Sugar alcohol-modified polyester nanoparticles for gene delivery via selective caveolae-mediated endocytosis†
Betsy
Reshma G
ab,
Chirag
Miglani
c,
Asish
Pal
 c and
Munia
Ganguli
c and
Munia
Ganguli
 *ab
*ab
aCSIR-Institute of Genomics and Integrative Biology, Mathura Road, New Delhi 110025, India. E-mail: mganguli@igib.in
bAcademy of Scientific and Innovative Research (AcSIR), Ghaziabad 201002, India
cChemical Biology Unit, Institute of Nanoscience and Technology, Sector 81, Mohali, Punjab 140306, India
First published on 30th January 2024
Abstract
Nucleic acid-based drugs are changing the scope of emerging medicine in preventing and treating diseases. Nanoparticle systems based on lipids and polymers developed to navigate tissue-level and cellular-level barriers are now emerging as vector systems that can be translated to clinical settings. A class of polymers, poly(β-amino esters) (PBAEs) known for their chemical flexibility and biodegradability, has been explored for gene delivery. These polymers are sensitive to changes in the monomer composition affecting transfection efficiency. Hence to add functionality to these polymers, we partially substituted ligands to an identified effective polymer chemistry. We report here a new series of statistical copolymers based on PBAEs where the backbone is modified with sugar alcohols to selectively facilitate the caveolae-mediated endocytosis pathway of cellular transport. These ligands are grafted at the polymer's backbone, thereby establishing a new strategy of modification in PBAEs. We demonstrate that these polymers form nanoparticles with DNA, show effective complexation and cargo release, enter the cell via selective caveolae-mediated endocytosis, exhibit low cytotoxicity, and increase transfection in neuronal cells.
Introduction
The advent of nucleic acid therapeutics has huge potential to treat diseases.1 Establishing such molecules for gene therapy or gene editing in clinical settings requires smart carriers that can efficiently complex nucleic acids to overcome various cellular and tissue level barriers. Viruses function as natural gene delivery agents and hence have been approved for the treatment of human diseases.2 However, the immunogenicity of viral vectors hinders redosing for therapeutic efficiencies.3 Nanoparticle-mediated strategies provide effective and safe solutions to overcome biological barriers for nucleic acid delivery.4Among the materials investigated for nucleic acid delivery, poly(β-amino esters) (PBAEs) have received special attention because of their chemical and structural diversity.5 PBAE polymers were first introduced by Lynn and Langer as gene delivery vectors.6 The polymers are formed in two steps by Michael's addition reaction with diacrylates and amines followed by end-capping.7 Over the years the polymer structure has been optimized for transfection efficiency and stability in vivo.8–10 The chemical flexibility of the polymer has been explored for improving its transfection efficiency7–9,11,12 and overcoming physical barriers for organ- and cellular-level tropism.13–15 More recently, combinatorial libraries emphasizing the physiochemical diversity of the choice of amines and divinyl monomers have been employed for organ tropism of the liver, spleen, and lungs.16,17 Apart from the chemical flexibility, these polymers self-assemble with diverse cargoes. PBAE polymers carry plasmid and mRNA13,18 for gene expression, siRNA19 for gene silencing, and the ribonucleoprotein complex for gene editing.20
The search for efficient polymers has been made by synthesizing libraries of polymers. However, this approach is expensive and exhaustible with less predictability from in vitro to in vivo conditions.21 Very few unique chemical spaces have been reported from combinatorial library screening to exhibit extrahepatic delivery. More targeted approaches involve protein corona modulation22,23 and antibody or peptide-mediated targeting.24,25 Surface engineering with antibodies and peptides increases the complexity of the delivery system, limiting clinical translation.26–28 Hence, there is a need for rational functional design of these polymers emerging from the biological identity of the target cell.
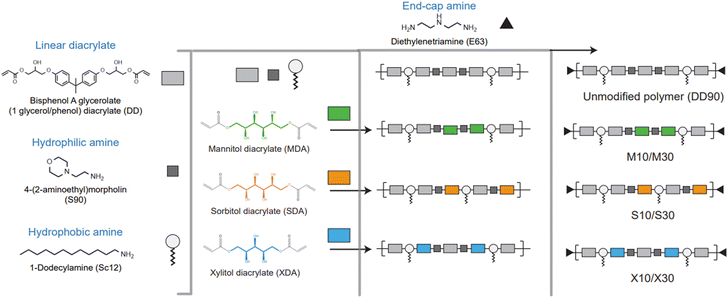
Stable polymers are synthesized as a four-component terpolymer, where each component enables efficient transfection in vitro and in vivo. We hypothesized that if functionality has to arise from the polymer it has to be partially substituted. To implement partial substitution-based functionality, we opted for modifying the PBAE polymer backbone using sugar alcohols, namely mannitol, sorbitol, and xylitol. Sugar alcohols have found their way to overcome the physical and biological barriers of the brain29–31 and lungs.32 Additionally, they have been reported to exhibit selective caveolae-mediated endocytosis.33
Caveolae-mediated endocytosis has recently gained recognition as a significant pathway for internalization and transport within various tissues. A large number of caveolae are found in the endothelial cells of peripheral organs and muscle cells.34 The dysregulation of caveolae in the brain microvascular endothelium of aged mice has opened up opportunities for the delivery of drugs to the brain via this endocytosis route.35 New strategies that employ caveolae-mediated transcytosis as a means to deliver drugs to the brain using both chemical and physical techniques have been developed.36–38 Caveolae, which are preformed vesicles ranging from 80 to 100 nm, are invaginations found in the plasma membrane and play a role in size-dependent endocytosis.39 However, sugar alcohol modification leads to an increase in the size of the nanoparticles as reported with plasmid DNA (pDNA).40–42 We hypothesized that using PBAEs we could control the size of the nanoparticles and also enable more chemical versatility to these modifications.
Herein, for the first time, we have reported the incorporation of sugar alcohols into the backbone of PBAE polymers, thereby making it a multicomponent statistical copolymer design strategy. These modifications have random integration of the sugar alcohols, whose doping is controlled by the feed ratios in the synthesis step. Sugar alcohol-modified polymers were able to effectively complex and release pDNA, improving transfection and cellular viability in neuronal cells. The backbone modification retained the osmotic properties, thereby polarizing the nanoparticles from nonspecific endocytosis to selective caveolae-mediated endocytosis. The high transfection efficiency in neuronal cells indicates that the system can be taken forward to enhance gene delivery to the brain.
Results and discussion
Design, synthesis, and characterization of modified polymers
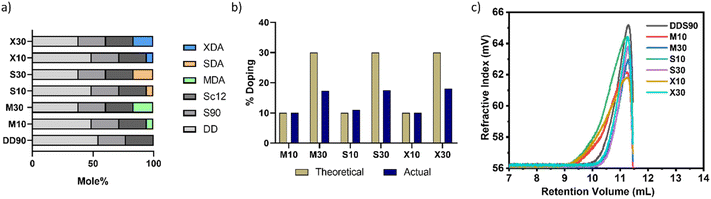
Among other polymer chemistries, PBAE was chosen because of its structural diversity and the ability to overcome gene delivery hurdles.5 Sugar alcohol monomers, such as mannitol, sorbitol, and xylitol, were known to selectively target caveolae-mediated endocytosis.41,42 Even though the three sugar alcohols are very similar in structure, their physical and chemical properties vary significantly. To dope sugar alcohols, we selected a combination of monomers reported for effective nucleic acid delivery.16 DD90 has been a very successful polymer backbone chemistry as reported by different groups.6,13,17 Starting with the hydrophobic DD90 as the foundational structure, we introduced sugar-alcohol in the backbone as a fourth element in the base polymer. Diacrylates of mannitol, sorbitol, and xylitol were synthesized with two-mole equivalents of acryloyl chloride to form mannitol diacrylate (MDA), sorbitol diacrylate (SDA), and xylitol diacrylate (XDA), respectively (Fig. S1†). The unmodified DD90 polymer was synthesized with diacrylate (B7), hydrophilic amine (S90), and hydrophobic amine (Sc12) in one pot Michael's addition reaction. For sugar alcohol incorporation, B7 diacrylate was partially substituted with sugar alcohol diacrylates (MDA, SDA, and XDA) at mole percentages of 10 and 30. This substitution was carried out alongside S90 and Sc12 to synthesize sugar alcohol-doped PBAE base polymers. Finally, the polymers were end-capped with diethylene triamine (E63). A schematic representation of the design strategy is shown in Fig. 1 and Fig. S1, S2† show the synthesis steps and structures. Doping sugar alcohols at higher percentages (more than 30%) resulted in a particle size over 150 nm. These particles did not show transfection and hence were excluded.The mole percentages of monomers used in the polymer synthesis reaction are given in Fig. 2a. The unmodified polymer DD90 and sugar alcohol-modified polymers were characterized by proton nuclear magnetic resonance (1H-NMR) spectroscopy and gel permeation chromatography (GPC). The percentage of grafting of mannitol (M10, M30), sorbitol (S10, S30), and xylitol (X10, X30) were calculated from the 1H-NMR spectra (400 MHz for 1H-NMR) with DD90 as a reference (Fig. S3†). 1H-NMR confirmed the doping percentage of sugar alcohols in the backbone of the polymer: 10 and 17.3% (M10 and M30) (Fig. S4 and S5†), 11 and 17.5% (S10 and S30) (Fig. S6 and S7†), and 10 and 18% (X10 and X30) (Fig. S8 and S9†). Sugar alcohol incorporation was found to be less with M30, S30, and X30, while the theoretical and actual doping percentages were identical in the case of lower doping ratios (Fig. 2b). The polymer molecular weight was between 4.5 and 7 kDa with PDI less than 2 (Table S2†). Multicomponent sugar alcohol-doped polymers M30, S30, and X30 formed uniform polymers with PDIs of 1.2–1.3 (Fig. 2c). These polymers were taken up for further studies without any additional purification.
Effects of sugar alcohol modification on polymer properties
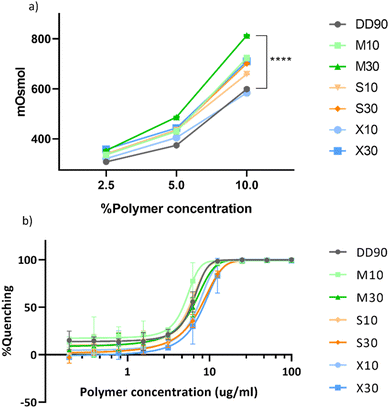
For the backbone modification of sugar alcohol to enable functionality, the osmotic properties of the sugar alcohols should be retained in the modified PBAE. The osmolarity was measured at 2.5, 5, and 10% polymer concentrations in 100 mM sodium acetate (pH 5.2) (Fig. 3a). DD diacrylate has two OH functional groups per monomer, hence the unmodified polymer in itself has osmotic properties. The addition of sugar alcohols into the backbone enhanced the osmolarity significantly. Among the modified polymers, M30 has the highest osmolarity of about 800 mOsmol, which is about 200 mOsmol higher than the unmodified polymer DD90 at 10% polymer concentration. All modified polymers had a positive effect on osmolarity in comparison with the unmodified polymer.Doping ligands into the backbone of the polymer can change polymer–DNA interactions. To analyze the nucleic acid binding efficiency of these modifications, we performed a RiboGreen nucleic acid binding assay. The polymers were serially diluted in sodium acetate buffer. pDNA with the RiboGreen dye was added to the polymer solution to allow binding. Fluorescence was plotted as percentage quenching against the polymer concentration (Fig. 3b). The IC50 values of binding can be calculated at 50% quenching, where higher values indicate weak interaction and vice versa. The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50 values increased with higher doping percentages but all polymers showed effective binding to DNA (Table S3†). The RiboGreen nucleic acid binding assay established efficient binding of modified polymers to DNA with a trend of 10% doped polymer binding better than those with 30% doping.
IC50 values increased with higher doping percentages but all polymers showed effective binding to DNA (Table S3†). The RiboGreen nucleic acid binding assay established efficient binding of modified polymers to DNA with a trend of 10% doped polymer binding better than those with 30% doping.
Nucleic acid binding is required but not a sufficient condition to predict the success of a delivery system. For a delivery system to work in vitro and in vivo, the nucleic acid should also be unpacked inside the cell.2,43,44 Complexation and release of DNA from nanoparticles is a balancing act.45 The efficient complexation as well as the efficient release of DNA from the nanoparticle can dictate the transfection efficiency.46 To study the complexation and release kinetics of the polymer-DNA complexes, we used gel electrophoresis. All polymers complexed plasmid DNA at weight ratios from 10 to 60 (Fig. S10a†). In the presence of heparin (anionic challenge), complete release was observed in M30, S30, and X30. DD90, M10, S10, and X10 exhibited partial release profiles even at the highest heparin ratio (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 32 w/w of polymer to heparin) (as depicted in Fig. S10b†) might be ascribed to the inherent hydrophobic nature of these polymers. This indicates that despite M30, S30, and X30 having good nucleic acid binding, they can release DNA completely and can achieve a balance between complexation and release which is an important aspect to consider when designing gene delivery agents. The complete release of pDNA with heparin in the case of M30, S30, and X30 raises a stability concern against nucleases. To check for nuclease protection, we treated the nanoparticles formed with pDNA and the polymer at 1
32 w/w of polymer to heparin) (as depicted in Fig. S10b†) might be ascribed to the inherent hydrophobic nature of these polymers. This indicates that despite M30, S30, and X30 having good nucleic acid binding, they can release DNA completely and can achieve a balance between complexation and release which is an important aspect to consider when designing gene delivery agents. The complete release of pDNA with heparin in the case of M30, S30, and X30 raises a stability concern against nucleases. To check for nuclease protection, we treated the nanoparticles formed with pDNA and the polymer at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 (w/w) with DNase I for 30 min and quantitatively measured free DNA with the RiboGreen dye. We observed that the nanoparticles formed enabled a high level of protection against DNase treatment, suggesting the formation of stable nanoparticles with complete encapsulation of pDNA (Fig. S11†).
60 (w/w) with DNase I for 30 min and quantitatively measured free DNA with the RiboGreen dye. We observed that the nanoparticles formed enabled a high level of protection against DNase treatment, suggesting the formation of stable nanoparticles with complete encapsulation of pDNA (Fig. S11†).
Preparation and characterization of polymer DNA nanoparticles
All polymers were rapidly self-assembled with pDNA in sodium acetate buffer at a DNA to polymer ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
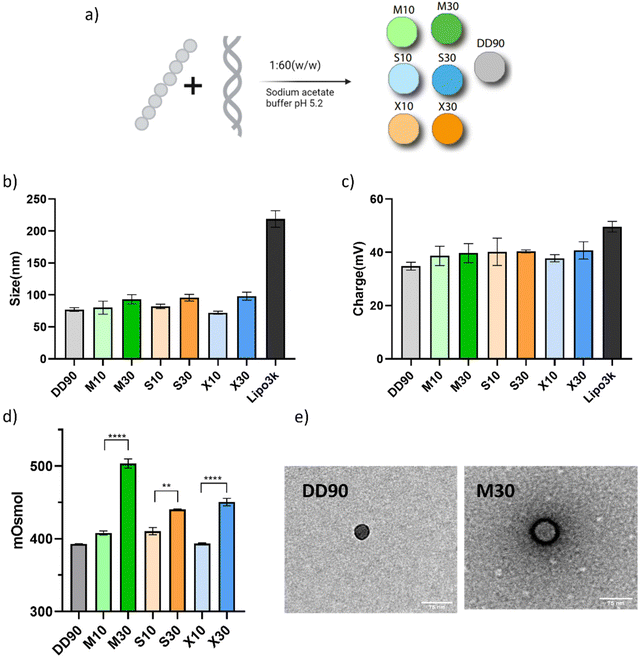
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 (w/w) (Fig. 4a). The weight ratio was taken from a previously reported work.16 The size of all the nanoparticles, estimated by dynamic light scattering, was below 100 nm (Fig. 4b). The zeta potential of the nanoparticles was estimated to be between 35 and 40 mV (Fig. 4c). The PDI of all the nanoparticles was below 0.25, indicating a uniform particle size (Fig. S12†). The nanoparticles formed with sugar alcohol-doped polymers did not vary significantly with the size and charge. We postulate that polyols might be counterproductive for nucleic acid binding, but the hydrophobic amines in PBAE enable efficient binding to pDNA. This interaction is crucial to keep the size of sugar alcohol modifications below 100 nm. Fig. 4b and c depict the size and charge of the particles formed with the commercial transfection agent Lipofectamine 3000 as a control.
60 (w/w) (Fig. 4a). The weight ratio was taken from a previously reported work.16 The size of all the nanoparticles, estimated by dynamic light scattering, was below 100 nm (Fig. 4b). The zeta potential of the nanoparticles was estimated to be between 35 and 40 mV (Fig. 4c). The PDI of all the nanoparticles was below 0.25, indicating a uniform particle size (Fig. S12†). The nanoparticles formed with sugar alcohol-doped polymers did not vary significantly with the size and charge. We postulate that polyols might be counterproductive for nucleic acid binding, but the hydrophobic amines in PBAE enable efficient binding to pDNA. This interaction is crucial to keep the size of sugar alcohol modifications below 100 nm. Fig. 4b and c depict the size and charge of the particles formed with the commercial transfection agent Lipofectamine 3000 as a control.
Osmotic stimuli are responsible for caveolae-mediated endocytosis.47 We measured the osmolarity of assembled nanoparticles to confirm that their surface properties reflect the osmolality function. In Fig. 4d, the sugar alcohol-doped polymeric nanoparticles demonstrated increased osmolarity, with approximately a 100 mOsmol difference for M30 and around 50 mOsmol for S30 and X30 in comparison with DD90. M10 and S10 showed marginally higher osmolarity while X10 showed no changes. Experimental findings demonstrate the enhanced osmolarity in sugar alcohol-modified nanoparticles, indicating increased functionality with higher doping percentages.
To gain structural insights into the nanoparticles, we performed transmission electron microscopy for DD90 and M30. We observed that the compositional changes in the polymer lead to different localizations of DNA within the nanoparticles as observed after positively staining DNA with uranyl acetate. In the case of DD90, the DNA seems to be compartmentalized throughout the structure of the nanoparticle, while M30 showed a prominent ring of DNA with a hollow interior (Fig. 4e). While the implication of the DNA arrangement within the nanoparticles remains elusive, it might be contributing to the complete release of the pDNA from the nanoparticles as seen with the release studies. We observed that the packaging of DNA is starkly different between DD90 and M30; however, there have been reports of the hollow center morphology with hydrophilic PBAE polymers.48
Cellular uptake and the mechanism of uptake
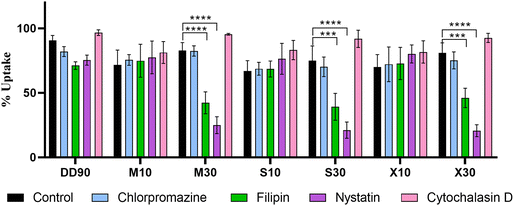
Modulating genes responsible for uptake, such as caveolin 1, have been shown to alter delivery in a cell type-specific manner.49 Caveolin 1 is upregulated under various disease conditions, enabling targeting opportunities.35,38,50 Apart from differential targeting opportunities with caveolae, it is attributed to an efficient endocytosis pathway for gene transfection.51 To confirm whether sugar alcohol doping in the backbone of the polymer facilitates selective caveolae-mediated endocytosis, we checked the uptake of nanoparticles in the presence of pharmacological endocytosis inhibitors. Four inhibitors, chlorpromazine (clathrin-mediated endocytosis), cytochalasin D (macropinocytosis), nystatin, and filipin (caveolae/lipid raft-mediated endocytosis), were used to access the mechanism of uptake. To enable this experiment, we labeled pDNA with Atto 488 as detailed in the Materials and Methods section. Polymeric nanoparticles showed good uptake of nanoparticles of about 70–90%. The total fluorescence intensity, however, was the highest for M30 with the sugar alcohol-modified PBAE (Fig. S13†). In the presence of inhibitors, M30, S30, and X30 showed a significant decrease from about 90% to about 50% with filipin and 30% with nystatin while other inhibitors did not affect uptake (Fig. 5). DD90, M10, S10, and X10 did not show entry by any specific pathway as there was no considerable decrease in uptake with the inhibitors. This confirms that the 30 percent molar doping of sugar alcohol polarized the nanoparticles to a specific caveolae/lipid raft-mediated endocytosis pathway.Cellular viability and transfection efficiency of sugar alcohol modification
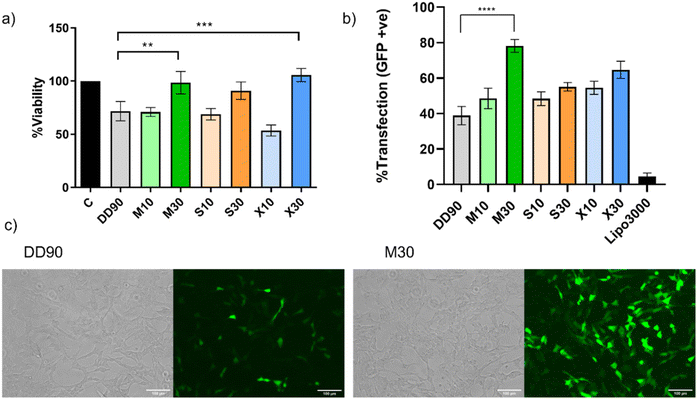
Ideal nucleic acid delivery systems should possess high efficiency and minimal toxicity.52 Recent work has reported a strong positive correlation of hydrophobicity with dose-independent transfection.16,53 The cell viability of the polymeric nanoparticles was estimated by MTT assay in SHSY5Y cell line. Cells were treated with nanoparticles in complete media with 300 ng of plasmid DNA in a 24-well plate for 24 h as detailed in the Materials and Methods section. It was observed that DD90, M10, S10, and X10 showed about 70% cell viability, which could be because of the hydrophobicity of the polymer (Fig. 6a). However, the sugar alcohols with higher doping (M30, S30, and X30) showed excellent cell viability. This indicates that doping with sugar alcohols can reduce the toxicity of the hydrophobic PBAE. All polymers performed significantly better when compared to the commercial agent Lipofectamine 3000 (Fig. S14†). The modified polymers showed higher transfection in comparison with unmodified DD90 with over 50% transfection efficiency in difficult-to-transfect SHSY5Y (Fig. 6b). M30 emerged as the lead candidate inducing superior transfection and exhibiting about 80% efficiency, nearly a two-fold increase compared with DD90 (Fig. 6c). The polyols in the sugar alcohols will reduce the hydrophobicity of the polymer. Interestingly, hydrophobicity changes with sugar alcohol modification did not affect the low-dose transfection efficiencies of the sugar alcohol-modified PBAE. The reduced hydrophobicity likely contributed to the enhanced cellular compatibility observed in the study. Interestingly, despite the decrease in hydrophobicity, the transfection efficiencies of these modified polymers were found to increase in serum-containing media with low doses of DNA. We found no colocalization of the nanoparticles with the lysosomes, ruling out endosomal escape for the differences in the transfection observed (Fig. S15†).M30 and caveolae-mediated endocytosis
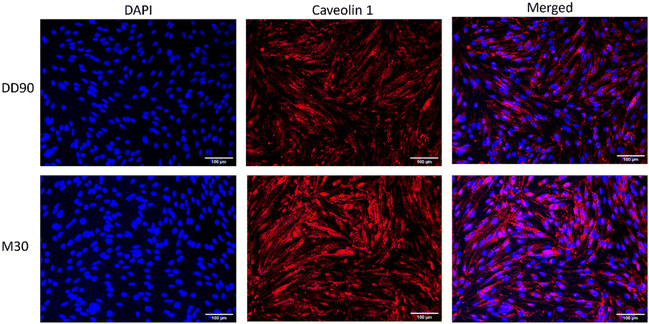
To corroborate our uptake study results for the involvement of caveolae-mediated endocytosis, we performed fluorescence microscopy studies with caveolae-specific markers.39 Cellular localization of the modified nanoparticles was observed by confocal microscopy using a caveolae-specific marker, FITC-labeled cholera toxin. After 4 h of incubation with DD90 and M30, we see the colocalization of Atto 488-labeled plasmid DNA containing nanoparticles with cholera toxin in the case of M30 (Fig. 7) confirming caveolae/lipid raft-mediated endocytosis. In previous studies, it has been reported that osmotically active mannitol-modified polyethyleneimine stimulated caveolin 1 in response to its hyperosmotic properties.30 To present a case for M30's use in material transport across the blood–brain barrier, we used microvascular brain endothelial cells – bEND.3. Cells were grown in a confluent monolayer over a coverslip to check for caveolin 1 expression with immunocytochemistry. The cells were fixed and permeabilized after 30 min of incubation with DD90 and M30 for caveolin 1 staining. Fig. 8 shows the abundance of caveolin 1 in M30 in comparison with DD90, indicating caveolae induction. For the prospect of gene delivery across the BBB, we reckon the particles that could be used in combination with agonist-induced models of caveolae induction36 or with physical methods like low-dose ultrasound.38Conclusion
We have developed highly efficient sugar alcohol-modified PBAE polymers from mannitol, sorbitol, and xylitol. To enable this modification, we have worked on the backbone incorporation of sugar alcohols in PBAE to overcome challenges with ligand conjugation complexity. We were able to synthesize 6 modified polymers doped at 10 and 30% doping ratios to the diacrylate in the polymer. These polymers showed an increase in osmolarity properties as expected from sugar alcohols in the backbone. More importantly, the modified polymers retained nucleic acid binding. The sugar alcohols at 30% doping got the balance right with complete complexation and complete release. The modification also led to selective caveolae-mediated endocytosis. Thus, we believe that the combination of efficient release and endocytosis mechanisms led to the efficient transfection of sugar alcohol-modified nanoparticles in the neuronal cell line SHSY5Y. The cellular viability profile also increased significantly with 30% doping of sugar alcohols. M30 was our lead polymer with a high transfection of about 80% transfection efficiency, twice that of DD90. There was concluding proof for caveolae-mediated endocytosis and stimulation with colocalization with a caveolae marker (cholera toxin) in neuronal cells and an increase in caveolin 1 expression in brain microvascular endothelial cells with M30. By successfully doping sugar alcohols into the backbone of the PBAE polymer and achieving efficient transfection, we have laid the foundation for exploring the potential of these modified nanoparticles for difficult-to-target organs. The reported polymer system has a less toxic composition and the alkyl side chains in the polymer can enable PEG lipid incorporation for its use in vivo. Through iterative optimization and exploration, this strategy has the potential to open up significant possibilities for the development of advanced gene delivery systems with enhanced transfection efficiency and selectivity, particularly for challenging targets and organs.Materials and methods
Bisphenol A glycerolate (1 glycerol/phenol) diacrylate (B7; CAS 4687949), 4-(2-aminoethyl) morpholine (S90; CAS 2038-031), 1-dodecylamine (Sc12; CAS 124-22-1), diethylenetriamine (E63; CAS 111400), mannitol (CAS 69658), sorbitol (CAS 50704), xylitol (CAS 87990), pyridine (CAS 110861), chlorpromazine hydrochloride (C0982), nystatin (N9150), filipin III (SAE0087) cytochalasin D (C8273), Atto 488 amine (74417), Atto 647N amine (95349), SPB (succinimidyl-[4-(psoralen-8-yloxy)] butyrate) (803545), DMSO-D6, dimethylformamide (CAS 68-12-2), diethyl ether (CAS 60-29-7), thiazolyl blue tetrazolium bromide (M5655), and the cholera toxin B subunit FITC conjugate (C1655) were purchased from Sigma-Aldrich. Acryloyl chloride (CAS 2123990) was purchased from Alfa Aesar. The pEGFP-C1 plasmid (4.7 kb) was amplified in Escherichia coli DH-5α and pDNA isolated with GenEluteTM from Sigma-Aldrich. The primary antibody against caveolin-1 (ab2910) was purchased from Abcam. The secondary antibody Alexa Fluor 594 goat anti-rabbit IgG (H + L) (A11012), ProLong Gold Antifade with DAPI (P3693), Lipofectamine 3000 (L3000-008), Lysotracker Red (L7528), and TURBO DNase (2238G2) were sourced from Thermo Fischer Scientific. Uranyl acetate was procured from CDH biochemicals.Synthesis of diacrylate sugar alcohol monomers
Diacrylate monomers of the sugar alcohols were synthesized according to previous reports.30 1 g of mannitol, sorbitol, or xylitol was weighed and dissolved in 20 mL of DMF. The solution was reacted with 10 mL of pyridine under stirring for 30 min at room temperature. 2 moles of acryloyl chloride in DMF were added and reacted at 4 °C overnight. The sugar alcohol diacrylates were precipitated with diethyl ether and dried in a speed vac.Polymer synthesis
Polymers were synthesized using a previously reported protocol.16 Specifically, the DD90 polymer was synthesized with bisphenol A glycerolate diacrylate, 4-(2-amino methyl) morpholine, and dodecylamine to form the base polymer. The modified base polymers M10, S10, X10, and M30, S30, and X30 were synthesized with 10% and 30% molar doping concentrations of MDA, SDA, and XDA, respectively, to bisphenol A glycerolate diacrylate. The detailed stoichiometric molar ratio is given in Table S1.† The diacrylates and amines were reacted at 90 °C for 48 h in DMF under stirring. Following this reaction, the polymers were capped with 1.5 moles of diethylenetriamine for 2 h at room temperature in DMSO. The polymers were precipitated, washed twice with diethyl ether, and dried under vacuum. The polymers were dissolved in DMSO at 100 mg mL−1 concentration and stored in single-use aliquots at −20 °C with a desiccant.NMR
NMR spectra were recorded on a 400 MHz Bruker (400 MHz for 1H-NMR). Polymers were dissolved in DMSO-d6 at a concentration of 20 mg/0.6 mL. The chemical shifts are reported in ppm and appear downfield to tetramethylsilane using the resonance of the deuterated solvent as an internal standard. Splitting patterns are designated as singlet (s), doublet (d), triplet (t), and multiplet (m).Gel permeation chromatography
GPC measurements were performed on a Malvern Omnisec instrument having an RI detector and a Shodex KD-806 M column with DMF as an eluent with a flow rate of 0.7 mL min−1 at a temperature of 35 °C with 0.01 M LiBr and PMMA as standards for all the samples. The results were analyzed by using Omnisec software. The sample peaks were analyzed for obtaining Mn, Mw, and PDI using the conventional calibration method. Samples for the GPC measurement was prepared (6 mg mL−1) by filtering solutions through a 0.2 μm nylon filter into a 2 mL GPC glass vial.Osmolarity measurement
Osmolarity values were measured in an aqueous buffer of 100 mM sodium acetate (pH 5.2). Polymers DD90, M10, M30, S10, S30, X10, and X30 were dissolved in 2.5, 5 and 10% concentrations and were measured as mOsmol using a cryoscopic osmometer (OSMOMAT 3000, GENOTEC).Nucleic acid binding assay
The sugar alcohol-modified polymers were evaluated for nucleic acid binding with the RiboGreen nucleic acid dye. The polymers were serially diluted from 100 μg μL−1 concentration in 25 mM sodium acetate buffer. The pDNA (pEGFP-C1) solution was prepared at a 1 μg mL−1 stock concentration with the RiboGreen dye in 25 mM sodium acetate buffer. 25 μL of the polymer solution was mixed with 75 μL of nucleic acid/RiboGreen solution in 96 well-black bottom plates. The samples were incubated for 20 min at 37 °C. The fluorescence reading was taken using a Tecan Infinite M200 Pro. Nucleic acid affinity with the polymers was characterized using the IC50 value of binding. The IC50 value was obtained by plotting the fluorescence quenching as a function of polymer concentration and fitting a sigmoid curve to the data. Lower IC50 values indicate higher binding and vice versa.Complexation and release by gel electrophoresis
Complexation and release studies were performed using the gel electrophoretic technique. Agarose gels were cast with 1% agarose dissolved in Tris-acetate-EDTA (TAE) buffer. 25 ng of plasmid was loaded per well. Polymers with different w/w ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, 1
0.5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, 1
5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 1
20, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, 1
30, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, and 1
40, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) were added to DNA to check the complexation variations between the polymers. For release studies, the DNA plasmid ratio was fixed to 1
60) were added to DNA to check the complexation variations between the polymers. For release studies, the DNA plasmid ratio was fixed to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 (w/w), and the heparin weights were set in gradients of 1
60 (w/w), and the heparin weights were set in gradients of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.25, 1
0.25, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, 1
0.5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, 1
4, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, 1
8, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16, and 1
16, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36 to the polymers and incubated for 30 min. Orange loading dye was added to the sample before loading the sample to the gel, electrophoresis was run at 100 V for 10 min and the gel was visualized using a gel doc system.
36 to the polymers and incubated for 30 min. Orange loading dye was added to the sample before loading the sample to the gel, electrophoresis was run at 100 V for 10 min and the gel was visualized using a gel doc system.
DNase assay
The ability of the nanoparticles to protect pDNA from nucleases was assayed with DNase treatment. Particles with a pDNA to polymer ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 (w/w) were formed using 2 μg of DNA in 25 mM sodium acetate (pH 5.2). Following incubation for 10 min, samples were split into two groups with and without DNase treatment. Nanoparticles were treated with 1 U μL−1 of DNase I at 37 °C for 30 min. Samples were transferred to 96 well-black bottom plates and 100 μL of RiboGreen solution (5 μg ml−1) was added to each sample to measure free DNA. The fluorescence intensity was determined using a TECAN plate reader. The percentage of pDNA protection was defined as (fluorescence with DNase)/(fluorescence without DNase) × 100.
60 (w/w) were formed using 2 μg of DNA in 25 mM sodium acetate (pH 5.2). Following incubation for 10 min, samples were split into two groups with and without DNase treatment. Nanoparticles were treated with 1 U μL−1 of DNase I at 37 °C for 30 min. Samples were transferred to 96 well-black bottom plates and 100 μL of RiboGreen solution (5 μg ml−1) was added to each sample to measure free DNA. The fluorescence intensity was determined using a TECAN plate reader. The percentage of pDNA protection was defined as (fluorescence with DNase)/(fluorescence without DNase) × 100.
Nanoparticle formation and characterization
The plasmid DNA and polymer at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 (w/w) were pipette mixed with 25 mM sodium acetate (pH 5.2) to formulate particles and incubated for 10 min at room temperature. For size measurements, 4 μg of plasmid was used in nanoparticle formation with polymers. Nanoparticles with Lipofectamine 3000 were formed as per the manufacturer's protocol. The nanoparticles were diluted in MilliQ at 0.002 μg μl−1 concentration and for zeta potential measurement, the nanoparticles were diluted in 10 mM NaCl and analyzed using a Zetasizer Nano ZS (Malvern Instruments, UK). The osmolarity of nanoparticles was measured using an Osmometer 3000 (GENEOTEC).
60 (w/w) were pipette mixed with 25 mM sodium acetate (pH 5.2) to formulate particles and incubated for 10 min at room temperature. For size measurements, 4 μg of plasmid was used in nanoparticle formation with polymers. Nanoparticles with Lipofectamine 3000 were formed as per the manufacturer's protocol. The nanoparticles were diluted in MilliQ at 0.002 μg μl−1 concentration and for zeta potential measurement, the nanoparticles were diluted in 10 mM NaCl and analyzed using a Zetasizer Nano ZS (Malvern Instruments, UK). The osmolarity of nanoparticles was measured using an Osmometer 3000 (GENEOTEC).
Transmission electron microscopy
TEM imaging was performed to get insight into the location of DNA within the nanoparticles. The formed nanoparticles were deposited on 200 mesh copper grids. The copper grids were washed with ultrapure water to remove buffer salts. The grids were then washed with 1% (w/v) uranyl acetate in ultrapure water and air-dried overnight. Samples were examined on a TECNAI G2 20 Twin electron microscope. 10–15 fields were captured for each sample.Cell culture
In vitro experiments were performed using human neuroblastoma cells (SHSY5Y) provided by Beena Pillai at CSIR-IGIB. Mouse brain microvascular endothelial cells (bEND.3) were procured from AddexBio. The cells were passaged with DMEM supplemented with 10% FBS from Gibco and were maintained and cultured using previously established conditions.Plasmid labeling
The plasmid was labeled as reported previously.54 100 μl (1 μg μL−1) of pDNA was incubated with 12.5 μL (1 μg μL−1) of NHS-psoralen in a round bottom 96-well plate. The reaction took place under a 365 nm UV lamp for 25 min to crosslink psoralen with plasmid DNA. The samples were kept in an ice bath to prevent denaturation. Following the crosslinking reaction, 40 μg of Atto 488 amine was added and incubated in the dark for 1 h incubation at room temperature. The labeled plasmid was purified by ethanol precipitation. DNA was resuspended in nuclease-free water and stored at −20 °C for further use.Mechanism of uptake with endocytosis inhibitors
The seeded cells in 24 well plates were incubated with inhibitors chlorpromazine (10 μg mL−1), filipin III (7.5 μM), cytochalasin D (500 nM), and nystatin (50 μg mL−1) for 30 min. Nanoparticles formed with Atto 488-labeled plasmid were added to the cells at 300 ng of DNA per well and incubated for 4 h at 37 °C. Any signal from the membrane-bound nanoparticles was quenched with 0.04% trypan blue. Flow cytometry was performed to estimate the uptake of nanoparticles. The results were recorded with an FL1 green channel. The percentage uptake was obtained using BD accuri software and plotted with the presence and absence of inhibitors for comparison.Transfection efficiency and cytotoxicity
The cells were plated at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per 24 well in 400 μL of complete media and allowed to adhere overnight. The nanoparticles were formulated with plasmid DNA (pEGFP-C1) as described earlier at 1
000 per 24 well in 400 μL of complete media and allowed to adhere overnight. The nanoparticles were formulated with plasmid DNA (pEGFP-C1) as described earlier at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 (v/v) of DNA to the polymer. The nanoparticles were incubated with the cells in complete media for 24 h at 300 ng per well. For transfections using commercially available reagents, Lipofectamine 3000 (Thermo Fisher Scientific) was used as instructed by the manufacturer. After 24 h, cytotoxicity was measured by the MTT assay and transfection was evaluated by flow cytometry. For viability assay, the MTT reagent was added in opti MEM to obtain a final concentration of 0.5 mg mL−1 and incubated at 37 °C for 2 h. The medium was carefully aspirated and 200 μL of DMSO was added to solubilize the formazan crystals and the absorbance was measured using a Tecan microplate reader. The cell viability of the treated cells was normalized to that of untreated controls. For the estimation of the gene expression of the GFP reporter, flow cytometry was performed on a BD Accuri C6. The percentage of positive GFP was calculated using BD Accuri C6 software and plotted.
60 (v/v) of DNA to the polymer. The nanoparticles were incubated with the cells in complete media for 24 h at 300 ng per well. For transfections using commercially available reagents, Lipofectamine 3000 (Thermo Fisher Scientific) was used as instructed by the manufacturer. After 24 h, cytotoxicity was measured by the MTT assay and transfection was evaluated by flow cytometry. For viability assay, the MTT reagent was added in opti MEM to obtain a final concentration of 0.5 mg mL−1 and incubated at 37 °C for 2 h. The medium was carefully aspirated and 200 μL of DMSO was added to solubilize the formazan crystals and the absorbance was measured using a Tecan microplate reader. The cell viability of the treated cells was normalized to that of untreated controls. For the estimation of the gene expression of the GFP reporter, flow cytometry was performed on a BD Accuri C6. The percentage of positive GFP was calculated using BD Accuri C6 software and plotted.
Lysosome colocalization
SHSY5Y cells were seeded in 24 well plates at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well density. At 60–70% confluency, the cells were incubated with nanoparticles of DD90, M30, S30, and X30 carrying Atto 488-labeled plasmid DNA. After 4 h, the medium was removed and Lysotracker Red (50 nM) was added in Fluorobrite medium and incubated for 10 min. Later, the cells were washed with PBS and imaged in Fluorobrite medium with EVOS M5000.
000 cells per well density. At 60–70% confluency, the cells were incubated with nanoparticles of DD90, M30, S30, and X30 carrying Atto 488-labeled plasmid DNA. After 4 h, the medium was removed and Lysotracker Red (50 nM) was added in Fluorobrite medium and incubated for 10 min. Later, the cells were washed with PBS and imaged in Fluorobrite medium with EVOS M5000.
Colocalization with cholera toxin
The cells were seeded at 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per 12-well plate on a coverslip to adhere overnight. The nanoparticles DD90, M30, S30, and X30 formed with Atto 647-labeled plasmid were incubated with the cells in fresh media for 3 h. Cholera toxin labeled with FITC was added to the wells as per the manufacturer's instruction and incubated for 30 min. The coverslips were fixed with 4% paraformaldehyde for 15 min and mounted with ProLong Gold Antifade with DAPI on a glass slide and sealed. Images were captured using a Leica SP8 confocal microscope.
000 per 12-well plate on a coverslip to adhere overnight. The nanoparticles DD90, M30, S30, and X30 formed with Atto 647-labeled plasmid were incubated with the cells in fresh media for 3 h. Cholera toxin labeled with FITC was added to the wells as per the manufacturer's instruction and incubated for 30 min. The coverslips were fixed with 4% paraformaldehyde for 15 min and mounted with ProLong Gold Antifade with DAPI on a glass slide and sealed. Images were captured using a Leica SP8 confocal microscope.
Immunocytochemistry in bEND.3
bEND.3 cells were seeded in a 6-well plate with a coverslip at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well to adhere overnight. After 4 h of transfection with the nanoparticles, the cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. After fixation, the cells were washed gently with PBS, followed by permeabilization with washing buffer (0.1% Triton-X100 in PBS for 5 min). The cells were kept submerged in a blocking buffer (5% BSA in PBS with 0.1% Tween 20) for 60 min at room temperature. Subsequently, the blocking buffer was switched with the primary antibody solution (anti-caveolin 1 antibody diluted to 1
000 cells per well to adhere overnight. After 4 h of transfection with the nanoparticles, the cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. After fixation, the cells were washed gently with PBS, followed by permeabilization with washing buffer (0.1% Triton-X100 in PBS for 5 min). The cells were kept submerged in a blocking buffer (5% BSA in PBS with 0.1% Tween 20) for 60 min at room temperature. Subsequently, the blocking buffer was switched with the primary antibody solution (anti-caveolin 1 antibody diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in blocking buffer), and the cells were kept submerged in it overnight at 4 °C. The following day, the primary antibody solution was discarded, and the cells were washed with washing buffer before treatment with the secondary antibody solution (goat anti-rabbit IgG (H + L) secondary antibody Alexa Fluor 594 diluted to 1
500 in blocking buffer), and the cells were kept submerged in it overnight at 4 °C. The following day, the primary antibody solution was discarded, and the cells were washed with washing buffer before treatment with the secondary antibody solution (goat anti-rabbit IgG (H + L) secondary antibody Alexa Fluor 594 diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 in blocking buffer) for 60 min. Next, the secondary antibody solution was discarded, and the cells were washed with washing buffer. The glass coverslip was mounted on top of the cells using ProLong Gold Antifade (with DAPI) and images were visualized using a Life Technologies Floid Cell Imaging Station.
800 in blocking buffer) for 60 min. Next, the secondary antibody solution was discarded, and the cells were washed with washing buffer. The glass coverslip was mounted on top of the cells using ProLong Gold Antifade (with DAPI) and images were visualized using a Life Technologies Floid Cell Imaging Station.
Statistics
Statistical analyses were performed using GraphPad Prism software (GraphPad Software, Inc.). The results are depicted as the mean and standard with three repeats if not stated otherwise. Significance was calculated with two-way ANOVA. Statistical significance was denoted as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.Author contributions
B. R. G.: conceptualization, methodology, investigation, validation, visualization, and writing – original draft. C. M.: investigation and validation. A. P.: validation, writing – review and editing, resources, and supervision. M. G.: conceptualization, methodology, validation, supervision, writing – review and editing, and funding acquisition.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was funded by the Council of Scientific and Industrial Research (MLP2011). B. R. G. acknowledges funding under the CSIR-GATE for a fellowship. We thank Rakesh Jha for confocal microscopy and Aseem Chaphalkar for the illustrations from CSIR-IGIB. We thank the technical staff and TEM facility at the National Institute of Immunology, Delhi for TEM image acquisition. Figures were created with Adobe Illustrator.References
- B. B. Mendes, J. Conniot, A. Avital, D. Yao, X. Jiang, X. Zhou, N. Sharf-Pauker, Y. Xiao, O. Adir, H. Liang, J. Shi, A. Schroeder and J. Conde, Nat. Rev. Methods Primers, 2022, 2, 24 CrossRef CAS PubMed.
- I. Lostalé-Seijo and J. Montenegro, Nat. Rev. Chem., 2018, 2, 258–277 CrossRef.
- N. F. Nidetz, M. C. McGee, L. V. Tse, C. Li, L. Cong, Y. Li and W. Huang, Pharmacol. Ther., 2020, 207, 119142 CrossRef PubMed.
- S. A. Dilliard and D. J. Siegwart, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed.
- J. Karlsson, K. R. Rhodes, J. J. Green and S. Y. Tzeng, Expert Opin. Drug Delivery, 2020, 17, 1395–1410 CrossRef CAS PubMed.
- D. M. Lynn and R. Langer, J. Am. Chem. Soc., 2000, 122, 10761–10768 CrossRef CAS.
- J. J. Green, R. Langer and D. G. Anderson, Acc. Chem. Res., 2008, 41, 749–759 CrossRef CAS PubMed.
- A. A. Eltoukhy, D. Chen, C. A. Alabi, R. Langer and D. G. Anderson, Adv. Mater., 2013, 25, 1487–1493 CrossRef CAS PubMed.
- W. Wang, D. Zhou, L. Cutlar, Y. Gao, W. Wang, J. O'Keeffe-Ahern, S. McMahon, B. Duarte, F. Larcher, B. J. Rodriguez and U. Greiser, Sci. Adv., 2016, 2, 1600102 CrossRef PubMed.
- Y. Li, X. Wang, Z. He, M. Johnson, S. A., I. Lara-Sáez, J. Lyu and W. Wang, J. Am. Chem. Soc., 2023, 145, 17187–17200 CrossRef CAS PubMed.
- A. A. Eltoukhy, D. J. Siegwart, C. A. Alabi, J. S. Rajan, R. Langer and D. G. Anderson, Biomaterials, 2012, 33, 3594–3603 CrossRef CAS PubMed.
- Y. Deng, J. Zhang, X. Sun, L. Li, M. Zhou, S. Liu, F. Chen, C. Pan, Z. Yu, M. Li, W. Zhong and M. Zeng, J. Controlled Release, 2023, 363, 597–605 CrossRef CAS PubMed.
- L. Rotolo, D. Vanover, N. C. Bruno, H. E. Peck, C. Zurla, J. Murray, R. K. Noel, L. O'Farrell, M. Araínga, N. Orr-Burks, J. Y. Joo, L. C. S. Chaves, Y. Jung, J. Beyersdorf, S. Gumber, R. Guerrero-Ferreira, S. Cornejo, M. Thoresen, A. K. Olivier, K. M. Kuo, J. C. Gumbart, A. R. Woolums, F. Villinger, E. R. Lafontaine, R. J. Hogan, M. G. Finn and P. J. Santangelo, Nat. Mater., 2023, 22, 369–379 CrossRef CAS PubMed.
- A. F. Rodrigues, C. Rebelo, S. Simões, C. Paulo, S. Pinho, V. Francisco and L. Ferreira, Adv. Sci., 2023, 10, 2205475 CrossRef CAS PubMed.
- Z. Deng, W. Gao, F. Kohram, E. Li, T. V. Kalin, D. Shi and V. V. Kalinichenko, Bioact. Mater., 2024, 31, 1–17 CAS.
- Y. Rui, D. R. Wilson, S. Y. Tzeng, H. M. Yamagata, D. Sudhakar, M. Conge, C. A. Berlinicke, D. J. Zack, A. Tuesca and J. J. Green, Sci. Adv., 2022, 8, 2855 CrossRef PubMed.
- J. C. Kaczmarek, A. K. Patel, L. H. Rhym, U. C. Palmiero, B. Bhat, M. W. Heartlein, F. DeRosa and D. G. Anderson, Biomaterials, 2021, 275, 120966 CrossRef CAS PubMed.
- P. Mastorakos, C. Zhang, E. Song, Y. E. Kim, H. W. Park, S. Berry, W. K. Choi, J. Hanes and J. S. Suk, J. Controlled Release, 2017, 262, 37–46 CrossRef CAS PubMed.
- K. L. Kozielski, A. Ruiz-Valls, S. Y. Tzeng, H. Guerrero-Cázares, Y. Rui, Y. Li, H. J. Vaughan, M. Gionet-Gonzales, C. Vantucci, J. Kim, P. Schiapparelli, R. Al-Kharboosh, A. Quiñones-Hinojosa and J. J. Green, Biomaterials, 2019, 209, 79–87 CrossRef CAS PubMed.
- Y. Rui, D. R. Wilson, J. Choi, M. Varanasi, K. Sanders, J. Karlsson, M. Lim and J. J. Green, Sci. Adv., 2019, 5, 3255 CrossRef PubMed.
- K. Paunovska, C. D. Sago, C. M. Monaco, W. H. Hudson, M. G. Castro, T. G. Rudoltz, S. Kalathoor, D. A. Vanover, P. J. Santangelo, R. Ahmed, A. V. Bryksin and J. E. Dahlman, Nano Lett., 2018, 18, 2148–2157 CrossRef CAS PubMed.
- C. Fornaguera, M. Guerra-Rebollo, M. Á. Lázaro, A. Cascante, N. Rubio, J. Blanco and S. Borrós, Adv. Healthc. Mater., 2019, 8, 1900849 CrossRef CAS PubMed.
- T. J. Harris, J. J. Green, P. W. Fung, R. Langer, D. G. Anderson and S. N. Bhatia, Biomaterials, 2010, 31, 998–1006 CrossRef CAS PubMed.
- T. T. Smith, S. B. Stephan, H. F. Moffett, L. E. McKnight, W. Ji, D. Reiman, E. Bonagofski, M. E. Wohlfahrt, S. P. S. Pillai and M. T. Stephan, Nat. Nanotechnol., 2017, 12, 813–822 CrossRef CAS PubMed.
- N. Distasio, F. Dierick, T. Ebrahimian, M. Tabrizian and S. Lehoux, Acta Biomater., 2022, 143, 356–371 CrossRef CAS PubMed.
- D. Rosenblum, N. Joshi, W. Tao, J. M. Karp and D. Peer, Nat. Commun., 2018, 9, 1410 CrossRef PubMed.
- T. Liang, Z. Xing, L. Jiang and J. J. Zhu, VIEW, 2021, 2, 20200131 CrossRef CAS.
- G. Lin and M. Zhang, Acc. Chem. Res., 2023, 2007166–2007169 Search PubMed.
- S. Pandey, M. C. Lee, J. Woon Lim, Y. H. Choung, K. J. Jang, S. B. Park, J. E. Kim, J. H. Chung and P. Garg, Biomaterials, 2022, 281, 121359 CrossRef CAS PubMed.
- T. E. Park, B. Singh, H. Li, J. Y. Lee, S. K. Kang, Y. J. Choi and C. S. Cho, Biomaterials, 2015, 38, 61–71 CrossRef CAS PubMed.
- P. Garg, S. Pandey, H. Seonwoo, S. Yeom, Y. H. Choung, C. S. Cho, P. H. Choung and J. H. Chung, Chem. Commun., 2015, 51, 3645–3648 RSC.
- A. Kumari, S. Pal, B. Reshma G., F. P. Mohny, N. Gupta, C. Miglani, B. Pattnaik, A. Pal and M. Ganguli, Mol. Pharm., 2022, 19, 1309–1324 CrossRef CAS PubMed.
- S. J. Hong, M. H. Ahn, J. Sangshetti and R. B. Arote, Acta Biomater., 2019, 97, 105–115 CrossRef CAS PubMed.
- C. Matthaeus and J. W. Taraska, Front. Cell Dev. Biol., 2021, 8, 614472 CrossRef PubMed.
- A. C. Yang, M. Y. Stevens, M. B. Chen, D. P. Lee, D. Stähli, D. Gate, K. Contrepois, W. Chen, T. Iram, L. Zhang, R. T. Vest, A. Chaney, B. Lehallier, N. Olsson, H. Du Bois, R. Hsieh, H. C. Cropper, D. Berdnik, L. Li, E. Y. Wang, G. M. Traber, C. R. Bertozzi, J. Luo, M. P. Snyder, J. E. Elias, S. R. Quake, M. L. James and T. Wyss-Coray, Nature, 2020, 583, 425–430 CrossRef CAS PubMed.
- J. H. Chang, C. Greene, K. Frudd, L. Araujo dos Santos, C. Futter, B. J. Nichols, M. Campbell and P. Turowski, Cell Rep. Med., 2022, 3, 100497 CrossRef CAS PubMed.
- D. E. Tylawsky, H. Kiguchi, J. Vaynshteyn, J. Gerwin, J. Shah, T. Islam, J. A. Boyer, D. R. Boué, M. Snuderl, M. B. Greenblatt, Y. Shamay, G. P. Raju and D. A. Heller, Nat. Mater., 2023, 22, 391–399 CrossRef CAS PubMed.
- R. Pandit, W. K. Koh, R. K. P. Sullivan, T. Palliyaguru, R. G. Parton and J. Götz, J. Controlled Release, 2020, 327, 667–675 CrossRef CAS PubMed.
- M. S. de Almeida, E. Susnik, B. Drasler, P. Taladriz-Blanco, A. Petri-Fink and B. Rothen-Rutishauser, Chem. Soc. Rev., 2021, 50, 5397–5434 RSC.
- W. S. Lee, Y. K. Kim, Q. Zhang, T. E. Park, S. K. Kang, D. W. Kim, C. S. Cho and Y. J. Choi, Nanomedicine, 2014, 10, 525–534 CrossRef CAS PubMed.
- M. A. Islam, C. H. Yun, Y. J. Choi, J. Y. Shin, R. Arote, H. L. Jiang, S. K. Kang, J. W. Nah, I. K. Park, M. H. Cho and C. S. Cho, Biomaterials, 2011, 32, 9908–9924 CrossRef CAS PubMed.
- T. E. Park, B. Kang, Y. K. Kim, Q. Zhang, W. S. Lee, M. A. Islam, S. K. Kang, M. H. Cho, Y. J. Choi and C. S. Cho, Biomaterials, 2012, 33, 7272–7281 CrossRef CAS PubMed.
- A. L. J. Marschall, BioDrugs, 2021, 35, 643–671 CrossRef CAS PubMed.
- P. Roy, N. W. Kreofsky, M. E. Brown, C. Van Bruggen and T. M. Reineke, JACS Au, 2023, 3, 1876–1889 CrossRef CAS PubMed.
- C. L. Grigsby and K. W. Leong, J. R. Soc., Interface, 2010, 7, 67–82 CrossRef PubMed.
- D. V. Schaffer, N. A. Fidelman, N. Dan and D. A. Lauffenburger, Biotechnol. Bioeng., 2000, 67, 598–606 CrossRef CAS PubMed.
- M. A. Islam, T. E. Park, J. Firdous, H. S. Li, Z. Jimenez, M. Lim, J. W. Choi, C. H. Yun and C. S. Cho, Prog. Mater. Sci., 2022, 128, 100961 CrossRef.
- M. Keeney, S. G. Ong, A. Padilla, Z. Yao, S. Goodman, J. C. Wu and F. Yang, ACS Nano, 2013, 7, 7241–7250 CrossRef CAS PubMed.
- C. D. Sago, M. P. Lokugamage, G. N. Lando, N. Djeddar, N. N. Shah, C. Syed, A. V. Bryksin and J. E. Dahlman, Nano Lett., 2018, 18, 7590–7600 CrossRef CAS PubMed.
- X. L. Qian, Y. H. Pan, Q. Y. Huang, Y. B. Shi, Q. Y. Huang, Z. Z. Hu and L. X. Xiong, OncoTargets Ther., 2019, 12, 1539–1552 CrossRef CAS PubMed.
- Y. Y. Won, R. Sharma and S. F. Konieczny, J. Controlled Release, 2009, 139, 88–93 CrossRef CAS PubMed.
- M. Breunig, U. Lungwitz, R. Liebl and A. Goepferich, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 14454–14459 CrossRef CAS PubMed.
- Z. Zhang, N. Qiu, S. Wu, X. Liu, Z. Zhou, J. Tang, Y. Liu, R. Zhou and Y. Shen, Adv. Mater., 2021, 33, 2102219 CrossRef CAS PubMed.
- D. R. Wilson, A. Mosenia, M. P. Suprenant, R. Upadhya, D. Routkevitch, R. A. Meyer, A. Quinones-Hinojosa and J. J. Green, J. Biomed. Mater. Res., Part A, 2017, 105, 1813–1825 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Polymer characterization, DNA polymer complexation release studies, DNase protection, nanoparticle PDI, nanoparticle uptake, transfection, and fluorescence microscopy images of lysosomal colocalization. See DOI: https://doi.org/10.1039/d3nr05300h |
| This journal is © The Royal Society of Chemistry 2024 |