/q to 26/4 gave monolayered strips with a width of ∼121.0 nm and lengths ranging between 2 and 6 μm (Fig. S14, ESI†), further to 28/4 and 29/5 only generated tile-oligomer fragments (Fig. S15, ESI†).
With c44R serving as the scaffold strands, we also succeeded in assembling pure RNA:RNA DAE-Ep/q nanotubes and DAE-KL hexagonal nanogrids (Fig. 5). We designed five DAE-Ep/q variants with p/q perturbed at 22/4, 22/6, 23/5, 23/7, and 24/8. Among these variants, DAE-E22/6 performed best to provide ripe, rigid RNA nanotubes with a diameter of ∼39.0 nm, a height of ∼3.6 nm, and lengths ranging between 2 and 7 μm (Fig. 5B and Fig. S16, ESI†); DAE-E22/4 and DAE-E23/5 formed similar but low-yield nanotubes with relatively shorter lengths ranging between 2 and 5 μm, which were accompanied by densely distributed tile-oligomer fragments (Fig. S17 and S18, ESI†); finally, DAE-E23/7 and DAE-E24/8 only generated much shorter nanofibers with lengths ranging between 0.5 and 2 μm (Fig. S19, ESI†). The above results presented a similar assembling tendency but differentiated slightly in detail with those reported in the reference using the linear RNA strand for scaffolding.34 For example, in the reference, DAE-E22/6 cannot form nanotubes but lengthy monolayered strips, while both DAE-E23/7 and DAE-E24/8 designs were optimal for assembling nanotubes, among which a few exceeded 10 μm in length. The reason for the assembling differences might be mainly attributed to the topology difference of the scaffold strands; however, the heterogeneity of the in vitro transcribed RNA strands, especially the considerable 3′ terminal heterogeneity, should also be accounted for.8,44,45
Inspired by the cotranscriptional, one-strand DX origami technology via kissing-loop base pairing interactions,35 we designed a two-strand DAE-KL system for self-assembly (Fig. 5C). With the c44R as the core scaffold, a transcriptional ssRNA complements to c44R and forms the DAE core domain, and the rest of the ssRNA strand self-associates at the four corners of the DAE-KL unit to generate two peripheral 120° KL pairs. The one-tile DAE-KL units assembled mainly to hexagonal nanogrids with a linear lattice constant at 23.7 nm (71 nm/3), very close to the theoretical value of 23.6 nm. Additional deformed polygons such as diamonds, pentagons, elongated hexagons, heptagons, and even octagons also occurred minorly. The above observations can be accounted for the formation of different vertex joints: regularly three-branched (statistically estimated at ∼70%), and minorly occurred two-branched and four-branched (Fig. 5D and S20).
Conclusions
In summary, using a newly established dumbbell splinting plus T4 DNA ligation strategy, we successfully converted two short, linear ssRNAs, miR-16 at 22 nt and a 44 nt strand adapted from the literature,34 to their respective monomeric circRNAs in near-quantitative yields. We screened the experimental conditions, including the concentrations of four components, RNA substrate, T4 DNA ligase, ATP, and Mg2+, and temperature plus incubation time for optimisation of monomeric cyclisation ligations. Based on this, we further applied the c44R strand to construct both hybrid and pure RNA DAE tiles and their DAE-Ep/q, DAE-Op/q, and DAE-KL assemblies with perturbed p/q variants, which appear as nanotubes, flat nanoarrays, and wireframe nanogrid patterns.Author contributions
SL, CM, and SJX developed the concept for this work through discussion (conceptualisation). SL conducted experiments and analysis including material synthesis, denaturing PAGE assay, RNA purification, nanostructure assembling and AFM imaging (investigation and formal analysis). YC and XG supported the experiments in transcription and AFM imaging, respectively (investigation). SL gathered and trimmed the data and constructed figures for visualisation and presentation (data curation and visualisation). SL and SJX wrote the original draft (writing). CM and SJX reviewed and edited the manuscript (writing and editing).Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
SJX acknowledges the financial support from the National Natural Science Foundation of China (91753134 and 21571100). CM acknowledges the financial support from the NSF (2025187 and 2107393).Notes and references
- Y. M. Demirci and M. D. Sacar Demirci, J. Integr. Bioinform., 2021, 18, 45–50 CrossRef PubMed.
- X. Liu, W. Xiong, M. Ye, T. Lu, K. Yuan, S. Chang, Y. Han, Y. Wang, L. Lu and Y. Bao, Signal Transduction Targeted Ther., 2023, 8, 441 CrossRef CAS PubMed.
- S. M. Nur, M. A. Hasan, M. A. Amin, M. Hossain and T. Sharmin, Interdiscip. Sci., 2015, 7, 257–265 CrossRef CAS PubMed.
- S. S. Sohrab, S. A. El-Kafrawy, Z. Mirza, M. A. Kamal and E. I. Azhar, Curr. Pharm. Des., 2018, 24, 62–77 CrossRef CAS PubMed.
- Y. Enuka, M. Lauriola, M. E. Feldman, A. Sas-Chen, I. Ulitsky and Y. Yarden, Nucleic Acids Res., 2016, 44, 1370–1383 CrossRef CAS PubMed.
- S. Umekage and Y. Kikuchi, J. Biotechnol., 2009, 139, 265–272 CrossRef CAS PubMed.
- V. E. Paluzzi, C. Zhang and C. Mao, Angew. Chem., Int. Ed., 2023, 62, e202218443 CrossRef CAS PubMed.
- P. Obi and Y. G. Chen, Methods, 2021, 196, 85–103 CrossRef CAS PubMed.
- S. Muller and B. Appel, RNA Biol., 2017, 14, 1018–1027 CrossRef PubMed.
- M. J. Moore and C. C. Query, Methods Enzymol., 2000, 317, 109–123 CAS.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2019, 119, 6459–6506 CrossRef CAS PubMed.
- S. Nummelin, J. Kommeri, M. A. Kostiainen and V. Linko, Adv. Mater., 2018, 30, e1703721 CrossRef PubMed.
- Y. He, T. Ye, M. Su, C. Zhang, A. E. Ribbe, W. Jiang and C. Mao, Nature, 2008, 452, 198–201 CrossRef CAS PubMed.
- P. Yin, R. F. Hariadi, S. Sahu, H. M. Choi, S. H. Park, T. H. Labean and J. H. Reif, Science, 2008, 321, 824–826 CrossRef CAS PubMed.
- P. W. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- D. Jasinski, F. Haque, D. W. Binzel and P. Guo, ACS Nano, 2017, 11, 1142–1164 CrossRef CAS PubMed.
- E. Poppleton, N. Urbanek, T. Chakraborty, A. Griffo, L. Monari and K. Gopfrich, RNA Biol., 2023, 20, 510–524 CrossRef CAS PubMed.
- D. Han, X. Qi, C. Myhrvold, B. Wang, M. Dai, S. Jiang, M. Bates, Y. Liu, B. An, F. Zhang, H. Yan and P. Yin, Science, 2017, 358, eaao2648 CrossRef PubMed.
- W. Grabow and L. Jaeger, F1000Prime Rep., 2013, 5, 46 Search PubMed.
- E. Westhof, B. Masquida and L. Jaeger, Fold Des., 1996, 1, R78–88 CrossRef CAS PubMed.
- W. W. Grabow and L. Jaeger, Acc. Chem. Res., 2014, 47, 1871–1880 CrossRef CAS PubMed.
- C. Geary, A. Chworos, E. Verzemnieks, N. R. Voss and L. Jaeger, Nano Lett., 2017, 17, 7095–7101 CrossRef CAS PubMed.
- N. B. Leontis and E. Westhof, Science, 2014, 345, 732–733 CrossRef CAS PubMed.
- A. S. Abu Almakarem, A. I. Petrov, J. Stombaugh, C. L. Zirbel and N. B. Leontis, Nucleic Acids Res., 2012, 40, 1407–1423 CrossRef CAS PubMed.
- K. A. Afonin, M. Viard, A. Y. Koyfman, A. N. Martins, W. K. Kasprzak, M. Panigaj, R. Desai, A. Santhanam, W. W. Grabow, L. Jaeger, E. Heldman, J. Reiser, W. Chiu, E. O. Freed and B. A. Shapiro, Nano Lett., 2014, 14, 5662–5671 CrossRef CAS PubMed.
- G. C. Shukla, F. Haque, Y. Tor, L. M. Wilhelmsson, J. J. Toulme, H. Isambert, P. Guo, J. J. Rossi, S. A. Tenenbaum and B. A. Shapiro, ACS Nano, 2011, 5, 3405–3418 CrossRef CAS PubMed.
- K. A. Afonin, M. Viard, I. Kagiampakis, C. L. Case, M. A. Dobrovolskaia, J. Hofmann, A. Vrzak, M. Kireeva, W. K. Kasprzak, V. N. KewalRamani and B. A. Shapiro, ACS Nano, 2015, 9, 251–259 CrossRef CAS PubMed.
- L. Rolband, D. Beasock, Y. Wang, Y. G. Shu, J. D. Dinman, T. Schlick, Y. Zhou, J. S. Kieft, S. J. Chen, G. Bussi, A. Oukhaled, X. Gao, P. Sulc, D. Binzel, A. S. Bhullar, C. Liang, P. Guo and K. A. Afonin, Comput. Struct. Biotechnol. J., 2022, 20, 6120–6137 CrossRef CAS PubMed.
- M. Chandler, T. Lyalina, J. Halman, L. Rackley, L. Lee, D. Dang, W. Ke, S. Sajja, S. Woods, S. Acharya, E. Baumgarten, J. Christopher, E. Elshalia, G. Hrebien, K. Kublank, S. Saleh, B. Stallings, M. Tafere, C. Striplin and K. A. Afonin, Molecules, 2018, 23, 3178 CrossRef PubMed.
- J. M. Stewart, M. Viard, H. K. Subramanian, B. K. Roark, K. A. Afonin and E. Franco, Nanoscale, 2016, 8, 17542–17550 RSC.
- M. F. Parsons, M. F. Allan, S. Li, T. R. Shepherd, S. Ratanalert, K. Zhang, K. M. Pullen, W. Chiu, S. Rouskin and M. Bathe, Nat. Commun., 2023, 14, 382 CrossRef CAS PubMed.
- E. C. Wamhoff, L. Ronsard, J. Feldman, G. A. Knappe, B. M. Hauser, A. Romanov, J. B. Case, S. Sanapala, E. C. Lam, K. J. S. Denis, J. Boucau, A. K. Barczak, A. B. Balazs, M. S. Diamond, A. G. Schmidt, D. Lingwood and M. Bathe, Nat. Commun., 2024, 15, 795 CrossRef CAS PubMed.
- S. H. Ko, M. Su, C. Zhang, A. E. Ribbe, W. Jiang and C. Mao, Nat. Chem., 2010, 2, 1050–1055 CrossRef CAS PubMed.
- J. M. Stewart, C. Geary and E. Franco, ACS Nano, 2019, 13, 5214–5221 CrossRef CAS PubMed.
- C. Geary, P. W. Rothemund and E. S. Andersen, Science, 2014, 345, 799–804 CrossRef CAS PubMed.
- M. Wang, H. Huang, Z. Zhang and S. J. Xiao, Nanoscale, 2016, 8, 18870–18875 RSC.
- M. Ali, N. Afshan, C. Jiang, H. Zheng and S. J. Xiao, Nanoscale, 2019, 11, 22216–22221 RSC.
- C. Jiang, B. Lu, W. Zhang, Y. P. Ohayon, F. Feng, S. Li, N. C. Seeman and S.-J. Xiao, J. Am. Chem. Soc., 2022, 144, 6759–6769 CrossRef CAS PubMed.
- Y. Wang, W. Ge, B. Lu, J. J. Zhu and S. J. Xiao, Nanoscale, 2020, 12, 19597–19603 RSC.
- R. An, Q. Li, Y. Fan, J. Li, X. Pan, M. Komiyama and X. Liang, Nucleic Acids Res., 2017, 45, e139 CrossRef PubMed.
- K. Shi, T. E. Bohl, J. Park, A. Zasada, S. Malik, S. Banerjee, V. Tran, N. Li, Z. Yin, F. Kurniawan, K. Orellana and H. Aihara, Nucleic Acids Res., 2018, 46, 10474–10488 CrossRef CAS PubMed.
- S. Ghafouri-Fard, T. Khoshbakht, B. M. Hussen, S. T. Abdullah, M. Taheri and M. Samadian, Cancer Cell Int., 2022, 22, 342 CrossRef CAS PubMed.
- H. Yan, S. H. Park, G. Finkelstein, J. H. Reif and T. H. LaBean, Science, 2003, 301, 1882–1884 CrossRef CAS PubMed.
- J. F. Milligan, D. R. Groebe, G. W. Witherell and O. C. Uhlenbeck, Nucleic Acids Res., 1987, 15, 8783–8798 CrossRef CAS PubMed.
- Y. Gholamalipour, A. Karunanayake Mudiyanselage and C. T. Martin, Nucleic Acids Res., 2018, 46, 9253–9263 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and experimental methods, additional dPAGE photographs, additional AFM images and necessary sectional profiling data, and nucleic acid sequences. See DOI: https://doi.org/10.1039/d4nh00236a |
| This journal is © The Royal Society of Chemistry 2024 |