Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bio-gel nanoarchitectonics in tissue engineering
Jingwen
Song
 *a,
Wenyan
Lyu
*a,
Wenyan
Lyu
 bc,
Kohsaku
Kawakami
bc,
Kohsaku
Kawakami
 ad and
Katsuhiko
Ariga
ad and
Katsuhiko
Ariga
 *bc
*bc
aResearch Center for Functional Materials, National Institute for Materials Science (NIMS), 1-1 Namiki, Tsukuba 305-0044, Ibaraki, Japan. E-mail: SONG.Jingwen@nims.go.jp
bGraduate School of Frontier Sciences, The University of Tokyo, 5-1-5 Kashiwa-no-ha, Kashiwa 277-8561, Japan
cResearch Center for Materials Nanoarchitectonics, National Institute for Materials Science (NIMS), 1-1 Namiki, Tsukuba 305-0044, Japan. E-mail: ARIGA.Katsuhiko@nims.go.jp
dGraduate School of Pure and Applied Sciences, University of Tsukuba, 1-1-1 Tennodai, Tsukuba 305-8577, Ibaraki, Japan
First published on 13th June 2024
Abstract
Given the creation of materials based on nanoscale science, nanotechnology must be combined with other disciplines. This role is played by the new concept of nanoarchitectonics, the process of constructing functional materials from nanocomponents. Nanoarchitectonics may be highly compatible with applications in biological systems. Conversely, it would be meaningful to consider nanoarchitectonics research oriented toward biological applications with a focus on materials systems. Perhaps, hydrogels are promising as a model medium to realize nanoarchitectonics in biofunctional materials science. In this review, we will provide an overview of some of the defined targets, especially for tissue engineering. Specifically, we will discuss (i) hydrogel bio-inks for 3D bioprinting, (ii) dynamic hydrogels as an artificial extracellular matrix (ECM), and (iii) topographical hydrogels for tissue organization. Based on these backgrounds and conceptual evolution, the construction strategies and functions of bio-gel nanoarchitectonics in medical applications and tissue engineering will be discussed.
1. Introduction
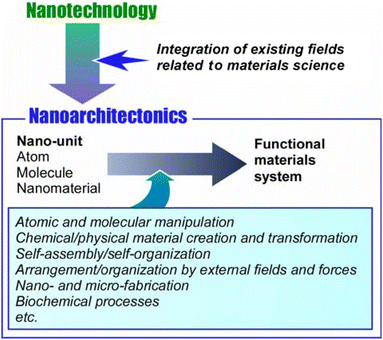
The development of mankind has been accompanied by the development of available functional materials. As human social life has become larger and more diverse, social demands have increased dramatically. For example, functional materials have been developed to confront many problems such as energy production,1 energy storage,2 various sensing,3 environmental protection,4 carbon neutrality,5 device development,6 drug delivery,7 regenerative medicine,8 pathogen control,9 and cancer treatment.10 Most of them do not rely on the inherent performance of the material itself. The key to the expression of advanced functions is the control of sophisticated structures that extend to the nanoscale. The development of the concept of nanotechnology has clarified this trend. It has made it possible to observe11 and manipulate12 structures at the atomic and molecular levels. Nanotechnology has allowed us to elucidate scientific phenomena and principles at the nanoscale. Given the creation of materials based on nanoscale science, nanotechnology must be combined with other disciplines. This role is played by the new concept of nanoarchitectonics,13 the process of constructing functional materials from nanocomponents. It can be called a post-nanotechnology concept.14 The ultimate goal of nanoarchitectonics is to construct functional materials systems with many functional units, such as biological systems.15While Richard Feynman proposed nanotechnology in the mid-20th century,16 Masakazu Aono initiated nanoarchitectonics at the beginning of the 21st century.17 Nanoarchitectonics combines various processes that have been mainly formulated in the field of materials science, with the aim of architecting functional materials systems from nano-units such as atoms and molecules (Fig. 1).18 It selects and combines atomic and molecular manipulation, chemical/physical material creation and transformation, self-assembly/self-organization, arrangement/organization by external fields and forces, nano- and micro-fabrication, biochemical processes, etc. to architect functional materials.19 Thus, unlike self-assembly,20 which is based on simple equilibrium, nanoarchitectonics is suited to creating asymmetric and hierarchical materials structures.21 Nanoscale processes are subject to uncertainties such as thermal fluctuations, stochastic distributions, and quantum effects. Therefore, functional materials systems are built in such a way that effects are harmonized.22 The form in which functional molecules are organized hierarchically and the mode in which they work symbiotically with thermal fluctuations is similar to those of biofunctional systems.23 Therefore, the ultimate goal of nanoarchitectonics is to construct advanced functional systems like biological systems and to architect materials systems that interact with biofunctional systems. For example, exploration of functional systems based on bio-gels would be a good research target.
Nanoarchitectonics is a methodology for architecting functional materials systems from atoms and molecules. Since all materials are in principle made of atoms and molecules, the methodology of nanoarchitectonics applies to all materials. It is an integrative concept that can be likened to the theory of everything in physics24 and can be called the method for everything in materials science.25 The endeavor of nanoarchitectonics will provide new insights and deeper thinkings in the field of materials chemistry in general. In fact, research studies advocating nanoarchitectonics are being conducted in a wide range of fields, not only in fields related to chemistry26 and physics,27 but also many related to biology and bio-chemistry.28 Nanoarchitectonics is expected to contribute to many areas, including those related to basic bio-chemistry,29 biosensing,30 drug delivery,31 cell control,32 and therapeutic areas.33 Nanoarchitectonics may be highly compatible with applications in biological systems. Conversely, it would be meaningful to consider nanoarchitectonics research oriented toward biological applications with a focus on materials systems.
The methodology of nanoarchitectonics is not about launching an entirely new concept. Rather, it is an integration of existing fields. Therefore, there are many examples of research that have characteristics of nanoarchitectonics even if they do not advocate nanoarchitectonics. From this perspective, we will consider how conventional technologies can contribute to nanoarchitectonics. The point is to assemble rational three-dimensional (3D) structures from nano-units such as atoms, molecules, and ions. One methodology is to organize two-dimensional (2D) structures and then assemble them into an organized 3D functional structure.34 Functional integration in biological systems is the rational integration of functional molecules based on various membrane structures.35 Therefore, organizing functional molecules intrinsically into 2D systems such as thin films may be one of the best strategies of nanoarchitectonics for creating highly functional materials. On this basis, building 3D structures is a suitable way to create materials for bio-applications. Typical methodologies are the Langmuir–Blodgett (LB) method36 and layer-by-layer (LbL) assembly.37 These methods are rational methods for building functional structures from two to three dimensions, but they only synthesize structures at the thin-film level. They are not always suitable as methods for building macroscopic materials.
A methodology for creating well-defined structures in two or three dimensions from molecules or ions is the synthesis of metal–organic frameworks (MOFs)38 and covalent organic frameworks (COFs).39 In this case, crystal structures with precise pore structures are formed. 3D precise structures can be made, but with a few exceptions,40 most are rigid structures. Also, large structures at the level of animal experiments are not always possible. In addition, there are many other methodologies to create 3D structures by supramolecular assembly. Among them, gel creation is a methodology that incorporates a large amount of solvent molecules to form macroscopic structures using only a small amount of constituent molecules.41 The characteristic feature of this method is that it produces very soft structures due to the large amount of solvent. Hydrogels in which the solvent material is water molecules are particularly suitable for biological applications. When considering biological applications through 3D nanoarchitectonics, it is useful to consider gels as target materials.
First, a brief look at some recent examples of gel research shows that gels, which are already widely studied materials, are also being investigated for controlling fundamental physical properties, such as mechanical properties are controlled through structural design, while the order and disorder characteristics are regulated using physicochemical procedures.42 This indicates that there remains ample room for nanoarchitectonics to contribute to the field of gels as well. Mayumi, Ito, and co-workers are investigating gels with sliding ring structures.43 Recently, they found that moderate cross-linking of poly(ethylene glycol) hydrogels led to parallel orientation of the chains during elongation and rapid and reversible strain-induced crystallization.44 The resulting toughness was an order of magnitude greater than that of covalently crosslinked poly(ethylene glycol) homogeneous gels. Sekine and Nankawa examined carboxymethylcellulose nanofiber hydrogels formed by solid–liquid phase separation.45 Kubota discussed supramolecular-polymer composite hydrogels as multi-network hydrogels in his recent review.46 The design of this composite hydrogel can rationally integrate the stimulus-response of the supramolecular gel with the stiffness of the polymeric gel. Yamanaka and co-workers investigated the adsorption of silica, polystyrene, and titania particles onto polyacrylamide and polydimethylacrylamide hydrogels.47 The van der Waals force was again found to be a strong enough driving force for particles absorbed on the surface of polymer hydrogels in water. Supramolecular polymer-based hydrogels typically exhibit highly ordered structures, which are stable and in a thermodynamic equilibrium state. Recently, Xing, Yan, and their co-workers prepared long-range disordered biomolecular glasses using amino acid and peptide nanoarchitectonics. These glasses demonstrate biocompatibility, biodegradability, and biorecyclability, surpassing the properties of currently used commercial glasses and plastic materials.48
In addition to the basic studies of physical properties, several advanced applications have been reported. Li, Gao, and co-workers discussed in a recent review flexible sensors based on biohydrogels of bacterial cellulose as natural biomass.49 Potential applications include human–machine interfaces, wearable medicine, and bionic intelligent robotics. Masud, Hossain, Kaneti, and co-workers developed κ-carrageenan hydrogel-coated mesoporous gold electrodes.50 These were useful for the chronocoulometric detection of microRNA. Lu, Gu, and co-workers developed a conductive polymer hydrogel strain sensor that exhibited both extreme strain and negligible hysteresis.51 It was created by a micro-phase semi-separated network design of the polymer and a fabrication technique that combined 3D printing and continuous freeze–thawing. The technology could be directed toward stretchable electronic skin and intelligent robotic systems. Hydrogels are ideal materials for human–machine interfaces because of their mechanical and chemical similarity with biological tissues. A recent review reported by Yuk et al. provided a comprehensive discussion of functional modes, design principles, and future applications of hydrogel interfaces for human–machine integration.52 Kai, Yu, Huang, and co-workers also discussed the correlation between hydrogel properties and device performances in their recent review.53 This is to provide a perspective on current challenges and future directions for the development of flexible electronics using environmentally responsive hydrogels.
For example, properties such as stiffness, pore size, viscoelasticity, nano- and microarchitecture, degradability, ligand presentation, and stimulus responsiveness can be widely modulated in hydrogels. Therefore, applications in the biomedical field are quite promising. The review by Cao, Zhang, and co-workers discussed the rational structural and functional design of hydrogels utilizing materials engineering to influence cell signaling cascades and fates.54 Cheng, Yue, and co-workers proposed an antibiotic-free strategy to combat implant-related infections by using biodegradable and cytocompatible hydrogel coatings.55 Duan, Guo, and co-workers developed a photo-crosslinked multifunctional antimicrobial adhesive hemostatic hydrogel dressing based on polyethylene glycol monomethyl ether modified glycidyl methacrylate functionalized chitosan, methacrylamide dopamine, and zinc ions.56 This is intended to kill drug-resistant bacteria and promote wound healing. Guo and co-workers developed a self-healing hydrogel based on quaternized chitosan, oxidized dextran, tobramycin, and polydopamine-coated polypyrrole nanowires, with good electrical conductivity and antioxidant activity.57 This is aimed at burn wound repairment. Qi, Shen, Hu, and co-workers developed a hydrogel with melanin nanoparticles loaded on a polysaccharide matrix composed of biguanide chitosan and oxidized β-glucan for efficient healing of bacterially infected diabetic wounds.58 This wound dressing material could be a new option for the treatment of diabetic wounds. Fu, Zhu, Fan, and co-workers developed a gel that is conducive to diabetic chronic wounds.59 They encapsulated gelatin microspheres containing insulin and celecoxib. The gel was prepared using phenylborate-grafted polyvinyl alcohol and chitosan. Showing dual-response temperature-sensitive shape self-adaptation to glucose and matrix metalloproteinase-9, Zhao, Yang, and co-workers hybridized collagen-based natural hydrogels with protocatechuic acid aldehyde for their ability to regulate macrophage heterogeneity.60 Useful for promoting angiogenesis and diabetic wound healing, He, Shen, and co-workers developed a novel glycyrrhizic acid-based hybrid hydrogel dressing with intrinsic immunomodulatory properties.61 This hybrid hydrogel, which also focuses on promoting rapid healing of diabetic wounds, is composed of an interpenetrating polymer network with excellent injectability and mechanical strength. Zhu, Fan, and co-workers developed an injectable hydrogel based on Ti3C2 MXene nanosheets coated with hyaluronic acid-grafted-dopamine and polydopamine.62 This could be applied to diabetic wound healing in combination with mild photothermal stimulation. Guo and co-workers designed a cross-linked multifunctional adhesive hydrogel based on sodium alginate, gelatin, and protocatechuic aldehyde, with ferric ions added.63 The developed reversible adhesive hydrogel dressing can be served as a versatile tissue sealant.
As described above, gels, especially hydrogels, have been studied in many fields. They can be used for various purposes because they can incorporate multiple components of nanoarchitectonics. They have many advantages, such as the ability to freely control their mechanical properties through solvent content and internal nano- and micro-structures. As can be seen from the examples, many efforts are being made for practical applications in the biomedical field. Perhaps, hydrogels are promising as a model medium to realize nanoarchitectonics in biofunctional materials science. In this review, we will provide a more detailed overview of some of the more narrowly defined targets, especially for tissue engineering, and look at the utility of hydrogel design and synthesis (nanoarchitectonics from molecules and polymers). Specifically, we will discuss (i) hydrogel bio-inks for 3D bioprinting, (ii) dynamic hydrogels as an artificial extracellular matrix (ECM), and (iii) topographical hydrogels for tissue organization. In detail, hydrogel bio-inks stand out for the rheological properties of initial extrudability and printability as inks, as well as their self-supporting and shape maintenance abilities after being used to construct hydrogel scaffolds. Additionally, by regulating the printing formulation or programs, hydrogel bio-inks loaded with cells or growth factors of interest can be readily customized to accomplish the spatiotemporal distribution which has great potential for artificial 3D tissues or organs, etc. In dynamic hydrogels, the emphasis shifts to mechanical properties that dynamically respond to environmental stimuli such as pH, temperature, light, or biochemical signals. Dynamic hydrogels undergo reversible changes in stiffness or degradation rates to provide a supportive matrix that can remodel and adapt to cell activity, facilitating cell migration, proliferation, and differentiation. Topographical hydrogels rely on surface patterning and nano/micro-scale features to influence cell behaviors. They have precise spatial cues that mimic the native tissue microenvironment and guide cell adhesion, alignment, and organization. Based on these backgrounds and conceptual evolutions, the construction strategies and functions of bio-gel nanoarchitectonics in medical applications and tissue engineering will be discussed.
2. Hydrogel bio-inks for 3D bioprinting
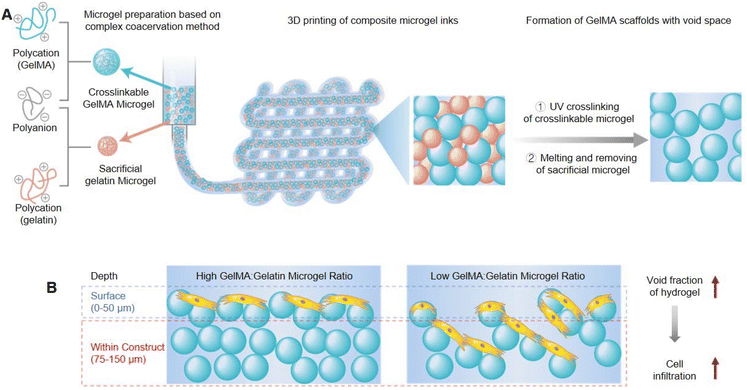
3D bioprinting technology is expected to achieve scalable biofabrication of structurally complex and functionally powerful tissue simulations for the study and amelioration of human diseases. Bio-ink materials for 3D printing must present two main roles: first, as raw materials, they can be used to manufacture structured constructs and second, as downstream cell induction niches, they can support cell growth, proliferation, and function. Hydrogels are ideal bio-ink materials that can maintain high cell viability and can also combine with traditional biological growth factors strategies to guide cell phenotype and fate.Conventional hydrogels exhibit a homogeneous structure, which as bio-inks for 3D printing will restrict cell growth and mobility as well as impede the diffusion of oxygen and nutrients, thus hindering cell viability, infiltration, and proliferation. Seymour et al. constructed microgel-based composite granular 3D printing inks composed of UV-cross-linkable gelatin methacryloyl (GelMA) microgels and sacrificial gelatin microgels (Fig. 2A).64 After UV cross-linking and heat melting, crosslinked GelMA scaffolds remained, but gelatin was removed and left behind continuous void space. The void fraction could be readily controlled by the proportion of the gelatin microgel within total composite inks. Experiments of the rheological characteristics assessed that the total concentration of microgels impacted the printability and shape maintenance, which were independent of the ratio of GelMA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) gelatin microgel inks, so the printability was decoupled from the void fraction. Furthermore, it also demonstrated that the interstitial void space of printed scaffolds was beneficial for human umbilical vein endothelial cell (HUVEC) growth and infiltration, which was positively associated with the void fraction (Fig. 2B). Therefore, this work could provide a new insights into bioprinting and tissue engineering applications through fabricating microporous GelMA microgel inks with advanced printability and shape maintenance.
gelatin microgel inks, so the printability was decoupled from the void fraction. Furthermore, it also demonstrated that the interstitial void space of printed scaffolds was beneficial for human umbilical vein endothelial cell (HUVEC) growth and infiltration, which was positively associated with the void fraction (Fig. 2B). Therefore, this work could provide a new insights into bioprinting and tissue engineering applications through fabricating microporous GelMA microgel inks with advanced printability and shape maintenance.
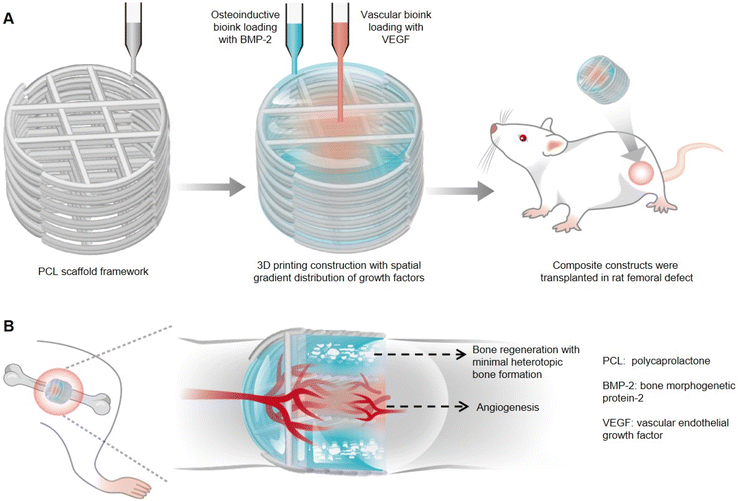
Therapeutic growth factors delivery often requires higher than normal doses, which can cause unwanted side effects. Freeman et al. established a novel 3D printed composite scaffold with distinct spatiotemporally distributed growth factors, which could be used as an implant for angiogenesis and bone defect healing.65 Two kinds of alginate/methylcellulose-based bioinks respectively containing vascular endothelial growth factor (VEGF) and bone morphogenetic protein-2 (BMP-2) were incorporated to fabricate the composite printed construct to achieve the above-mentioned tissue regeneration (Fig. 3A). It had been demonstrated that the spatial distribution of VEGF with decreasing load from the center to periphery within the implant was more conducive to vascular infiltration than the implant loading with homogeneously distributed VEGF, which could be credited to the cell migration chemotactic effect. Moreover, peripherally distributed BMP-2 with a slow-release profile, achieved by the addition of nanoparticles LAPONITE® due to electrostatic attraction, was assessed as more favorable for new bone formation than BMP-2 with fast release in implants. Furthermore, the whole was greater than the sum of the parts, and thereby the composite growth factor delivery system with both VEGF and BMP-2 was also applied to bone defect healing. It had been proved that the composite implant could not only enhance angiogenesis more significantly compared to the VEGF gradient-only loaded hydrogel but also promote bone regeneration and maturation with little heterotopic bone formation (Fig. 3B), which was mainly attributed to appropriate growth factor release kinetics and non-supraphysiological dosages. Therefore, the composite growth factor loaded system displayed in this work provides some new inspirations in biohydrogel designs and precisely controlled tissue regeneration, such as the spatiotemporal synergetic technique and controlled release strategies of multiple growth factors under the auxiliary of nanoparticles.
In addition to the above examples, there are diverse applications of hydrogels for 3D bioprinting. Properties such as biocompatibility with shear viscosity reduction, adequate yield strength, and fast self-healing are desired in hydrogel inks for 3D bioprinting. The searches of gel structures for this purpose are underway from various viewpoints. Zhang et al. developed novel self-healing pre-crosslinked hydrogel microparticles of chitosan methacrylate/poly(vinyl alcohol) hybrid hydrogels.66 Using this ink, they directly printed a series of biomimetic constructs with very high aspect ratios and delicate microstructures. It was possible to fabricate constructs easily and versatilely with excellent shape fidelity at organ-related scales, such as blood vessels, human ears, and rat femurs. Bioprinting by digital light processing (DLP) is advantageous for the biofabrication of structurally complex tissues. Wang et al. used a method of selective enzymatic digestion of hyaluronan methacrylate for DLP bioprinting.67 The molecular cleavage approach provided a tissue-matched structure without loss of structural complexity and fidelity. With this method, stem cell-derived NGN2-accelerated progenitor cells (SNaPs) differentiated into neurons and astrocytes. They exhibited strong electrophysiological and other brain-like properties. With further optimization, the molecular cleavage strategy was expected to be applied to tissue engineering, which has been difficult to realize.
Kim et al. used bioink made from silk fibroin for DLP-3D bioprinting in tissue engineering applications.68 Mechanical and rheological properties could be adjusted by varying the content of the silk fibroin-based bioink. The bioink allowed the construction of very complex organ structures such as the heart, blood vessels, brain, trachea, and ears with excellent structural stability and reliable biocompatibility. It was particularly noteworthy that this approach only used naturally derived polymers. Recently, four-dimensional (4D) printing has emerged as a next generation biofabrication technology. Kim et al. reported a 4D bioprinting system based on DLP and a photocurable silk fibroin hydrogel, containing two or more cell types.69 Using this 4D bioprinting system, tracheal mimetic tissues consisting of two types of cells were produced. When implanted into a damaged rabbit trachea, the implant spontaneously integrated with the host trachea. Both epithelium and cartilage formed at the predicted site. This raised the possibility of using this system for tissue engineering and biomedical applications.
The above illustrates the usefulness of hydrogels as bioinks for 3D bioprinting. Their mechanical and biocompatibility properties are important parameters. The target organs and biological tissues are diverse. The phenomena are intricate and complex, but the underlying polymer chemistry and physical chemistry are based on prior knowledge. In other words, the design and synthesis of optimal hydrogels using such knowledge leave a variety of grounds for the contribution of nanoarchitectonics.
3. Dynamic hydrogels as an artificial ECM
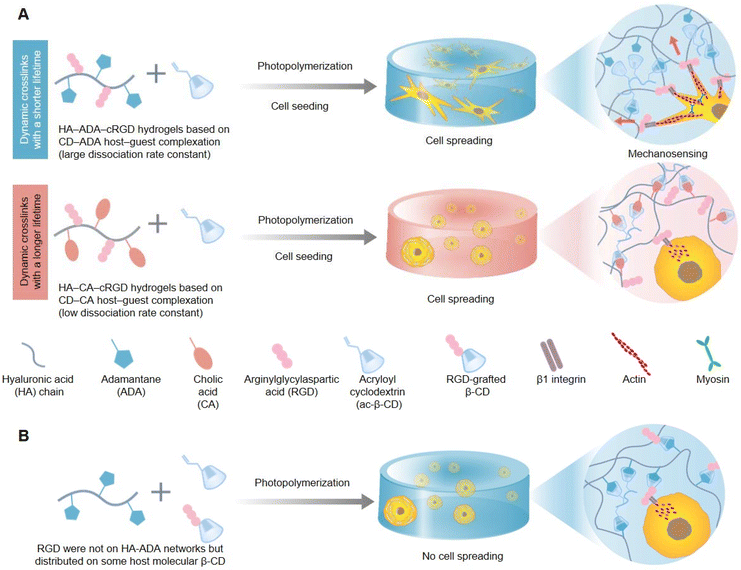
In living organisms, cells constantly interact with the highly dynamic ECM and remodel it. This process promotes various cell behaviors and fates, including growth, proliferation, migration, differentiation, and apoptosis. In addition to intrinsic thermodynamic remodeling, the degradation of biological polymers and the dissociation/recombination of force-induced physical crosslinks, which are two orthogonal external sources, also play roles in the dynamic characteristics of biological polymer networks in the natural ECM. Hydrogels are widely used as tunable biomimetic 3D cell culture matrices. Dynamic hydrogels can change their structures and properties under environmental stimuli. The formation of these hydrogels is usually achieved through physical interaction or chemical cross-linking, building a 3D network nanoarchitectonics that has the capacity to swell in water and respond to external cues. Moreover, dynamic hydrogels comprise reversible bonds like hydrogen bonds, ionic interactions, or dynamic covalent bonds, so that the hydrogel can have reversible structural changes, which provides it the applications for self-healing, adaptation and responsiveness to environmental cues. Some hydrogels show fast responses to stimuli such as temperature, pH, light, and ionic strength. As a result, dynamic hydrogels can be assembled from building blocks that adapt to cell activities when living cells are introduced. However, the natural ECM is very complex, and designing the physical and chemical properties of hydrogels to precisely manipulate their dynamic interactions with cells remains challenging. Therefore, designing a 3D hydrogel matrix with adjustable dynamic characteristics to reproduce the spatiotemporal hierarchy of ECM dynamics is of great significance in comprehending the relationship between the natural ECM and cell behaviors.To explore the dynamic correlation between the natural ECM and cell behaviors, researchers are committed to developing dynamic cell-adaptive hydrogels cross-linked by dynamic reversible bonds. Yang et al. investigated the indispensable role played by the dynamic property of 3D hydrogels based on supramolecular crosslinks in guiding cell behaviors and fates (Fig. 4).70 Hydrogels with similar reaction equilibrium constants but distinct kinetic constants were prepared through host–guest reversible interactions between the mono-acryloyl β-cyclodextrin (ac-β-CD) and hyaluronic acid chains (HA) equally grafted with adamantane (ADA) or cholic acid (CA) prior to photo-initiated polymerization, and were termed HA-ADA and HA-CA, respectively. Consistent with the higher dissociation kinetic constants of the HA-ADA hydrogel, it also exhibited faster stress relaxation than the HA-CA hydrogel as verified through rheological experiments. In addition, human Mesenchymal Stem Cells (hMSCs) cultured within the HA-ADA-cRGD (RGD is the abbreviation for arginylglycylaspartic acid) hydrogel presented extensive stellate spreading and intensive cellular mechanotransduction and osteogenic differentiation, while hMSCs within the HA-CA-cRGD hydrogel exhibited a round morphology and weak mechanosensing and adipogenic differentiation (Fig. 4A). In addition, the precise conjugated sites and robust binding strength of cell-adhesive ligands of RGD peptides also played a vital role in cellular spreading and mechanosensing (Fig. 4B). The above-mentioned cellular behaviors and multi-cell assembly impacted by the dynamics of hydrogels involved the concerted action of cell–matrix interactions, cell-adhesive ligand β1 integrin, and cytoskeletal mechanotransduction. Altogether, higher dynamics of the ECM could provide the cell membrane more local space freedom to explore and recruit cell-adhesive ligands of the ECM, and activate intracellular downstream mechanical signaling pathways better.
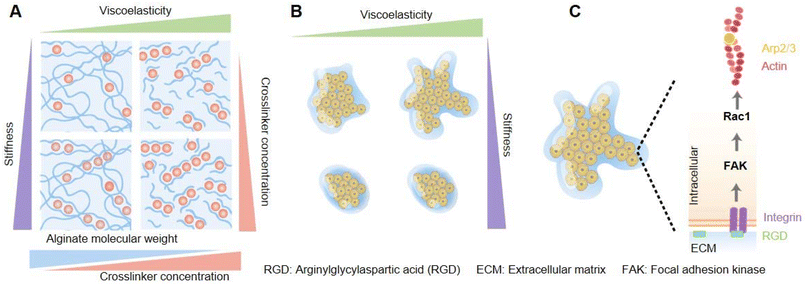
The collective behavior of cell and tissue patterns are also controlled by the mechanical properties of the ECM, for example, the viscosity, elasticity, viscoelasticity, fluidity, and so on. Among these features, how the viscoelasticity of the ECM affects the collective cell's organization from the spatial and temporal aspects remains not very well studied. Elosegui-Artola et al. explored and illustrated the effects driven by the passive viscoelasticity of the ECM on the spatiotemporal organization of encapsulated collective cells using alginate hydrogels (Fig. 5).71 Through coordinating the formula of alginate polymers and calcium crosslinker concentration, hydrogels with varying stress relaxation times were prepared, with independent control of the stiffness (Fig. 5A). Compared with elastic matrices, epithelial cell spheroids cultured within viscoelastic hydrogels were experimentally validated to show morphological instability (rapid growth, formation of finger-like protrusions, and spherical symmetry breaking) (Fig. 5B), and higher cellular mechanotransduction involved the activation of Rac1 signaling pathways, and facilitation of epithelial-to-mesenchymal transitions (EMTs), which was recognized to promote tumor growth and metastasis (Fig. 5C). Moreover, a computational simulation and prediction model was created. The calculation formulas of three variables of the matrix that could be used to recapitulate tissue organization within hydrogels were considered in the model – the scaled fluidity and passive mechanical relaxation time and the scaled cell proliferative capacity – and they were in concert to design a phase diagram to predict cell patterning. Based on formulations, the computational model simulated that decreased viscosity of the ECM could lead to tissue growth and instability, which was validated by the above-mentioned experiments. Besides, the model predicted cell motility, tissue proliferation, and increased stiffness of the matrix independent of viscoelasticity or elasticity, and all these also could contribute to tissue growth and instability. Definitely, these predictions were substantiated by related experiments. To prove the universal applicability of the model, the intestinal organoids culturing experiment was performed and indeed verified that the viscoelasticity of ECM promoted tissue growth and morphological changes.
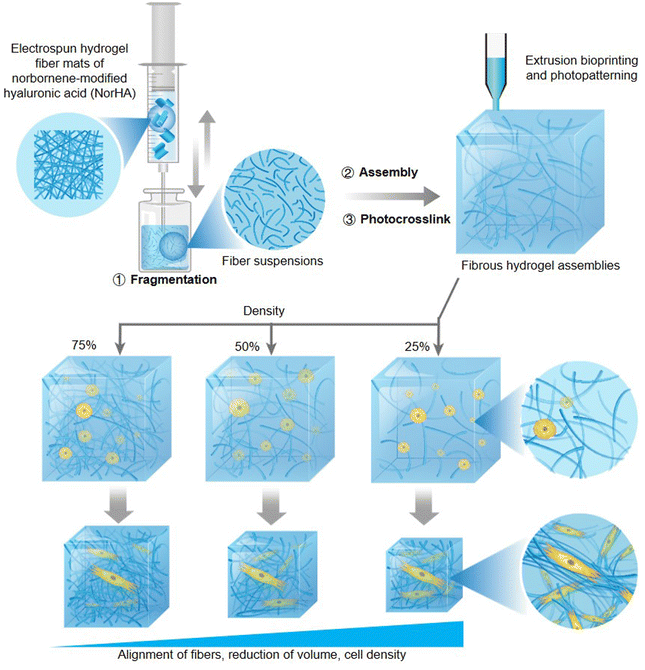
Natural fiber matrices are critical components of the ECM, and the effects of programmable and remodelable assembly of a 3D fibrous hydrogel scaffold on cellular behavior have yet to be investigated. Davidson et al. fabricated reassembled norbornene-modified hyaluronic acid (NorHA) fibrous hydrogel assemblies by photocrosslinking the resuspension solution of fragments produced from mechanical disruption of the preprepared electrospun fiber (Fig. 6).72 The hydrogels were found to possess excellent physical properties of shear-thinning and self-healing before photocrosslinking, which is conducive to the potential of application in extrusion printing constructs. Moreover, the features of strain-stiffening and strain-induced fiber alignment had been detected after photocrosslinking, and both features were dramatically demonstrated in the low-density (20%) fibrous assemblies compared to those of median (50%) and high (75%) density hydrogels. After encapsulating and culturing cells within varying fibre density hydrogels, the low-density fibrous assemblies were found to exhibit more extensive and more pericellular fiber recruitment, more reduction of volume, higher cell density, and compressive moduli under the cellular mechanical forces than median and high-density hydrogels. Due to the spatial distribution of matrix fibers being converted from the initial isotropic orientation into an organized alignment consistent with a micro-scale hydrogel geometry under cell contraction, the cell-laden fibrous assemblies were proposed to have the utility of microtissue fabrication. Furthermore, the hydrogel system could also be utilized to fabricate programmable biohydrogels via printing technology or photolithography methods, which were enabled by the shear-thinning induced extrudability and photocrosslinking of fibrous assemblies, respectively.
As seen in the examples above, the physical and structural cues of ECMs are diverse. Hydrogels are frequently used as synthetic ECMs for 3D cell culture. In addition to the examples described above, various other examples of studies have been reported. Lou et al. reported an interpenetrating network (IPN) hydrogel system based on dynamically covalently cross-linked HA and collagen I.73 The stress relaxation of IPN hydrogels can be tuned by the component HA. The modulation was based on the affinity of the HA cross-linker, the molecular weight, and the concentration of HA. The resulting viscoelastic hyaluronan-collagen IPN hydrogels mimicked well the microenvironment of the ECM, where mechanical cues in 3D cell culture could direct the function and fate of stem cells. Bone marrow-derived hMSCs are promising cells for regenerative therapies, and ex vivo expansion is necessary to obtain clinically useful cell numbers. Killaars et al. used hydrogels containing allyl sulfide cross-linkers and radical-mediated addition–fragmentation chain transfer processes at different time points.74 Hydrogels containing hMSCs were softened in situ. The effects of short- and long-term mechanical stimuli on epigenetic modification of hMSCs were quantified. Epigenetic remodeling was persistent, suggesting that memory may be retained in neural progenitor cell (NPC) culture in 3D hydrogels. This was an attractive strategy to increase the number of stem cells needed for therapeutic purposes. How the properties of the 3D material affect the maintenance of NPC stem cells in the absence of differentiation factors has been less well explored. Madl et al. found that stem cell maintenance did not correlate with initial hydrogel firmness in the range of physiologically appropriate firmness.75 Conversely, hydrogel degradation correlated with and was necessary for the maintenance of NPC stemness. This finding suggested that matrix remodeling without cytoskeletal tension generation was one strategy to maintain cellular stemness in 3D.
Zhang et al. investigated the mechanosensing behavior of human mesenchymal stem cells using a thermosensitive electrospun microfiber crosslinked hydrogel.76 The ability to switch in situ from a rigid (37 °C) to a soft (25 °C) state in a temperature-induced manner for a number of cycles was investigated. Cell proliferation, adhesion, mechanical threshold for nuclear migration of YAP signals, and osteogenic differentiation were enhanced compared to hMSCs grown under normal culture conditions. This might be due to enhanced mechanical feedbacks via dynamic mechanical interactions between cells and 3D fibrous constructs. The influence of early protein deposition on cell behavior in hydrogels had not been well explored. Loebel et al. used biological orthogonal labeling techniques to visualize early proteins within 1 day of culture in a variety of hydrogels.77 The role of nascent proteins in cell recognition of engineered materials had been reaffirmed and had implications for in vitro cell signaling studies and tissue repairment applications. However, there is still room for further investigations in complex feedback mechanisms. Intracellular biomacromolecules were dynamically reorganized in response to signals. This is dynamic nanoarchitectonics in living systems. Freeman et al. prepared hydrogels based on peptide amphiphiles, which can have DNA chains that degrade in response to chemical triggers, and investigated their superstructural assembly behaviors.78 The hydrogels had the property of degrading upon the addition of molecules or changes in charge density and organizing into a superstructure of entangled filaments. Experimental and simulation results indicated that reversible superstructures were formed when the large-scale dynamics of the supramolecular material was controlled by the formation of strong non-covalent bonds that could be broken from the outside. This dynamic supramolecular system allowed us to consider how changes in the structural features of the hydrogel network modulate important phenotypic changes in astrocytes associated with brain and spinal cord injury and neurological diseases.
The natural ECM has a decisive influence on cell behavior. Therefore, nanoarchitectonics of its artificial mimics is important for cellular tissue engineering and regenerative medicine. Hydrogels are very promising candidates, as seen in many of the examples above. The development of an artificial ECM is challenging due to the need for responses to the interaction/feedback from cells. Therefore, it is necessary to analyze the dynamic cell behavior and synthesize corresponding materials and structures. In such cases, the dynamic nanoarchitectonics approach is considered a powerful target.
4. Topographical hydrogels for tissue organization
Topographical scaffolding plays a crucial role in the development of epithelial organs and the solid vascular networks connecting them. In tissue engineering, the scaffold used is one of the most critical elements. Biocompatible hydrogel based topographical scaffold fabrications are expected to mimic from the microscale of the native ECM to macroscale of tissue topographical structures.The mechanism of fluid transport by multivascular networks and intravascular topographies has always been a very challenging research topic as such complex 3D network channels are difficult to construct. Grigoryan et al. constructed poly(ethylene glycol) diacrylate (PEGDA) hydrogels containing multivascular networks based on 3D stereolithography through bottom-up LBL photopolymerization under the assistance of innovative biocompatible photoabsorber additives, such as tartrazine (yellow food coloring FD&C Yellow 5, E102), curcumin (from turmeric), or anthocyanin (from blueberries).79 3D monolithic hydrogels with the vessel wall integrated with a functional bicuspid valve were fabricated, which could effectively spontaneously control the open-state of anterograde flows and the close-state of retrograde flows. Besides, entangled vascular networks comprising two separated and non-intersected channels also were established. When one of the channels was oxygen ventilation and another was perfused human red blood cells, respectively, vascularized alveolar and lung-mimetic models were created and validated to extremely recapitulate the functional intervascular oxygen transportation between the circulatory system and respiratory system. Furthermore, superior therapeutic effects of the transplantation of compositive hydrogel carriers containing vascular networks and encapsulated hepatocytes in chronic liver injury were determined, so it was revealed that the system had clinical application value.
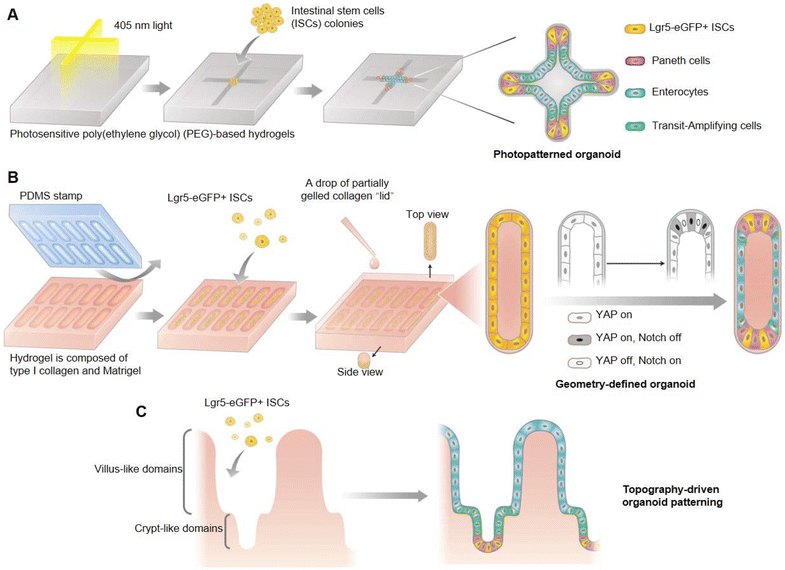
The stem cell self-organization process that is fundamental to organoid development has been difficult to control, which leads to the lack of repeatability in most existing technology for organoid culturing. Gjorevski et al. developed an external control strategy based on photosensitive poly(ethylene glycol) (PEG) hydrogels to confine the geometry of intestinal organoids, and demonstrated the indispensable influence of a pre-defined geometrical framework for the development of intestinal organoids (Fig. 7A).80 The crypt–villus axis of intestinal organoids mostly occurred at the localized position of cell accumulation within the shape-confined organoids, which involved greater spatial distribution in the activation of YAP and Notch signaling pathways, thereby influencing the stem cell fate by regulating Paneth cell localization and differentiation (Fig. 7B), and ultimately mediating the formation of crypt and villu domains (Fig. 7C). This pre-setting of organoid geometry could make the whole development process of organoids highly conservative, and there was no need to introduce external biochemical gradients. Furthermore, it was also demonstrated that the natural physiological structure of the intestinal epithelium in vivo could be further simulated when intestinal organoids were cultivated within the shape-confined hydrogel scaffolds.
Similarly, in tissue engineering for muscle repair, scaffold design also plays a critical role. The topographical hydrogel scaffolds with conductivity that mimic the ECM are crucial for skeletal muscle repair. Xue et al. created an anisotropic and conductive hydrogel scaffold by using gelatin methacryloyl (GelMA) as the base hydrogel and silver nanowires (AgNW) as the conductive dopant, employing a directional freezing technique to achieve the desired topographical scaffold for muscle defect repair (Fig. 8).81 These topographical and mechanical properties closely resemble those of the native muscle ECM, enabling cell orientation through contact cues and electrical stimulation. When transplanted into rats with muscle defects, the electrically stimulated topographical hydrogel scaffold showed a marked improvement in muscle reconstruction, achieving muscle mass and strength restoration ratios of 95% and 99% of normal levels, respectively. This finding presents the potential of topographically optimized and conductive hydrogel scaffolds in advancing muscle repair applications.
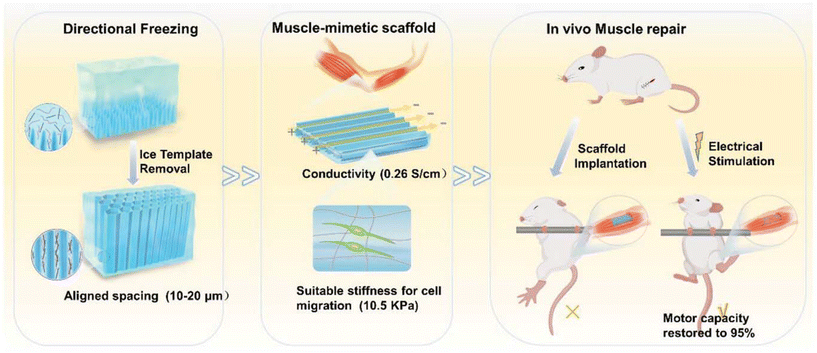 | ||
| Fig. 8 Schematic diagram of topographical scaffolds with conductivity for in vivo muscle repair under electrical stimulation. Reprinted with permission from ref. 46. Copyright 2024, Royal Society of Chemistry. | ||
As seen in the above examples, cells and their tissues do not have a uniform structure, and the corresponding gels require topographic structural control. Various attempts have been made, not limited to the above examples. Deshmukh et al. developed an acoustofluidic device that continuously patterned mammalian cells within hydrogel fibers.82 Photopolymerizable hydrogels were externally induced to undergo gelatinization by light when the cells were placed under the influence of an acoustic field. Muscle progenitor cells (myoblasts) could be patterned in parallel lines within the hydrogel, mimicking the structure of skeletal muscle. There, increased formation of myotubes and spontaneous twitching of myotubes could be observed. In addition, the anisotropy of natural tissues of other cell types, such as tendons, ligaments, and neurons, could be mimicked. Liu et al. reported a new technique called filament optical biofabrication.83 This technique allowed for the rapid fabrication of hydrogels consisting of a unidirectional network of microfilaments with a diameter on the length scale of a single cell. The formed microfilaments exhibit outstanding cell induction properties in fibroblasts, tendon cells, endothelial cells, and myoblasts. It is also possible to form multidirectional microfilaments within a single hydrogel construct. Potential applications of nanoarchitectonics in complex multicellular tissue engineering constructs would be considered more. Ma et al. reported an approach for versatile tissue engineering from tendon to bone, utilizing calcium silicate nanowires and alginate composite hydrogels as building blocks.84 3D printing technology and mechanical stretching post-processing were integrated to create structures with significantly enhanced mechanical properties ranging from nanoscale to submicron and microscale. The composite hydrogels significantly enhanced tissue regeneration of bone-to-tendon, especially fibrocartilage transition tissue in vivo.
As is the case with many mammalian tissues, organisms have specific cellular arrangements that allow for their unique functions. Correspondingly, topographic controls such as the patterning of hydrogels and their mixtures with the network become important. Biological systems universally possess hierarchical and topographic structural properties. Therefore, artificial structures such as hydrogels must have a topographic structure. The nanoarchitectonics methodology, which has advantages in creating hierarchical structures, is expected to be able to meet such demands.
5. Conclusions
Some of the above examples are reviewed and summarized. These examples illustrate the utility of the concept of nanoarchitectonics for research in the biological and medical fields, especially in tissue engineering, using gels. For hydrogels as bioinks for 3D bioprinting, mechanical and biocompatible properties are important parameters. The organs and biological tissues to which they are applied are complex systems, but fundamentally they are controlled upon a combination of known phenomena of polymer chemistry and physical chemistry. This is in common with the concept of nanoarchitectonics, which is based on the harmonization of fundamental processes and phenomena. Also, it is useful for cellular tissue engineering and regenerative medicine to construct artificial mimics of ECMs with nanoarchitectonics of gels, as they play a decisive role in the behaviors of cells. During this process, the interaction with cells is dynamic at the interfaces they come in contact with. Interfacial nanoarchitectonics emphasizing dynamic elements and nanoarchitectonics of soft materials surfaces will be key. As shown in the last few examples, it is not the cells themselves, but their specific arrangement and organization that play a major role in the expression of function. Patterning technology that corresponds to the hierarchical structure of living organisms is required. A nanoarchitectonics technique that favors the formation of hierarchical structures can meet these requirements.Cells that perform biofunctions, their tissue bodies, organs, and living organisms are structurally and functionally complex systems. As a material that is compatible with these complex systems, hydrogels are promising because of their ability to manipulate various components, structures, and mechanical properties. It is necessary to assemble unit molecules, polymers, ions, etc. into macroscopic structures comparable to those of cells and tissues. This is where the concept of nanoarchitectonics has many opportunities to demonstrate its power. Human beings have devised a variety of gels that can cope with such complex systems through accumulated experience and scientific knowledge. However, a more rational approach requires the introduction of data processing methodologies. The concepts of machine learning85 and materials informatics86 are being used to select, design, and develop optimal materials. There are also proposals to combine nanoarchitectonics and materials informatics.87 A rational approach to large-scale production is also needed. The development of such methodologies may also be achieved by supporting nanoarchitectonics with artificial intelligence. The fabrication of optimal gel structures from nanoscale structural elements by nanoarchitectonics will be useful for the development of tissue engineering biomedical materials at the practical level. The participation of new technologies such as machine learning will also promote it.
Author contributions
Jingwen Song: conceptualization, writing – original draft, and writing – review & editing. Wenyan Lyu: writing – original draft. Kohsaku Kawakami: project administration, review & editing, and funding acquisition. Katsuhiko Ariga: conceptualization, writing – original draft, writing – review & editing, and funding acquisition.Data availability
This is a review article and does not include any original data.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was partially supported by Japan Society for the Promotion of Science KAKENHI (Grant Numbers JP20H00392 and JP23H05459).References
- (a) D. Guo, R. Shibuya, C. Akiba, S. Saji, T. Kondo and J. Nakamura, Science, 2016, 351, 361–365 CrossRef CAS PubMed; (b) E. Zhang, Q. Zhu, J. Huang, J. Liu, G. Tan, C. Sun, T. Li, S. Liu, Y. Li, H. Wang, Xi. Wan, Z. Wen, F. Fan, J. Zhang and K. Ariga, Appl. Catal., B, 2021, 293, 120213 CrossRef CAS; (c) K. Maeda, F. Takeiri, G. Kobayashi, S. Matsuishi, H. Ogino, S. Ida, T. Mori, Y. Uchimoto, S. Tanabe, T. Hasegawa, N. Imanaka and H. Kageyama, Bull. Chem. Soc. Jpn., 2022, 95, 26–37 CrossRef CAS; (d) G. Chen, S. K. Singh, K. Takeyasu, J. P. Hill, J. Nakamura and K. Ariga, Sci. Technol. Adv. Mater., 2023, 23, 413–423 CrossRef PubMed; (e) H. Imahori, Bull. Chem. Soc. Jpn., 2023, 96, 339–352 CrossRef CAS.
- (a) A. H. Khan, S. Ghosh, B. Pradhan, A. Dalui, L. K. Shrestha, S. Acharya and K. Ariga, Bull. Chem. Soc. Jpn., 2017, 90, 627–648 CrossRef; (b) Md. S. Islam, Y. Shudo and S. Hayami, Bull. Chem. Soc. Jpn., 2022, 95, 1–25 CrossRef CAS; (c) A. Yoshino, Bull. Chem. Soc. Jpn., 2022, 95, 195 CrossRef CAS; (d) T. Hosaka and S. Komaba, Bull. Chem. Soc. Jpn., 2022, 95, 569–581 CrossRef CAS; (e) P. A. Shinde, Q. Abbas, N. R. Chodankar, K. Ariga, M. A. Abdelkareem and A. G. Olabi, J. Energy Chem., 2023, 79, 611–638 CrossRef CAS.
- (a) S. Ishihara, J. Labuta, W. V. Rossom, D. Ishikawa, K. Minami, J. P. Hill and K. Ariga, Phys. Chem. Chem. Phys., 2014, 16, 9713–9746 RSC; (b) A. Muranaka, H. Ban, M. Naito, S. Miyagawa, M. Ueda, S. Yamamoto, M. Harada, H. Takaya, M. Kimura, N. Kobayashi, M. Uchiyama and Y. Tokunaga, Bull. Chem. Soc. Jpn., 2022, 95, 1428–1437 CrossRef CAS; (c) T. Murata, K. Minami, T. Yamazaki, T. Sato, H. Koinuma, K. Ariga and N. Matsuki, Bull. Chem. Soc. Jpn., 2023, 96, 29–34 CrossRef CAS; (d) S. d. K. Sundaram, Md. M. Hossain, M. Rezki, K. Ariga and S. Tsujimura, Biosensors, 2023, 13, 1018 CrossRef PubMed; (e) Y. Sasaki and T. Minami, ChemNanoMat, 2024, 10, e202300335 CrossRef CAS.
- (a) M. Wen, G. Li, H. Liu, J. Chen, T. An and H. Yamashita, Environ. Sci.: Nano, 2019, 6, 1006–1025 RSC; (b) X. Han, S. Wang, M. Liu and L. Liu, Bull. Chem. Soc. Jpn., 2022, 95, 1445–1452 CrossRef CAS; (c) R. Li, D. Huang, S. Chen, L. Lei, Y. Chen, J. Tao, W. Zhouab and G. Wang, Nanoscale, 2022, 14, 10299–10320 RSC; (d) J. Wang, F. Matsuzawa, N. Sato, Y. Amano and M. Machida, Bull. Chem. Soc. Jpn., 2023, 96, 1088–1098 CrossRef CAS; (e) J. Yang, L. Huang, J. You and Y. Yamauchi, Small, 2023, 19, 2301044 CrossRef CAS PubMed.
- (a) R. Zhang and T. Hanaoka, Nat. Commun., 2022, 13, 3629 CrossRef CAS PubMed; (b) A. Chapman, E. Ertekin, M. Kubota, A. Nagao, K. Bertsch, A. Macadre, T. Tsuchiyama, T. Masamura, S. Takaki, R. Komoda, M. Dadfarnia, B. Somerday, A. T. Staykov, J. Sugimura, Y. Sawae, T. Morita, H. Tanaka, K. Yagi, V. Niste, P. Saravanan, S. Onitsuka, K.-S. Yoon, S. Ogo, T. Matsushima, G. Tumen-Ulzii, D. Klotz, D. H. Nguyen, G. Harrington, C. Adachi, H. Matsumoto, L. Kwati, Y. Takahashi, N. Kosem, T. Ishihara, M. Yamauchi, B. B. Saha, M. A. Islam, J. Miyawaki, H. Sivasankaran, M. Kohno, S. Fujikawa, R. Selyanchyn, T. Tsuji, Y. Higashi, R. Kirchheim and P. Sofronis, Bull. Chem. Soc. Jpn., 2022, 95, 73 CrossRef CAS; (c) S. Chen, J. Liu, Q. Zhang, F. Teng and B. C. McLellan, Renewable Sustainable Energy Rev., 2022, 167, 112537 CrossRef CAS; (d) N. Tsubaki, Y. Wang, G. Yan and Y. He, Bull. Chem. Soc. Jpn., 2023, 96, 291–302 CrossRef CAS; (e) T. Fukushima, M. Higashi and M. Yamauchi, Bull. Chem. Soc. Jpn., 2023, 96, 1209–1215 CrossRef CAS.
- (a) T. Tsuchiya, T. Nakayama and K. Ariga, Appl. Phys. Express, 2022, 15, 100101 CrossRef CAS; (b) Y. Saito, H. Sasabe, H. Tsuneyama, S. Abe, M. Matsuya, T. Kawano, Y. Kori, T. Hanayama and J. Kido, Bull. Chem. Soc. Jpn., 2023, 96, 24–28 CrossRef CAS; (c) M. Ishii, Y. Yamashita, S. Watanabe, K. Ariga and J. Takeya, Nature, 2023, 622, 285–291 CrossRef CAS PubMed; (d) M. Matsuya, H. Sasabe, S. Sumikoshi, K. Hoshi, K. Nakao, K. Kumada, R. Sugiyama, R. Sato and J. Kido, Bull. Chem. Soc. Jpn., 2023, 96, 183–189 CrossRef CAS; (e) Y. Yamamoto, H. Yano, S. Karashima, R. Uenishi, N. Orimo, J. Nishitani and T. Suzuki, Bull. Chem. Soc. Jpn., 2023, 96, 938–942 CrossRef CAS; (f) K. Ariga, S. Akakabe, R. Sekiguchi, M. L. Thomas, Y. Takeoka, M. Rikukawa and M. Yoshizawa-Fujita, ACS Omega, 2024, 9, 22203–22212 CrossRef CAS PubMed.
- (a) S. Quader, K. Kataoka and H. Cabral, Adv. Drug Delivery Rev., 2022, 182, 114115 CrossRef CAS PubMed; (b) Y. Ju, H. Liao, J. J. Richardson, J. Guo and F. Caruso, Chem. Soc. Rev., 2022, 51, 4287–4336 RSC; (c) Y. Han, P. Wen, J. Li and K. Kataoka, J. Controlled Release, 2022, 345, 709–720 CrossRef CAS PubMed; (d) L. T. B. Nguyen and M. Abe, Bull. Chem. Soc. Jpn., 2023, 96, 899–906 CrossRef CAS; (e) S. Mohanan, C. I. Sathish, T. J. Adams, S. Kan, M. Liang and A. Vinu, Bull. Chem. Soc. Jpn., 2023, 96, 1188–1195 CrossRef CAS.
- (a) H. Sekine, T. Shimizu, K. Sakaguchi, I. Dobashi, M. Wada, M. Yamato, E. Kobayashi, M. Umezu and T. Okano, Nat. Commun., 2013, 4, 1399 CrossRef PubMed; (b) L. Moroni, J. A. Burdick, C. Highley, S. J. Lee, Y. Morimoto, S. Takeuchi and J. J. Yoo, Nat. Rev. Mater., 2018, 3, 21–37 CrossRef CAS PubMed; (c) X. Fu, G. Liu, A. Halim, Y. Ju, Q. Luo and G. Song, Cells, 2019, 8, 784 CrossRef CAS PubMed; (d) Y. Shang, J. Zeng, Z. Xie, N. Sasaki and M. Matsusaki, Bull. Chem. Soc. Jpn., 2022, 95, 1163–1168 CrossRef CAS; (e) R. Hama, A. Ulziibayar, J. W. Reinhardt, T. Watanabe, I. Kelly and T. Shinoka, Biomolecules, 2023, 13, 280 CrossRef CAS PubMed.
- (a) Y. Hashimoto, T. Suzuki and K. Hashimoto, Mol. Psychiatry, 2022, 27, 1898–1907 CrossRef CAS PubMed; (b) M. Komiyama, Bull. Chem. Soc. Jpn., 2022, 95, 1308–1317 CrossRef CAS; (c) Y. Cui, A. Taguchi, H. Shida, S. Konno, K. Takayama, A. Taniguchi and Y. Hayashi, Bull. Chem. Soc. Jpn., 2022, 95, 1156–1162 CrossRef CAS; (d) A. Watanabe, M. Iwagami, J. Yasuhara, H. Takagi and T. Kuno, Vaccine, 2023, 41, 1783–1790 CrossRef CAS PubMed; (e) S. Sugita, H. Ishitani and S. Kobayashi, Bull. Chem. Soc. Jpn., 2023, 96, 744–751 CrossRef CAS.
- (a) A. Bieniek, A. P. Terzyk, M. Wiśniewski, K. Roszek, P. Kowalczyk, L. Sarkisov, S. Keskin and K. Kaneko, Prog. Mater. Sci., 2021, 117, 100743 CrossRef CAS; (b) A. R. Pradipta, H. Michiba, A. Kubo, M. Fujii, T. Tanei, K. Morimoto, K. Shimazu and K. Tanaka, Bull. Chem. Soc. Jpn., 2022, 95, 421–426 CrossRef CAS; (c) V. J. Sahayasheela, Z. Yu, Y. Hirose, G. N. Pandian, T. Bando and H. Sugiyama, Bull. Chem. Soc. Jpn., 2022, 95, 693–699 CrossRef CAS; (d) L. Sutrisno and K. Ariga, NPG Asia Mater., 2023, 15, 21 CrossRef CAS; (e) C. Tay, A. Tanaka and S. Sakaguchi, Cancer Cell, 2023, 41, 450–465 CrossRef CAS PubMed.
- (a) Y. Sugimoto, P. Pou, M. Abe, P. Jelinek, R. Pérez, S. Morita and Ó. Custance, Nature, 2007, 446, 64–67 CrossRef CAS PubMed; (b) A. Shiotari and Y. Sugimoto, Nat. Commun., 2017, 8, 14313 CrossRef CAS PubMed; (c) K. Kimura, K. Miwa, H. Imada, M. Imai-Imada, S. Kawahara, J. Takeya, M. Kawai, M. Galperin and Y. Kim, Nature, 2019, 570, 210–213 CrossRef CAS PubMed; (d) K. Tada, Y. Hinuma, S. Ichikawa and S. Tanaka, Bull. Chem. Soc. Jpn., 2023, 96, 373–380 CrossRef CAS; (e) H. Hoelzel, S. Lee, K. Y. Amsharov, N. Jux, K. Harano, E. Nakamura and D. Lungerich, Nat. Chem., 2023, 15, 1444–1451 CrossRef CAS PubMed.
- (a) Y. Okawa and M. Aono, Nature, 2001, 409, 683–684 CrossRef CAS PubMed; (b) P. Mishra, J. P. Hill, S. Vijayaraghavan, W. Van Rossom, S. Yoshizawa, M. Grisolia, J. Echeverria, T. Ono, K. Ariga, T. Nakayama, C. Joachim and T. Uchihashi, Nano Lett., 2015, 15, 4793–4798 CrossRef CAS PubMed; (c) G. J. Simpson, V. García-López, P. Petermeier, L. Grill and J. M. Tour, Nat. Nanotechnol., 2017, 12, 604–606 CrossRef CAS PubMed; (d) S. Kawai, O. Krejčí, T. Nishiuchi, K. Sahara, T. Kodama, R. Pawlak, E. Meyer, T. Kubo and A. S. Foster, Sci. Adv., 2020, 6, eaay8913 CrossRef CAS PubMed; (e) W.-H. Soe, M. Kleinwächter, C. Kammerer, G. Rapenne and C. Joachim, J. Phys. Chem. C, 2020, 124, 22625–22630 CrossRef CAS.
- (a) K. Ariga, Q. Ji, W. Nakanishi, J. P. Hill and M. Aono, Mater. Horiz., 2015, 2, 406–413 RSC; (b) L. Cao, Y. Huang, B. Parakhonskiy and A. G. Skirtach, Nanoscale, 2022, 14, 15964–16002 RSC.
- K. Ariga, Nanoscale Horiz., 2021, 6, 364–378 RSC.
- K. Ariga and Y. Yamauchi, Chem. - Asian J., 2020, 15, 718–728 CrossRef CAS PubMed.
- (a) R. P. Feynman, Eng. Sci., 1960, 23, 32–36 Search PubMed; (b) M. Roukes, Sci. Am., 2001, 285, 48–51 CrossRef CAS PubMed.
- K. Ariga, K. Minami, M. Ebara and J. Nakanishi, Polym. J., 2016, 48, 371–389 CrossRef CAS.
- (a) K. Ariga, Curr. Opin. Colloid Interface Sci., 2023, 63, 101656 CrossRef CAS; (b) O. Azzaroni, E. Piccinini, G. Fenoy, W. Marmisollé and K. Ariga, Nanotechnology, 2023, 34, 472001 CrossRef PubMed.
- K. Ariga, J. Li, J. Fei, Q. Ji and J. P. Hill, Adv. Mater., 2016, 28, 1251–1286 CrossRef CAS PubMed.
- (a) K. Ariga, M. Nishikawa, T. Mori, J. Takeya, L. K. Shrestha and J. P. Hill, Sci. Technol. Adv. Mater., 2020, 20, 51–95 CrossRef PubMed; (b) E. Mieda, Y. Morishima, T. Watanabe, H. Miyake and S. Shinoda, Bull. Chem. Soc. Jpn., 2023, 96, 538–544 CrossRef CAS; (c) K. Saito and Y. Yamamura, Bull. Chem. Soc. Jpn., 2023, 96, 607–613 CrossRef CAS.
- (a) K. Ariga, X. Jia, J. Song, J. P. Hill, D. T. Leong, Y. Jia and J. Li, Angew. Chem., Int. Ed., 2020, 59, 15424–15446 CrossRef CAS PubMed; (b) K. Ariga, J. Song and K. Kawakami, Chem. Commun., 2024, 60, 2152–2167 RSC.
- M. Aono and K. Ariga, Adv. Mater., 2016, 28, 989–992 CrossRef CAS PubMed.
- (a) M. Komiyama, K. Yoshimoto, M. Sisido and K. Ariga, Bull. Chem. Soc. Jpn., 2017, 90, 967–1004 CrossRef; (b) K. Ariga, Int. J. Mol. Sci., 2022, 23, 3577 CrossRef CAS PubMed.
- R. B. Laughlin and D. Pines, Proc. Natl. Acad. Sci., 2000, 97, 28–31 CrossRef CAS PubMed.
- (a) K. Ariga and R. Fakhrullin, Bull. Chem. Soc. Jpn., 2022, 95, 774–795 CrossRef CAS; (b) K. Ariga, Bull. Chem. Soc. Jpn., 2024, 97, uoad001 CrossRef.
- (a) T. Govindaraju and M. B. Avinash, Nanoscale, 2012, 4, 6102–6117 RSC; (b) W. Nakanishi, K. Minami, L. K. Shrestha, Q. Ji, J. P. Hill and K. Ariga, Nano Today, 2014, 9, 378–394 CrossRef CAS; (c) K. Ariga and M. Shionoya, Bull. Chem. Soc. Jpn., 2021, 94, 839–859 CrossRef CAS; (d) G. Chen, F. Sciortino, K. Takeyasu, J. Nakamura, J. P. Hill, L. K. Shrestha and K. Ariga, Chem. – Asian J., 2022, 17, e202200756 CrossRef CAS PubMed; (e) P. A. Shinde, N. R. Chodankar, H.-J. Kim, M. A. Abdelkareem, A. A. Ghaferi, Y.-K. Han, A. G. Olabi and K. Ariga, ACS Energy Lett., 2023, 8(10), 4474–4487 CrossRef CAS; (f) E. Ruiz-Hitzky and C. Ruiz-Garcia, Nanoscale, 2023, 15, 18959–18979 RSC.
- (a) M. Ramanathan, L. K. Shrestha, T. Mori, Q. Ji, J. P. Hill and K. Ariga, Phys. Chem. Chem. Phys., 2013, 15, 10580–10611 RSC; (b) J. Kim, J. H. Kim and K. Ariga, Joule, 2017, 1, 739–768 CrossRef CAS; (c) A. Nayak, S. Unayama, S. Tai, T. Tsuruoka, R. Waser, M. Aono, I. Valov and T. Hasegawa, Adv. Mater., 2018, 30, 1703261 CrossRef PubMed; (d) M. Eguchi, A. S. Nugraha, A. E. Rowan, J. Shapter and Y. Yamauchi, Adv. Sci., 2021, 8, 2100539 CrossRef CAS PubMed; (e) K. Lee, M. Han, G. Kwon, Y. Jeon, J. Kim and J. You, Appl. Surf. Sci., 2023, 613, 155955 CrossRef CAS.
- (a) K. Ariga, Q. Ji, T. Mori, M. Naito, Y. Yamauchi, H. Abe and J. P. Hill, Chem. Soc. Rev., 2013, 42, 6322–6345 RSC; (b) R. Chang, L. Zhao, R. Xing, J. Li and X. Yan, Chem. Soc. Rev., 2023, 52, 2688–2712 RSC; (c) Q. Liu, H. Li, B. Yu, Z. Meng, X. Zhang, J. Li and L. Zheng, Adv. Funct. Mater., 2022, 32, 2201196 CrossRef CAS; (d) Z. Li, F. Yu, X. Xu, T. Wang, J. Fei, J. Hao and J. Li, J. Am. Chem. Soc., 2023, 145, 20907–20912 CrossRef CAS PubMed.
- (a) K. Ariga, Small Struct., 2021, 2, 2100006 CrossRef CAS; (b) Y. Jia, X. Yan and J. Li, Angew. Chem., Int. Ed., 2022, 61, e202207752 CrossRef CAS PubMed; (c) D. Parbat, N. Jana, M. Dhar and U. Manna, ACS Appl. Mater. Interfaces, 2023, 15, 25232–25247 CrossRef CAS PubMed; (d) X. Shen, J. Song, C. Sevencan and D. T. Leong, Sci. Technol. Adv. Mater., 2023, 23, 199–224 CrossRef PubMed.
- (a) A. Juste-Dolz, M. Delgado-Pinar, M. Avella-Oliver, E. Fernández, J. L. Cruz, M. V. Andrés and Á. Maquieira, ACS Appl. Mater. Interfaces, 2022, 14, 41640–41648 CrossRef CAS PubMed; (b) J. Liu, R. Wang, H. Zhou, M. Mathesh, M. Dubey, W. Zhang, B. Wang and W. Yang, Nanoscale, 2022, 14, 10286–10298 RSC; (c) S. Kim, S. Baek, R. Sluyter, K. Konstantinov, J. H. Kim, S. Kim and Y. H. Kim, EcoMat, 2023, 5, e12356 CrossRef; (d) J. V. Vaghasiya, C. C. Mayorga-Martinez and M. Pumera, npj Flexible Electron., 2023, 7, 26 CrossRef PubMed; (e) H.-M. Deng, M.-J. Xiao, Y.-L. Yuan, R. Yuan and Y.-Q. Chai, Sens. Actuators, B, 2024, 398, 134715 CrossRef CAS.
- (a) A. R. Ferhan, S. Park, H. Park, H. Tae, J. A. Jackman and N.-J. Cho, Adv. Funct. Mater., 2022, 32, 2203669 CrossRef CAS; (b) M. Komiyama, Beilstein J. Nanotechnol., 2023, 14, 218–232 CrossRef CAS PubMed; (c) Y. N. Reddy, A. De, S. Paul, A. K. Pujari and J. Bhaumik, Biomacromolecules, 2023, 24, 1717–1730 CrossRef CAS PubMed; (d) S. Mohanan, X. Guan, M. Liang, A. Karakoti and A. Vinu, Small, 2023, 2301113 CrossRef PubMed; (e) W. Tian, C. Wang, R. Chu, H. Ge, X. Sun and M. Li, Biomater. Res., 2023, 27, 100 CrossRef CAS PubMed.
- (a) E. Psarra, U. König, Y. Ueda, C. Bellmann, A. Janke, E. Bittrich, K.-J. Eichhorn and P. Uhlmann, ACS Appl. Mater. Interfaces, 2015, 7, 12516–12529 CrossRef CAS PubMed; (b) B. Tian, J. Liu, S. Guo, A. Li and J.-B. Wan, Int. J. Biol. Macromol., 2023, 243, 125161 CrossRef CAS PubMed; (c) X. Jia, J. Chen, W. Lv, H. Li and K. Ariga, Int. J. Biol. Macromol., 2023, 4, 101251 CAS; (d) W. Hu, J. Shi, W. Lv, X. Jia and K. Ariga, Sci. Technol. Adv. Mater., 2023, 23, 393–412 CrossRef PubMed; (e) Z. Hajikhani, I. Haririan, M. Akrami and S. Hajikhani, Nanomedicine, 2023, 18, 1441–1458 CrossRef CAS PubMed.
- (a) Q. Ren, N. Yu, L. Wang, M. Wen, P. Geng, Q. Jiang, M. Li and Z. Chen, J. Colloid Interface Sci., 2022, 614, 147–159 CrossRef CAS PubMed; (b) H. Duan, F. Wang, W. Xu, G. Sheng, Z. Sun and H. Chu, Dalton Trans., 2023, 52, 16085–16102 RSC; (c) Y. Liu, J. Zhao, X. Xu, Y. Xu, W. Cui, Y. Yang and J. Li, Angew. Chem., Int. Ed., 2023, 62, e202308019 CrossRef CAS PubMed; (d) P. Jayachandran, S. Ilango, V. Suseela, R. Nirmaladevi, M. R. Shaik, M. Khan, M. Khan and B. Shaik, Biomedicines, 2023, 11, 217 CrossRef CAS PubMed; (e) N. Yang, X. Pan, X. Zhou, Z. Liu, J. Yang, J. Zhang, Z. Jia and Q. Shen, Adv. Healthcare Mater., 2023, 2302752 Search PubMed.
- (a) K. Ariga, Nanoscale, 2022, 14, 10610–11062 RSC; (b) K. Ariga, Chem. Mater., 2023, 35, 5233–5254 CrossRef CAS.
- (a) I. R. Vetter and A. Wittinghofer, Science, 2001, 294, 1299–1304 CrossRef CAS PubMed; (b) K. N. Ferreira, T. M. Iverson, K. Maghlaoui, J. Barber and S. Iwata, Science, 2004, 303, 1831–1838 CrossRef CAS PubMed; (c) D. A. Bryant and D. P. Canniffe, J. Phys. B: At., Mol. Opt. Phys., 2018, 51, 033001 CrossRef.
- (a) K. Ariga, Y. Yamauchi, T. Mori and J. P. Hill, Adv. Mater., 2013, 25, 6477–6512 CrossRef CAS PubMed; (b) K. Ariga, Langmuir, 2020, 36, 7158–7180 CrossRef CAS PubMed; (c) S. Negi, M. Hamori, H. Kitagishi and K. Kano, Bull. Chem. Soc. Jpn., 2022, 95, 1537–1545 CrossRef CAS; (d) S. Negi, M. Hamori, Y. Kubo, H. Kitagishi and K. Kano, Bull. Chem. Soc. Jpn., 2023, 96, 48–56 CrossRef CAS; (e) O. N. Oliveira Jr., L. Caseli and K. Ariga, Chem. Rev., 2022, 122, 6459–6513 CrossRef CAS PubMed; (f) J. Adachi, M. Naito, S. Sugiura, N. H.-T. Le, S. Nishimura, S. Huang, S. Suzuki, S. Kawamorita, N. Komiya, J. P. Hill, K. Ariga, T. Naota and T. Mori, Bull. Chem. Soc. Jpn., 2022, 95, 889–897 CrossRef CAS.
- (a) G. Decher, Science, 1997, 277, 1232–1237 CrossRef CAS; (b) K. Ariga, J. P. Hill and Q. Ji, Phys. Chem. Chem. Phys., 2007, 9, 2319–2340 RSC; (c) G. Rydzek, Q. Ji, M. Li, P. Schaaf, J. P. Hill, F. Boulmedais and K. Ariga, Nano Today, 2015, 10, 138–167 CrossRef CAS; (d) Z. Zhang, J. Zeng, J. Groll and M. Matsusaki, Biomater. Sci., 2022, 10, 4077–4094 RSC; (e) K. Ariga, Y. Lvov and G. Decher, Phys. Chem. Chem. Phys., 2022, 24, 4097–4115 RSC; (f) J. Borges, J. Zeng, X. Q. Liu, H. Chang, C. Monge, C. Garot, K. Ren, P. Machillot, N. E. Vrana, P. Lavalle, T. Akagi, M. Matsusaki, J. Ji, M. Akashi, J. F. Mano, V. Gribova and C. Picart, Adv. Healthcare Mater., 2024, 2302713 CrossRef CAS PubMed.
- (a) Y. Shan, G. Zhang, W. Yin, H. Pang and Q. Xu, Bull. Chem. Soc. Jpn., 2022, 95, 230–260 CrossRef CAS; (b) M. Daniel, G. Mathew, M. Anpo and B. Neppolian, Coord. Chem. Rev., 2022, 468, 214627 CrossRef CAS; (c) L. Larasati, W. W. Lestari and M. Firdaus, Bull. Chem. Soc. Jpn., 2022, 95, 1561–1577 CrossRef CAS; (d) T. Ohata, K. Tachimoto, K. J. Takeno, A. Nomoto, T. Watanabe, I. Hirosawa and R. Makiura, Bull. Chem. Soc. Jpn., 2023, 96, 274–282 CrossRef CAS; (e) Q. Yao, X. Zhang, Z.-H. Lu and Q. Xu, Coord. Chem. Rev., 2023, 493, 215302 CrossRef CAS.
- (a) Y. Hara and K. Sakaushi, Nanoscale, 2021, 13, 6341–6356 RSC; (b) Y. Charles-Blin, T. Kondo, Y. Wu, S. Bandow and K. Awaga, Bull. Chem. Soc. Jpn., 2022, 95, 972–977 CrossRef CAS; (c) L. Huang, J. Yang, Y. Asakura, Q. Shuai and Y. Yamauchi, ACS Nano, 2023, 17, 8918–8934 CrossRef CAS PubMed; (d) S. Zhang, L. Lombardo, M. Tsujimoto, Z. Fan, E. K. Berdichevsky, Y.-S. Wei, K. Kageyama, Y. Nishiyama and S. Horike, Angew. Chem., Int. Ed., 2023, 62, e202312095 CrossRef CAS PubMed; (e) Y. Zhao, T. Irie, J. Sakai, H. Mabuchi, S. Biswas, T. Sekine, S. Das, T. Ben and Y. Negishi, ACS Appl. Nano Mater., 2023, 6, 19210–19217 CrossRef CAS.
- (a) N. Ma and S. Horike, Chem. Rev., 2022, 122, 4163–4203 CrossRef CAS PubMed; (b) S. Horike, Bull. Chem. Soc. Jpn., 2023, 96, 887–898 CrossRef CAS.
- (a) C. Creton, Macromolecules, 2017, 50, 8297–8316 CrossRef CAS; (b) R. Yoshida, Polym. J., 2022, 54, 827–849 CrossRef CAS; (c) M. Z. I. Nizami, B. D. L. Campéon, A. Satoh and Y. Nishina, Bull. Chem. Soc. Jpn., 2022, 95, 713–720 CrossRef CAS; (d) H. Jia and T. Michinobu, ChemNanoMat, 2023, 9, e202300020 CrossRef CAS; (e) R. Tamate and T. Ueki, Chem. Rec., 2023, 23, e202300043 CrossRef CAS PubMed.
- R. Chang, C. Yuan, P. Zhou, R. Xing and X. Yan, Acc. Chem. Res., 2024, 57, 289–301 CrossRef CAS PubMed.
- (a) Y. Noda, Y. Hayashi and K. Ito, J. Appl. Polym. Sci., 2014, 131, 40509 CrossRef; (b) A. B. Imran, K. Esaki, H. Gotoh, T. Seki, K. Ito, Y. Sakai and Y. Takeoka, Nat. Commun., 2014, 5, 5124 CrossRef PubMed; (c) K. Mayumi, C. Liu, Y. Yasuda and K. Ito, Gels, 2021, 7, 91 CrossRef CAS PubMed.
- C. Liu, N. Morimoto, L. Jiang, S. Kawahara, T. Noritomi, H. Yokoyama, K. Mayumi and K. Ito, Science, 2021, 372, 1078–1081 CrossRef CAS PubMed.
- Y. Sekine and T. Nankawa, Bull. Chem. Soc. Jpn., 2023, 96, 1150–1155 CrossRef CAS.
- R. Kubota, Bull. Chem. Soc. Jpn., 2023, 96, 802–812 CrossRef CAS.
- Y. Aoyama, N. Sato, A. Toyotama, T. Okuzono and J. Yamanaka, Bull. Chem. Soc. Jpn., 2022, 95, 314–324 CrossRef CAS.
- R. Xing, C. Yuan, W. Fan, X. Ren and X. Yan, Sci. Adv., 2023, 9, eadd8105 CrossRef CAS PubMed.
- X. Pan, J. Li, N. Ma, X. Ma and M. Gao, Chem. Eng. J., 2023, 461, 142062 CrossRef CAS.
- B. Salahuddin, M. K. Masud, S. Aziz, C.-H. Liu, N. Amiralian, A. Ashok, S. M. A. Hossain, H. Park, M. A. Wahab, M. A. Amin, M. A. Chari, A. E. Rowan, Y. Yamauchi, M. S. A. Hossain and Y. V. Kaneti, Bull. Chem. Soc. Jpn., 2022, 95, 198–207 CrossRef CAS.
- Z. Shen, Z. Zhang, N. Zhang, J. Li, P. Zhou, F. Hu, Y. Rong, B. Lu and G. Gu, Adv. Mater., 2022, 34, 2203650 CrossRef CAS PubMed.
- H. Yuk, J. Wu and X. Zhao, Nat. Rev. Mater., 2022, 7, 935–952 CrossRef CAS.
- L. Hu, P. L. Chee, S. Sugiarto, Y. Yu, C. Shi, R. Yan, Z. Yao, X. Shi, J. Zhi, D. Kai, H.-D. Yu and W. Huang, Adv. Mater., 2023, 35, 2205326 CrossRef CAS PubMed.
- H. Cao, L. Duan, Y. Zhang, J. Cao and K. Zhang, Signal Transduction Targeted Ther., 2021, 6, 426 CrossRef CAS PubMed.
- Y. Liu, T. Dong, Y. Chen, N. Sun, Q. Liu, Z. Huang, Y. Yang, H. Cheng and K. Yue, ACS Appl. Mater. Interfaces, 2023, 15, 11507–11519 CrossRef CAS PubMed.
- Y. Yang, Y. Liang, J. Chen, X. Duan and B. Guo, Bioact. Mater., 2022, 8, 341–354 CAS.
- Y. Huang, L. Mu, X. Zhao, Y. Han and B. Guo, ACS Nano, 2022, 16, 13022–13036 CrossRef CAS PubMed.
- Y. Xiang, X. Qi, E. Cai, C. Zhang, J. Wang, Y. Lan, H. Deng, J. Shen and R. Hu, Chem. Eng. J., 2023, 460, 141852 CrossRef CAS.
- W. Zhou, Z. Duan, J. Zhao, R. Fu, C. Zhu and D. Fan, Bioact. Mater., 2022, 17, 1–17 CAS.
- Y.-J. Fu, Y.-F. Shi, L.-Y. Wang, Y.-F. Zhao, R.-K. Wang, K. Li, S.-T. Zhang, X.-J. Zha, W. Wang, X. Zhao and W. Yang, Adv. Sci., 2023, 10, 2206771 CrossRef CAS PubMed.
- Y. Qian, Y. Zheng, J. Jin, X. Wu, K. Xu, M. Dai, Q. Niu, H. Zheng, X. He and J. Shen, Adv. Mater., 2022, 34, 2200521 CrossRef CAS PubMed.
- Y. Li, R. Fu, Z. Duan, C. Zhu and D. Fan, ACS Nano, 2022, 16, 7486–7502 CrossRef CAS PubMed.
- Y. Liang, H. Xu, Z. Li, A. Zhangji and B. Guo, Nano-Micro Lett., 2022, 14, 185 CrossRef CAS PubMed.
- A. J. Seymour, S. Shin and S. C. Heilshorn, Adv. Healthcare Mater., 2021, 10, 2100644 CrossRef CAS PubMed.
- F. E. Freeman, P. Pitacco, L. H. A. van Dommelen, J. Nulty, D. C. Browe, J.-Y. Shin, E. Alsberg and D. J. Kelly, Sci. Adv., 2020, 6, eabb5093 CrossRef CAS PubMed.
- H. Zhang, Y. Cong, A. R. Osi, Y. Zhou, F. Huang, R. P. Zaccaria, J. Chen, R. Wang and J. Fu, Adv. Funct. Mater., 2020, 30, 1910573 CrossRef CAS.
- M. Wang, W. Li, J. Hao, A. Gonzales III, Z. Zhao, R. S. Flores, X. Kuang, X. Mu, T. Ching, G. Tang, Z. Luo, C. E. Garciamendez-Mijares, J. K. Sahoo, M. F. Wells, G. Niu, P. Agrawal, A. Q. Hinojosa, K. Eggan and Y. S. Zhang, Nat. Commun., 2022, 13, 3317 CrossRef PubMed.
- S. H. Kim, Y. K. Yeon, J. M. Lee, J. R. Chao, Y. J. Lee, Y. B. Seo, Md. T. Sultan, O. J. Lee, J. S. Lee, S.-i. Yoon, I.-S. Hong, G. Khang, S. J. Lee, J. J. Yoo and C. H. Park, Nat. Commun., 2018, 9, 1620 CrossRef PubMed.
- S. H. Kim, Y. B. Seo, Y. K. Yeon, Y. J. Lee, H. S. Park, Md. T. Sultan, J. M. Lee, J. S. Lee, O. J. Lee, H. Hong, H. Lee, O. Ajiteru, Y. J. Suh, S.-H. Song, K.-H. Lee and C. H. Park, Biomaterials, 2020, 260, 120281 CrossRef CAS PubMed.
- B. Yang, K. Wei, C. Loebel, K. Zhang, Q. Feng, R. Li, S. H. D. Wong, X. Xu, C. Lau, X. Chen, P. Zhao, C. Yin, J. A. Burdick, Y. Wang and L. Bian, Nat. Commun., 2021, 12, 3514 CrossRef CAS PubMed.
- A. Elosegui-Artola, A. Gupta, A. J. Najibi, B. R. Seo, R. Garry, C. M. Tringides, I. de Lázaro, M. Darnell, W. Gu, Q. Zhou, D. A. Weitz, L. Mahadevan and D. J. Mooney, Nat. Mater., 2023, 22, 117–127 CrossRef CAS PubMed.
- M. D. Davidson, M. E. Prendergast, E. Ban, K. L. Xu, G. Mickel, P. Mensah, A. Dhand, P. A. Janmey, V. B. Shenoy and J. A. Burdick, Sci. Adv., 2021, 7, eabi8157 CrossRef CAS PubMed.
- J. Lou, R. Stowers, S. Nam, Y. Xia and O. Chaudhuri, Biomaterials, 2018, 154, 213–222 CrossRef CAS PubMed.
- A. R. Killaars, J. C. Grim, C. J. Walker, E. A. Hushka, T. E. Brown and K. S. Anseth, Adv. Sci., 2019, 6, 1801483 CrossRef PubMed.
- C. M. Madl, B. L. LeSavage, R. E. Dewi, C. B. Dinh, R. S. Stowers, M. Khariton, K. J. Lampe, D. Nguyen, O. Chaudhuri, A. Enejder and S. C. Heilshorn, Nat. Mater., 2017, 16, 1233–1242 CrossRef CAS PubMed.
- J. G. Zhang, C. Cheng, J. L. Cuellar-Camacho, M. Li, Y. Xia, W. Li and R. Haag, Adv. Funct. Mater., 2018, 28, 1804773 CrossRef.
- C. Loebel, R. L. Mauck and J. A. Burdick, Nat. Mater., 2019, 18, 883–891 CrossRef CAS PubMed.
- R. Freeman, M. Han, Z. Álvarez, J. A. Lewis, J. R. Wester, N. Stephanopoulos, M. T. McClendon, C. Lynsky, J. M. Godbe, H. Sangji, E. Luijten and S. I. Stupp, Science, 2018, 362, 808–813 CrossRef CAS PubMed.
- B. Grigoryan, S. J. Paulsen, D. C. Corbett, D. W. Sazer, C. L. Fortin, A. J. Zaita, P. T. Greenfield, N. J. Calafat, J. P. Gounley, A. H. Ta, F. Johansson, A. Randles, J. E. Rosenkrantz, J. D. Louis-Rosenberg, P. A. Galie, K. R. Stevens and J. S. Miller, Science, 2019, 364, 458–464 CrossRef CAS PubMed.
- (a) N. Gjorevski, M. Nikolaev, T. E. Brown, O. Mitrofanova, N. Brandenberg, F. W. DelRio, F. M. Yavitt, P. Liberali, K. S. Anseth and M. P. Lutolf, Science, 2022, 375, eaaw9021 CrossRef CAS PubMed; (b) T. R. Huycke and Z. J. Gartner, Science, 2022, 375, 26–27 CrossRef CAS PubMed.
- Y. Xue, J. Li, T. Jiang, Q. Han, Y. Jing, S. Bai and X. Yan, Adv. Healthcare Mater., 2024, 13, 2302180 CrossRef CAS PubMed.
- D. V. Deshmukh, P. Reichert, J. Zvick, C. Labouesse, V. Künzli, O. Dudaryeva, O. Bar-Nur, M. W. Tibbitt and J. Dual, Adv. Funct. Mater., 2022, 32, 2113038 CrossRef CAS.
- H. Liu, P. Chansoria, P. Delrot, E. Angelidakis, R. Rizzo, D. Rütsche, L. A. Applegate, D. Loterie and M. Zenobi-Wong, Adv. Mater., 2022, 34, 2204301 CrossRef CAS PubMed.
- H. Ma, C. Yang, Z. Ma, X. Wei, M. R. Younis, H. Wang, W. Li, Z. Wang, W. Wang, Y. Luo, P. Huang and J. Wang, Adv. Healthcare Mater., 2022, 11, 2102837 CrossRef CAS PubMed.
- (a) Y. Takagiwa, Z. Hou, K. Tsuda, T. Ikeda and H. Kojima, ACS Appl. Mater. Interfaces, 2021, 13, 53346–53354 CrossRef CAS PubMed; (b) Y. Liang, C. Jiao, P. Zhou, W. Li, Y. Zang, Y. Liu, G. Yang, L. Liu, J. Cheng, G. Liang, J. Wang, Z. Zhong and W. Yan, Bull. Chem. Soc. Jpn., 2023, 96, 148–155 CrossRef CAS; (c) Y. Komoto, J. Ryu and M. Taniguchi, Chem. Commun., 2023, 59, 6796–6810 RSC; (d) N. Saito, A. Nawachi, Y. Kondo, J. Choi, H. Morimoto and T. Ohshima, Bull. Chem. Soc. Jpn., 2023, 96, 465–474 CrossRef CAS; (e) K. Nakaguro, Y. Mitsuta, S. Koseki, T. Oshiyama and T. Asada, Bull. Chem. Soc. Jpn., 2023, 96, 1099–1107 CrossRef CAS.
- (a) S. Ju, T. Shiga, L. Feng, Z. Hou, K. Tsuda and J. Shiomi, Phys. Rev. X, 2017, 7, 021024 Search PubMed; (b) L. Himanen, A. Geurts, A. S. Foster and P. Rinke, Adv. Sci., 2019, 6, 1900808 CrossRef PubMed; (c) S. Hashimura, Y. Yamaguchi, H. Takeda, N. Tanibata, M. Nakayama, N. Niizeki and T. Nakaya, J. Phys. Chem. C, 2023, 127, 21665–21674 CrossRef CAS; (d) K. Hatakeyama-Sato, Polym. J., 2023, 55, 117–131 CrossRef CAS; (e) X. Zheng, X. Zhang, T.-T. Chen and I. Watanabe, Adv. Mater., 2023, 35, 2302530 CrossRef CAS PubMed; (f) S. Hashimura, Y. Yamaguchi, H. Takeda, N. Tanibata, M. Nakayama, N. Niizeki and T. Nakaya, J. Phys. Chem. C, 2023, 127, 21665–21674 CrossRef CAS.
- (a) W. Chaikittisilp, Y. Yamauchi and K. Ariga, Adv. Mater., 2022, 34, 2107212 CrossRef CAS PubMed; (b) R. Hikichi, Y. Tokura, Y. Igarashi, H. Imai and Y. Oaki, Bull. Chem. Soc. Jpn., 2023, 96, 766–774 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |