Role of aptamer technology in extracellular vesicle biology and therapeutic applications
Rocky
Chowdhury†
 a,
Sadegh
Eslami†
a,
Sadegh
Eslami†
 b,
Cuong Viet
Pham
c,
Alin
Rai
b,
Cuong Viet
Pham
c,
Alin
Rai
 bf,
Jia
Lin
d,
Yingchu
Hou
bf,
Jia
Lin
d,
Yingchu
Hou
 e,
David W.
Greening
e,
David W.
Greening
 *bf and
Wei
Duan
*bf and
Wei
Duan
 *a
*a
aSchool of Medicine, Deakin University, and IMPACT Strategic Research Centre, Waurn Ponds, VIC 3216, Australia. E-mail: wei.duan@deakin.edu.au
bMolecular Proteomics Laboratory, Baker Heart and Diabetes Institute, Melbourne, Victoria, Australia. E-mail: david.greening@baker.edu.au
cMolecular Imaging and Theranostics Laboratory, Baker Heart and Diabetes Institute, Melbourne, VIC 3004, Australia
dDepartment of Biochemistry and Molecular Biology, West China School of Basic Medical Sciences & Forensic Medicine, Sichuan University, Chengdu, China
eLaboratory of Tumor Molecular and Cellular Biology College of Life Sciences, Shaanxi Normal University 620 West Chang'an Avenue, Xi'an, Shaanxi 710119, China
fDepartment of Cardiovascular Research, Translation and Implementation, and La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Australia
First published on 25th May 2024
Abstract
Extracellular vesicles (EVs) are cell-derived nanosized membrane-bound vesicles that are important intercellular signalling regulators in local cell-to-cell and distant cell-to-tissue communication. Their inherent capacity to transverse cell membranes and transfer complex bioactive cargo reflective of their cell source, as well as their ability to be modified through various engineering and modification strategies, have attracted significant therapeutic interest. Molecular bioengineering strategies are providing a new frontier for EV-based therapy, including novel mRNA vaccines, antigen cross-presentation and immunotherapy, organ delivery and repair, and cancer immune surveillance and targeted therapeutics. The revolution of EVs, their diversity as biocarriers and their potential to contribute to intercellular communication, is well understood and appreciated but is ultimately dependent on the development of methods and techniques for their isolation, characterization and enhanced targeting. As single-stranded oligonucleotides, aptamers, also known as chemical antibodies, offer significant biological, chemical, economic, and therapeutic advantages in terms of their size, selectivity, versatility, and multifunctional programming. Their integration into the field of EVs has been contributing to the development of isolation, detection, and analysis pipelines associated with bioengineering strategies for nano-meets-molecular biology, thus translating their use for therapeutic and diagnostic utility.
1. Introduction
Extracellular vesicles (EVs) are membranous vesicles (30–300 nm) released by all cellular organisms, including plants, fungi, and bacteria.1–4 They act as important signalling entities for the selective packaging and transfer of bioactive molecules such as proteins, lipids and nucleic acids between cells to induce a signalling response and regulate cellular processes.5 The production and dynamic release of EVs as biological messengers appear to be evolutionarily conserved across all kingdoms of life. Functionally, EVs are involved in different homoeostasis processes, such as metabolism,6 neuronal function,7 cell development and differentiation,8,9 cell proliferation, inflammation,10 embryo implantation and endometrial function,11–13 and tissue repair and regeneration.14–16 EV-mediated crosstalk may occur unidirectionally or bidirectionally between cells or via systemic communication, during which EVs traffic to various tissues and organs.17,18 This intercellular interaction may involve not only the release and delivery of EV cargo but also cell surface interactions and target-cell modulation (i.e., surface-mediated signalling).19,20 Besides the horizontal transfer of cargo molecules such as proteins and RNAs between cells, EVs can induce signaling in target cells upon interaction at the cell surface (juxtacrine signaling) and complex with and neutralize soluble factors through receptors on the EV surface.21 Thus, EV membrane proteins, such as proteoglycans, lectins, and integrins, as well as components of the extracellular matrix, mediate the EV–plasma membrane interaction. Moreover, EVs have intrinsic tropism at the tissue and cell levels as a result of their surface displayed molecules. The capacity of EVs to essentially capture a snapshot of selected molecular cargos from their parental cells in space and time and their ability to transfer complex biological information make them unique and promising next generation diagnostics and therapeutics.EVs may be classified based on a set of characteristics, including their biophysical properties (size and density), biochemical composition, process of cellular release, and surface charge. As heterogeneously sized (from ∼30 to more than 3000 nm in diameter) particles, EVs include small EVs (sEVs, including exosomes), microvesicles (MVs) (or large EVs (lEVs), microparticles and ectosomes), shed midbody remnants (sMBr), migrasomes and apoptotic bodies20,22–28 (Table 1). The nature and relative abundance of EV cargo are selectively determined during EV biogenesis,20,29,30 and depend on their cell of origin22,31–34 as well as source tissue.6 In addition, EV cargo loading is influenced by the different physiological, pathological, and stress conditions of the cell/tissue of origin. Over the past decade, including those driven by the international EV community, awareness of the need to rigorously isolate specific EV types and populations for research and therapeutic purposes has been a central focus in the field to better understand their fundamental form and composition, and further elucidate mechanisms of EV biogenesis and function. Currently, size, density, surface charge, membrane surface proteins, and lipid membranes have been explored for the isolation of EVs; specifically, these include differential ultracentrifugation, density gradient ultracentrifugation, polymer-based precipitation, charge-mediated electrostatic adsorption, ultrafiltration, size exclusion, lipid anchors, and antibody-based immunoisolation.22,46,47 However, there is a pressing need for bioaffinity-mediated EV isolation48 to capture a disease-subset of EVs, understand a subpopulation of EVs, or even capture cell-specific surface antigens.31,34,49,50 Innovation in the development of novel technologies for cell- or tissue-specific targeting of EV will accelerate the clinical translation of EV-based diagnostics and therapeutics. Recently, the development of aptamer technology has seen multidisciplinary applications and impacts on various aspects of the field of EVs, including enrichment and retrieval, biology and signalling, imaging, targeting, and therapeutic delivery. Their facile modification and programmability as well as their significant affinity recognition ability to target and interact make the feature of aptamers a single-stranded DNA sequence highly advantageous51 in promoting the understanding, targetability and therapeutic enhancement of EVs.
| Extracellular vesicles | Other types of extracellular vesicles | ||||||
|---|---|---|---|---|---|---|---|
| Subtype | Small EVs (sEVs)23,33,35–39 (incl. exosomes) | Large EVs (lEVs)23,29,40,41 (incl. ectosomes, microvesicles) | Shed midbody remnants (sMB-Rs)26 | Exopher42 | Large oncosome43 | Migrasomes28,44 | Apoptotic bodies45 |
| Biogenesis | Endosomal maturation pathway, intraluminal vesicles (various sorting mechanisms) | Plasma membrane budding or protrusions, membrane fission | Cytokinesis (cytokinetic abscission) | Plasma membrane budding | Plasma membrane budding | Plasma membrane budding | Plasma membrane budding |
| Markers (enrichment) | CD63, CD81, SDCBP, ESCRT complex proteins: ALIX/PDCD6IP and TSG101 | ANXA1, RPS7, ARF6 | MKLP1, RACGAP1 | Phosphatidylserine, LC3, Tom20 | CK18, GOT1 | TSPAN4, ITGA5 | Phosphatidylserine |
| Particle diameter range | 30–200 nm | ∼100–1000 nm | ∼200–600 nm | ≥1 μm | ≥1 μm | ≥1 μm | 50–5000 nm |
| Buoyant density | 1.08–1.14 g ml−1 | 1.08–1.14 g ml−1 | 1.22–1.30 g ml−1 | ||||
2. Therapeutic extracellular vesicles
EVs have inherent advantages as therapeutic mediators due to their size, membrane stability, non-toxicity and non-immunogenicity, and propensity for cellular uptake and transversion of biological barriers, including tissue barriers (blood–brain, blood–cerebrospinal fluid barriers), plasma membranes, and their ability to deliver cargo across endosomal membranes. In addition to these intrinsic properties, the ability to modify the surface and luminal cargo loading using various molecular biology and bioengineering approaches makes EVs an attractive vehicle for delivering a wide range of therapeutics.EV-associated bioactive cargo has been shown to promote several processes in the body that contribute to endogenous tissue remodelling and therapeutic tissue repair. Stem- and progenitor cell-derived EVs have been shown to mediate therapeutic effects in various preclinical disease models, which have resulted in the initiation of several early phase clinical trials.52,53 Moreover, pre-clinical studies on cardiac pathology highlight the regenerative capacity of EVs and engender their use as cell-free therapeutic tools.54 Clinical applications of EV-based therapeutics, derived from various immune cells (e.g., dendritic cells55–59 and engineered cell counterparts60–64), tumour cells,65–67 and bacteria,68 are being trialled in immunomodulation in the context of antigen cross-presentation, cancer immune surveillance and tumour escape, tissue regeneration and recovery, and as delivery vehicles for combination therapies.50
In addition to the delivery of bioactive cargo, EVs harbor various cell surfaces and integral membrane proteins based on their source, mediating associated cell recognition, adhesion, antigen uptake, and recipient cell function.69–74 In addition to target cell surface interactions, EVs can also bind complement factors and complement regulatory proteins,19,75 as well as cytokines,76 with such surface composition a functional determinant of EV–cell interactions, impacting the accumulation of EVs in specific cell subsets and organs.76 Immune (dendritic) cell-derived EVs, harbouring functional membrane-bound IL-15Rα (NKG2D ligand), were shown in vivo to promote receptor–ligand-dependent immune cell activation and proliferation, resulting in tumour suppression.58 EVs from monocyte-derived dendritic cells also carry functional NKG2D ligands and can promote the proliferation and activation of NK cells,58 suggesting their potential utility for tumour vaccination.
In the context of tissue regeneration and repair, stem cell-derived EVs can ameliorate the effects of various diseases in the liver (fibrosis, hepatitis, and inflammation), brain (stroke), heart (myocardial infarction and contractility), kidney (renal ischemia and stenosis), and immune system. Moreover, EVs secreted from differentiated induced pluripotent stem cells, cardiospheres or embryonic stem cells administered directly into infarcted hearts have been shown to attenuate ischaemic injury in both small and large animal models.77,78 Through direct administration to the heart, EVs are believed to signal directly to the myocardium and supporting cells, including fibroblasts and endothelial cells, altering their responses to ischaemic injury. The diverse beneficial effects of seemingly identical entities (i.e., mesenchymal stem cell (MSC)-EVs, and bone marrow-derived stem cell (BMDSC)-EVs79–84) suggest a complex repertoire of active cargo85 working synergistically, as opposed to a single molecular component. Applying a system-based understanding of EV form and effect, such as omic and multi-omic-based approaches, is currently a focus in the EV field.86,87
Current advances in the field of EV therapeutics have highlighted current and emerging strategies in engineering EVs (either through altered cell-based genetic engineering, or directly altering EV form/type through EV modification) as an effective means to tune a specified function for an intended disease phenotype. Various strategies to genetically engineer cells to produce EVs with a protein tag for the targeted delivery of biologics to immune cells without the need for chemical modifications have been shown.88 Furthermore, EVs provide a deliverable approach to ablate protein expression (i.e., through the delivery of the programmable nuclease Cas9); alternately, co-delivery of a homology-directed repair template can insert specified mutations, insertions or deletions into the genomes of target cells, or further through EV-mediated delivery of adeno-associated virus (AAV) vehicles to overcome anti-vector immunity and inherent limited tissue tropism. Cells genetically engineered to produce functionalized EVs may even be implanted to continuously generate such particles in situ. In contrast, although modification of EVs directly may confer cargo loading flexibility, this approach requires more extensive purification and introduces challenges from a manufacturing and regulatory standpoint.
Recently, aptamer technology was applied to the field of EVs based on specific interactions between aptamers and EVs; such studies have highlighted their potential associated with EV isolation, the capacity to modify and enhance various interactions between aptamers and EVs to understand specific EVs, and cargo packaging in EVs for delivery and gene editing.
3. Aptamer: a unique class of nucleic acid-based affinity ligands for biology and medicine
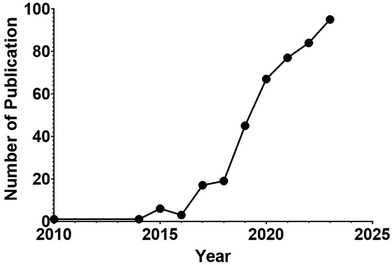
Aptamers are short single-stranded nucleic acid molecules with the ability to fold into defined three-dimensional structures and thus can bind specific targets.89 Also known as “chemical antibodies”, aptamers have a distinct molecular identity as well as a disparate route of selection from their protein counterparts, antibodies. Being DNA or RNA, aptamers are selected via an in vitro PCR-based procedure termed the systematic evolution of ligands by exponential enrichment (SELEX), which was developed in 1990.90,91 Unlike the production of antibodies, which rely on the use of animals or cultured cells, SELEX is conducted in a microtube without the involvement of animals or cells. Therefore, aptamers have been selected against small inorganic molecules, proteins, specific types of cells or organs, and even whole organisms with high specificity and affinity.92–95 Because of the in vitro nature of selection, aptamers can be developed with higher affinity for their cognate targets (KD of 10−12 M) than for antibodies96 with comparable specificity. Therefore, there is increased interest in adopting aptamer technology in both basic biomedical research and sustained efforts in its translation into the clinic (Fig. 1). | ||
| Fig. 1 (A) Counts of yearly publications on aptamer in PubMed. (B) Number of clinical trials per year in NIH clinical trial registry (https://clinicaltrials.gov/). | ||
As nucleic acid-based affinity ligands, aptamers have several advantages over antibodies in their applications in biomedical research and clinical translation.97 Being chemically synthesized, aptamers have very low and controllable batch-to-batch variations in terms of the quality and consistency of the synthesized DNA or RNA. They can also be rapidly and scalably generated in large quantities. The stability of DNA and chemically modified RNA aptamers is unrivalled by antibodies, as aptamers remain stable in low or high pH, in an organic solvent, over a sustained period of elevated temperature, and can withstand physical forces, e.g. shaking or vortexing. Pertaining to EV research and development, several distinct features make aptamers an invaluable class of affinity ligands. First, most aptamers possess a molecular mass of 10–15 kDa, which is one order of magnitude smaller than that of monoclonal antibodies. Being small, the motion of aptamers is much faster than antibodies in aqueous milieus in vitro and in vivo, as the speed for a molecule moving via Brownian motion is inversely related to its size. Second, a monoclonal antibody of 150 kDa has a coverage footprint area in the range of 38–88 nm2,98 while that for an aptamer of 15 kDa is in the range of ∼8–16 nm2.99 This translates to 10-fold more molecules of aptamer binding to the same area occupied by a single monoclonal antibody on the surface of the EV membrane. The mechanism underlying the advantage of more aptamers engaging an area occupied by a single antibody has been elucidated in an elegant study on the selectivity of targeted drug delivery, where Prins and co-workers have shown that multivalent weak interactions between DNA ligand and its receptor on particles lead to enhanced selectivity.100 Consequently, the small size of aptamers affords enhanced binding avidity and specificity of aptamers to their targets on the surface of EVs. Third, aptamers bind to their targets via their properly folded three-dimensional structure via induced fitting101 without the involvement of covalent bonding. As shown in Fig. 2, the binding motif of aptamers is generated by folding the linear DNA or RNA strand via internal hydrogen bonding into a defined 3D structure. Unlike antibody-based affinity chromatography in which harsh conditions are employed to elute the bound antigen, e.g. extreme pH and high salt, the bound antigen or EV-associated (surface) ligand can be released readily from aptamer via the disruption of the 3-D confirmation of the aptamer under physiological conditions. Finally, as per molecule, a DNA aptamer is generally 50–100 times cheaper than a monoclonal antibody. Thus, it is commercially uncompetitive, if not unviable, to use antibody-based chromatography to manufacture large quantities of biotherapeutics, such as EVs. In addition, the pharmaceutical industry routinely uses alkaline solutions, e.g. 0.1 M NaOH, to clean or sterile chromatography vessels. These alkaline solutions totally destroy the antibody's ability to bind to their targets. In contrast, DNA aptamers can withstand 30 cycles of treatment with 1 M NaOH, followed by washing with saline without compromising their binding capacity.102 Needless to say, extreme pH and high salt inevitably damage the structural integrity and functional competence of EVs isolated via affinity chromatography. Since 2010,103 aptamers have been applied as a frontier technology for EV biology and research, including their application in isolation, detection, labelling/tracking and EV-based diagnostics and therapeutics (Fig. 3, Tables 2 and 3).51,89
 | ||
| Fig. 2 Schematic representation of folding of an aptamer from the initial linear strand into a defined 3D structure followed by target binding. | ||
| Target | Aptamer | Source of EVs | Affinity (KD) | Applications | Ref. | |
|---|---|---|---|---|---|---|
| Isolation | CD63 | CD63 aptamer | MDA-MB-231, A549 cell culture medium | 38.71 nM to CD63 | Magnetic bead-based exosome immuno-separation system | 104 |
| HepG2 | LZH8 | HepG2 cell culture medium | 96 nM to HepG2 | A nanotetrahedron (NTH)-assisted aptasensor for direct capture and detection of hepatocellular exosome | 105 and 106 | |
| MUC-1 | MUC-1 aptamer | Plasma from patients with cancer or healthy subjects | NA | aptamer-based magnetic isolation | 107 | |
| PTK7 | Sgc8 | A549 cell culture medium | NA | Microfluidic system | 108 | |
| Vascular endothelial growth factor | VEGF aptamer | EV from serum of patients with prostate cancer | 400 nM | Using nano-flocytometry-based thermophoretic profiling of EVs for the early detection and classification of prostate cancer | 106 and 109 | |
| PSMA | PSMA | EV from SKBR3 and LNCaP cells and blood of cancer patients | NA | Multiplex detection of cancer EVs via surface enhanced Raman scattering (SERS) | 110 | |
| Detection | MUC-1 | S1.3/S2.2 | Exosome-derived from MCF-7, SGC7901 cells, and serum from patients with breast and gastric cancer | 0.135 nM to MUC-1 peptides | Electrochemical, fluorescence, colorimetric, and SERS based aptasensors | 111 and 112 |
| HER2 | HER2 aptamer | Exosome-derived from HCC1954, P0403, HEK-293, A549, HCT116, SK-BR3, HeLa, MCF-7 cells, and human plasma | NA | Colorimetric, SPR, and SERS based aptasensor for specific HER2 + exosomes detection | 111 | |
| αvβ6 | αvβ6 aptamer | Exosome-derived from HCC1954, P0403, HEK-293, A549, HCT116, SK-BR3, HeLa, MCF-7 cells, and human plasma | NA | Colorimetric, aptasensor for specific αvβ6 + exosomes detection | 113 | |
| PD-L1 | MJ5C | Exosome-derived from A375 and k562 cells and human plasma. | 91 ± 12 nM to PD-L1 positive cell | HOLMES-ExoPD-L1 fluorescent sensor for detection of glycosylated PD-L1 exosomes | 114 | |
| ALPPL-2 | SQ2 | Exosome-derived from PANC-1+, Capan-1, cells, and human serum. | 22.5 ± 4.5 nM to ALPPL-2 | Colorimetric sandwich-based sensor | 115 | |
| CEA | CEA aptamer | Exosome-derived from MCF-7, SK-BR-3, MDA-MB-231, BT47 cells and human serum. | NA | Magneto-ediated electrochemical and SERS based sensor | 111 | |
| PSA | AS2 | Exosome-derived from PC3, HeLa cells, and prostate cancerous and human plasma. | 0.7 nM to PSA protein | Label free qualitive and quantitative aptasensor | 116 | |
| PDGF | Group 36 | Exosome-derived from HepG2, HeLa, MCF-7, MCF-10A cells, and human blood. | 0.58–78 nM to different isoforms of PDGF | ExpoAPP-fluorescence-based aptasensor | 117 | |
| PSA | PSap4#5 | Human serum | 40 nM | Using fluorescent aptamers to discriminate prostate cancer from benign prostate enlargement via thermophoretic profiling | 106 and 118 | |
| EpCAM | CF-7 cell-derived exosomes in bovine serum | NA | A colorimetric aptasensor to detect and quantify cancer cell line-derived sEVs | 119 and 120 | ||
| Mycobacterial EV membrane | Aptamer-21 | Mycobacterial EV | 65 nM | Visualizing the release of mycobacterial membrane-derived EVs in infected macrophages using confocal microscopy | 121 | |
| Alkaline phosphatase placental-like 2 | SQ2 | Blood EV from cancer patients | 23 nM | Using ALPPL2 directed sandwich aptamer-linked immobilized sorbent assay for the detection of pancreatic cancer-derived EV | 115 and 122 | |
| Tracking and labeling | PD-L1 | MJ5C | Exosome derived A375 human melanoma cells culture medium | 91 ± 12 nM to PD-L1 positive cell | Aptamer-based metabolic glycan labeling for in situ visualizing and functional study of exosomes | 123 |
| ATP | ATP aptamer | Exosome derived HepG2 cells culture medium | NA | Aptamer-based probing of exosome internalizing pathways via confocal microscopy | 124 | |
| CEM cells | Sgc8 | Exosome derived immature dendritic cells culture medium | 0.8 ± 0.09 nM to CEM cells | Tracking of cellular uptake pathway of aptamer-functionalized exosomes | 125 | |
| PD-L1 | Ex-50.T | EV from HeLa, Cal27 and 293FT cells | NA | Detecting PD-L1 positive sEV on magnetic chips | 126 |
| Aptamer | Target and disease | Therapeutic cargo | Source of EVs | Ref. |
|---|---|---|---|---|
| AS1411 | Nucleolin. Breast cancer, leukemia, and colorectal cancer models | cel-miR-67 and let-7 miRNA, miRNA-21 Sponge and doxorubicin | Immature dendritic cells, recombinant 293T cells, HEK-293 cells | 127 and 128 |
| A9g | PSMA. Prostate cancer cell lines and mouse model of prostate cancer | siRNA against survivin | HEK-293 cells | 129 |
| EGFRapt | EGFR. Breast cancer cell lines and mouse model of breast cancer | siRNA against survivin | HEK-293 cells | 129 |
| sgc8 | PTK7. T-leukemia cell line | Doxorubicin | Immature dendritic cells | 125 |
| 5TR1 | MUC-1. Breast cancer cell line and mouse model of colorectal adenocarcinoma | Doxorubicin | Mesenchymal stem cells | 130 |
| A9g | PSMA (KD 130 nM). Prostate cancer xenograft | survivin siRNA | HEK293T | 129 |
| LJM-3064 | Myelin. Suppression of inflammatory response as well as lowered demyelination lesion region in CNS. | MSC-derived exosomes | MSC-derived exosomes | 131 and 132 |
| BMSC-specific aptamer | Bone marrow stromal cells. Promoting bone regeneration in a mouse model of ovariectomy-induced postmenopausal osteoporosis. | Bone marrow stromal cell-derived exosomes | Bone marrow stromal cell | 133 and 134 |
The transition from bench to clinic presents significant challenges, with the efficacy of aptamers reliant on understanding the role of their targets in disease pathology. Moreover, the decision to utilize aptamers as either standalone treatments (monotherapy) or as part of a multi-agent regimen in clinical trials profoundly influences the outcome. Table 4 provides a summary of the clinical trials involving 15 classes of aptamers over the past decade, sourced from ClinicalTrials.gov. Among them, ten aptamers successfully completed Phase 1 trials but did not progress to Phase 2. Additionally, three aptamers concluded Phase 2 trials without advancing to Phase 3, which is primarily attributed to their inability to meet the primary endpoints. Finally, one aptamer's Phase 3 trial failed due to its lack of efficacy. Remarkably, the anti-complement C5 RNA aptamer, avacincaptad pegol, has persevered through a 15-year clinical development journey. It transitioned from multi-agent combination trials to a single-agent monotherapy trial, ultimately achieving FDA approval in September 2023. This milestone came 15 years after its initial Phase 1 trial in 2008.135 Avacincaptad pegol now stands as the second aptamer approved by the FDA, offering treatment for geographic atrophy secondary to age-related macular degeneration.136
| Aptamer | Target | NCT number | Conditions | Year started | Study status |
|---|---|---|---|---|---|
| AON-D21 | Complement component C5a | NCT05962606 | Community-acquired pneumonia | 2023 | Recruiting (phase 2) |
| AON-D21 | Complement component C5a | NCT05018403 | Phase 1, first-in-man single ascending intravenous doses | 2021 | Completed (phase 1) |
| AON-D21 | Complement component C5a | NCT05343819 | Phase 1, multiple ascending intravenous doses | 2022 | Completed (phase 1) |
| X-aptamer | Biomarkers for hepatocellular carcinoma patients treated with Lipiodol TACE | NCT04459468 | To identify proteomic biomarkers for outcome prediction of TACE Treatment in liver cancer | 2022 | Recruiting |
| ApTOLL | Toll-like Receptor 4 | NCT05293236 | COVID-19 | 2022 | Phase 1 terminated due to lack of patients |
| AptameX | S Protein of SARS-CoV-2 | NCT04974203 | Saliva-based COVID-19 test, | 2021 | Unknown |
| Tenofovir (TFV) aptamer | Tenofovir (TFV) | NCT04870671 | Biosensor for TFV in biological fluid | 2021 | Completed (phase 1) |
| BT200 | A1 Domain of Von Willebrand Factor (VWF) | NCT04677803 | Von Willebrand diseases; Hemophilia A | 2020 | Completed (phase 2) |
| Aptamer against antiretroviral drugs | Dolutegravir, emtricitabine, lamivudine, tenofovir | NCT04302896 | Urine test for adherence to antiretroviral drugs | 2020 | Completed (early phase 1) |
| Zimura®/Izervay® (Avacincaptad pegol) | Complement component C5 | NCT04435366 | Geographic atrophy; macular degeneration | 2020 | Phase 3, completed in 2023, FDA approved |
| ApTOLL | Toll-like Receptor 4 | NCT04742062 | Stroke | 2019 | Completed (phase 1) |
| 68Ga-Sgc8 | Protein tyrosine kinase-7 (PTK7) | NCT03385148 | Colorectal cancer | 2017 | Unknown (early phase 1) |
| Zimura® (Avacincaptad pegol) | complement component C5 | NCT05571267 | Macular degeneration | 2016 | Phase 2, completed |
| Aptamer against atrial fibrillation biomarkers | Atrialf fibrillation biomarkers | NCT03188484 | To Identify proteins differentially expressed in atrial fibrillation using SOMAscan® platform | 2016 | Phse 1 completed in 2019 |
| Fovista® (E10030) | Platelet-derived Growth factor (PDGF) | NCT02387957 | Age-related macular degeneration (AMD) | 2016 | Phase 2 terminated in 2017, primary endpoint not achieved |
| Zimura®/Izervay® (Avacincaptad pegol) | Complement Component C5 | NCT02686658 | Geographic atrophy; macular degeneration | 2016 | Phase 2; completed in 2019 |
| Aptamer against bladder cancer biomarkers | Bladder cancer biomarkers | NCT02957370 | Aptamer sensors for clinical staging and monitoring of bladder cancer | 2015 | Completed in 2022 |
| Zimura® (Avacincaptad pegol) | Complement component C5 | NCT02397954 | Idiopathic Polypoidal Choroidal Vasculopathy | 2015 | Phase 2 completed in 2019 |
| Fovista® (E10030) | Platelet-derived growth factor (PDGF) | NCT02214628 | Age-related macular degeneration (AMD) | 2014 | Phase 2 terminated in 2019, primary endpoint not achieved |
| Fovista® (E10030) | Platelet-derived growth factor (PDGF) | NCT01940887 | Age-related macular degeneration (AMD) | 2014 | Phase 3 terminated in 2017, primary endpoint not achieved |
| Spiegelmer lexaptepid pegol (NOX-H94) | Hepcidin | NCT02079896 | Anemia in end stage renal Disease | 2014 | Phase 1 and 2 completed in 2015 |
| Fovista® (E10030) | Platelet-derived growth factor (PDGF) | NCT02591914 | Neovascular age-related macular degeneration (AMD) | 2014 | Completed (phase 1) |
4. Aptamer-based sensor for the detection and analysis of EVs
Many aptamers developed previously for cell biology and cancer biology have been utilized for the detection and analysis of EVs,89,137 including aptamers against PSA, HER2, PD-L1, CEA, VEGF, PSMA, and EGFR. One of the most frequently utilized aptamers in the EV field is a 32-nucleotide DNA aptamer against human CD63 with a KD of 40 nM developed by Base Pair Biotechnologies, Inc. (#ATW0056).138 Various aptamer-based biosensors (aptasensors) have been developed with modes of signal transduction, including fluorescent, electrochemical, colorimetric and surface-enhanced Raman spectroscopic detection.139 The signal of aptamer-based sensors can be further enhanced via direct amplification in which nucleic acid amplification products are generated on the EV surface or competitive amplification; aptamers competitively bound on EVs can release trigger DNA upon interaction between aptamers and their targets on EVs for subsequent amplification. The progress on aptasensors over the past decades has been extensively covered in recent reviews.139–141 Here, we discuss new developments since 2023. Li and co-workers developed an aptamer-based method for the detection and classification of ovarian cancers by profiling tumor markers on small EVs.142 They isolated EVs from plasma by ultrafiltration, followed by the analysis of a set of seven EV surface cancer biomarkers, i.e. CA125, STIP1, CD24, EpCAM, EGFR, MUC1, and HER2, using a nanoflow cytometer, a flow cytometer purposely developed for the analysis of EVs from 40 to 1000 nm in size and equipped with single-photon counting capacity.143,144 By molecular profiling plasma sEVs from 54 patients with untreated ovarian cancer, they were able to identify patients with cancer with an accuracy of 94.2%. Huang et al. developed an electrochemical biosensor with divalent aptamer-functionalized nanochannels by simultaneously conjugating CD63 and EpCAM aptamers.145 The target EVs can be recognized and selectively captured in a divalent collaborative manner, resulting in a varied ionic transport behaviour corresponding to the abundance of captured EVs. This method achieves an impressive limit of detection (LOD) of 4.43 × 103 particles per mL with HeLa cell-derived EVs in 10 mm KCL (pH 7.4). A similar dual aptamer recognition strategy was employed by Guo and colleagues who constructed a fluorescent biosensor using DNA aptamers against CD63 and PTK7, achieving a limit of detection (LOD) of 2.2 × 107 particles per mL using EVs from MCF7 breast cancer cells.146 Li and colleagues also employed a CD63 aptamer to develop a molybdenum disulfide-integrated iron organic framework.147 This is a colorimetric aptasensor with a mimetic nanozyme (MoS2-MIL-101(Fe) with superior peroxidase enzymatic activity to generate a change in colour in the TMB-H2O2 system to report the detection of EV, with an LOD of 3.37 × 106 particles per mL when studied using EVs derived from the HGC-7901 cell line. Furthermore, Cheng and colleagues developed a clove-like gold nanocluster aptasensor in which a CD63 aptamer-modified DNA tetrahedron was attached to the surface of a gold electrode. Following EV-CD63 aptamer interaction, a sequential DNA aptamer against mucin 1 (MUC1) in a clove configuration was used to generate a strong electrochemical signal, resulting in an LOD of 1.58 × 105 particles per mL using MCF7 cell-derived EVs.148In terms of clinically relevant sensitivity, many aptasensors possess an LOD sufficient for detecting the lower end of the concentration range of the analyte in the targeted sample matrix. For example, the LOD for SARS-CoV-2 detection using an electrochemical aptasensor is <1100 copies per mL,149 while for a capacitive laser-induced graphene-based aptasensor, it stands at 1790 copies per mL.150 These LODs translate to a detection capability that is 2 orders of magnitude beyond the acceptable diagnostic sensitivity for SARS-Cov-2 (1.0 × 106 genome copies per mL) according to the World Health Organization.151 As for protein detection, an on-chip integrated graphene aptasensor achieves an LOD of 160 aM for COVID-19 neutralizing antibodies in serum,152 which is 5 orders of magnitude lower than the minimum target concentration (30 pM) in patients.153 Thus, the current generation of aptasensors can attain the sensitivity and specificity required for real-world applications.
5. Aptamer as the next generation affinity ligand for EV isolation
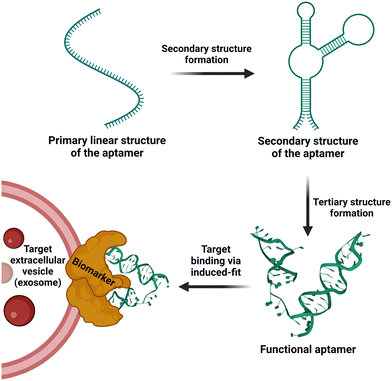
Over the past five decades, the technologies used for the isolation and separation of EVs have harnessed the biophysical properties of EVs, including charge, buoyant density, solubility and size.48 However, the emergence of biochemical interaction-based affinity isolation methods provides enhanced isolation of EV from cells, biofluids, and tissue sources.154,155 Interestingly, as revealed by a survey of methods used for the isolation of EV in the last two decades, affinity isolation was the least utilized method employed by the EV field; for example, 8% of published papers used affinity isolation compared with 46% of publications that used ultracentrifugation and 18% papers that used size exclusion chromatography.156 Despite unparalleled purity and high EV recovery rates, the adoption of affinity chromatography for EVs is hampered by several inherent limitations, including high cost in the use of monoclonal antibodies and limited scalability. As antibody-focused approaches are the most widely used class of ligands in EV affinity chromatography, the harsh elution conditions (non-physiological pH and high salt) required to release EVs from the antibody matrix are key factors that discourage the utilization of affinity-based methods for EV isolation.157–159It is in this context that aptamers emerge as the next generation of alternative affinity ligands for the isolation, concentration and purification of EVs under physiological conditions. The fundamental biochemical and structural differences between antibodies and aptamers lie in the fact that the former consists of 20 monomer units (amino acids) with a very stable 3D structure, while the latter comprises only 4 monomer units (nucleotides) with a 3D structure that can be facilely altered after the capture of EVs to release EVs under physiological conditions. This inconspicuous attribute of aptamers is best exemplified by the pioneering work of rapid capture, followed by the non-destructive release of EVs.107 Zhang and co-workers conjugated a biotinylated CD63 DNA aptamer to streptavidin-coated magnetic beads. Upon capture of EVs derived from the conditioned culture medium of MCF-7 cells, the EVs were released in phosphate-buffered saline at 37 °C for over 15 min by the addition of 1.5 μM full-length antisense oligonucleotide. This approach altered the 3D structure of the folded CD63 aptamer, abolishing its ability to bind to CD63 on the surface of EVs (Fig. 4).70 More innovative aptamer-based affinity EV isolation strategies have been developed in subsequent years. Travas-Sejdic and colleagues developed an electrochemically switchable cloth by conjugating a thiolated CD63 aptamer to AuCl4− on a microporous cloth. The captured EVs were released with up to 92% efficiency under −1.2 V voltage in 5 min (ref. 160) although the CD63 aptamer remains attached to the surface of the EVs. Alternatively, Qian et al. developed a dual aptamer-based microfluidic approach to isolate EVs by conjugating the desthiobiotin-labelled CD63 aptamer and PTK7 aptamer onto the surface of a microfluidic chip functionalized with streptavidin. Because the binding affinity of desthiobiotin to streptavidin is five orders of magnitude lower than that for biotin,161 Qian and colleagues were able to elute up to 108 EVs per ml in 20 min with a large amount of free biotin. Once again, both the CD63 and PTK7 aptamers remained on the surface of the EVs. In an attempt to further enhance the efficiency of EV-aptamer-based affinity isolation, a multivalent DNA flower strategy has been pursued.162,163 To achieve a multivalent interaction between the capture aptamer and target proteins on the EV surface, a single copy of the DNA aptamer against EpCAM or PTK7 was converted to a long single-stranded aptamer with up to 800 repeated units by enzyme-catalysed rolling circle amplification. The biotinylated long aptamer repeats were then self-assembled into microspheres in a one-pot reaction. The DNA flower-captured EVs were released via the digestion of the DNA flower with deoxyribonuclease I. This multivalent affinity isolation method is robust and highly efficient, with a small caveat of contamination of the eluted EVs with deoxyribonuclease I (Fig. 5).
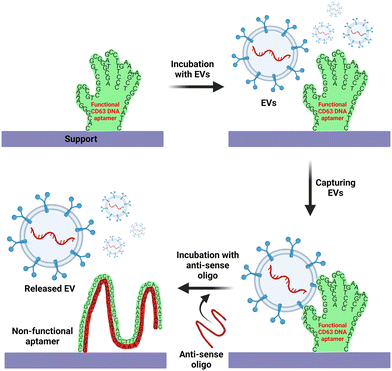 | ||
| Fig. 4 Schematic illustration of affinity isolation of EV using CD63 DNA aptamer followed by the releasing of EVs in physiological conditions. Adapted from ref. 107. | ||
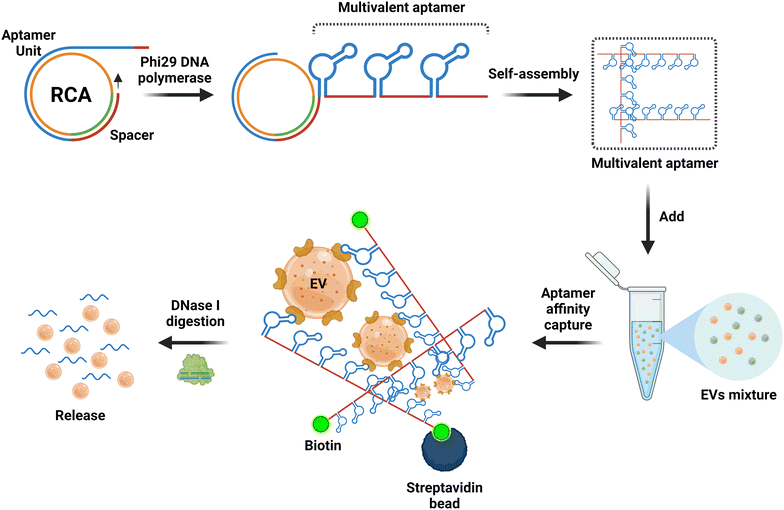 | ||
| Fig. 5 Schematic of the multivalent aptamers prepared by an isothermal enzymatic techniques known as rolling circle amplification (RCA) for aptamer-based EV isolation. Biotin-modified multivalent aptamers with up to 800 repeated aptamer units are prepared by RCA, followed by the formation of a 3D aptamer network in a suspension for the aptamer-based affinity isolation of EVs. The EVs are retrieved by degradation of the aptamers with dna nucleases. Adapted from ref. 161 and 163. | ||
One of the limitations of aptamer-based affinity isolation of EV is the presence of co-eluent in the isolated EV, i.e. antisense oligonucleotides in the case of CD63 aptamer-based EV isolation or DNase I and biotin-oligonucleotide in the case of isothermal enzymatic isolation. Depending on the nature of downstream applications, there is flexibility in how one handles isolated EVs. For instance, in omic-based studies, one may choose to utilize EVs with co-eluted contaminants. Alternatively, one can implement an extra purification step to eliminate these co-eluted substances efficiently. This may be accomplished by employing either streptavidin magnetic beads/columns or size exclusion chromatography to ensure the preservation of the integrity of the isolated EVs.
6. Application of aptamer technology in EV-based translational medicine
6.1 Aptamer-functionalized EV therapeutics for nanomedicine
Since the first approval of PEGylated liposomal doxorubicin (Doxil®) as a nanomedicine by the US Food and Drug Administration in 1995, many new classes of nanoparticle-based nanodrug systems have been developed for improving treatment efficacy and reducing toxicity via improved drug solubility, optimized biodistribution and controlled drug release.164 Moreover, the utility and advantage of nanomedicine as nanosized formulations permit pharmacokinetic and biodistributions that are different from those possible for free drugs and hence can confer considerable therapeutic benefits.18,165 In particular, functionalized nanodrug systems can offer enhanced bioavailability of free drugs, prolonged half-life of systemically administered drugs, reduced immunogenicity, and targeted delivery to specific tissues.165To complement these systems, EVs have emerged as a promising nano-sized drug delivery platform due to their structural stability, low immunogenicity, human compatibility and degradability and enhanced safety emanating from their acellular nature18 (Table 5). Therapeutic cargos, e.g. nucleic acid, protein, small molecular drugs, can be loaded into (or surface modified on) EVs by pre-loading methods in which cells are co-incubated with cargos or transfected with target genes, followed by the production of EVs or post-loading where isolated EVs are loaded with cargos via electroporation, sonication, freeze-thawing or chemical induction.166,167 However, EVs cannot generally target specific types of cells or tissues; thus, it is desirable to modify and enhance the intended therapeutic effect of EVs by targeting moieties for precision medicine.51
| Characteristic | Extracellular vesicles | Liposome/synthetic nanoparticles | Polymer | Free drug |
|---|---|---|---|---|
| Adapted from ref. 171–173. | ||||
| Size | 40–150 nm (dependent on subtype) | 25–1000 nm | 50–1000 nm | 1–5 nm |
| Source | Natural cells | Liposome synthesis | Chemical synthesis | Chemical, peptide synthesis |
| Structure | Phospholipid bilayer, comprise proteins, lipids, and nucleic acids | Double-layer membrane, comprise fat-soluble molecules | Polymerization, comprise polymers | Peptide, enzymes, small molecules |
| Complexity | ++++ | ++ | ++ | + |
| Toxicity | Low | Low | High | Low |
| Stability | High | Low | High | Varied |
| Multifunctionality (modified, diverse purpose) | +++ | +++ | + | + |
| Immunogenicity | + | ++ | ++++ | ++ |
| Targeting capacity | Inherent/modifiable | Modifiable | Modifiable | Limited |
| Circulation time | ++ | +++ | +++ | + |
The aptamer is an ideal class of targeting moiety for EV-based therapeutics because it is vastly superior to its antibody counterpart in tissue penetration and can deliver cargos selectively to target cells in vivo.168,169 Aiming at the isolation of aptamers that specifically recognize EVs from breast cancer cells, Esposito and colleagues developed an Exo-SELEC procedure in which EVs from breast cancer cells were used to select aptamers targeting breast cancer EVs.170 The group isolated a 2′-fluoro-pyrimidine modified RNA aptamer termed ex-50.T of 33 nucleotides in length with a KD of 0.8 nM towards EVs from breast cancer cells. This ex-50.T aptamer was able to inhibit EV uptake by MDA-MB-231 cells by ∼50% and inhibited MCF-7 cell migration induced by MDA-MB-231-derived EV by 30%. Thus, this novel aptamer against breast cancer-derived EV provides a strategy for cancer specificity and EV targeting.
The presence of mRNAs and miRNAs in EVs was first reported by Valadi and co-workers in 2007.174 Since then, EV-based delivery of therapeutic RNA into target tissues has been an area of intense investigation in translational medicine.175 However, most efforts have focused on EV-mediated delivery of miRNA because the RNA cargos in native EVs are largely small RNAs. The size of mRNA is much larger (1000–1500 nt); therefore, it remains a major challenge for mRNA as a cargo for EV-mediated therapeutic delivery. In an attempt to develop an intradermally delivered mRNA for collagen-replacement therapy, You and co-workers loaded collagen mRNA into EVs from human dermal fibroblasts via cellular nanoporation for the treatment of photoaged skin.176 In this brute de force approach, transient nanometric pores are created on the surface of cells to allow for the large-scale loading of full-transcript mRNAs into secreted EVs by applying transient electrical pulses of ∼200 V.177 However, a more elegant and potentially widely applicable strategy for loading EVs with mRNA of interest is derived from the utilization of our knowledge of the biogenesis pathways of EV for therapeutic cargo loading.178 Recently, Zhang and colleagues developed an EV biogenesis pathway-based molecular engineering strategy to load long mRNA into EVs using a DNA aptamer for gene therapy.179 Based on an unorthodox hypothesis that translation inhibition of target mRNA is beneficial for sorting into EVs, Zhang et al. designed a special DNA aptamer consisting of a single-strand segment matching the region surrounding the start codon AUG of the target mRNA and a double-stranded segment that could be recognized by zinc finger motifs (Fig. 6). They first engineered host HEK293 cells by overexpressing a recombinant protein via the fusion of CD9 with a zinc finger motif. By design, the single-stranded segment of the aptamer prevents ribosome assembly in the AUG and thus arrests the translation of the target mRNA. At the other end of the aptamer, the double-stranded segment binds to the zinc finger fused with CD9, thus shuttling the target mRNA into the endosome to generate EVs loaded with the mRNA. They demonstrated that large mRNA with 2382 nt could be successfully loaded into EVs using this approach. To explore the potential of therapeutic applications, Zhang and co-workers further demonstrated that EVs loaded with Pgc1α mRNA induced adipose tissue browning in vivo, while EVs loaded with interlukin-10 mRNA suppressed intestinal inflammation in a mouse model of inflammatory bowel disease.179 With further refinement, this EV biogenesis-based mRNA loading strategy will accelerate a new frontier for EV-based mRNA therapy, including novel mRNA vaccines.
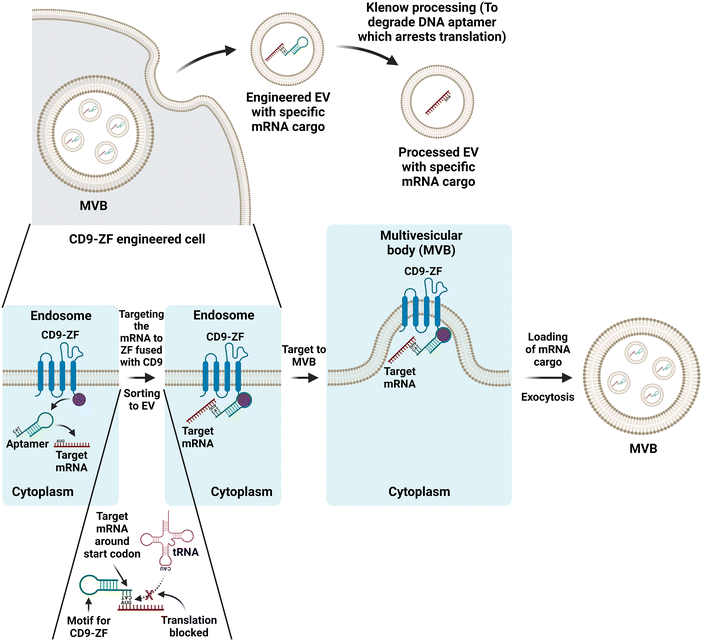 | ||
| Fig. 6 Schematic illustration of selective sorting of target mRNA into CD9-zinger finger (ZF) engineered EV by a dna aptamer. Top: the therapeutic mRNA is targeted to MVB followed by the loading of the mRNA into EVs by the transfected DNA aptamer within cells engineered by overexpression of the recombinant CD9-ZF. Finally, the DNA aptamers in the released EVs are enzymatically removed. Bottom: the translation of the target mRNA is arrested by the annealing of the single-stranded segment of the DNA aptamer to the regions surrounding the start codon AUG of the target mRNA, followed by the targeting of the mRNA to the endosome membrane via the interaction between the double-stranded motif on the DNA aptamer and the ZF in the recombinant CD9-ZF protein. Eventually, the EVs loaded with the target mRNA are formed at MVB and released by exocytosis. Adapted from ref. 179. | ||
6.2 Aptamer-guided EV drug delivery into the brain
Gliomas make up virtually 80% of all lethal primary brain tumours, of which glioblastoma multiform (GBM) is the most malignant type with a 5-year survival rate of <10%. The 15-month average life expectancy of GBM patients has remained unchanged over the past several decades largely due to the invasive nature of tumour growth and the hindrance of the blood–brain barrier (BBB) in treatment efficacy.180 Compared to synthetic drug delivery systems, e.g. polymeric nanoparticles and liposomes, EVs have low toxicity, excellent biocompatibility, high stability and favourable penetration of the BBB.181 Aptamers are often employed as a targeting moiety for targeted delivery into the brain. For example, EVs from naïve macrophage can cross the BBB via the interaction between integrin lymphocyte function-associated antigen 1 (LFA-1), intercellular adhesion molecule 1 (ICAM-1) and the carbohydrate-binding C-type lectin receptors expressed on endothelial cells on the BBB.182 Therefore, Liu and co-workers surface-functionalized EVs derived from macrophages with a cholesterol-modified AS1411 DNA aptamer against nucleolin, a protein that is overexpressed on the plasma membrane of some tumor cells. The AS1411 aptamer-decorated EVs were then loaded with glutathione-responsive biodegradable silica nanoparticles and injected intravenously. After crossing the BBB and entering the tumour cells via endocytosis, the EVs produced large amounts of O2 to relieve tumour hypoxia using an orthotopic GBM xenograft model. Such aptamer-EV mediated sonodynamic therapy efficiently inhibited the GBM metastasis via the activation of HIF-α1. A different approach was adopted by Ye and colleagues, who generated aptamer-guided EVS encapsulating two chemotherapy drugs.183 They initially loaded EVs derived from the human leukemia monocytic cell line THP-1 with temozolomide and O6-benzylguanine, followed by the conjugation of two targeting ligands to the surface of drug-loaded EVs, an angiopep-2 peptide for enhanced BBB penetration and a CD133 RNA aptamer for targeting the cancer stem cells of GMB. After intravenous administration, the enhanced BBB penetration and superior tumor accumulation of the engineered EVs were prominent in a U87MG-bearing glioma mouse xenograft model, with a therapeutic efficacy 60 fold higher than that of the free drugs.Parkinson's disease is a progressive neurodegenerative disease that affects a person's control of their body movements.184 The vast majority of PD subtypes and most cases of sporadic PD share Lewy bodies as a characteristic pathological hallmark, with α-synuclein aggregates being the main component.185 In pursuing a reduction in neuropathological deficits, Zheng and co-workers developed DNA aptamers targeting α-synuclein aggregates.186 These aptamers can effectively reduce α-synuclein aggregation in a neuronal cell line and primary neurons and reduce mitochondrial dysfunction and cell defects induced by α-synuclein overexpression. Subsequently, the DNA aptamer F5R1 with a KD of 2.4 nM was found to bind preferably to fibrillar rather than monomeric α-synuclein.187 Subsequently, the therapeutic potential of F5R1 was investigated by encapsulating the aptamer into EVs from HEK293 cells following polyethylenimine-assisted transfection. To facilitate the penetration of the BBB, they modified the aptamer-loaded EVs by fusing the neuron-specific rabies viral glycoprotein (RVG) peptide to the extra-EV N terminus of Lamp2b, an abundant membrane protein of EV, to allow facile entry into the brain. Intraperitoneal administration of the RVG-decorated EVs containing the F5R1 aptamer to C57BL/6J female mice led to retrograde transport and transsynaptic transmission into the central nervous system through the axons and synapses of peripheral neurons, circumventing the BBB.187 Such EV-aptamer treatment decreased α-synuclein aggregation in the substantia nigra and ameliorated motor dysfunction in the mouse α-synuclein preformed fibril model of the sporadic Parkinson's disease. This pioneering work provides valuable insight into the potential for the application of EV-DNA aptamer therapy delivered into the brain for the future treatment of neurodegenerative diseases.
6.3 Aptamer-targeted EVs in regenerative medicine
Regenerative medicine applies engineering and life science principles to repair or replace damaged tissues, promote cell survival and tissue/organ renewal and restore normal functions.188 Due to their ability to self-renewal and differentiate into specific cells, stem cells are frequently employed to regenerate and repair tissues that have been damaged or affected by the disease.189 Mesenchymal stem cells or mesenchymal stromal cells (MSCs) are adult stem cells that encompass a broad differentiation potential. In the past few decades, MSCs have been isolated and characterized from various human tissues (e.g., bone marrow, fat, placenta, and umbilical cord) and have been applied in regenerative medicine.190 Various key studies have highlighted that the paracrine signaling factors of stem cells (and not the cells directly) reduce injury resulting from myocardial ischemia/reperfusion;191 this foundation has now indicated that many, if not most, of the reported effects of MSC are actually mediated by soluble factors released by these cells, in particular the EVs released by MSCs.192,193 Intense interest and efforts have expanded this field using EVs from MSC in the development of novel therapeutic strategies for regenerative medicine. However, the risk of an off-target effect due to the low efficiency of EV delivery to the site of injury, in particular the specific type of cells underlying the pathology, remains a major barrier to the successful translation of EV-based therapy. The employment of aptamers to guide EVs loaded with therapeutic cargos to the site of tissue/organ injury has become an increasingly attractive strategy for developing translational strategies for regenerative medicine.194The critical importance of targeting MSC-derived EVs by an aptamer to the intended sites of clinical intervention was demonstrated by Luo and colleagues.134 These researchers found that EVs from bone marrow stromal cells could effectively augment the osteoblastic differentiation ability of bone marrow mesenchymal stem cells (BMSCs) in vitro. However, they failed to prevent ovariectomy-induced postmenopausal osteoporosis using a mouse model. To mitigate this issue, a 40 nt DNA aptamer targeting BMSC133 was conjugated on the surface of EVs. As expected, such aptamer-guided EVs were able to be efficiently internalized into BMSCs in vitro and accumulate in bone marrow in vivo. Furthermore, in the bone, the aptamer-targeted EVs could promote bone formation and enhance bone mass in mouse models of postmenopausal osteoporosis and femur fracture in vivo.134
The indispensable role of aptamers in targeting EVs in the development of efficient strategies for the prevention and treatment of osteoporosis and fracture was further independently confirmed by Shou and colleagues.195 Zhou et al. utilized a three-way junction RNA nanoparticle to engineer EVs functionalized by the same 40 nt DNA aptamer used by Xie et al. for bone-specific targeting. As shown in Fig. 7A, they began by linking the 40 nt DNA aptamer against BMSCs to one of the arms of the three-way RNA scaffold,196 followed by the conjugation of a cholesterol molecule to the 2nd arm of the RNA junction with a triethylene glycol (TEG) spacer for anchoring onto the membrane of EVs. To enable tracking of the aptamer-functionalized EV in vivo, a fluorescent reporter (Alexa Fluor 647) was conjugated to the last arm. This BMSC aptamer was used to functionalize the surface of EVs derived from M2 macrophages that had been shown to attenuate inflammation and promote tissue repair, most likely via EV cargos of miR-690 and miR-378a-3p.197,198 The targeting of aptamer-EV to BMSCs in vitro was confirmed by flow cytometry and confocal microscopy analysis. The DNA aptamer-guided EVs were then systematically administered through tail vein injection to mice from a femur fracture model. These BMSC aptamer-functionalized EVs were found to accumulate mainly at the site of bone fracture with a slow release of cargos, thereby significantly accelerating the healing processes compared with that of the control EVs without BMSC aptamer (Fig. 7B). This work provides an elegant illustration of how aptamer-guided EVs can circumvent the limitation of unmodified EVs, as the latter are rapidly accumulated in and removed by reticuloendothelial organs, e.g. the liver and spleen, upon systemic administration.199 Notably, the utilization of the three-way junction approach for functionalizing EVs with a targeting aptamer exemplifies the versatility of the aptamer armamentarium, as such an approach can be adapted for inserting any targeting aptamer of interest into the membrane of any type of EVs relevant to the intended basic research or pre-clinical purpose.
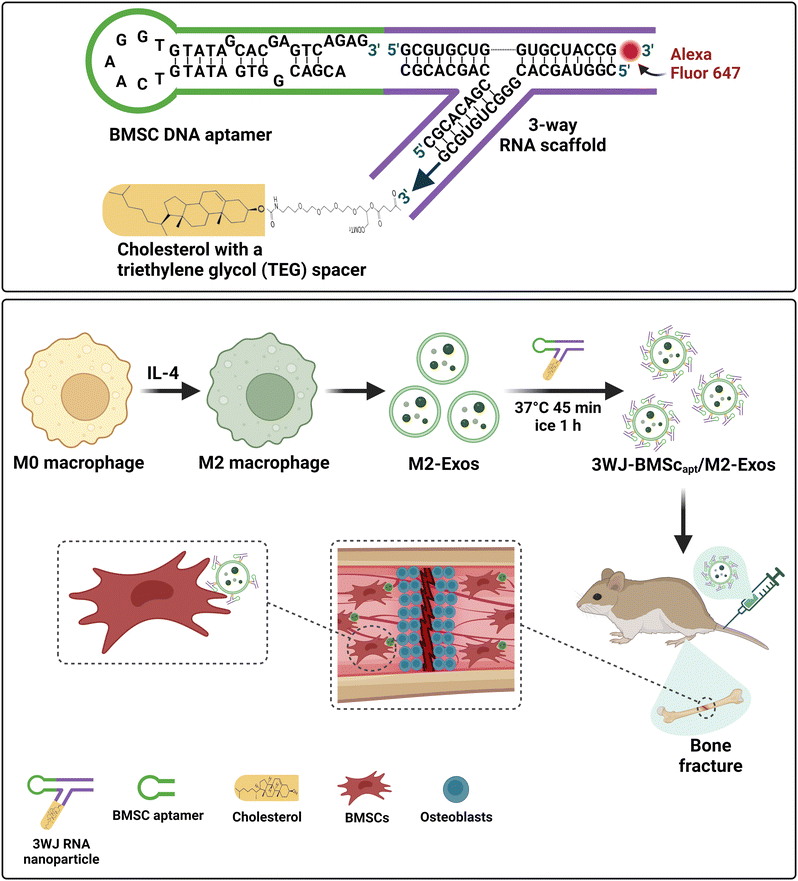 | ||
| Fig. 7 Schematic illustration of promotion of the healing of bone fracture by 3-way junction RNA nanoparticles-DNA aptamer functionalized EVs. (A) The formation of the 3-way junction RNA nanoparticles by linking of the DNA aptamer targeting BMSC to one arm of the 3-wyar RNA junction and the conjugation cholesterol to the other end of the RNA junction for the insertion of the nanoparticles onto the membrane of the EVs derived from M2 macrophages. A fluorophore is conjugated on the third end to enable in vivo tracking. (B) The DNA aptamer-guided EVs are injected intravenously to mice from a femur fracture model. These BMSC aptamer functionalized EVs are targeted to the site of bone fracture followed by a slow release of miRNA cargos, thereby significantly accelerating the bone regeneration processes. Adapted from ref. 195 and 196. | ||
An alternative way to conjugate aptamers to the surface of EVs is to covalently conjugate a carboxylic acid-functionalized aptamer to the amine groups on EVs through 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)/N-hydroxysuccinimide (NHS) chemistry. Using this approach, Shamili and co-workers functionalized EVs from MSC with a 40-nr DNA aptamer LJM-3064.132 Because the LJM-3064 aptamer has a high affinity for myelin and the ability to promote remyelination, it was employed as both a targeting ligand and a therapeutic agent. When intravenously injected into mice with experimental autoimmune encephalomyelitis, the LJM-3064 aptamer-EV produced a robust suppression of the inflammatory response as well as a lowered demyelination lesion region in the central nervous system, resulting in reduced severity of the disease in vivo.
6.4 Aptamer-guided EVs as a novel therapeutic modality for cardiovascular diseases
Cardiovascular diseases, including hypertension, heart failure, atherosclerosis, stroke, and ischemic heart disease, are the leading causes of death worldwide.200 EVs have emerged as a promising treatment strategy for cardiovascular disease. As a vehicle for signalling molecules, EV-mediated communication between different heart cells has been shown to play a crucial role in the maintenance of cardiac homeostasis and the progress of cardiac diseases.201,202 With the advantages of stability, minimal immunogenicity and the ease of pre- and post-isolation modification, several MSC-derived EV therapeutic studies have been developed with therapeutic cargos consisting mainly of microRNAs as well as long non-coding RNA and covalently closed RNA.203 EV-mediated delivery of miRNAs, including miR-19, miR-210, miR 133a, miR 30d-5p, miR-214-3p, let-7i-5p, let-7g-5p, miR-129-5p, miR-320a, miR-1246, miR19a, miR-29b, miR-455, miR-17 and miR-210, have been explored for EV-mediated treatment of coronary artery disease, heart failure, cardiomyopathy and atrial fibrillation.204 Other approaches have used EV-mimetics and their bioactive protein cargo from stem cells for cardiac cell repair. Lozano et al.205 integrated quantitative proteomics to understand cell remodelling following treatment to reveal the upregulation of angiogenic proteins (MFGE8, MYH10, VDAC2) in endothelial cells and pro-survival proteins (CNN2, THBS1, IGF2R) in cardiomyocytes, while attenuation of cardiac fibrosis and extracellular matrix remodelling capacity in cardiac fibroblasts (ACTN1, COL1A1/2/4A2/12A1, ITGA1/11, THBS1).205 This study presents a scalable approach to generating functional EV-mimetics to deliver protein cargo and remodel the cell and signaling landscape for cardiac repair.A critical challenge of EV-based therapy in the management of cardiovascular diseases is the lack of a robust systemic administration regime to deliver sufficient amounts of EVs to the site of the diseased cells/tissue, followed by the meaningful therapeutic retention of the EVs. Aptamers are ideal candidates for targeted EV therapeutics for cardiovascular diseases due to their high specificity, ease of cellular internalization and rapid tissue penetration and accumulation.206
Two groups of researchers have made inroads into the development of aptamers targeting cardiac muscle cells. Catalucci and colleagues were the first to report the use of an internalizing aptamer for the selective delivery of a small therapeutic peptide to cardiac cells.207 These researchers generated an aptamer-peptide chimera by conjugating the PDGFRβ-targeting Gint4.T aptamer to a small mimetic peptide (MP). The MP has been shown to restore myocardial function in pathological heart conditions associated with defective L-type calcium channel (LTCC) function peptide via targeting of the Cavβ2 subunit of the LTCC. When applied to HL-1cardiac muscle cells that contract and retain phenotypic characteristics of the adult cardiomyocyte,208 the Gint4.T-MP aptamer-peptide chimera was successfully internalized in cells and was able to not only restore LTCC protein levels but also recover LTCC-dependent calcium fluxes in the cardiac cells.207 Nonetheless, PDGFRβ is expressed in many tissues/organs outside the heart.209 The development of aptamers with excellent cardio-specificity will advance the development of effective new strategies for the treatment of cardiovascular disease. The heart remains a dauting challenge for developing therapies directed specifically at only one cell type, as cardiomyocytes constitute most of the cell mass but only account for 30% of the total cell number in the heart.210 It was only until 2023 that cardiac muscle-specific aptamers were developed.211 To develop targeted therapy for dilated cardiomyopathy, a type of heart muscle disease that causes the heart ventricles to thin and weaken, with very poor five-year survival rates,212 Phylactou and colleagues isolated a 2′-fluoropyrimidine-modified RNA aptamer, Aptamer-10478.211 This RNA aptamer has high selectivity towards cardiomyocytes as well as good stability in the serum. Future endeavours in isolating aptamers specific towards cells/tissues underlying cardiovascular pathogenesis will advance the clinical translation of this novel cell-free therapeutics for cardiovascular diseases.
7. Conclusion and perspectives
Oligonucleotide drugs, including aptamers, have gained exceptional scientific and pre-clinical interest, with numerous drugs being approved or used in clinical trials.213 To date, there are more than 18 marketed products based on antisense oligonucleotides, aptamers and small interfering RNAs, and many others are in the pipeline for both academia and industry.214 A major technological trigger for this development has been progress in oligonucleotide chemistry to overcome inherent limitations in the pharmacokinetics of free drugs and reduce the cost of manufacturing. Such strategies focus on enhancing the stability of native RNA and DNA aptamers to minimize degradation by nucleases.215 Although extensive oligonucleotide libraries of sufficient sequence complexity exist, searching only for sequence space often yields aptamers that are limited in specificity and particularly in affinity, with the field moving towards incorporating chemical and structure space to identify mature functional aptamers.216 Because the main hurdle for pharmacotherapy is delivery to target tissues, the adoption of delivery technologies, such as nanoparticles and, recently, EVs, is a game changer for many therapeutic indications.As discussed, EVs as cell-derived lipid-based nanocarriers are remarkable in their biocompatibility and potential for highly specific active targeting through the surface display of endogenous cellular ligands. Moreover, owing in large part to the complex, multimodal biogenesis pathways responsible for their formation and release, EVs contain a diverse molecular cargo that overcomes redundancy in intercellular signalling. Their inherent capacity to transverse cell membranes and transfer such complex bioactive cargo, as well as their ability to be modified through various engineering and modification strategies have attracted significant therapeutic interest.221,222 With further refinement, molecular bioengineering strategies are providing a new frontier for EV-based therapy, including novel mRNA vaccines, antigen cross-presentation and immunotherapy, organ delivery and repair, and cancer immune surveillance and targeted therapeutics.50,221 The main challenges for using EVs as delivery systems are (i) reproducible, large–scale production,217 (ii) effective loading of drugs or reprogramming of the EV cargo directly (through genetic manipulation), and (iii) target selectivity to enhance delivery and retention at the site of the disease. Despite the prolonged circulatory half-life of cargo encapsulated within EVs, most therapeutic EVs tend to accumulate in the liver, with only a small fraction of the injected dose reaching the intended target cells or tissues.218 Additional breakthroughs are required to improve cell- or tissue-specific delivery. Functional and tissue-targeted modification of EVs from cells used in cell therapy, such as cardiosphere-derived cells (in Phase III clinical trial NCT05126758 (https://clinicaltrials.gov/ct2/show/NCT05126758)) or other stem cells, would further increase efficacy in target tissues.
As aptamers provide numerous biological, chemical, and therapeutic advantages, including their size, selectivity, versatility, and multifunctional programming, their integration into the field of EVs contributes to the development of isolation, detection, and analysis pipelines for therapeutic and diagnostic utility. Aptamers have largely improved the sensitivity in EV detection coupled with some flexible technologies, such as fluorescence signal amplification, energy transfer, nanozymes catalysis, conversion to ssDNA detection, and sandwich format sensors. Although there are still some crucial technological issues and challenges to be addressed, these center on the stability of aptamers in biological settings in vivo and the specificity of aptamer interactions, compounded by the molecular diversity and abundance within and on the surface of EVs. An in-depth analysis of EVs as drug delivery entities may generate valuable insights that can be applied to synthetic particles. However, there are inherent pharmacokinetic challenges with such approaches. The protein corona, an absorption surrounding particles when circulating in vivo, makes nanocarriers prone to be recognized by the innate immune system and eliminated by phagocytic cells in the lungs, liver, and spleen.82 Therefore, merely coating cell-derived EVs, artificial EVs, or hybrid EV systems with aptamers may not adequately fulfill the intended targeting purpose in vivo. Innovative approaches, such as modifying the EV membrane and its associated protein corona with active peptides,219,220 promise to enhance EV targeting and therapeutic efficacy.
The successful translation of aptamer technology into EV-based medical applications depends on the advances in resolving several key limitations of aptamers: (i) the lack of the repeatability of aptamer-target binding due to inherent structural flexibility of oligonucleotides; (ii) the vulnerability of aptamer to its reaction environment in the absence of a standard protocol; (iii) fabricating procedures for many aptamer-based platforms are tedious, making them unfavorable for mass production; and (iv) given the uniqueness of each aptamer, transitioning from one aptamer to another on a particular sensing or isolation platform can be complex and challenging.
There is a growing interest in and convergence between aptamer technology and the EV field. Sequence-specific libraries are overcoming some of the inherent challenges with aptamer targeting and specificity to address fundamental questions in the EV field, namely the origin, form and function of EVs. Recent efforts have focused on identifying novel aptamer targets and stable EV signatures associated with a particular cell, disease or condition. Such technical developments will provide a new understanding of EV biology and application and ultimately translate our rapidly expanding understanding of the diversity of EVs from cells, organs and organisms to novel diagnostics and therapeutics.221,222
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was funded by the National Health and Medical Research Council (NHMRC) (D. W. G.), National Heart Foundation (NHF) (D. W. G.) (105072). D. W. G. is supported as the Hains Fellow, J. B. is the recipient of postgraduate scholarships from the NHMRC and National Heart Foundation (NHF). S. E. is supported through an Australian RFP Scholarship, in addition to Bright Sparks Foundation. All figures were made in BioRender.com.References
- M. Toyofuku, N. Nomura and L. Eberl, Types and origins of bacterial membrane vesicles, Nat. Rev. Microbiol., 2019, 17(1), 13–24 CrossRef CAS PubMed.
- J. Tulkens, O. D. Wever and A. Hendrix, Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization, Nat. Protoc., 2020, 15(1), 40–67 CrossRef CAS PubMed.
- Y. Jia, et al., Exosomes secreted from sonic hedgehog-modified bone mesenchymal stem cells facilitate the repair of rat spinal cord injuries, Acta Neurochir., 2021, 163(8), 2297–2306 CrossRef PubMed.
- J. Rizzo, M. L. Rodrigues and G. Janbon, Extracellular Vesicles in Fungi: Past, Present, and Future Perspectives, Front. Cell. Infect. Microbiol., 2020, 10, 346 CrossRef CAS PubMed.
- G. van Niel, et al., Challenges and directions in studying cell-cell communication by extracellular vesicles, Nat. Rev. Mol. Cell Biol., 2022, 23(5), 369–382 CrossRef CAS PubMed.
- R. Garcia-Martin, et al., Tissue differences in the exosomal/small extracellular vesicle proteome and their potential as indicators of altered tissue metabolism, Cell Rep., 2022, 38(3), 110277 CrossRef CAS PubMed.
- P. Sharma, et al., Exosomes regulate neurogenesis and circuit assembly, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(32), 16086–16094 CrossRef CAS PubMed.
- J. C. Gross, et al., Active Wnt proteins are secreted on exosomes, Nat. Cell Biol., 2012, 14(10), 1036–1045 CrossRef CAS PubMed.
- Y. J. Jung, et al., Cell reprogramming using extracellular vesicles from differentiating stem cells into white/beige adipocytes, Sci. Adv., 2020, 6(13), eaay6721 CrossRef CAS PubMed.
- W. C. Zhang, et al., Adipose-Derived Stromal Cells Attenuate Adipose Inflammation in Obesity through Adipocyte Browning and Polarization of M2 Macrophages, Mediators Inflammation, 2019. 2019, 1731540 Search PubMed.
- S. Gurung, et al., Exosomes and soluble secretome from hormone-treated endometrial epithelial cells direct embryo implantation, Mol. Hum. Reprod., 2020, 26(7), 510–520 CrossRef CAS PubMed.
- Q. H. Poh, et al., Proteome reprogramming of endometrial epithelial cells by human trophectodermal small extracellular vesicles reveals key insights into embryo implantation, Proteomics, 2021, 21(13–14), e2000210 CrossRef PubMed.
- A. Rai, et al., Proteomic profiling of human uterine extracellular vesicles reveal dynamic regulation of key players of embryo implantation and fertility during menstrual cycle, Proteomics, 2021, 21(13–14), e2000211 CrossRef PubMed.
- M. T. Roefs, J. P. G. Sluijter and P. Vader, Extracellular Vesicle-Associated Proteins in Tissue Repair, Trends Cell Biol., 2020, 30(12), 990–1013 CrossRef CAS PubMed.
- B. Chen, et al., Stem Cell-Derived Extracellular Vesicles as a Novel Potential Therapeutic Tool for Tissue Repair, Stem Cells Transl. Med., 2017, 6(9), 1753–1758 CrossRef PubMed.
- J. Yao, et al., A Minimally Invasive Exosome Spray Repairs Heart after Myocardial Infarction, ACS Nano, 2021, 15(7), 11099–11111 CrossRef CAS PubMed.
- C. Crewe, et al., Extracellular vesicle-based interorgan transport of mitochondria from energetically stressed adipocytes, Cell Metab., 2021, 33(9), 1853–1868 CrossRef CAS PubMed.
- I. K. Herrmann, M. J. A. Wood and G. Fuhrmann, Extracellular vesicles as a next-generation drug delivery platform, Nat. Nanotechnol., 2021, 16(7), 748–759 CrossRef CAS PubMed.
- A. Rai, et al., Proteomic dissection of large extracellular vesicle surfaceome unravels interactive surface platform, J. Extracell. Vesicles, 2021, 10(13), e12164 CrossRef CAS PubMed.
- G. van Niel, G. D'Angelo and G. Raposo, Shedding light on the cell biology of extracellular vesicles, Nat. Rev. Mol. Cell Biol., 2018, 19(4), 213–228 CrossRef CAS PubMed.
- D. Gupta, et al., Amelioration of systemic inflammation via the display of two different decoy protein receptors on extracellular vesicles, Nat. Biomed. Eng., 2021, 5(9), 1084–1098 CrossRef CAS PubMed.
- R. Xu, et al., Extracellular vesicle isolation and characterization: toward clinical application, J. Clin. Invest., 2016, 126(4), 1152–1162 CrossRef PubMed.
- D. K. Jeppesen, et al., Reassessment of Exosome Composition, Cell, 2019, 177(2), 428–445 CrossRef CAS PubMed.
- J. A. Martinez-Greene, et al., Quantitative proteomic analysis of extracellular vesicle subgroups isolated by an optimized method combining polymer-based precipitation and size exclusion chromatography, J. Extracell. Vesicles, 2021, 10(6), e12087 CrossRef PubMed.
- K. Al-Nedawi, et al., Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells, Nat. Cell Biol., 2008, 10(5), 619–624 CrossRef CAS PubMed.
- A. Rai, et al., Secreted midbody remnants are a class of extracellular vesicles molecularly distinct from exosomes and microparticles, Commun. Biol., 2021, 4(1), 400 CrossRef CAS PubMed.
- E. Willms, et al., Cells release subpopulations of exosomes with distinct molecular and biological properties, Sci. Rep., 2016, 6, 22519 CrossRef CAS PubMed.
- H. Jiao, et al., Mitocytosis, a migrasome-mediated mitochondrial quality-control process, Cell, 2021, 184(11), 2896–2910 CrossRef CAS PubMed.
- J. W. Clancy, M. Schmidtmann and C. D'Souza-Schorey, The ins and outs of microvesicles, FASEB Bioadv., 2021, 3(6), 399–406 CrossRef PubMed.
- R. Palmulli and G. van Niel, To be or not to be… secreted as exosomes, a balance finely tuned by the mechanisms of biogenesis, Essays Biochem., 2018, 62(2), 177–191 CrossRef PubMed.
- D. W. Greening and R. J. Simpson, Understanding extracellular vesicle diversity - current status, Expert Rev. Proteomics, 2018, 15(11), 887–910 CrossRef CAS PubMed.
- D. Zabeo, et al., Exosomes purified from a single cell type have diverse morphology, J. Extracell. Vesicles, 2017, 6(1), 1329476 CrossRef PubMed.
- J. Kowal, et al., Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(8), E968–E977 CrossRef CAS PubMed.
- L. Martin-Jaular, et al., Unbiased proteomic profiling of host cell extracellular vesicle composition and dynamics upon HIV-1 infection, EMBO J., 2021, 40(8), e105492 CrossRef CAS PubMed.
- F. G. Kugeratski, et al., Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker, Nat. Cell Biol., 2021, 23(6), 631–641 CrossRef CAS PubMed.
- M. Ostrowski, et al., Rab27a and Rab27b control different steps of the exosome secretion pathway, Nat. Cell Biol., 2010, 12(1), 19–30 CrossRef CAS PubMed.
- D. W. Greening, et al., Proteomic insights into extracellular vesicle biology–defining exosomes and shed microvesicles, Expert Rev. Proteomics, 2017, 14(1), 69–95 CrossRef CAS PubMed.
- S. B. Arya, et al., Ceramide-rich microdomains facilitate nuclear envelope budding for non-conventional exosome formation, Nat. Cell Biol., 2022, 24(7), 1019–1028 CrossRef CAS PubMed.
- T. Matsui, et al., ALIX and ceramide differentially control polarized small extracellular vesicle release from epithelial cells, EMBO Rep., 2021, 22(5), e51475 CrossRef CAS PubMed.
- M. Mathieu, et al., Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9, Nat. Commun., 2021, 12(1), 4389 CrossRef CAS PubMed.
- A. Lischnig, et al., Quantitative Proteomics Identifies Proteins Enriched in Large and Small Extracellular Vesicles, Mol. Cell. Proteomics, 2022, 21(9), 100273 CrossRef CAS PubMed.
- J. A. Nicolas-Avila, et al., A Network of Macrophages Supports Mitochondrial Homeostasis in the Heart, Cell, 2020, 183(1), 94–109 CrossRef CAS PubMed.
- V. R. Minciacchi, et al., Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles, Oncotarget, 2015, 6(13), 11327–11341 CrossRef PubMed.
- L. Ma, et al., Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration, Cell Res., 2015, 25(1), 24–38 CrossRef CAS PubMed.
- G. K. Atkin-Smith, et al., A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure, Nat. Commun., 2015, 6, 7439 CrossRef PubMed.
- Q. Zhang, et al., Comprehensive isolation of extracellular vesicles and nanoparticles, Nat. Protoc., 2023, 18(5), 1462–1487 CrossRef CAS PubMed.
- K. S. Visan, et al., Comparative analysis of tangential flow filtration and ultracentrifugation, both combined with subsequent size exclusion chromatography, for the isolation of small extracellular vesicles, J. Extracell. Vesicles, 2022, 11(9), e12266 CrossRef PubMed.
- T. Liangsupree, E. Multia and M. L. Riekkola, Modern isolation and separation techniques for extracellular vesicles, J. Chromatogr. A, 2021, 1636, 461773 CrossRef CAS PubMed.
- R. Xu, et al., Extracellular vesicles in cancer - implications for future improvements in cancer care, Nat. Rev. Clin Oncol., 2018, 15(10), 617–638 CrossRef CAS PubMed.
- D. W. Greening, et al., Extracellular vesicles as next generation immunotherapeutics, Semin. Cancer Biol., 2023, 90, 73–100 CrossRef CAS PubMed.
- P. H. Tran, et al., Aptamer-guided extracellular vesicle theranostics in oncology, Theranostics, 2020, 10(9), 3849–3866 CrossRef CAS PubMed.
- D. Athauda, et al., Utility of Neuronal-Derived Exosomes to Examine Molecular Mechanisms That Affect Motor Function in Patients With Parkinson Disease: A Secondary Analysis of the Exenatide-PD Trial, JAMA Neurol., 2019, 76(4), 420–429 CrossRef PubMed.
- G. Camussi and J. Lötvall, The importance of controlled clinical trials with extracellular vesicles, J. Extracell. Vesicles, 2023, 12(7), e12347 CrossRef PubMed.
- C. Beltrami, et al., Human Pericardial Fluid Contains Exosomes Enriched with Cardiovascular-Expressed MicroRNAs and Promotes Therapeutic Angiogenesis, Mol. Ther., 2017, 25(3), 679–693 CrossRef CAS PubMed.
- L. Zitvogel, et al., Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes, Nat. Med., 1998, 4(5), 594–600 CrossRef CAS PubMed.
- B. Escudier, et al., Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial, J. Transl. Med., 2005, 3(1), 10 CrossRef PubMed.
- M. A. Morse, et al., A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer, J. Transl. Med., 2005, 3(1), 9 CrossRef PubMed.
- S. Viaud, et al., Dendritic cell-derived exosomes promote natural killer cell activation and proliferation: a role for NKG2D ligands and IL-15Ralpha, PLoS One, 2009, 4(3), e4942 CrossRef PubMed.
- S. Viaud, et al., Updated technology to produce highly immunogenic dendritic cell-derived exosomes of clinical grade: a critical role of interferon-gamma, J. Immunother., 2011, 34(1), 65–75 CrossRef PubMed.
- Z. Lu, et al., Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models, J. Hepatol., 2017, 67(4), 739–748 CrossRef CAS PubMed.
- B. Zuo, et al., Universal immunotherapeutic strategy for hepatocellular carcinoma with exosome vaccines that engage adaptive and innate immune responses, J. Hematol. Oncol., 2022, 15(1), 46 CrossRef CAS PubMed.
- C. D. Phung, et al., Anti-CTLA-4 antibody-functionalized dendritic cell-derived exosomes targeting tumor-draining lymph nodes for effective induction of antitumor T-cell responses, Acta Biomater., 2020, 115, 371–382 CrossRef CAS PubMed.
- H. Zhu, et al., An efficient and safe MUC1-dendritic cell-derived exosome conjugate vaccine elicits potent cellular and humoral immunity and tumor inhibition in vivo, Acta Biomater., 2022, 138, 491–504 CrossRef CAS PubMed.
- Z. Du, et al., Modified dendritic cell-derived exosomes activate both NK cells and T cells through the NKG2D/NKG2D-L pathway to kill CML cells with or without T315I mutation, Exp. Hematol. Oncol., 2022, 11(1), 36 CrossRef CAS PubMed.
- S. Hu, et al., Engineered exosome-like nanovesicles suppress tumor growth by reprogramming tumor microenvironment and promoting tumor ferroptosis, Acta Biomater., 2021, 135, 567–581 CrossRef CAS PubMed.
- M. Yildirim, et al., TLR ligand loaded exosome mediated immunotherapy of established mammary Tumor in mice, Immunol. Lett., 2021, 239, 32–41 CrossRef CAS PubMed.
- B. Zuo, et al., Alarmin-painted exosomes elicit persistent antitumor immunity in large established tumors in mice, Nat. Commun., 2020, 11(1), 1790 CrossRef CAS PubMed.
- C. W. Obiero, et al., A Phase 2a Randomized Study to Evaluate the Safety and Immunogenicity of the 1790GAHB Generalized Modules for Membrane Antigen Vaccine against Shigella sonnei Administered Intramuscularly to Adults from a Shigellosis-Endemic Country, Front. Immunol., 2017, 8, 1884 CrossRef PubMed.
- I. Parolini, et al., Microenvironmental pH is a key factor for exosome traffic in tumor cells, J. Biol. Chem., 2009, 284(49), 34211–34222 CrossRef CAS PubMed.
- A. H. Hutagalung and P. J. Novick, Role of Rab GTPases in membrane traffic and cell physiology, Physiol. Rev., 2011, 91(1), 119–149 CrossRef CAS PubMed.
- D. Feng, et al., Cellular internalization of exosomes occurs through phagocytosis, Traffic, 2010, 11(5), 675–687 CrossRef CAS PubMed.
- C. Thery, M. Ostrowski and E. Segura, Membrane vesicles as conveyors of immune responses, Nat. Rev. Immunol., 2009, 9(8), 581–593 CrossRef CAS PubMed.
- J. L. Hood, R. S. San and S. A. Wickline, Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis, Cancer Res., 2011, 71(11), 3792–3801 CrossRef CAS PubMed.
- J. Skog, et al., Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers, Nat. Cell Biol., 2008, 10(12), 1470–1476 CrossRef CAS PubMed.
- E. I. Buzas, et al., Molecular interactions at the surface of extracellular vesicles, Semin. Immunopathol., 2018, 40(5), 453–464 CrossRef CAS PubMed.
- L. G. Lima, et al., Tumor microenvironmental cytokines bound to cancer exosomes determine uptake by cytokine receptor-expressing cells and biodistribution, Nat. Commun., 2021, 12(1), 3543 CrossRef CAS PubMed.
- W. T. Meng and H. D. Guo, Small Extracellular Vesicles Derived from Induced Pluripotent Stem Cells in the Treatment of Myocardial Injury, Int. J. Mol. Sci., 2023, 24(5) Search PubMed.
- B. Sun, et al., New treatment methods for myocardial infarction, Front. Cardiovasc. Med., 2023, 10, 1251669 CrossRef CAS PubMed.
- Y. Han, et al., Exosomes from hypoxia-treated human adipose-derived mesenchymal stem cells enhance angiogenesis through VEGF/VEGF-R, Int. J. Biochem. Cell Biol., 2019, 109, 59–68 CrossRef CAS PubMed.
- C. Xue, et al., Exosomes Derived from Hypoxia-Treated Human Adipose Mesenchymal Stem Cells Enhance Angiogenesis Through the PKA Signaling Pathway, Stem Cells Dev., 2018, 27(7), 456–465 CrossRef CAS PubMed.
- B. Lukomska, et al., Challenges and Controversies in Human Mesenchymal Stem Cell Therapy, Stem Cells Int., 2019. 2019, 9628536 Search PubMed.
- L. Kordelas, et al., MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease, Leukemia, 2014, 28(4), 970–973 CrossRef CAS PubMed.
- A. M. Williams, et al., Mesenchymal Stem Cell-Derived Exosomes Provide Neuroprotection and Improve Long-Term Neurologic Outcomes in a Swine Model of Traumatic Brain Injury and Hemorrhagic Shock, J. Neurotrauma, 2019, 36(1), 54–60 CrossRef PubMed.
- G. de Couto, et al., Exosomal MicroRNA Transfer Into Macrophages Mediates Cellular Postconditioning, Circulation, 2017, 136(2), 200–214 CrossRef CAS PubMed.
- W. S. Toh, et al., MSC exosome works through a protein-based mechanism of action, Biochem. Soc. Trans., 2018, 46(4), 843–853 CrossRef CAS PubMed.
- A. Rai, et al., A Protocol for Isolation, Purification, Characterization, and Functional Dissection of Exosomes, Methods Mol. Biol., 2021, 2261, 105–149 CrossRef CAS PubMed.
- E. Shaba, et al., Multi-Omics Integrative Approach of Extracellular Vesicles: A Future Challenging Milestone, Proteomes, 2022, 10(2) CrossRef CAS PubMed.
- D. M. Stranford, et al., Genetically encoding multiple functionalities into extracellular vesicles for the targeted delivery of biologics to T cells, Nat. Biomed. Eng., 2023 Search PubMed.
- S. M. Nimjee, et al., Aptamers as Therapeutics, Annu. Rev. Pharmacol. Toxicol., 2017, 57, 61–79 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, In vitro selection of RNA molecules that bind specific ligands, Nature, 1990, 346(6287), 818–822 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase, Science, 1990, 249(4968), 505–510 CrossRef CAS PubMed.
- P. Burgstaller, A. Girod and M. Blind, Aptamers as tools for target prioritization and lead identification, Drug Discovery Today, 2002, 7(24), 1221–1228 CrossRef CAS PubMed.
- D. Proske, et al., Aptamers–basic research, drug development, and clinical applications, Appl. Microbiol. Biotechnol., 2005, 69(4), 367–374 CrossRef CAS PubMed.
- S. M. Nimjee, C. P. Rusconi and B. A. Sullenger, Aptamers: an emerging class of therapeutics, Annu. Rev. Med., 2005, 56, 555–583 CrossRef CAS PubMed.
- M. Kohlberger and G. Gadermaier, SELEX: Critical factors and optimization strategies for successful aptamer selection, Biotechnol. Appl. Biochem., 2022, 69(5), 1771–1792 CrossRef CAS PubMed.
- M. Kimoto, Y. W. Shermane Lim and I. Hirao, Molecular affinity rulers: systematic evaluation of DNA aptamers for their applicabilities in ELISA, Nucleic Acids Res., 2019, 47(16), 8362–8374 CrossRef CAS PubMed.
- R. Thevendran, et al., Strategies to bioengineer aptamer-driven nanovehicles as exceptional molecular tools for targeted therapeutics: A review, J. Controlled Release, 2020, 323, 530–548 CrossRef CAS PubMed.
- M. Reth, Matching cellular dimensions with molecular sizes, Nat. Immunol., 2013, 14(8), 765–767 CrossRef CAS PubMed.
- M. Z. Sabri, et al., The assessment of three dimensional modelling design for single strand DNA aptamers for computational chemistry application, Biophys. Chem., 2020, 267, 106492 CrossRef CAS PubMed.
- M. R. W. Scheepers, I. L. J. van and M. W. J. Prins, Multivalent weak interactions enhance selectivity of interparticle binding, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(37), 22690–22697 CrossRef CAS PubMed.
- S. Cai, et al., Investigations on the interface of nucleic acid aptamers and binding targets, Analyst, 2018, 143(22), 5317–5338 RSC.
- C. Forier, et al., DNA aptamer affinity ligands for highly selective purification of human plasma-related proteins from multiple sources, J. Chromatogr. A, 2017, 1489, 39–50 CrossRef CAS PubMed.
- D. Ishimaru, et al., Mechanism of regulation of bcl-2 mRNA by nucleolin and A + U-rich element-binding factor 1 (AUF1), J. Biol. Chem., 2010, 285(35), 27182–27191 CrossRef CAS PubMed.
- Z. Zhou, Y. Chen and X. Qian, Target-Specific Exosome Isolation through Aptamer-Based Microfluidics, Biosensors, 2022, 12(4) Search PubMed.
- S. Wang, et al., Aptasensor with Expanded Nucleotide Using DNA Nanotetrahedra for Electrochemical Detection of Cancerous Exosomes, ACS Nano, 2017, 11(4), 3943–3949 CrossRef CAS PubMed.
- C. Liu, et al., Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers, Nat. Biomed. Eng., 2019, 3(3), 183–193 CrossRef CAS PubMed.
- K. Zhang, et al., Rapid Capture and Nondestructive Release of Extracellular Vesicles Using Aptamer-Based Magnetic Isolation, ACS Sens., 2019, 4(5), 1245–1251 CrossRef CAS PubMed.
- D. Shangguan, et al., Aptamers evolved from live cells as effective molecular probes for cancer study, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(32), 11838–11843 CrossRef CAS PubMed.
- A. S. Potty, et al., Biophysical characterization of DNA aptamer interactions with vascular endothelial growth factor, Biopolymers, 2009, 91(2), 145–156 CrossRef CAS PubMed.
- Z. Wang, et al., Screening and multiple detection of cancer exosomes using an SERS-based method, Nanoscale, 2018, 10(19), 9053–9062 RSC.
- X. Su, et al., Integrated SERS-Vertical Flow Biosensor Enabling Multiplexed Quantitative Profiling of Serological Exosomal Proteins in Patients for Accurate Breast Cancer Subtyping, ACS Nano, 2023, 17(4), 4077–4088 CrossRef CAS PubMed.
- Y. Zhou, et al., Detection of breast cancer-derived exosomes using the horseradish peroxidase-mimicking DNAzyme as an aptasensor, Analyst, 2019, 145(1), 107–114 RSC.
- L. Xu, et al., Development of a simple, sensitive and selective colorimetric aptasensor for the detection of cancer-derived exosomes, Biosens. Bioelectron., 2020, 169, 112576 CrossRef CAS PubMed.
- L. Zhu, et al., Quantification-Promoted Discovery of Glycosylated Exosomal PD-L1 as a Potential Tumor Biomarker, Small Methods, 2022, 6(9), e2200549 CrossRef PubMed.
- H. S. Shin, et al., ALPPL2 Is a Potential Diagnostic Biomarker for Pancreatic Cancer-Derived Extracellular Vesicles, Mol. Ther.–Methods Clin. Dev., 2019, 15, 204–210 CrossRef CAS PubMed.
- J. Chen, et al., Isolation and Visible Detection of Tumor-Derived Exosomes from Plasma, Anal. Chem., 2018, 90(24), 14207–14215 CrossRef CAS PubMed.
- M. Lee, et al., Recent Advances in Biological Applications of Aptamer-Based Fluorescent Biosensors, Molecules, 2023, 28(21), 7327 CrossRef CAS PubMed.
- N. Savory, et al., Selection of DNA aptamer against prostate specific antigen using a genetic algorithm and application to sensing, Biosens. Bioelectron., 2010, 26(4), 1386–1391 CrossRef CAS PubMed.
- C. Liu, et al., λ-DNA- and Aptamer-Mediated Sorting and Analysis of Extracellular Vesicles, J. Am. Chem. Soc., 2019, 141(9), 3817–3821 CrossRef CAS PubMed.
- J. Kuang, et al., A colorimetric aptasensor based on a hemin/EpCAM aptamer DNAzyme for sensitive exosome detection, Analyst, 2022, 147(22), 5054–5061 RSC.
- S. Das, et al., Development of DNA Aptamers to Visualize Release of Mycobacterial Membrane-Derived Extracellular Vesicles in Infected Macrophages, Pharmaceuticals, 2021, 15(1) Search PubMed.
- P. Dua, et al., Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein, Cancer Res., 2013, 73(6), 1934–1945 CrossRef CAS PubMed.
- L. Zhu, et al., Coupling Aptamer-based Protein Tagging with Metabolic Glycan Labeling for In Situ Visualization and Biological Function Study of Exosomal Protein-Specific Glycosylation, Angew. Chem., Int. Ed., 2021, 60(33), 18111–18115 CrossRef CAS PubMed.
- F. He, et al., Probing exosome internalization pathways through confocal microscopy imaging, Chem. Commun., 2019, 55(93), 14015–14018 RSC.
- J. Zou, et al., Aptamer-Functionalized Exosomes: Elucidating the Cellular Uptake Mechanism and the Potential for Cancer-Targeted Chemotherapy, Anal. Chem., 2019, 91(3), 2425–2430 CrossRef CAS PubMed.
- H. Y. Dong, et al., Precise selection of aptamers targeting PD-L1 positive small extracellular vesicles on magnetic chips, Chem. Commun., 2021, 57(29), 3555–3558 RSC.
- N. F. Hosseini, et al., AS1411 aptamer-functionalized exosomes in the targeted delivery of doxorubicin in fighting colorectal cancer, Biomed. Pharmacother., 2022, 155, 113690 CrossRef CAS PubMed.
- X. Tong, et al., Progress in cancer drug delivery based on AS1411 oriented nanomaterials, J. Nanobiotechnol., 2022, 20(1), 57 CrossRef CAS PubMed.
- F. Pi, et al., Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression, Nat. Nanotechnol., 2018, 13(1), 82–89 CrossRef CAS PubMed.
- E. Bagheri, et al., Targeted doxorubicin-loaded mesenchymal stem cells-derived exosomes as a versatile platform for fighting against colorectal cancer, Life Sci., 2020, 261, 118369 CrossRef CAS PubMed.
- B. Nastasijevic, et al., Remyelination induced by a DNA aptamer in a mouse model of multiple sclerosis, PLoS One, 2012, 7(6), e39595 CrossRef CAS PubMed.
- F. Hosseini Shamili, et al., Immunomodulatory properties of MSC-derived exosomes armed with high affinity aptamer toward mylein as a platform for reducing multiple sclerosis clinical score, J. Controlled Release, 2019, 299, 149–164 CrossRef CAS PubMed.
- C. J. Li, et al., MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation, J. Clin. Invest., 2015, 125(4), 1509–1522 CrossRef PubMed.
- Z. W. Luo, et al., Aptamer-functionalized exosomes from bone marrow stromal cells target bone to promote bone regeneration, Nanoscale, 2019, 11(43), 20884–20892 RSC.
- D. Al Shaer, et al., 2023 FDA TIDES (Peptides and Oligonucleotides) Harvest, Pharmaceuticals, 2024, 17(2), 243 CrossRef CAS PubMed.
- O. A. Halawa, et al., A Review of Completed and Ongoing Complement Inhibitor Trials for Geographic Atrophy Secondary to Age-Related Macular Degeneration, J. Clin. Med., 2021, 10(12), 2580 CrossRef CAS PubMed.
- Y. Zhang, B. S. Lai and M. Juhas, Recent Advances in Aptamer Discovery and Applications, Molecules, 2019, 24(5), 941 CrossRef PubMed.
- Q. Zhou, et al., Development of an aptasensor for electrochemical detection of exosomes, Methods, 2016, 97, 88–93 CrossRef CAS PubMed.
- L. Wu, et al., Aptamer-Based Liquid Biopsy, ACS Appl. Bio Mater., 2020, 3(5), 2743–2764 CrossRef CAS PubMed.
- D. Sun, et al., Recent progress in aptamer-based microfluidics for the detection of circulating tumor cells and extracellular vesicles, J. Pharm. Anal., 2023, 13(4), 340–354 CrossRef PubMed.
- L. Wu, et al., Aptamer-Based Detection of Circulating Targets for Precision Medicine, Chem. Rev., 2021, 121(19), 12035–12105 CrossRef CAS PubMed.
- J. Li, et al., An Aptamer-Based Nanoflow Cytometry Method for the Molecular Detection and Classification of Ovarian Cancers through Profiling of Tumor Markers on Small Extracellular Vesicles, Angew. Chem., Int. Ed., 2023, e202314262 Search PubMed.
- Y. Tian, et al., Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry, J. Extracell. Vesicles, 2020, 9(1), 1697028 CrossRef CAS PubMed.
- H. Liu, et al., Analysis of extracellular vesicle DNA at the single-vesicle level by nano-flow cytometry, J. Extracell. Vesicles, 2022, 11(4), e12206 CrossRef CAS PubMed.
- Y. Huang, et al., Divalent Aptamer-Functionalized Nanochannels for Facile Detection of Cancer Cell-Derived Exosomes, Sensors, 2023, 23(22), 9139 CrossRef CAS PubMed.
- F. Guo, et al., A dual aptamer recognition-based fluorescent biosensor for extracellular vesicles assays with high sensitivity and specificity, Sens. Actuators, B, 2023, 389, 133890 CrossRef CAS.
- C. Li, et al., Molybdenum Disulfide-Integrated Iron Organic Framework Hybrid Nanozyme-Based Aptasensor for Colorimetric Detection of Exosomes, Biosensors, 2023, 13(8), 800 CrossRef CAS PubMed.
- W. Cheng, et al., Highly Sensitive Aptasensor for Detecting Cancerous Exosomes Based on Clover-like Gold Nanoclusters, Anal. Chem., 2023, 95(7), 3606–3612 CrossRef CAS PubMed.
- P. Sen, et al., High-Precision Viral Detection Using Electrochemical Kinetic Profiling of Aptamer-Antigen Recognition in Clinical Samples and Machine Learning, Angew. Chem., Int. Ed., 2024, 63(20), e202400413 CrossRef CAS PubMed.
- G. Moreira, et al., A capacitive laser-induced graphene based aptasensor for SARS-CoV-2 detection in human saliva, PLoS One, 2023, 18(8), e0290256 CrossRef CAS PubMed.
- A. I. Cubas-Atienzar, et al., Limit of detection in different matrices of 19 commercially available rapid antigen tests for the detection of SARS-CoV-2, Sci. Rep., 2021, 11(1), 18313 CrossRef CAS PubMed.
- L. Xu, et al., On-chip integrated graphene aptasensor with portable readout for fast and label-free COVID-19 detection in virus transport medium, Sens. Diagn., 2022, 1(4), 719–730 RSC.
- R. Zhuo, et al., Comparison of SARS-CoV-2 spike antibody quantitative titer reporting using the World Health Organization International Standard Units by four commercial assays, J. Clin. Virol., 2022, 156, 105292 CrossRef CAS PubMed.
- B. J. Tauro, et al., Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes, Methods, 2012, 56(2), 293–304 CrossRef CAS PubMed.
- M. Holcar, M. Kandušer and M. Lenassi, Blood Nanoparticles - Influence on Extracellular Vesicle Isolation and Characterization, Front. Pharmacol., 2021, 12, 773844 CrossRef CAS PubMed.
- S. Giancaterino and C. Boi, Alternative biological sources for extracellular vesicles production and purification strategies for process scale-up, Biotechnol. Adv., 2023, 63, 108092 CrossRef CAS PubMed.
- N. Kruljec and T. Bratkovič, Alternative Affinity Ligands for Immunoglobulins, Bioconjugate Chem., 2017, 28(8), 2009–2030 CrossRef CAS PubMed.
- G. Perret and E. Boschetti, Aptamer-Based Affinity Chromatography for Protein Extraction and Purification, Adv. Biochem. Eng./Biotechnol., 2020, 174, 93–139 CrossRef CAS PubMed.
- L. F. Yang, et al., Aptamers 101: aptamer discovery and in vitro applications in biosensors and separations, Chem. Sci., 2023, 14(19), 4961–4978 RSC.
- A. Akbarinejad, et al., Novel Electrochemically Switchable, Flexible, Microporous Cloth that Selectively Captures, Releases, and Concentrates Intact Extracellular Vesicles, ACS Appl. Mater. Interfaces, 2020, 12(35), 39005–39013 CrossRef CAS PubMed.
- J. D. Hirsch, et al., Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation, Anal. Biochem., 2002, 308(2), 343–357 CrossRef CAS PubMed.
- F. Xue, et al., Isolation of extracellular vesicles with multivalent aptamers, Analyst, 2021, 146(1), 253–261 RSC.
- Y. Ren, et al., Multivalent DNA Flowers for High-Performance Isolation, Detection, and Release of Tumor-Derived Extracellular Vesicles, ACS Appl. Mater. Interfaces, 2023, 15(48), 55358–55368 CrossRef CAS PubMed.
- Y. Jia, et al., Approved Nanomedicine against Diseases, Pharmaceutics, 2023, 15(3), 774 CrossRef CAS PubMed.
- M. J. Mitchell, et al., Engineering precision nanoparticles for drug delivery, Nat. Rev. Drug Discovery, 2021, 20(2), 101–124 CrossRef CAS PubMed.
- M. Zhang, et al., Engineered exosomes from different sources for cancer-targeted therapy, Signal Transduction Targeted Ther., 2023, 8(1), 124 CrossRef PubMed.
- Q. Wu, et al., Advances in Extracellular Vesicle Nanotechnology for Precision Theranostics, Adv. Sci., 2023, 10(3), e2204814 CrossRef PubMed.
- D. Xiang, et al., Superior Performance of Aptamer in Tumor Penetration over Antibody: Implication of Aptamer-Based Theranostics in Solid Tumors, Theranostics, 2015, 5(10), 1083–1097 CrossRef CAS PubMed.
- D. Xiang, et al., Transforming doxorubicin into a cancer stem cell killer via EpCAM aptamer-mediated delivery, Theranostics, 2017, 7(17), 4071–4086 CrossRef CAS PubMed.
- C. L. Esposito, et al., Identification of a novel RNA aptamer that selectively targets breast cancer exosomes, Mol. Ther.–Nucleic Acids, 2021, 23, 982–994 CrossRef CAS PubMed.
- L. Wang, et al., Engineering Extracellular Vesicles as Delivery Systems in Therapeutic Applications, Adv. Sci., 2023, 10(17), e2300552 CrossRef PubMed.
- S. Yadav, A. K. Sharma and P. Kumar, Nanoscale Self-Assembly for Therapeutic Delivery, Front. Bioeng. Biotechnol., 2020, 8, 127 CrossRef PubMed.
- K. W. Witwer and J. Wolfram, Extracellular vesicles versus synthetic nanoparticles for drug delivery, Nat. Rev. Mater., 2021, 6(2), 103–106 CrossRef CAS PubMed.
- H. Valadi, et al., Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells, Nat. Cell Biol., 2007, 9(6), 654–659 CrossRef CAS PubMed.
- K. O'Brien, et al., RNA delivery by extracellular vesicles in mammalian cells and its applications, Nat. Rev. Mol. Cell Biol., 2020, 21(10), 585–606 CrossRef PubMed.
- Y. You, et al., Intradermally delivered mRNA-encapsulating extracellular vesicles for collagen-replacement therapy, Nat. Biomed. Eng., 2023, 7(7), 887–900 CrossRef CAS PubMed.
- Z. Yang, et al., Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation, Nat. Biomed. Eng., 2020, 4(1), 69–83 CrossRef CAS PubMed.
- J. Radler, et al., Exploiting the biogenesis of extracellular vesicles for bioengineering and therapeutic cargo loading, Mol. Ther., 2023, 31(5), 1231–1250 CrossRef PubMed.
- S. Zhang, et al., Selective Encapsulation of Therapeutic mRNA in Engineered Extracellular Vesicles by DNA Aptamer, Nano Lett., 2021, 21(20), 8563–8570 CrossRef CAS PubMed.
- D. Qi, et al., Assessment and prediction of glioblastoma therapy response: challenges and opportunities, Brain, 2023, 146(4), 1281–1298 CrossRef PubMed.
- M. Abdelsalam, et al., Insights into Exosome Transport through the Blood-Brain Barrier and the Potential Therapeutical Applications in Brain Diseases, Pharmaceuticals, 2023, 16(4), 571 CrossRef CAS PubMed.
- D. Yuan, et al., Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain, Biomaterials, 2017, 142, 1–12 CrossRef CAS PubMed.
- S. Liang, H. Xu and B. C. Ye, Membrane-Decorated Exosomes for Combination Drug Delivery and Improved Glioma Therapy, Langmuir, 2022, 38(1), 299–308 CrossRef CAS PubMed.
- M. T. Hayes, Parkinson's Disease and Parkinsonism, Am. J. Med., 2019, 132(7), 802–807 CrossRef PubMed.
- P. Riederer, et al., Lewy bodies, iron, inflammation and neuromelanin: pathological aspects underlying Parkinson's disease, J. Neural Transm., 2023, 130(5), 627–646 CrossRef PubMed.
- Y. Zheng, et al., Novel DNA Aptamers for Parkinson's Disease Treatment Inhibit α-Synuclein Aggregation and Facilitate its Degradation, Mol. Ther.–Nucleic Acids, 2018, 11, 228–242 CrossRef CAS PubMed.
- X. Ren, et al., Exosomal DNA Aptamer Targeting α-Synuclein Aggregates Reduced Neuropathological Deficits in a Mouse Parkinson's Disease Model, Mol. Ther.–Nucleic Acids, 2019, 17, 726–740 CrossRef CAS PubMed.
- A. E. Altyar, et al., Future regenerative medicine developments and their therapeutic applications, Biomed. Pharmacother., 2023, 158, 114131 CrossRef CAS PubMed.
- Z. Liang, et al., Dedifferentiated fat cells: current applications and future directions in regenerative medicine, Stem Cell Res. Ther., 2023, 14(1), 207 CrossRef PubMed.
- R. Margiana, et al., Clinical application of mesenchymal stem cell in regenerative medicine: a narrative review, Stem Cell Res. Ther., 2022, 13(1), 366 CrossRef CAS PubMed.
- R. C. Lai, et al., Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury, Stem Cell Res., 2010, 4(3), 214–222 CrossRef CAS PubMed.
- K. W. Witwer, et al., Defining mesenchymal stromal cell (MSC)-derived small extracellular vesicles for therapeutic applications, J. Extracell. Vesicles, 2019, 8(1), 1609206 CrossRef CAS PubMed.
- S. I. van de Wakker, et al., Extracellular Vesicle Heterogeneity and Its Impact for Regenerative Medicine Applications, Pharmacol. Rev., 2023, 75(5), 1043–1061 CrossRef CAS PubMed.
- Z. Luo, et al., Application of aptamers in regenerative medicine, Front. Bioeng. Biotechnol., 2022, 10, 976960 CrossRef PubMed.
- J. Shou, et al., 3WJ RNA Nanoparticles-Aptamer Functionalized Exosomes From M2 Macrophages Target BMSCs to Promote the Healing of Bone Fractures, Stem Cells Transl. Med., 2023, 12(11), 758–774 CrossRef PubMed.
- P. Guo, et al., Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation, Mol. Cell, 1998, 2(1), 149–155 CrossRef CAS PubMed.
- Z. Li, et al., Exosomes Derived From M2 Macrophages Facilitate Osteogenesis and Reduce Adipogenesis of BMSCs, Front. Endocrinol., 2021, 12, 680328 CrossRef PubMed.
- M. Kang, et al., Bone regeneration is mediated by macrophage extracellular vesicles, Bone, 2020, 141, 115627 CrossRef CAS PubMed.
- M. Kang, et al., Biodistribution of extracellular vesicles following administration into animals: A systematic review, J. Extracell. Vesicles, 2021, 10(8), e12085 CrossRef PubMed.
- M. Vaduganathan, et al., The Global Burden of Cardiovascular Diseases and Risk: A Compass for Future Health, J. Am. Coll. Cardiol., 2022, 80(25), 2361–2371 CrossRef PubMed.
- A. Okamura, et al., Can Extracellular Vesicles as Drug Delivery Systems Be a Game Changer in Cardiac Disease?, Pharm. Res., 2023, 40(4), 889–908 CrossRef CAS PubMed.
- B. Claridge, et al., Proteome characterisation of extracellular vesicles isolated from heart, Proteomics, 2021, 21(13–14), e2100026 CrossRef PubMed.
- B. You, et al., Extracellular Vesicles: A New Frontier for Cardiac Repair, Pharmaceutics, 2022, 14(9), 1848 CrossRef CAS PubMed.
- Y. Xu, et al., Exosomes and their derivatives as biomarkers and therapeutic delivery agents for cardiovascular diseases: Situations and challenges, J. Transl. Int. Med., 2023, 11(4), 341–354 CrossRef PubMed.
- J. Lozano, et al., Scalable Generation of Nanovesicles from Human-Induced Pluripotent Stem Cells for Cardiac Repair, Int. J. Mol. Sci., 2022, 23(22), 14334 CrossRef CAS PubMed.
- X. Chen, et al., Aptamer-based applications for cardiovascular disease, Front. Bioeng. Biotechnol., 2022, 10, 1002285 CrossRef PubMed.
- A. Romanelli, et al., An anti-PDGFRβ aptamer for selective delivery of small therapeutic peptide to cardiac cells, PLoS One, 2018, 13(3), e0193392 CrossRef PubMed.
- W. C. Claycomb, et al., HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte, Proc. Natl. Acad. Sci. U. S. A., 1998, 95(6), 2979–2984 CrossRef CAS PubMed.
- J. Shen, et al., PDGFR-β as a positive regulator of tissue repair in a mouse model of focal cerebral ischemia, J. Cereb. Blood Flow Metab., 2012, 32(2), 353–367 CrossRef CAS PubMed.
- N. Anto Michel, et al., Cellular Heterogeneity of the Heart, Front. Cardiovasc. Med., 2022, 9, 868466 CrossRef PubMed.
- S. Philippou, et al., Selective Delivery to Cardiac Muscle Cells Using Cell-Specific Aptamers, Pharmaceuticals, 2023, 16(9) Search PubMed.
- X. R. Xu, et al., Fifteen-year mortality and prognostic factors in patients with dilated cardiomyopathy: persistent standardized application of drug therapy and strengthened management may bring about encouraging change in an aging society, J. Geriatr. Cardiol., 2022, 19(5), 335–342 CAS.
- J. A. Kulkarni, et al., The current landscape of nucleic acid therapeutics, Nat. Nanotechnol., 2021, 16(6), 630–643 CrossRef CAS PubMed.
- M. Egli and M. Manoharan, Chemistry, structure and function of approved oligonucleotide therapeutics, Nucleic Acids Res., 2023, 51(6), 2529–2573 CrossRef CAS PubMed.
- M. R. Dunn, et al., Generating Biologically Stable TNA Aptamers that Function with High Affinity and Thermal Stability, J. Am. Chem. Soc., 2020, 142(17), 7721–7724 CrossRef CAS PubMed.
- R. Dolot, et al., Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity, Nucleic Acids Res., 2018, 46(9), 4819–4830 CrossRef CAS PubMed.
- R. C. de Abreu, et al., Native and bioengineered extracellular vesicles for cardiovascular therapeutics, Nat. Rev. Cardiol., 2020, 17(11), 685–697 CrossRef PubMed.
- G. Han, et al., Generalizable anchor aptamer strategy for loading nucleic acid therapeutics on exosomes, EMBO Mol. Med., 2024, 16(4), 1027–1045 CrossRef PubMed.
- R. Liam-Or, et al., Cellular uptake and in vivo distribution of mesenchymal-stem-cell-derived extracellular vesicles are protein corona dependent, Nat. Nanotechnol., 2024 DOI:10.1038/s41565-023-01585-y.
- P. Singh, et al., Removal and identification of external protein corona members from RBC-derived extracellular vesicles by surface manipulating antimicrobial peptides, J. Extracell. Biol., 2023, 2(3), e78 CrossRef CAS.
- A. Rai, B. Claridge, J. Lozano and D. W. Greening, The discovery of extracellular vesicles and their emergence as a next-generation therapy, Circ. Res., 2024, 135 DOI:10.1161/CIRCRESAHA.123.323054.
- B. Claridge, J. Lozano, Q. H. Poh and D. W. Greening, Development of extracellular vesicle therapeutics: Challenges, considerations, and opportunities, Front. Cell Dev. Biology, 2021, 9, 734720 CrossRef PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |