Relative biomembrane fusogenicities of the tumor-selective liposomes of RGDK- and CGKRK-lipopeptides†
Wahida
Rahaman
and
Arabinda
Chaudhuri
 *
*
Department of Chemical Sciences, Indian Institute of Science Education and Research Kolkata, Mohanpur, Nadia-741246, West Bengal, India. E-mail: arabinda.chaudhuri@iiserkol.ac.in
First published on 17th April 2024
Abstract
Cancer is the second leading cause of death globally after heart diseases. Currently used highly cytotoxic anti-cancer drugs not only kill cancer cells but also often kill non-cancerous healthy body cells, causing adverse side effects. Efforts are now being directed towards developing tumor-selective chemotherapy. Tumor/tumor endothelial cell selective peptide ligands are being covalently grafted onto the exo-surfaces of drug carriers such as liposomes, polymers, etc. A number of prior studies used conjugation of tumor/tumor endothelial cell-selective RGDK- or CGKRK-peptide ligands on the outer surfaces of liposomes, metal-based nanoparticles, single walled carbon nanotubes (SWNTs), etc. However, studies aimed at examining the relative cell membrane fusogenicities and the relative degrees of cellular uptake for the RGDK- and CGKRK-ligand-grafted nanometric drug carriers have not yet been undertaken. Herein, using the widely used liposomes of DOPC, DOPE, DOPS and cholesterol (45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, w/w ratio) as the model biomembranes and the fluorescence resonance energy transfer (FRET) assay for measuring membrane fusogenicities, we show that the liposomes of the RGDK-lipopeptide are more biomembrane fusogenic than the liposomes of the CGKRK-lipopeptide. Notably, such FRET assay-derived relative biomembrane fusogenicities of the liposomes of RGDK- and CGKRK-lipopeptides were found to be consistent with their relative degrees of cellular uptake in cultured cancer cells. The present findings open the door for undertaking in-depth in vivo studies aimed at evaluating the relative therapeutic potential of different nanocarriers of drugs/genes/siRNA having tumor-targeting RGDK- and CGKRK-peptides on their exo-surfaces.
15, w/w ratio) as the model biomembranes and the fluorescence resonance energy transfer (FRET) assay for measuring membrane fusogenicities, we show that the liposomes of the RGDK-lipopeptide are more biomembrane fusogenic than the liposomes of the CGKRK-lipopeptide. Notably, such FRET assay-derived relative biomembrane fusogenicities of the liposomes of RGDK- and CGKRK-lipopeptides were found to be consistent with their relative degrees of cellular uptake in cultured cancer cells. The present findings open the door for undertaking in-depth in vivo studies aimed at evaluating the relative therapeutic potential of different nanocarriers of drugs/genes/siRNA having tumor-targeting RGDK- and CGKRK-peptides on their exo-surfaces.
Introduction
Cells internalize macromolecules and therapeutic drug/gene-loaded nanometric particles majorly through endocytotic cellular uptake mechanisms in which a small part of the plasma membrane is utilized in engulfing the particles.1 Drugs/genes are delivered to target cells often using liposomes and polymer-based nanocarriers, and such nanocarrier-associated therapeutic payloads also enter cells majorly via endocytosis.2 Aimed at accomplishing tumor-selective delivery of anticancer drugs/genes/siRNAs, several receptors on the tumor cell surfaces are being exploited. For instance, integrins, αβ-heterodimeric transmembrane glycoprotein receptors,3 heparan sulphate receptors,4 folate receptors,5 and transferrin receptors6 overexpressed on several cancer cells and tumour endothelial cells (tumour vasculatures) are increasingly exploited for selective targeting of nanocarrier-bound drugs to tumours and tumour vasculatures. Three integrin receptors in particular (among the existing 26 integrin receptors), namely, αvβ3, αvβ5, and α5β1 integrins, have found widespread use.7–20 Among the high-affinity peptide ligands of these integrin receptors, RGDK ligands have shown significant therapeutic promise for selectively delivering anticancer drugs to tumors and tumor endothelial cells.7,14–20A phage display study21 identified another therapeutically promising heparin sulphate receptor-selective CGKRK-ligand which has also been widely exploited for targeting anticancer drugs/genes/siRNAs to tumors and tumor endothelial cells in both in vitro and in vivo settings.22–27 As mentioned above, both RGDK- and CGKRK-peptides have been widely used for tumor-selective chemotherapy. However, studies aimed at examining the relative biomembrane fusogenicities and the relative degrees of cellular uptake of these two tumor targeting peptide-grafted nanocarriers have not yet been undertaken. Herein, exploiting widely used liposomes of DOPC/DOPE/DOPS/cholesterol (45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, w/w) as model biomembranes and the fluorescence resonance energy transfer (FRET) assay for measuring biomembrane fusogenicity, we show that the liposomes of RGDK-lipopeptide/DOPC/cholesterol (1
15, w/w) as model biomembranes and the fluorescence resonance energy transfer (FRET) assay for measuring biomembrane fusogenicity, we show that the liposomes of RGDK-lipopeptide/DOPC/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 mole ratio) are more fusogenic than the liposomes of CGKRK-lipopeptide/DOPC/cholesterol (1
0.5 mole ratio) are more fusogenic than the liposomes of CGKRK-lipopeptide/DOPC/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 mole ratio). Notably, consistent with the observed higher biomembrane fusogenicity of the liposomes of the RGDK-lipopeptide (Fig. 1), the degree of cellular uptake for the liposomes of the RGDK-lipopeptide was found to be higher than that for the liposomes of the CGKRK-lipopeptide (Fig. 1) in four cultured animal cells. The findings reported herein open the door for undertaking in-depth in vivo studies aimed at evaluating the relative therapeutic potential of drug/gene/siRNA-loaded nanocarriers (including liposomes, polymers, polymeric micelles, carbon nanotubes, etc.) having tumor-targeting RGDK- and CGKRK-peptides on their exo-surfaces.
0.5 mole ratio). Notably, consistent with the observed higher biomembrane fusogenicity of the liposomes of the RGDK-lipopeptide (Fig. 1), the degree of cellular uptake for the liposomes of the RGDK-lipopeptide was found to be higher than that for the liposomes of the CGKRK-lipopeptide (Fig. 1) in four cultured animal cells. The findings reported herein open the door for undertaking in-depth in vivo studies aimed at evaluating the relative therapeutic potential of drug/gene/siRNA-loaded nanocarriers (including liposomes, polymers, polymeric micelles, carbon nanotubes, etc.) having tumor-targeting RGDK- and CGKRK-peptides on their exo-surfaces.
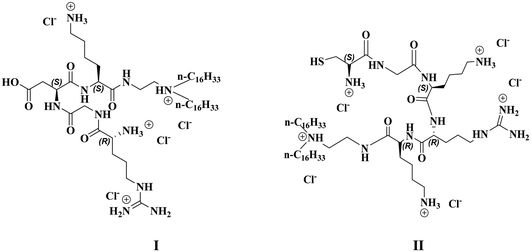 | ||
| Fig. 1 Chemical structures of the RGDK-lipopeptide (I) and the CGKRK-lipopeptide (II) used in the present study. | ||
Materials and methods
General reagents and methods
H-Lys(Boc)-2-Cl-Trtresin, Fmoc-Asp(O-tBu)-OH, Boc-Arg(pbf)-OH, Fmoc-Gly-OH, Fmoc-Arg(pbf)-OH, Boc-Cys(Trt)-OH, Amberlite IR 400 Cl− ion exchange resin, piperidine, DOPC, DOPS, DOPE, NBD-PE, triisopropylsilane and Rho-PE were purchased from Sigma Aldrich. Hexafluorophosphate azabenzotriazole tetramethyl uronium (HATU), 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), N,N-diisopropylethylamine (DIPEA), cholesterol, acetic acid (CH3COOH),1-(3-dimethylaminopropyl)-3-ethyl carbodiimide hydrochloride (EDC·HCl), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), sodium bicarbonate (NaHCO3), sodium sulfate anhydrous (Na2SO4), ammonium chloride (NH4Cl), Triton-X 100 and dimethyl sulfoxide (DMSO) were purchased from SRL. Hydroxybenzotriazole (HOBT) was purchased from Spectrochem. Dimethylformamide (DMF), dichloromethane (DCM), methanol (MeOH), trifluoroacetic acid (TFA) and chloroform (CHCl3) were purchased from Merck. Nuclease (DNase and RNase)-free water (H2O) was purchased from Invitrogen. MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) was purchased from Puregene. Cu-grids were purchased from Allied Scientific.Cells and culture media
RAW 264.7 (murine mouse macrophage), HEK-293, MDAMB-231, and B16F10 cells were purchased from ATCC. RPMI 1640, PBS, trypsin-EDTA, penicillin, streptomycin, FBS, DMEM, MEM and sodium pyruvate were purchased from Gibco. MEM (minimum essential medium) non-essential amino acids solution (100×) was purchased from STEMCELL Technologies. 96-well plates and confocal dishes were purchased from SPL Life Sciences.Synthesis
The synthesis of the RGDK-lipopeptide (I, Fig. 1) and the CGKRK-lipopeptide (II, Fig. 1) involved the following steps. The amphiphilic lipopeptides I and II were synthesized using the synthetic schemes (shown in Fig. S1 and Fig. S2,† respectively) as reported previously.25,28 >95% purity of the compounds I and II was confirmed by reversed phase analytical HPLC using two different mobile phases (100% methanol and 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v, methanol
5, v/v, methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water). ESI-MS was used to confirm the structure of the purified RGDK- and CGKRK-lipopeptides. Spectral details and HPLC profiles of the purified RGDK- and CGKRK-lipopeptides are provided in the ESI (Fig. S3–S10).†
water). ESI-MS was used to confirm the structure of the purified RGDK- and CGKRK-lipopeptides. Spectral details and HPLC profiles of the purified RGDK- and CGKRK-lipopeptides are provided in the ESI (Fig. S3–S10).†
ESI-MS data of the final RGDK-lipopeptide ( I ): [M + 1]+ = 965.9; [M/2 + 1]+ = 483.5.
ESI-MS data of the final CGKRK-lipopeptide ( II ): [M + 1]+ = 1081; [M/2 + 1]+ = 541.
Preparation of liposomes
The conventional thin film hydration method was followed to prepare two different liposomal formulations. The first one contained RGDK-lipopeptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC
DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol and second one contained CGKRK-lipopeptide
Chol and second one contained CGKRK-lipopeptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC
DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol using a 1
Chol using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 mol ratio. Briefly, the required volume of each lipid from their corresponding stock solutions (made in 3
0.5 mol ratio. Briefly, the required volume of each lipid from their corresponding stock solutions (made in 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v, chloroform
1, v/v, chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol) was taken in a glass vial to make 1 mL liposomal solutions (containing 1 mM total lipids). The organic solvents were removed using a gentle nitrogen flow and the resultant dry lipid film was kept under high vacuum for 6 h for complete removal of any trace of organic solvents. As previously described,28 the dried thin lipid films were allowed to swell overnight after adding nuclease-free water (1 mL) and vortexed. The resulting multi-lamellar liposomal solutions were sonicated (using a Ti-probe in an SKL-250D Ultrasonic Processor, Ningbo SjiaLab Equipment Company Ltd) in an ice bath for 20 min using 2 s pulse ON cycles, followed by 30 s of no sonication (pulse OFF cycles), during which any excess heat generated in the previous 2 s pulse ON cycle was dissipated in the surrounding ice bath. The solution was centrifuged at 11
methanol) was taken in a glass vial to make 1 mL liposomal solutions (containing 1 mM total lipids). The organic solvents were removed using a gentle nitrogen flow and the resultant dry lipid film was kept under high vacuum for 6 h for complete removal of any trace of organic solvents. As previously described,28 the dried thin lipid films were allowed to swell overnight after adding nuclease-free water (1 mL) and vortexed. The resulting multi-lamellar liposomal solutions were sonicated (using a Ti-probe in an SKL-250D Ultrasonic Processor, Ningbo SjiaLab Equipment Company Ltd) in an ice bath for 20 min using 2 s pulse ON cycles, followed by 30 s of no sonication (pulse OFF cycles), during which any excess heat generated in the previous 2 s pulse ON cycle was dissipated in the surrounding ice bath. The solution was centrifuged at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and the supernatant was extruded in a mini-extruder (Avanti Polar) using first a 0.2 μm size membrane filter and, thereafter, a 0.1 μm size membrane filter (each 20 times).
000 rpm for 10 min and the supernatant was extruded in a mini-extruder (Avanti Polar) using first a 0.2 μm size membrane filter and, thereafter, a 0.1 μm size membrane filter (each 20 times).
Hydrodynamic size and zeta potential measurements using dynamic light scattering
The hydrodynamic diameters and the zeta potential of the liposomes of RGDK -and CGKRK-lipopeptides were measured using the dynamic light scattering technique with a Malvern Nano ZS Zetasizer instrument equipped with a He–Ne laser (wavelength: 633 nm). 10 μL of each sample were diluted to 200 μL of water before measuring their sizes at room temperature.TEM sample preparation
First, 10 μL of 1 mM concentrated samples of the liposomes of RGDK- and CGKRK-lipopeptides were diluted to 200 μL with nuclease-free water, and a small amount of the diluted samples was drop-cast on a Cu-grid. Uranyl acetate was used to stain the grid once the samples had adhered to it. The Cu-grid was vacuumed for 8–10 h to ensure thorough drying of the samples. Finally, JEOL JEM-2100F microscopes were used to capture the pictures.FRET assay
A fluorescence resonance energy transfer (FRET) assay was used to determine the relative biomembrane fusogenecities of the liposomes of the CGKRK- and RGDK-lipopeptides, DOPC and cholesterol (co-lipids). The widely used model biomembrane consists of liposomes containing DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE
DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPS
DOPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol in a ratio of 45
Chol in a ratio of 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 (w/w) along with 1 mol% (with respect to total lipids) donor NBD-PE (excitation wavelength 485 nm) and acceptor Rho-PE (emission wavelength 595 nm). When this fluorophore-labelled model biomembrane was excited at 485 nm, fluorescence energy transfer occurred from NBD-PE to Rho-PE due to their spatial proximity and emission was observed at 595 nm. In a 1 mL cuvette, 500 μL of 1 mM liposomes of RGDK-lipopeptide I or CGKRK-lipopeptide II and 500 μL of the model biomembrane (containing both the donor and acceptor fluorophores) were mixed and incubated for 5 min. The percentage of membrane fusion was measured for 20 min in a Hitachi F-7000 spectrofluorometer using the following equation:
15 (w/w) along with 1 mol% (with respect to total lipids) donor NBD-PE (excitation wavelength 485 nm) and acceptor Rho-PE (emission wavelength 595 nm). When this fluorophore-labelled model biomembrane was excited at 485 nm, fluorescence energy transfer occurred from NBD-PE to Rho-PE due to their spatial proximity and emission was observed at 595 nm. In a 1 mL cuvette, 500 μL of 1 mM liposomes of RGDK-lipopeptide I or CGKRK-lipopeptide II and 500 μL of the model biomembrane (containing both the donor and acceptor fluorophores) were mixed and incubated for 5 min. The percentage of membrane fusion was measured for 20 min in a Hitachi F-7000 spectrofluorometer using the following equation:| % of membrane fusion = (F0 − Ft)/(F0 − Fα) × 100; |
MTT assay of the liposomes of the RGDK- and CGKRK-lipopeptide
HEK 293, RAW 264.7 and MDA-MB-231 cells (∼5 × 103 cell per well) were seeded in 96-well plates and cultured in complete MEM. Cells were kept in a CO2 incubator at 37 °C for 18 h. 1 mM stock liposomes of both RGDK- and CGKRK-lipopeptide were diluted with 100 μL of complete MEM with increasing liposome concentration from 0.1 to 50 μM. The diluted liposomes were added to the cells and incubated for 24 h. The medium was removed, and cells were washed with 100 μL of PBS. Then, 10 μL of MTT (final concentration – 0.5 mg mL−1) was added to each well, and the cells were incubated for 4 h in a CO2 incubator at 37 °C in the dark. Finally, MTT was discarded and formazan was dissolved in 100 μL of DMSO. Plates were kept under shaking conditions for 15 min in the dark and the absorbance was measured at 595 nm using a plate reader (BIO-RAD, iMark, Microplate Reader). The same amount of reagents (without cells) was added and used as a blank. Cells without any lipid were considered as untreated/control. The percentage of cell viability was calculated according to the equation below: Cell viability (%) = [A595(treated cells) − blank/A595(untreated cells) − blank] × 100.Measurements of the cellular uptake of the liposomes of RGDK- and CGKRK-lipopeptide at different time intervals in four different cell lines using an epifluorescence microscope
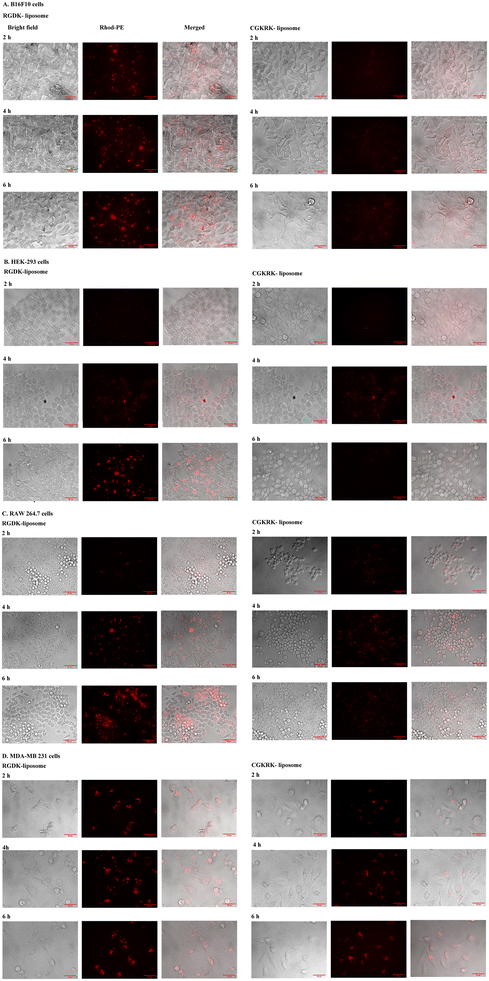
Cellular uptake studies were carried out by labelling the liposomes of RGDK- and CGKRK-lipopeptide with Rh-PE labelled dye (0.1 mol% with respect to lipopeptide) in four different cultured animal cell lines (two healthy cells HEK-293 and RAW-264.7 and two cancer cells B16F10 and MDAMB 231). Cells were seeded at a density of ∼104 in 96-well plates using 200 μL of complete media and kept in a CO2 incubator at 37 °C for 24 h. Next day, completely adhered cells were treated with 6.8 μL of Rho-PE (0.2 μM)-labelled liposomes of RGDK-lipopeptide (containing 0.2 mM DOPC, 0.2 mM RGDK-lipopeptide and 0.1 mM chol) and CGKRK-lipopeptide (containing 0.2 mM DOPC, 0.2 mM CGKRK-lipopeptide, and 0.1 mM cholesterol). The total volume of each well was made up to 200 μL with complete media. Cells were incubated with fluorescently labelled liposomes for 2 h, 4 h, and 6 h. Thereafter, the cell medium was removed, cells were washed with PBS solution and 200 μL of complete medium was added to the washed cells. Live cells were viewed under an epifluorescence microscope (Ix81, Olympus, Japan, equipped with a Cool Snap Myo-Photometrics camera).Measurements of the cellular uptake of the liposomes of RGDK- and CGKRK-lipopeptide in four different cell lines using a confocal microscope
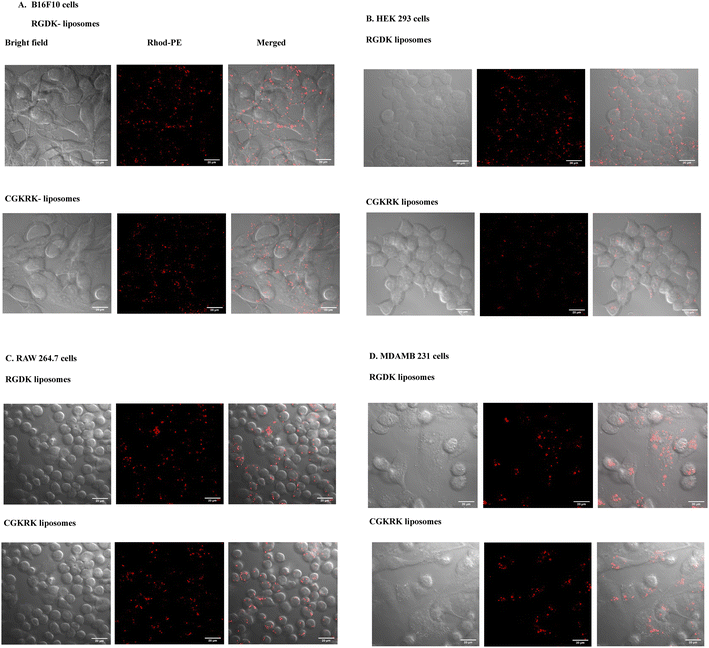
The cellular internalizations of Rh-PE labelled (0.1 mol% with respect to lipopeptide) liposomes of RGDK- and CGKRK-lipopeptide were also examined in HEK-293, RAW-264.7, B16F10 and MDAMB 231 using a confocal microscope, LSM 710 (Axio Observer Z1, Carl Zeiss). Cells were seeded at a density of ∼105 in a confocal dish using 300 μL of the complete medium and kept in a CO2 incubator at 37 °C for 24 h. Next day, completely adhered cells were treated with 6 μL of Rho-PE (0.2 μM) labelled liposomes of RGDK-lipopeptide (containing 0.2 mM DOPC, 0.2 mM RGDK-lipopeptide and 0.1 mM Chol) and CGKRK-lipopeptide (containing 0.2 mM DOPC, 0.2 mM CGKRK-lipopeptide, and 0.1 mM cholesterol). The total volume of each well was made up to 300 μL with complete medium. Cells were incubated with fluorescently labelled liposomes for 4 h. Thereafter, the cell medium was removed, cells were washed with PBS solution and 300 μL of the complete medium was added to the washed cells. Live cells were viewed under a confocal microscope, LSM 710 (Axio Observer Z1, Carl Zeiss).Results and discussion
Physicochemical properties of liposomal formulations
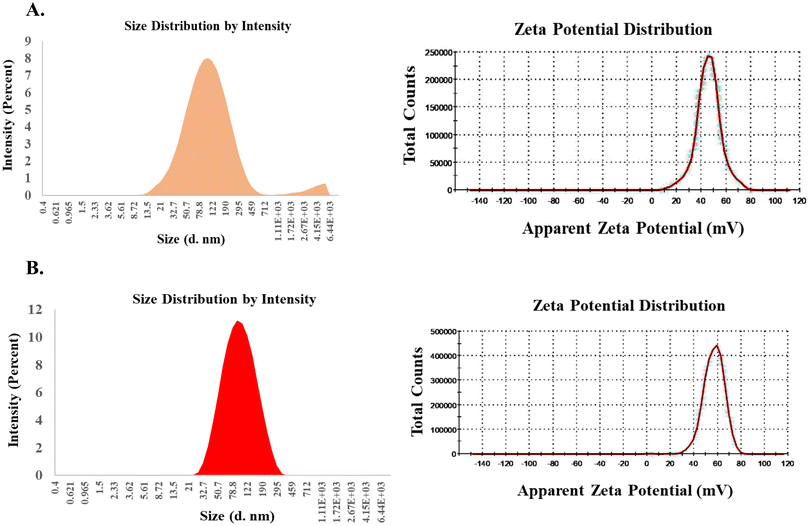
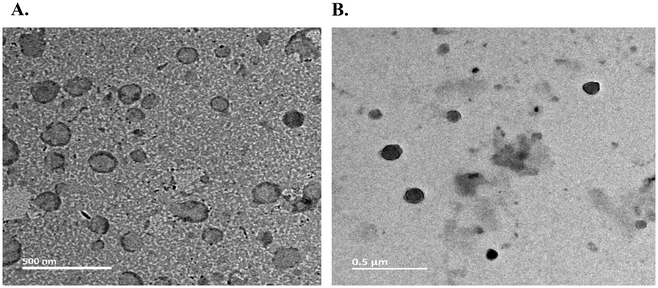
The hydrodynamic diameters and the surface potentials (ζ) of the liposomes of RGDK-lipopeptide I and CGKRK-lipopeptide II were measured using a Malvern Nano ZS Zetasizer instrument equipped with a He–Ne laser (wavelength 633 nm) using a previously reported protocol.28 The sizes and zeta potentials of the liposomes containing the RGDK-lipopeptide, DOPC (co-lipid) and cholesterol (co-lipid) and those containing the CGKRK-lipopeptide, DOPC (co-lipid) and cholesterol (co-lipid) were found to vary within the range 90–94 nm and 33–44 mV, respectively (Table 1 and Fig. 2). TEM images (Fig. 3, acquired using JEOL JEM-2100F microscopes) of both the liposomes were recorded as described previously.29| Liposomes | Hydrodynamic diameters (nm) | Zeta potentials (mV) | PDI |
|---|---|---|---|
| a Data are the mean SD (n = 3). | |||
Liposomes of the RGDK-lipopeptide, DOPC and Chol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5) mmol 0.5) mmol |
90.4 ± 6 | 33.2 ± 14 | 0.173 |
Liposomes of the CGKRK-lipopeptide, DOPC and Chol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5) mmol 0.5) mmol |
93.4 ± 5 | 43.8 ± 15 | 0.161 |
Measurement of biomembrane fusogenicity using the FRET assay
The widely used model biomembrane (liposomes of DOPC/DOPE/DOPS/Chol) was used in this FRET study as described previously.28,30 The findings summarized in Fig. 4 are consistent with higher biomembrane fusogenicities of the liposomes of RGDK-lipopeptide I than those of the liposomes of CGKRK-lipopeptide II. An important issue is worth mentioning here. Since the liposomes of the RGDK-lipopeptide were found to be more fusogenic with the model biomembrane than the liposomes of the CGKRK-lipopeptide, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 mole ratio of lipopeptide
0.5 mole ratio of lipopeptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC
DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol before fusion with model biomembranes is not likely to remain the same after fusion with the model membranes for both the liposomes.
cholesterol before fusion with model biomembranes is not likely to remain the same after fusion with the model membranes for both the liposomes.
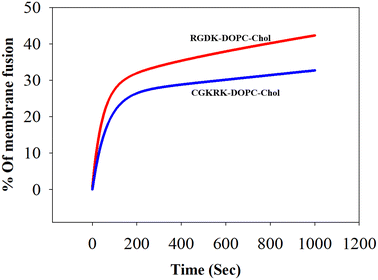 | ||
Fig. 4 Biomembrane fusogenicities of the liposomes of RGDK- and CGKRK-lipopeptides. Fusion was induced by adding the liposomes of RGDK-lipopeptide I (red) and CGKRK-lipopeptide II (blue) to the double fluorophore labelled model biomembrane, namely liposomes of DOPC/DOPE/DOPS/cholesterol (45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, w/w), as described in the text. The error bar profile is provided in the ESI (Fig. S11).† 15, w/w), as described in the text. The error bar profile is provided in the ESI (Fig. S11).† | ||
Cellular uptake study
The observed higher biomembrane fusogenicity of the liposomes of RGDK-lipopeptide I (Fig. 4) prompted us to next carry out cellular uptake experiments for 2 h, 4 h and 6 h in four different cultured animal cell lines (B16F10, HEK-293, RAW 264.7 and MDAMB 231) using epifluorescence microscopy and confocal microscopy as described previously.20,28 Notably, consistent with the findings in the FRET assay (Fig. 4), the degree of cellular uptake for the Rh-PE labeled liposomes of RGDK-lipopeptide I was observed to be higher than that for the liposomes of CGKRK-lipopeptides (Fig. 5). The degree of cellular internalization was higher for the liposomes of the RGDK-lipopeptide than that for the liposomes of the CGKRK-lipopeptide. The confocal microscopy images (Fig. 6) revealed the exact location of the Rh-PE dye in four different cell lines for both sets of liposomes.Compared to hormone receptor-positive normal breast cancers, triple-negative breast cancers (TNBCs) are typically more aggressive, more difficult to treat, and more likely to recur. Currently, surgery, chemotherapy, radiation therapy, and immunotherapy are used to treat TNBCs.31–35 The significantly high cellular uptake observed in triple-negative cancer cells (MDA-MB 231 cells) for the liposomes of RGDK-lipopeptide I indicates the possibility of their future use in combating hard-to-treat TNBCs.
Cytotoxicity assay
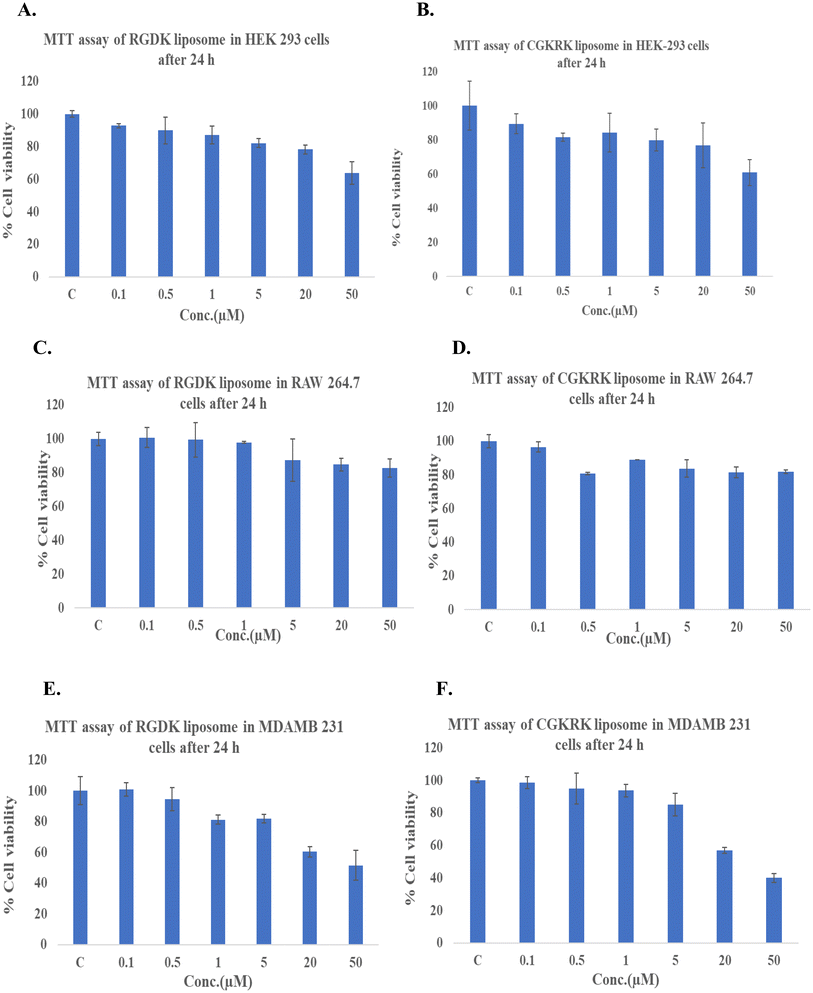
We measured the cellular toxicities of the presently described liposomal formulations in three different cell lines including HEK 293, RAW 264.7 and MDA-MB-231 cells using the MTT assay as reported earlier.28 Importantly, ∼76% HEK 293 cells were found to be viable upon incubating the cells for 24 h with the liposomes of RGDK- and CGKRK-lipopeptides containing 20 μM total lipid (Fig. 7A and B) and ∼80% of RAW 264.7 and MDA-MB-231 cells were viable after 24 hours of incubation with RGDK- and CGKRK-liposomes containing 50 μM total lipid and 5 μM total lipid (Fig. 7C–F).Many therapeutic drugs/genes/siRNAs, after endosomal cellular entry, need to be released from the endosomes into the cell cytoplasm before exhibiting their activities.36,37 To this end, fusogenic liposomes are widely used for cell tracking, nonviral gene transfer, and drug administration.38–40 The inclusion of such additional fusogenic lipid components in the presently described liposomal formulations of RGDK-lipopeptides may further enhance their biomembrane fusogenicities. The detrimental environment inside the endosomes leads to the degradation of the drugs/genes. Efforts are now being directed at designing a novel drug delivery strategy that can selectively deliver anticancer drugs/genes to the cell cytoplasm, avoiding endocytotic cellular uptake and subsequent degradation.41 An important point needs to be emphasized at this point of discussion. The present study showed that liposomes of RGDK-lipopeptides exhibit cellular uptake in both cancer and non-cancerous RAW264.7 cells. This finding clearly raises safety concerns regarding the use of integrin receptor-selective liposomes of RGDK-lipopeptides in cancer therapy. In antiangiogenic cancer therapy, the formation of new blood vessels from existing blood vessels (angiogenesis) is prevented by delivering anticancer drugs/genes selectively to tumor and tumor endothelial cells. Since the expression levels of integrin receptors in both tumor and tumor endothelial cells are remarkably high compared to their expression levels in healthy non-cancerous cells,42,43 under in vivo therapeutic settings, anticancer drugs associated with the integrin receptor-selective RGDK-lipopeptide are likely to be selectively delivered to both tumor and tumor endothelial cells in tumor tissues. For instance, prior studies showed tumor-selective accumulation of intraveneously administered doxorubicin encapsulated within the aqueous core of pegylated RGDK-lipopeptide.17 It needs to be investigated in the future whether grafting RGDK-ligands on the exo-surfaces of other nanocarriers of drugs/genes/siRNA (including polymeric micelles, carbon nanotubes, etc.) enhances their therapeutic outcome in tumor-selective chemotherapy. Studies aimed at examining the relative efficacies of RGDK-coated liposomal and polymeric drug carriers in killing cancer cells are worth undertaking.
Conclusions
In summary, by using widely used liposomes of DOPC/DOPE/DOPS/cholesterol (45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, w/w) as model biomembranes and the fluorescence resonance energy transfer (FRET) assay for measuring biomembrane fusogenicity, we have shown that the liposomes of RGDK-lipopeptide/DOPC/cholesterol (1
15, w/w) as model biomembranes and the fluorescence resonance energy transfer (FRET) assay for measuring biomembrane fusogenicity, we have shown that the liposomes of RGDK-lipopeptide/DOPC/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 mole ratio) are more fusogenic than the liposomes of CGKRK-lipopeptide/DOPC/cholesterol (1
0.5 mole ratio) are more fusogenic than the liposomes of CGKRK-lipopeptide/DOPC/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 mole ratio). Consistent with the observed higher biomembrane fusogenicity of the liposomes of the RGDK-lipopeptide, the degree of cellular uptake for the liposomes of the RGDK-lipopeptide was found to be higher than that for the liposomes of the CGKRK-lipopeptide in four cultured animal cell lines. The findings reported herein open the door for undertaking in-depth in vivo studies aimed at evaluating the relative therapeutic potential of drug/gene/siRNA-loaded nanocarriers (including liposomes, polymeric micelles, self-assembled lipid nanoparticles, carbon nanotubes, etc.) having tumor-targeting RGDK-peptides and CGKRK-lipopeptides on their exo-surfaces.
0.5 mole ratio). Consistent with the observed higher biomembrane fusogenicity of the liposomes of the RGDK-lipopeptide, the degree of cellular uptake for the liposomes of the RGDK-lipopeptide was found to be higher than that for the liposomes of the CGKRK-lipopeptide in four cultured animal cell lines. The findings reported herein open the door for undertaking in-depth in vivo studies aimed at evaluating the relative therapeutic potential of drug/gene/siRNA-loaded nanocarriers (including liposomes, polymeric micelles, self-assembled lipid nanoparticles, carbon nanotubes, etc.) having tumor-targeting RGDK-peptides and CGKRK-lipopeptides on their exo-surfaces.
Abbreviations
| DLS | Dynamic light scattering |
| RPMI | Roswell Park Memorial Institute |
| DMEM | Dulbecco's modified Eagle's medium |
| MEM | Minimum essential medium |
| PBS | Phosphate-buffered saline |
| FBS | Fetal bovine serum |
| HEK-293 | Human embryonic kidney cell line |
| B16F10 | Mouse melanoma cell line |
| nm | Nanometre |
| Fmoc | Fluorenyl methoxycarbonyl |
| DOPE | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine |
| DOPC | 1,2-Dioleoyl-3-sn-phosphatidylcholine |
| DOPS | 1,2-Dioleoyl-sn-glycero-3-phospho-L-serine |
| FRET | Förster resonance energy transfer |
| NBD-PE | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (ammonium salt) |
| Rhod-PE | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) |
| TEM | Transmission electron microscopy |
| ESI-MS | Electrospray ionization mass spectrometry |
| HRMS | High-resolution mass spectrometry |
| HPLC | High-performance liquid chromatography |
| MTT | (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide) |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. C. thanks the Indian Council of Medical Research (ICMR), Government of India, for awarding him an ICMR Emeritus Scientist position. W. R. thanks the Indian Institute of Science Education and Research Kolkata for an Institute Fellowship and infrastructural support to pursue doctoral research. We thank the Director, Indian Institute of Science Education and Research Kolkata for funding this research. We sincerely thank Dr Pradip K. Tarafdar and Dr Avijit Sardar for helping us with the FRET experiments in their laboratory. We thank Mr. Sumanta Moi, Mr. Uday Bhanu Sen, Ms. Tania Roy and Mr. Ritabrata Ghosh for helping us in executing TEM study, HRMS, ESI-MS and epifluorescence microscopy based cellular uptake experiments, respectively.References
- S. D. Conner and S. L. Schmid, Nature, 2003, 422, 37–44 CrossRef CAS PubMed.
- I. M. S. Degors, C. Wang, Z. U. Rehman and I. S. Zuhorn, Acc. Chem. Res., 2019, 52, 1750–1760 CrossRef CAS PubMed.
- D. J. Worm, S. Els-Heindl and A. G. Beck-Sickinger, Pept. Sci., 2020, 112, e24171 CrossRef CAS.
- L. Lv, et al. , Front. Pharmacol., 2020, 11, 558 CrossRef CAS PubMed.
- Y. H. Dinakar, A. Karole, S. Parvez, V. Jain and S. L. Mudavath, Biochim. Biophys. Acta, Gen. Subj., 2023, 1867, 130396 CrossRef CAS PubMed.
- S. Mojarad-Jabali, et al. , Expert Opin. Drug Delivery, 2022, 19, 685–705 CrossRef CAS PubMed.
- G. Mondal, S. Barui and A. Chaudhuri, Biomaterials, 2013, 34, 6249–6260 CrossRef CAS PubMed.
- E. Ruoslahti, Adv. Mater., 2012, 24, 3747–3756 CrossRef CAS PubMed.
- M. A. Dechantsreiter, et al. , J. Med. Chem., 1999, 42, 3033–3040 CrossRef CAS PubMed.
- D. Heckmann, A. Meyer, L. Marinelli, G. Zahn, R. Stragies and H. Kessle, Angew. Chem., Int. Ed., 2007, 46, 3571–3574 CrossRef PubMed.
- R. Haubner, R. Gratias, B. Diefenbach, S. L. Goodman, A. Jonczyk and H. Kessler, J. Am. Chem. Soc., 1996, 118, 7461–7472 CrossRef CAS.
- K. Chen and X. Chen, Theranostics, 2011, 1, 189–200 CrossRef CAS PubMed.
- K. N. Sugahara, et al. , Cancer Cell, 2009, 16, 510–520 CrossRef CAS PubMed.
- S. Samanta, R. Sistla and A. Chaudhuri, Biomaterials, 2010, 31, 1787–1797 CrossRef CAS PubMed.
- D. Pramanik, et al. , J. Med. Chem., 2008, 51, 7298–7302 CrossRef CAS PubMed.
- G. Mondal, S. Barui, S. Saha and A. Chaudhuri, J. Controlled Release, 2013, 172, 832–840 CrossRef CAS PubMed.
- S. Barui, S. Saha, G. Mondal, S. Haseena and A. Chaudhuri, Biomaterials, 2014, 35, 1643–1656 CrossRef CAS PubMed.
- P. Majumder, S. Bhunia, J. Bhattacharyya and A. Chaudhuri, J. Controlled Release, 2014, 180, 100–108 CrossRef CAS PubMed.
- K. Das, S. Nimushakavi, A. Chaudhuri and P. K. Das, ChemMedChem, 2017, 12, 738–750 CrossRef CAS PubMed.
- V. Vangala, N. V. Nimmu, S. Khalid, M. Kuncha, R. Sistla, R. Banerjee and A. Chaudhuri, Mol. Pharm., 2020, 17, 1859–1874 CrossRef CAS PubMed.
- J. A. Hoffman, et al. , Cancer Cell, 2003, 4, 383–391 CrossRef CAS PubMed.
- J. I. Griffin, et al. , Nanomedicine, 2017, 13, 1925–1932 CrossRef CAS PubMed.
- J. Zhang, et al. , J. Biomed. Nanotechnol., 2019, 15, 1546–1555 CrossRef CAS PubMed.
- S. K. Gulla, et al. , ACS Omega, 2018, 3, 8663–8676 CrossRef CAS PubMed.
- S. Barui, S. Saha, Y. Venu, G. K. Moku and A. Chaudhuri, Biomater. Sci., 2023, 11, 6135 RSC.
- L. Treps, J. Pathol., 2018, 246, 3–6 CrossRef CAS PubMed.
- L. Lv, et al. , Mol. Pharmaceutics, 2016, 13, 3506–3517 CrossRef CAS PubMed.
- W. Rahaman and A. Chaudhuri, Biomed. Mater., 2024, 19, 015004 CrossRef PubMed.
- B. Hazra, et al. , Langmuir, 2023, 39(48), 17031–17042 CrossRef CAS PubMed.
- M. Rajesh, J. Sen, M. Srujan, K. Mukherjee, B. Sreedhar and A. Chaudhuri, J. Am. Chem. Soc., 2007, 129, 11408–11420 CrossRef CAS PubMed.
- Y. Li, et al. , J. Hematol. Oncol., 2022, 15, 121 CrossRef CAS PubMed.
- G. Bianchini, C. D. Angelis, L. Licata and L. Gianni, Nat. Rev. Clin. Oncol., 2022, 19, 91–113 CrossRef CAS PubMed.
- H. A. Wahba and H. A. El-Hadaad, Cancer Biol. Med., 2015, 12, 106–116 CAS.
- O. Gulz, et al. , Ann. Oncol., 2009, 20, 1913–1927 CrossRef PubMed.
- R. A. Leon-Ferre and M. P. Goetz, Br. Med. J., 2023, 381, e071674 CrossRef PubMed.
- A. Wittrup, A. Ai, X. Liu, P. Hamar, R. Trifonova, K. Charisse, M. Manoharan, T. Kirchhausen and J. Lieberman, Nat. Biotechnol., 2015, 33, 870–876 CrossRef CAS PubMed.
- S. Senapati, A. K. Mahanta, S. Kumar and P. Maiti, Signal Transduction Targeted Ther., 2018, 3, 7 CrossRef PubMed.
- J. Kunisawa, et al. , J. Controlled Release, 2005, 105, 344–353 CrossRef CAS PubMed.
- M. Hoffmann, et al. , Pharmaceutics, 2023, 15, 1210 CrossRef CAS PubMed.
- S. Kube, et al. , Langmuir, 2017, 33, 1051–1059 CrossRef CAS PubMed.
- J. Yang, A. Bahreman, G. Daudey, J. Bussmann, R. C. L. Olsthoorn and A. Kros, ACS Cent. Sci., 2016, 2, 621–630 CrossRef CAS PubMed.
- S. M. Weis and D. A. Cheresh, Nat. Med., 2011, 17, 1359–1370 CrossRef CAS PubMed.
- J. S. Desgrosellier and D. A. Cheresh, Nat. Rev. Cancer, 2010, 10, 9–22 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr00450g |
| This journal is © The Royal Society of Chemistry 2024 |