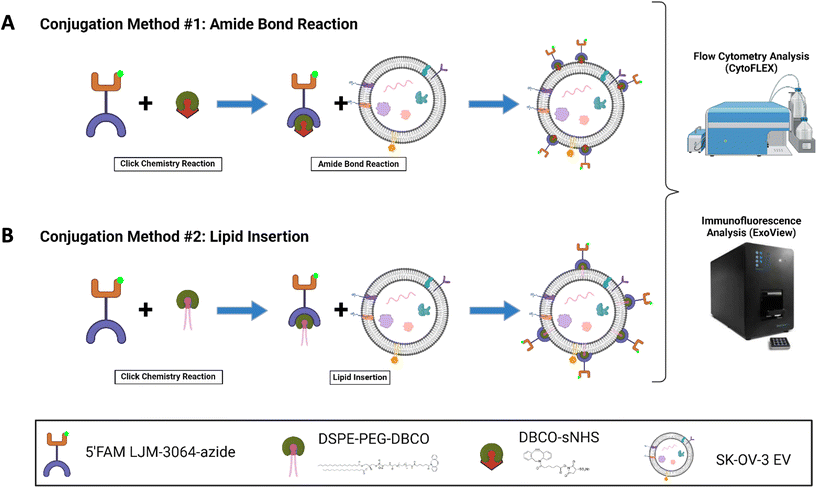
Assessing the conjugation efficiency of surface-modified extracellular vesicles using single nanovesicle analysis technologies
Leora
Goldbloom-Helzner
abc,
Harjn
Bains
c,
Emma G.
Loll
 ab,
Tanner
Henson
ac,
Rachel R.
Mizenko
c,
Priyadarsini
Kumar
ab,
Cheemeng
Tan
c,
Diana L.
Farmer
ab,
Randy P.
Carney
ab,
Tanner
Henson
ac,
Rachel R.
Mizenko
c,
Priyadarsini
Kumar
ab,
Cheemeng
Tan
c,
Diana L.
Farmer
ab,
Randy P.
Carney
 c and
Aijun
Wang
c and
Aijun
Wang
 *abc
*abc
aCenter for Surgical Bioengineering, Department of Surgery, School of Medicine, University of California-Davis, Sacramento, CA 95817, USA. E-mail: aawang@ucdavis.edu
bInstitute for Pediatric Regenerative Medicine, Shriners Children's, Sacramento, CA 95817, USA
cDepartment of Biomedical Engineering, University of California-Davis, Davis, CA 95616, USA
First published on 23rd September 2024
Abstract
Extracellular vesicles (EVs) are cell-secreted nanoscale vesicles with important roles in cell–cell communication and drug delivery. Although EVs pose a promising alternative to cell-based therapy, targeted delivery in vivo is lacking. Their surface is often modified to endow them with active targeting molecules to enable specific cell uptake and tailor EV biodistribution. A dominant paradigm has been to evaluate the EV surface functionalization using bulk analysis assays, such as western blotting and bead-based flow cytometry. Yet, the heterogeneity of EVs is now recognized as a major bottleneck for their clinical translation. Here, we engineer the EV surface at the single-vesicle level. We applied orthogonal platforms with single vesicle resolution to determine and optimize the efficiency of conjugating the myelin-targeting aptamer LJM-3064 to single EVs (Apt-EVs). The aptamers were conjugated using either lipid insertion or covalent protein modification, followed by an assessment of single-EV integrity and stability. We observed unique aptamer conjugation to single EVs that depends on EV size. Our study underscores the importance of single vesicle analysis for engineering EVs and provides a novel single-EV-based framework for modifying EV surfaces.
Introduction
Although cell-based therapy represents a promising approach in modern therapeutics, many studies suggest that cells exert their therapeutic effects via a paracrine mechanism.1,2 Cells secrete numerous factors, including a significant number of extracellular vesicles (EVs) to orchestrate key biological functions and cell behavior.3,4 The term “EV” is used as an umbrella term to broadly encompass nanoscale vesicles and related particles, ranging from ∼30 nm to more than 200 nm in diameter. EVs are derived from (among other mechanisms) the cell plasma membrane or endosomal machinery (canonical exosomes) and are recognized to play an important role in long-range cell–cell communication, transporting various functional molecules, including proteins, lipids, nucleic acids, glycans and metabolites.4–6 Compared to cell-based therapies, EVs are considered more shelf-stable, can exhibit lower immunogenicity in vivo,7 and can cross biological barriers, such as the blood brain barrier (BBB)8 (albeit the exact mechanisms are currently not fully elucidated), a major obstacle for most cell-based and drug therapies for central nervous system (CNS) diseases and injuries.5,9 Therefore, EVs represent a promising alternative approach to cell-based regenerative therapy.8,10 However, biodistribution studies examining EVs from different cell sources, such as the HEK293T, C2C12, B16-F10, and OLN-93 cell lines11 and umbilical cord mesenchymal stromal cells,12 and different common delivery routes (intravenous, intraperitoneal, and subcutaneous injection) have revealed that a significant amount of EVs accumulate in the lungs, spleen, and liver, while few naturally target the CNS.11–13 Therefore, there remains an unmet need to effectively deliver a sufficient dose of therapeutic EVs to specific cell types, such as neurons and oligodendrocytes, at desired injury sites in the brain and spinal cord.Several studies have explored the use of various surface modification methods to endow EVs with active targeting molecules, and thus boost adequate cell uptake in vitro and tailor EV biodistribution in vivo.14,15 For example, two chemical reactions, amine reactions (relying on binding to amino groups on EV proteins)16 and click chemistry,17,18 were validated by the addition of azide-fluor 545 onto EV surfaces.19 This study used high-performance liquid chromatography (HPLC) analysis of azide-fluor 545-conjugated liposomes to determine the number of binding groups on EVs for the estimation of EV conjugation efficiency. A subsequent study further explored the conjugation of a neuropilin-1 (NRP-1)-targeted peptide, RGE, to macrophage-derived EVs.20 Super-resolution microscopy confirmed the colocalization of the fluorescent RGE peptide with EVs with single vesicle resolution. Other studies have used conjugation methods that rely on the insertion of lipids into EV membranes. Previously, EV surfaces were modified with α-D-mannose or biotin through DSPE-incorporation, resulting in increased dendritic cell uptake. This study confirmed EV conjugation using a bulk biotin quantification kit.21 Another study used similar DSPE-based lipid insertion methods to PEGylate EV surfaces for improved tumor accumulation in vivo.22 Most recently, a study reported conjugation efficiency of a collagen-binding peptide, SILY, onto placental-derived mesenchymal stromal cell EVs (PMSC-EVs) using immunofluorescence image analysis technology, ExoView.23 To our knowledge, this is the only study that has assessed surface modification efficiency onto EVs via this technology. Although many studies have confirmed the conjugation of targeting molecules onto EV surfaces, there is not yet a universally established protocol to inform the optimization of EV surface conjugation nor is there a standard practice for comparing EV surface conjugation strategies.24 With this efficiency, researchers would have a standardized way to determine the degree of EV conjugation required to see the therapeutic effects of targeting molecules in vivo.
There is currently a lack of standard practices that can fully characterize and quantify the degree of EV surface conjugation (i.e., the conjugation efficiency) with single particle resolution over the size range of EVs (∼30–200 nm in diameter).25,26 Bulk analyses (such as bead-based flow cytometry27–30) have been used to assess the extent of EV labeling but do not address particle-to-particle differences within a mixture. This limitation may lead to inadequate surface modification, resulting in inaccurate conclusions in downstream dosing and functional assays. Here we applied two techniques with single EV resolution to assess EV conjugation efficiency: (1) a hybrid interferometry/immunofluorescence platform based on antibody chip capture31 and (2) single particle flow cytometry (EV-FC).32,33 In this study, we applied these two orthogonal platforms to optimize the conjugation efficiency of the aptamer LJM-3064, which binds with high affinity and high specificity to myelin,34–39 to modify a scalable EV source derived from a human ovarian cell line (SK-OV-3) as a proof of concept for engineering EVs for active targeting. To our knowledge, this is also the first study to optimize surface conjugation protocols and explore differential EV labeling based on size using immunofluorescence analysis (ExoView) technology. We also engineered PMSC-EVs, which have demonstrated neuroprotective and regenerative properties,23,40 with surface-conjugated LJM-3064 (PMSC-Apt-EVs) to assess its in vitro targeting potential to myelin-expressing cells. Finally, we will discuss differences between the two single vesicle analysis technologies to highlight method biases that could lead to differing results in efficiency calculation.
Experimental
EV isolation
The CELLine 1000AD bioreactor was utilized for the isolation of SK-OV-3 EVs as described previously.41,42 A total of 2.5 × 107 SK-OV-3 cells were seeded in the cell compartment of the bioreactor. These cells were maintained in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (complete growth medium). One liter of complete growth medium was added to the media compartment. After 24 h, to ensure full adherence of the cells to the matrix in the cell compartment, the cell compartment media was replaced with EV-depleted complete growth media, which was prepared by supplementing HyClone Medium high glucose with 1% penicillin/streptomycin and 10% FBS that had been centrifuged overnight at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g to deplete the bovine EVs. After 7 days of culture, the cell compartment media was extracted to isolate EVs and replaced with EV-depleted complete growth media. The media in the media compartment was also replaced. This process was repeated each week for a total of 8 weeks. PMSCs were isolated from early gestation placental chorionic villus tissue as previously described.4 The cells were expanded in D5 medium, including high-glucose DMEM, 5% FBS, 20 ng mL−1 recombinant human basic fibroblast growth factor (bFGF), 20 ng mL−1 recombinant human epidermal growth factor (EGF), and 1% penicillin/streptomycin, and incubated at 37 °C, 5% CO2. To isolate EVs, we followed optimized ultracentrifugation isolation procedures, as described previously.43 Briefly, 15 mL of cell culture supernatant was centrifuged at 300g for 10 min at 4 °C. The supernatant was collected and centrifuged at 2000g for 15 min at 4 °C. Then, the resulting supernatant was vacuum filtered through a 0.22 μM PES membrane. The filtered supernatant was transferred to thickwall polypropylene tubes (355462, Beckman Coulter) and centrifuged at 10
000g to deplete the bovine EVs. After 7 days of culture, the cell compartment media was extracted to isolate EVs and replaced with EV-depleted complete growth media. The media in the media compartment was also replaced. This process was repeated each week for a total of 8 weeks. PMSCs were isolated from early gestation placental chorionic villus tissue as previously described.4 The cells were expanded in D5 medium, including high-glucose DMEM, 5% FBS, 20 ng mL−1 recombinant human basic fibroblast growth factor (bFGF), 20 ng mL−1 recombinant human epidermal growth factor (EGF), and 1% penicillin/streptomycin, and incubated at 37 °C, 5% CO2. To isolate EVs, we followed optimized ultracentrifugation isolation procedures, as described previously.43 Briefly, 15 mL of cell culture supernatant was centrifuged at 300g for 10 min at 4 °C. The supernatant was collected and centrifuged at 2000g for 15 min at 4 °C. Then, the resulting supernatant was vacuum filtered through a 0.22 μM PES membrane. The filtered supernatant was transferred to thickwall polypropylene tubes (355462, Beckman Coulter) and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min (SW-28 rotor, Optima XL-100, Beckman Coulter) at 4 °C. This supernatant was spun twice at 120
000g for 30 min (SW-28 rotor, Optima XL-100, Beckman Coulter) at 4 °C. This supernatant was spun twice at 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 90 min at 4 °C, and the pellet was resuspended in triple-filtered PBS between spins. The final supernatant was discarded, and the pellet was resuspended in PBS. This solution was aliquoted to avoid freeze–thaw cycles and frozen at −80 °C until use.
000g for 90 min at 4 °C, and the pellet was resuspended in triple-filtered PBS between spins. The final supernatant was discarded, and the pellet was resuspended in PBS. This solution was aliquoted to avoid freeze–thaw cycles and frozen at −80 °C until use.
Conjugation methods
The LJM-3064 aptamer (5′-GGGTCGGCGGGTGGGGTGGGAGGTGGTC TTGTCTCTGGGT-3′) with a 5′ carboxyfluorescein modification and a 3′ azide modification was purchased from Integrated DNA Technologies.34 The folding process of LJM-3064 involved heating it to 95 °C for 5 minutes to denature secondary structures, followed by the addition of 1 mM MgCl2 to improve stability of the formed tertiary structures for functional application. Subsequently, the solution was cooled on ice for 8 minutes and further allowed to equilibrate at room temperature for 15 minutes before further use. Two conjugation methods, the amide bond reaction method and the lipid insertion method are explored in this study. The workflow schematic illustration of both methods is summarized in Fig. 1.Nanoparticle tracking analysis (NTA)
The EVs were diluted in ultrapure water for analysis by ZetaView (Particle Metrix) to determine their size, concentration, and zeta potential. The ZetaView tubing was rinsed with filtered ultrapure water prior to use. In accordance with the ZetaView manual, polystyrene 100 nm beads (Particle Metrix) were used for the daily calibration and autofocusing of particles within the cell. The samples were pre-diluted to an optimal concentration to allow for approximately 150 particles per frame. For each measurement, 11 positions were scanned for two cycles using the following parameters: camera sensitivity: 88, shutter: 150, frame rate: 30, and cell temperature: 25 °C.Cryogenic electron microscopy (CryoEM)
Aptamer-conjugated and native EV solutions were vitrified following standard protocols described elsewhere.44 Quantifoil grids (Orthogonal Array of 1.2 μm Diameter Holes – 1.3 μm Separation, mounted on a 200 M Cu grid #658-200-CU, Ted Pella) were vitrified in liquid ethane using a Leica EM GP2 Plunge Freezer after negative glow-discharge treatment of the grids and deposition of 4 μL of each sample. A Thermo Fisher Glacios electron microscope equipped with X-FEG optics, a Gatan K3 direct electron detector, and automation software (SerialEM) was used. The samples were vitrified during electron microscopy analysis, and cryoEM images were obtained at different magnifications (45k and 11k).Western blot
Western blotting of EV samples was performed according to previously established protocols.45 Thirteen microliters of EVs (per lane) were treated with NuPAGE LDS Sample Buffer (Thermo Fisher Scientific) containing the reducing agent DTT. The samples were heated to 90 °C for 5 min and centrifuged using a Sorvall Biofuge Pico Centrifuge (Heraeus #3325B rotor) at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 2 min. The samples were loaded on a 4–12% graded Tris-glycine SDS-polyacrylamide gel and run for 90 min at 150 V. The proteins were transferred to a 0.2 μm nitrocellulose membrane (Life Technologies) for 45 min at 100 V. The membranes were blocked in 5% nonfat dry milk in PBS with 0.05% Tween-20 for 1 h at room temperature with rotation, and then incubated with primary antibody in blocking buffer at 4 °C overnight. Antibodies against ALIX (Sigma Aldrich), tumor susceptibility gene 101 (TSG101) (Millipore Sigma), and calnexin (Cell Signaling Technology) were used at a 1
000g for 2 min. The samples were loaded on a 4–12% graded Tris-glycine SDS-polyacrylamide gel and run for 90 min at 150 V. The proteins were transferred to a 0.2 μm nitrocellulose membrane (Life Technologies) for 45 min at 100 V. The membranes were blocked in 5% nonfat dry milk in PBS with 0.05% Tween-20 for 1 h at room temperature with rotation, and then incubated with primary antibody in blocking buffer at 4 °C overnight. Antibodies against ALIX (Sigma Aldrich), tumor susceptibility gene 101 (TSG101) (Millipore Sigma), and calnexin (Cell Signaling Technology) were used at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution. Blots were then probed with their respective secondary antibodies. Secondary antibodies (goat anti-rabbit) modified with HRP were diluted at 1
500 dilution. Blots were then probed with their respective secondary antibodies. Secondary antibodies (goat anti-rabbit) modified with HRP were diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2500 in blocking buffer and incubated for 1 h at room temperature (23 °C) with rotation. Proteins were visualized with Supersignal West Pico PLUS chemiluminescent substrate (Thermo Fisher) using a ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA, USA).
2500 in blocking buffer and incubated for 1 h at room temperature (23 °C) with rotation. Proteins were visualized with Supersignal West Pico PLUS chemiluminescent substrate (Thermo Fisher) using a ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA, USA).
ExoView tetraspanin kit assay
Leprechaun Kits were used as purchased (Unchained Labs – previously NanoView Biosciences) to probe EVs at the single EV level. Chips were pre-scanned using the provided protocol to identify any previously adhered particles or scratches during manufacturing. Chips were placed in a 24 well plate (provided by Unchained Labs), avoiding contact with the sides of the well to prevent loss of sample from the chip. Apt-EV samples were diluted to 3 × 109 particles per mL in the incubation buffer provided in the kit. Then, 35–50 μL of the diluted Apt-EV solution was pipetted directly onto each chip. The plate was then covered with a plate sealer and aluminum foil to be incubated overnight at room temperature (23 °C). The chips in the 24 well plate were then washed to remove unbound particles utilizing the ExoView CW100 Plate Washer with MilliQ water and two buffer solutions (Solution A and Solution B) provided by the kit. After being prompted by the plate washer, the detection antibody solution was prepared by adding 0.6 μL of the provided tetraspanin panel (CF647-anti-CD63 and CF555-anti-CD81) to 300 μL of blocking solution per chip. After incubation and rinsing cycles with a plate washer, each chip was placed on the tilted side of its well and the plate was placed back into the plate washer for the remainder of the buffer to be drained completely. The chips were then placed on a Kim wipe to dry any residual wetness taking care not to disturb the top face of the chip. The chips were then transferred to the chuck and scanned for interferometric and fluorescence imaging. During data analysis, fluorescence cut-offs were chosen by limiting the number of detected particles on mouse IgG (MIgG) capture spots to fewer than 100 events.Single EV flow cytometry (EV-FC)
Flow cytometry (CytoFLEX, Beckman Coulter) was also used to assess EV surface modifications. Previously filtered PBS (through a Whatman Anotop 0.02 μm syringe filter) was run at the lowest flow rate of 10 μL min−1 and 405 nm SSC was used to trigger EV particles above background noise with a threshold of 2500 (a.u.). The blue laser channel (525 nm) was used to gate the LJM-3064+ EVs using unmodified SK-OV-EVs. The gate was drawn to reach at most 1% conjugation on unmodified SK-OV-3 EVs before application to Apt-EV samples. All samples were diluted such that the event rate remained between 2000 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per second and then run for 120 s at the lowest flow rate of 10 μL min−1. Each sample was recorded until the total number of events reached 500
000 events per second and then run for 120 s at the lowest flow rate of 10 μL min−1. Each sample was recorded until the total number of events reached 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Standard beads and FCMPASS software were used to calibrate the size and fluorescence of the Apt-EVs.46 For size calibration, we combined NIST traceable polystyrene beads (Thermo Fisher) of the following sizes: 81, 100, 152, 203, 269, 345, 401, and 453 nm. The beads were diluted in 0.2 μm-filtered MilliQ water to reach a total event rate of approximately 10
000. Standard beads and FCMPASS software were used to calibrate the size and fluorescence of the Apt-EVs.46 For size calibration, we combined NIST traceable polystyrene beads (Thermo Fisher) of the following sizes: 81, 100, 152, 203, 269, 345, 401, and 453 nm. The beads were diluted in 0.2 μm-filtered MilliQ water to reach a total event rate of approximately 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per s and recorded for 120 s. These data were analyzed in FCMPASS software to convert the arbitrary scatter intensity to the nominal diameter of the particles. For fluorescence calibration for the 525 nm laser, beads of 5 specified populations of FITC using Molecules of Equivalent Soluble Fluorophores (MESF) values were run similarly to the NIST beads on the CytoFLEX. These data were also analyzed in FCMPASS software to convert the arbitrary fluorescence units to MESF values.47
000 events per s and recorded for 120 s. These data were analyzed in FCMPASS software to convert the arbitrary scatter intensity to the nominal diameter of the particles. For fluorescence calibration for the 525 nm laser, beads of 5 specified populations of FITC using Molecules of Equivalent Soluble Fluorophores (MESF) values were run similarly to the NIST beads on the CytoFLEX. These data were also analyzed in FCMPASS software to convert the arbitrary fluorescence units to MESF values.47
Oligodendroglioma uptake assay
PMSC-EVs and PMSC-Apt-EVs were labeled with CellTrace™ CFSE dye (Thermo Fisher Scientific) at a concentration of 20 μM. CFSE was incubated with 1 × 1010 PMSC-EVs or PMSC-Apt-EVs for 1 h at 37 °C on rotation. Excess CFSE was removed using SEC. Human oligodendroglioma cells (HOG, Millipore) were cultured in D10 medium, including high-glucose DMEM, 10% FBS, 100 UI mL−1 of penicillin and 100 μg mL−1 of streptomycin and incubated at 37 °C, 5% CO2. HOGs were seeded onto 48 well plates (CELLTREAT) at 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 per well. CFSE-PMSC-EVs or CFSE-PMSC-Apt-EVs were added to each well and incubated with the HOGs at a ratio of 1 × 109 per 60k cells for 4 h. In the control groups, CFSE free dye or CFSE-labeled PMSC-EVs with a scrambled aptamer (CSFE-PMSC-ScrApt-EVs) were incubated with the HOGs for 4 h. Media with CFSE-PMSC-EVs or CFSE-PMSC-Apt-EVs was aspirated, and the cells were fixed in 10% Formalin (Epredia) for 30 min. After washing with PBS, the cells were blocked in 1% Bovine Serum Albumin (BSA, Thermo Fisher Scientific) for 1 h at room temperature and membranes were permeabilized by incubation with 0.5% Triton X-100 in PBS. Cells were then incubated overnight with a primary antibody against myelin basic protein (Rb anti-MBP, Abcam, ab216668) at 1
000 cells per cm2 per well. CFSE-PMSC-EVs or CFSE-PMSC-Apt-EVs were added to each well and incubated with the HOGs at a ratio of 1 × 109 per 60k cells for 4 h. In the control groups, CFSE free dye or CFSE-labeled PMSC-EVs with a scrambled aptamer (CSFE-PMSC-ScrApt-EVs) were incubated with the HOGs for 4 h. Media with CFSE-PMSC-EVs or CFSE-PMSC-Apt-EVs was aspirated, and the cells were fixed in 10% Formalin (Epredia) for 30 min. After washing with PBS, the cells were blocked in 1% Bovine Serum Albumin (BSA, Thermo Fisher Scientific) for 1 h at room temperature and membranes were permeabilized by incubation with 0.5% Triton X-100 in PBS. Cells were then incubated overnight with a primary antibody against myelin basic protein (Rb anti-MBP, Abcam, ab216668) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 dilution in 1% BSA at 4 °C. After being washed with PBS 3 times, the cells were incubated with the relevant secondary antibody (Invitrogen) at 1
250 dilution in 1% BSA at 4 °C. After being washed with PBS 3 times, the cells were incubated with the relevant secondary antibody (Invitrogen) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution in PBS for 1 h at room temperature, and then nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific). After being washed with PBS 3 times, the 20× images were captured using the FV1200 Fluoview confocal laser scanning microscope. Quantification of the number of cells with internalized CFSE-PMSC-EVs or CFSE-PMSC-Apt-EVs was performed using an ImageJ coded program (NIH) reliant on pixel intensity threshold settings.
500 dilution in PBS for 1 h at room temperature, and then nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific). After being washed with PBS 3 times, the 20× images were captured using the FV1200 Fluoview confocal laser scanning microscope. Quantification of the number of cells with internalized CFSE-PMSC-EVs or CFSE-PMSC-Apt-EVs was performed using an ImageJ coded program (NIH) reliant on pixel intensity threshold settings.
Statistical methods
All experiments were performed in triplicate, and the error bars represent one standard deviation (s.d.) from the mean. Statistical significance was assessed using the built-in functions of GraphPad Prism 7 software. For comparisons of two populations, Student's t-test was used, while ANOVA was used for comparisons of three or more populations. Data is presented as mean and standard error. ns = not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.Results
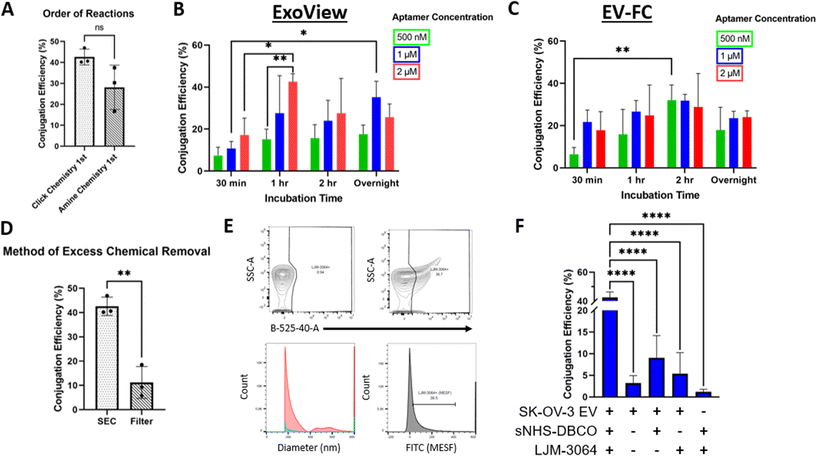
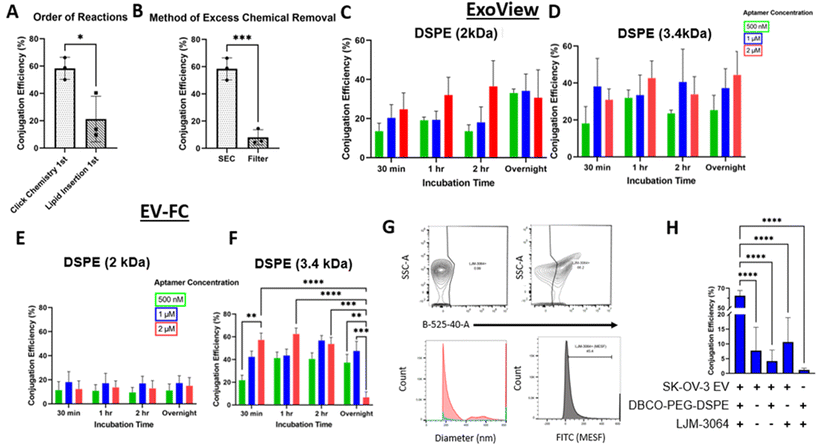
The conjugation of LJM-3064 to SK-OV-3 EVs was optimized for two conjugation methods: (1) amide bond reaction method (Fig. 1A) and (2) lipid insertion method (Fig. 1B) – both of which employed click chemistry as part of the reactions. In both conjugations, LJM-3064 was modified with a 3′ azide group and a 5′ carboxyfluorescein. Both conjugation methods were optimized by varying the following parameters: order of reactions (click chemistry 1st followed by amide bond reactions or lipid insertion vs. amide bond reactions or lipid insertion 1st followed by click chemistry), method of excess chemical removal (ultrafilter vs. SEC), length of PEG spacer (2 kDa vs. 3.4 kDa), incubation times (30 min, 1 h, 2 h, or overnight), and aptamer concentration (500 nM, 1 μM, or 2 μM). The various incubation steps allowed either sNHS to react with amine groups on the proteins present on EV surfaces in an amide bond reaction or DSPE to incorporate into the EV lipid bilayer in the lipid insertion method. EV conjugation efficiency was calculated at the single EV level to determine exact conjugation efficiencies using two technologies: one based on both a single-particle interferometric reflectance imaging sensor and immunofluorescence, and the other based on EV-FC. We investigated all parameters previously described to maximize the percentage of LJM-3064+ EVs within both modification strategies. We then characterized Apt-EVs post-conjugation for preserved physical and biochemical characteristics and explored the patterns of LJM-3064 conjugation within different EV subpopulations based on size.Calculation of Apt-EV conjugation efficiency
Apt-EVs were incubated on ExoView® chips, captured by 3 tetraspanin capture antibodies (CD9, CD63, and CD81), and fluorescently labeled for CD63 and CD81. The detected conjugation efficiency of Apt-EVs was calculated by dividing the number of LJM-3064+ EVs (488 nm) by the total number of immobilized EVs (detected by fluorescently-tagged CD63 and CD81 antibodies). Apt-EVs were also run as samples on the CytoFLEX. The detected conjugation efficiency of Apt-EVs was calculated by the percentage of EV particles included in the LJM-3064+ gate.Informed optimization of Apt-EVs by varying protocol parameters
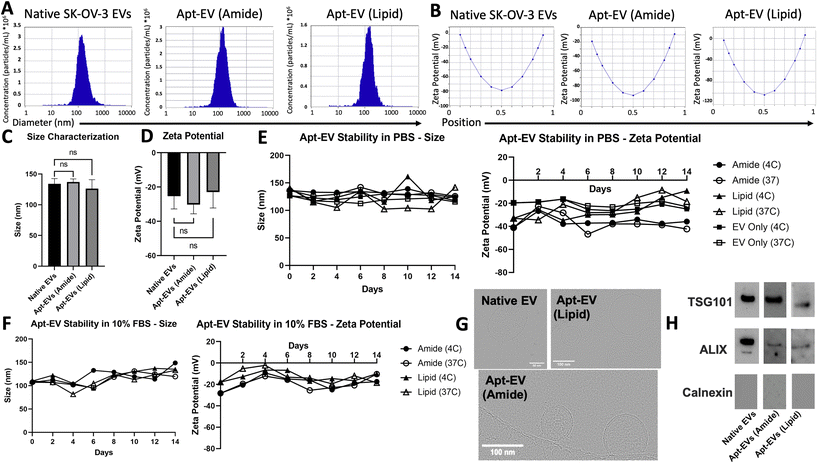
Apt-EV physical and biochemical properties are preserved post-conjugation
In accordance with the Minimum Information for Studies of Extracellular Vesicles 2018 (MISEV2018) guidelines,48 physical characterization, including size, surface charge, morphological analysis, and biomarker expression, was conducted to demonstrate that conjugation did not compromise EV integrity. NTA was used to confirm the retained size profiles and surface charge of the Apt-EVs. Representative size profiles identified EVs – both modified and unmodified – at 100–200 nm in diameter, which is down to the effective limit of detection of the ZetaView instrument (Fig. 4A). Zeta potential was also measured for native SK-OV-3 EVs as well as both Apt-EVs. The surface charge ranged from −30 to −20 mV (Fig. 4B). We compared the size distributions of native SK-OV-3 EVs, amide bond Apt-EVs and lipid insertion Apt-EVs. EV size (Fig. 4C) and surface charge (Fig. 4D) did not change significantly due to either surface modification method. Apt-EVs stored in PBS or 10% FBS were also observed over a two-week period under both 4 °C and 37 °C storage conditions to determine their stability under short-term shelf storage and clinically relevant conditions. NTA measurements of EVs over this period revealed stable size profiles at 100–150 nm. NTA revealed that the surface charge of the amide bond Apt-EVs’ in PBS remained stable over the two-week period, while the surface charge of lipid insertion Apt-EVs’ began to increase around day 10 (Fig. 4E). When stored in 10% FBS, the surface charge of the amide bond and lipid insertion Apt-EVs remained relatively stable at both temperatures, with fluctuations between −28 mV and −2 mV (Fig. 4F). CryoEM confirmed that EV integrity was not significantly affected post-conjugation. The EV membrane remained intact and most EV particles maintained a spherical shape (Fig. 4G). Western blotting analysis confirmed the preservation of the typical EV markers TSG101 and ALIX and the absence of the endoplasmic reticulum marker calnexin on Apt-EVs post-conjugation compared to native SK-OV-3 EVs (Fig. 4H).Differential labeling of LJM-3064 on EVs based on size
Analysis of both conjugation methods by immunofluorescence revealed differential LJM-3064 labeling of EVs for both amide bond reactions (Fig. 5A) and lipid insertion (Fig. 5B) methods based on size breakdown. The Apt-EV conjugation efficiency was calculated after the sample data were broken down into 10 nm fragments ranging from 50–150 nm (50–60 nm, 60–70 nm, etc.). It was apparent that for both conjugation methods, the total EV size distributions were similar. Additionally, nearly 50% of the total Apt-EVs from both conjugation methods were between 50 and 60 nm in diameter. Additionally, Apt-EVs with a diameter of 100 nm or more made up less than 15% of the total population of EVs captured and detected on the single particle interferometric reflectance imaging sensor (SP-IRIS) chip. Analysis of the Apt-EVs for both conjugation methods revealed that larger EVs (>80 nm) were more efficiently labeled than smaller EVs (50–80 nm), even though larger EVs made up a smaller portion of the total captured EV sample. For instance, for Apt-EV samples that fell within a size range of 50–60 nm (which made up ∼50% of the total captured EV sample), the average conjugation efficiency was less than 5%.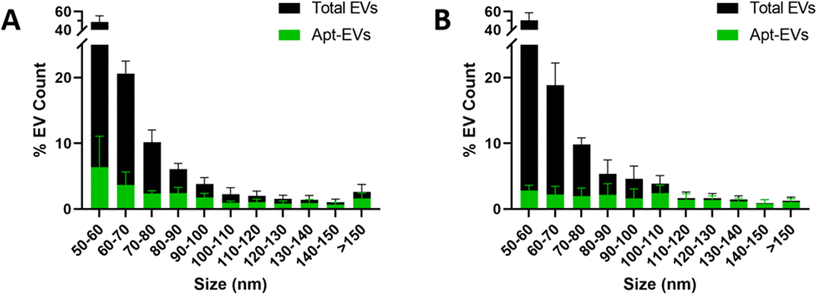 | ||
| Fig. 5 Analysis of differential labeling of Apt-EVs from amide bond reactions (A) and lipid insertion (B) based on size. | ||
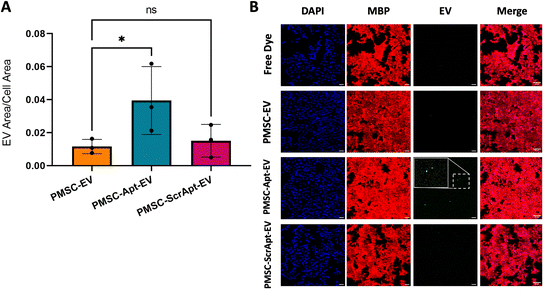
PMSC-Apt-EVs showed significantly increased uptake in oligodendrocytes in vitro
To assess for improved uptake of Apt-EVs in myelin-expressing cells, fluorescently-labeled PMSC-EVs, PMSC-Apt-EVs, and a scrambled aptamer-conjugated PMSC-EV control (PMSC-ScrApt-EVs) were added to HOG cells for 4 hours. The lipid insertion conjugation method was used with click chemistry 1st, 2 μM LJM-3064, 3.4 kDa DSPE-PEG-DBCO, EV incubation for 1 h, and SEC instead of ultrafiltration. The quantification of EV area within the cell area demonstrated significantly increased uptake of PMSC-Apt-EVs compared to native PMSC-EVs (p < 0.05). After employing the PMSC-ScrApt-EVs, no statistically significant enhancement in cell uptake was observed, suggesting that the observed improvement in cell uptake of EVs can be attributed to the functional LJM-3064 aptamer (Fig. 6A and B).Discussion
This study aimed to fully characterize and inform optimization of surface-modified EVs via platforms with single vesicle resolution. We examined the differential conjugation efficiencies of two common conjugation approaches, either based on protein conjugation or lipid insertion. To our knowledge, this is the first study to assess conjugation efficiency via immunofluorescence imaging and EV-FC. The advantages, disadvantages, and limits of detection for each platform are discussed in the following sections, ultimately demonstrating the need for orthogonal measures to increase the accuracy and comprehensive characterization of engineered EVs.Single EV analysis technologies fundamentally measure different particle populations, leading to inconsistencies in the calculation of conjugation efficiency
NTA relies on Brownian motion to determine EV size and concentration. A camera captures the Brownian motions of EVs and uses a laser to track the scattered light of the particles. The ZetaView limit of detection is approximately 85 nm, and most of the detected particles are between 90 and 300 nm in diameter.49 ExoView uses SP-IRIS to determine the size of EVs by measuring the contrast enhancement in scattered light from nanoparticles bound to a layered silicon substrate.50 The limit of detection of SP-IRIS has been reported in some studies to be 50 nm (ref. 49 and 50) and SP-IRIS can detect particles up to 200 nm in diameter. EV-FC, measured in this study by a Beckman Coulter CytoFLEX, uses the side scatter of the 405 nm violet laser as a trigger channel to detect EVs from noise.51,52 FC size measurements were calibrated using standard beads and FCMPASS software.47 Based on this calibration, the limit of detection of this flow cytometer was measured to be ∼150 nm. It is clear from these limitations that no single platform is suitable for assessing the full-size range of EVs. The limits of detection of many technologies vary so widely that each reported measurement may describe different subpopulations of the same sample.49 Moreover, the precise size range of EVs is not known. Typical studies have reported a lower range of EVs of ∼50 nm, with much variance around that figure in recently published literature.53–57 Ultimately, the different limits of detection lead to inconsistencies in directly comparing conjugation efficiencies across various platforms.The different limits of detection are further confounded in immunofluorescence imaging, which relies on captured tetraspanin-expressing particles. Instead, FC relies on thresholding to count any particle above background noise and is not biased by tetraspanin capture. In terms of detecting the number of LJM-3064 positive particles, immunofluorescence imaging only counts Apt-EVs or potentially adsorbed residual aptamer onto the chip, whereas the FC counts any particle positive for the aptamer, which could be aggregates of aptamer or other components (e.g., DSPE aggregates, EV fragments). These potential impurities could also influence NTA counts and size/surface charge distributions. This finding highlights the importance of using orthogonal technologies to fully describe the characteristics of Apt-EV samples.
The Apt-EV conjugation efficiencies reported in this paper differed between the results of immunofluorescence imaging and EV-FC calculations. For instance, the conjugation efficiency was greater when using immunofluorescence imaging than when using EV-FC with the same parameters (e.g., 2 kDa DSPE) in lipid insertion. This could be explained by ExoView's tendency to detect a lower number of total particles based on the equipment's discrimination of non-tetraspanin-expressing particles. This may also be due to the higher limit of detection EV-FC, which could lead to misrepresentation of Apt-EV subpopulations based on size. Further characterization of Apt-EV size distribution and tetraspanin expression (in the case of EV-FC) will be needed to better equate these two technologies.
Differential conjugation efficiencies based on EV size emerge across protein- or lipid-based conjugates
To the best of our knowledge, we are the first to explicitly investigate the dependence of EV conjugation efficiency on EV size.19 Upon analysis, it was clear that the conjugation efficiencies of Apt-EVs for both conjugation methods were different based on their size breakdown. In both amide bond reactions and lipid insertion methods, Apt-EVs were disproportionately labeled such that as the EV size increased, the efficiency of conjugation also increased (as detected by immunofluorescence). This differential labeling of LJM-3064 on EVs has implications for understanding differential conjugation efficiency based on EV size, including further investigation of differential functions of EVs based on size in terms of EV surface protein expression and native cargo. This phenomenon may be attributed to the conjugation method. In amide bond reactions, potential differences in surface protein density could influence conjugation efficiency. If protein density – i.e., the number of proteins per surface area (nm2) – is consistent regardless of EV size, larger EVs (100–200 nm) would have a greater absolute number of surface proteins and therefore would have more amine groups available for protein-aided conjugation (i.e., amide bond reactions). Literature has also shown that NHS hydrolysis is dependent on the pH of the buffer solution.58 A reduction in available reactive amine groups could explain why protein-based conjugation does not reach maximum efficiency.59,60 In this study, we used neutral buffer PBS for click chemistry reactions to support the stability of EVs, as vesicle aggregation may occur in slightly acidic or alkaline pH conditions.61 Future studies could evaluate different pH conditions as an additional parameter to further optimize conjugation efficiency and test the other approaches that have been used to modify cell membrane with NHS-PEG-maleimide and thiolated peptides or proteins.62,63 For lipid insertion, the difference in LJM-3064 labeling could be due to an increase in surface area and a potential increase in lipid insertion locations within the larger EVs. Changes in membrane curvature as well as steric hindrance of smaller EVs may also reveal interesting explanations for these observations.EV biogenesis could also be a factor that affects EV surface conjugation efficiency. For instance, smaller EVs (30–100 nm) are largely derived from the invagination of endosomes (via multivesicular bodies) and downstream fusion with the plasma membrane for release into the extracellular space; these vesicles are canonically referred to as “exosomes”. Larger EVs are largely derived from the plasma membrane through membrane blebbing and pinching off into the extracellular space. Both of these simplified vesicle populations overlap greatly, and no unique marker has been found to indicate biogenesis pathways.50 However, some investigations are underway to examine proteomic differences across sizes,64 although surface protein density remains elusive.
These theories for both conjugation methods, however, would have to be further investigated through fractionation of the EV population based on size and careful characterization of total protein content and lipid content, a technological feat that does not yet exist. Although this study investigated two direct EV conjugation methods for comparison, additional conjugations are warranted to fully describe the nature of conjugation efficiencies based on EV size distribution.
Uptake of PMSC-Apt-EVs in HOG cells reveals the targeting potential of LJM-3064 in vitro
We engineered PMSC-EVs by surface-conjugation with the LJM-3064 aptamer and applied PMSC-Apt-EVs to HOG cells to assess the targeting potential of LJM-3064 to myelin-expressing cells. After quantifying the EV area within the cell area, there was no significant difference in uptake of PMSC-EVs compared to PMSC-ScrApt-EVs. This data supports the notion that the observed improvement in cell uptake of PMSC-Apt-EVs can be attributed to the functional LJM-3064 aptamer. While this study confirmed the in vitro targeting potential of LJM-3064, further studies should be conducted to assess the aptamer's targeting potential to the CNS in other ex vivo or in vivo animal models.Conclusions
This study aimed to standardize the process of optimizing and fully describing surface-conjugated EVs using single EV analysis technologies. This work shows how immunofluorescence imaging (ExoView) and EV-FC (CytoFLEX) technologies can help inform EV surface conjugation protocols through variation of method parameters and calculation of the resultant conjugation efficiency. The optimization of both amide bond reaction and lipid insertion conjugations explored how the incubation time, aptamer concentration, method of excess chemical removal, and order of reactions affected the conjugation efficiency. In both conjugations, running click chemistry 1st before either amide bond reactions or lipid insertion, and using SEC instead of an ultrafilter resulted in the highest conjugations. Immunofluorescence imaging and EV-FC were used to measure different percentages of LJM-3064+ EVs, but further evaluation of the limits of detection of each instrument revealed that the conjugation efficiency reported by each instrument may describe a different subpopulation of EVs based on size. Physical and biomarker characterization, including NTA, cryoEM, and western blot analysis was performed to successfully confirm the preservation of EV integrity and stability after modification. In addition, further inspection of both conjugation methods via ExoView revealed differential LJM-3064 labeling of EVs via both the amide bond reaction method and the lipid insertion method based on EV size. In vitro uptake in oligodendrocytes demonstrated significantly increased PMSC-Apt-EV uptake compared to native PMSC-EVs and PMSC-ScrApt-EVs suggesting that EV surface conjugation with targeting ligands can effectively enhance the EV targeting potential. These observations will enhance researchers’ understanding of the differential conjugation efficiency in their own EV samples and encourage them to evaluate the targeting and functional properties of EVs based on size to confirm that they are delivering functional EVs to their desired tissue target in vivo. This overall engineering strategy was a necessary step in preparing EVs for improved targeting while maintaining the physical and biomolecular properties of native EVs. Few studies have quantified the degree of EV conjugation needed to observe a significant improvement in targeting and functional effects in vitro and in vivo. The practices established in this study are the first step in determining this important correlation.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. L. G. H.: conceptualization, methodology, data collection and analysis, writing—original draft preparation, writing—review and editing. H. B.: data collection and analysis, writing—review and editing. E. G. L.: data collection and analysis, writing—review and editing. T. H.: data collection and analysis. R. R. M.: data collection and analysis, writing—review and editing. P. K.: conceptualization, writing—review and editing. C. T.: supervision. D. L. F.: supervision. R. P. C.: supervision, conceptualization, writing—review and editing. A. W.: supervision, conceptualization, methodology, data collection and analysis, writing—review and editing, funding acquisition.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported in part by the UC Davis School of Medicine Cultivating Team Science Award, the UC Davis Health Ventures Office, Venture Catalyst's Science Translation and Innovative Research (STAIR™) proof-of-concept grant program, the UC Davis College of Engineering Next Level Vision Award, National Institutes of Health (NIH) grants (1R01NS115860-01A1, 1R01EB034279-01, 1R01EB033389-01A1, and 1R01NS131538-01A1), and Shriners Children's research grants (85135-NCA-21, 70012-NCA-23, 85400-NCA-24, and 85220-NCA-24). L. G. H. was supported by a Predoctoral Fellowship in Drug Delivery from the PhRMA Foundation. T. H. was supported by the Department of Defense (DOD) through the National Defense Science and Engineering Graduate (NDSEG) Fellowship program. The funding sources had no involvement in the study design, collection, data analysis, data interpretation, writing of the manuscript, or the decision to submit the manuscript for publication. This project was supported by the UC Davis Flow Cytometry Shared Resource Laboratory with funding from UC Davis Comprehensive Cancer Center Support Grant (CCSG) awarded by the National Cancer Institute (NCI P30 CA093373 and S10 OD018223) with technical assistance from Bridget McLaughlin. Furthermore, this work was supported by the UC Davis BioEM Facility (funded by user fees, the Department of Molecular and Cellular Biology, the College of Biological Sciences, the Office of Research, and the Provost's Office) and led by the technical director, Dr Fei Guo (who is supported by discretionary funds provided by Professor Jodi Nunnari (MCB)). The authors would also like to acknowledge BioRender for providing the software used to generate Fig. 1.References
- X. Liang, Y. Ding, Y. Zhang, H. F. Tse and Q. Lian, Paracrine mechanisms of mesenchymal stem cell-based therapy: Current status and perspectives, Cell Transplant., 2014, 23(9), 1045–1059, DOI:10.3727/096368913X667709/ASSET/IMAGES/LARGE/10.3727_096368913X667709-FIG1.JPEG.
- S. Fukushima, N. G. Campbell and S. R. Coppen, et al., Quantitative assessment of initial retention of bone marrow mononuclear cells injected into the coronary arteries, J. Heart Lung Transplant., 2011, 30(2), 227–233, DOI:10.1016/J.HEALUN.2010.09.002.
- Y. J. Chen, K. Chung and C. Pivetti, et al., Fetal surgical repair with placenta-derived mesenchymal stromal cell engineered patch in a rodent model of myelomeningocele, J. Pediatr. Surg., 2018, 53(1), 183–188, DOI:10.1016/j.jpedsurg.2017.10.040.
- L. Lankford, Y. J. Chen and Z. Saenz, et al., Manufacture and preparation of human placenta-derived mesenchymal stromal cells for local tissue delivery, Cytotherapy, 2017, 19(6), 680–688, DOI:10.1016/j.jcyt.2017.03.003.
- L. Margolis and Y. Sadovsky, The biology of extracellular vesicles: The known unknowns, PLos Biol., 2019, 17(7), e3000363, DOI:10.1371/journal.pbio.3000363.
- J. Phan, P. Kumar, D. Hao, K. Gao, D. Farmer and A. Wang, Engineering mesenchymal stem cells to improve their exosome efficacy and yield for cell-free therapy, J. Extracell. Vesicles, 2018, 7(1), 1522236, DOI:10.1080/20013078.2018.1522236.
- J. Zeng, C. Gu, Y. Sun and X. Chen, Engineering of M2 Macrophages-Derived Exosomes via Click Chemistry for Spinal Cord Injury Repair, Adv. Healthc. Mater., 2023, 12(11), 2203391, DOI:10.1002/ADHM.202203391.
- Z. Ren, Y. Qi, S. Sun, Y. Tao and R. Shi, Mesenchymal Stem Cell-Derived Exosomes: Hope for Spinal Cord Injury Repair, Stem Cells Dev., 2020, 29(23), 1467–1478, DOI:10.1089/scd.2020.0133.
- J. Saint-Pol, F. Gosselet, S. Duban-Deweer, G. Pottiez and Y. Karamanos, Targeting and Crossing the Blood-Brain Barrier with Extracellular Vesicles, Cells, 2020, 9(4), 851, DOI:10.3390/cells9040851.
- W. Liu, Y. Wang and F. Gong, et al., Exosomes derived from bone mesenchymal stem cells repair traumatic spinal cord injury by suppressing the activation of a1 neurotoxic reactive astrocytes, J. Neurotrauma, 2019, 36(3), 469–484, DOI:10.1089/neu.2018.5835.
- O. P. B. Wiklander, J. Z. Nordin and A. O'Loughlin, et al., Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting, J. Extracell. Vesicles, 2015, 4(2015), 1–13, DOI:10.3402/jev.v4.26316.
- A. M. Tolomeo, G. Zuccolotto and R. Malvicini, et al., Biodistribution of Intratracheal, Intranasal, and Intravenous Injections of Human Mesenchymal Stromal Cell-Derived Extracellular Vesicles in a Mouse Model for Drug Delivery Studies, Pharmaceutics, 2023, 15(2), 548, DOI:10.3390/PHARMACEUTICS15020548/S1.
- C. P. Lai, O. Mardini and M. Ericsson, et al., Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter, ACS Nano, 2014, 8(1), 483–494, DOI:10.1021/nn404945r.
- L. Ramasubramanian, P. Kumar and A. Wang, Engineering Extracellular Vesicles as Nanotherapeutics for Regenerative Medicine, Biomolecules, 2019, 10(1), 48, DOI:10.3390/biom10010048.
- L. Goldbloom-Helzner, D. Hao and A. Wang, Developing Regenerative Treatments for Developmental Defects, Injuries, and Diseases Using Extracellular Matrix Collagen-Targeting Peptides, Int. J. Mol. Sci., 2019, 20(17), 4072, DOI:10.3390/ijms20174072.
- T. Tian, H. Zhang and C. He, et al., Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy, Biomaterials, 2018, 150, 137–149, DOI:10.1016/j.biomaterials.2017.10.012.
- S. I. Presolski, V. P. Hong and M. G. Finn, Copper-Catalyzed Azide–Alkyne Click Chemistry for Bioconjugation, Curr. Protoc. Chem. Biol., 2011, 3(4), 153–162, DOI:10.1002/9780470559277.CH110148.
- S. Mack, M. F. Fouz, S. K. Dey and S. R. Das, Pseudo-Ligandless Click Chemistry for Oligonucleotide Conjugation, Curr. Protoc. Chem. Biol., 2016, 8(2), 83–95, DOI:10.1002/CPCH.1.
- T. Smyth, K. Petrova and N. M. Payton, et al., Surface functionalization of exosomes using click chemistry, Bioconjugate Chem., 2014, 25(10), 1777–1784, DOI:10.1021/bc500291r.
- G. Jia, Y. Han and Y. An, et al., NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo, Biomaterials, 2018, 178, 302–316, DOI:10.1016/J.BIOMATERIALS.2018.06.029.
- E. S. Choi, J. Song, Y. Y. Kang and H. Mok, Mannose-Modified Serum Exosomes for the Elevated Uptake to Murine Dendritic Cells and Lymphatic Accumulation, Macromol. Biosci., 2019, 19(7), e1900042, DOI:10.1002/MABI.201900042.
- S. A. A. Kooijmans, L. A. L. Fliervoet and R. Van Der Meel, et al., PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time, J. Controlled Release, 2016, 224, 77–85, DOI:10.1016/J.JCONREL.2016.01.009.
- D. Hao, L. Lu and H. Song, et al., Engineered extracellular vesicles with high collagen-binding affinity present superior in situ retention and therapeutic efficacy in tissue repair, Theranostics, 2022, 12(13), 6021–6037, DOI:10.7150/THNO.70448.
- L. Goldbloom-Helzner, H. Bains and A. Wang, Approaches to Characterize and Quantify Extracellular Vesicle Surface Conjugation Efficiency, Life, 2024, 14(4), 511, DOI:10.3390/LIFE14040511.
- R. R. Mizenko, T. Brostoff and T. Rojalin, et al., Tetraspanins are unevenly distributed across single extracellular vesicles and bias sensitivity to multiplexed cancer biomarkers, J. Nanobiotechnol., 2021, 19(1), 1–17, DOI:10.1186/S12951-021-00987-1/FIGURES/7.
- S. Rayamajhi and S. Aryal, Surface functionalization strategies of extracellular vesicles, J. Mater. Chem. B, 2020, 8(21), 4552–4569, 10.1039/D0TB00744G.
- X. Gao, N. Ran and X. Dong, et al., Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy, Sci. Transl. Med., 2018, 10(444), 195, DOI:10.1126/SCITRANSLMED.AAT0195/SUPPL_FILE/AAT0195_.PDF.
- R. Tamura, S. Uemoto and Y. Tabata, Augmented liver targeting of exosomes by surface modification with cationized pullulan, Acta Biomater., 2017, 57, 274–284, DOI:10.1016/J.ACTBIO.2017.05.013.
- H. Suárez, A. Gámez-Valero and R. Reyes, et al., A bead-assisted flow cytometry method for the semi-quantitative analysis of Extracellular Vesicles, Sci. Rep., 2017, 7(1), 11271, DOI:10.1038/s41598-017-11249-2.
- O. P. B. Wiklander, R. B. Bostancioglu and J. A. Welsh, et al., Systematic Methodological Evaluation of a Multiplex Bead-Based Flow Cytometry Assay for Detection of Extracellular Vesicle Surface Signatures, Front. Immunol., 2018, 9, 1326, DOI:10.3389/FIMMU.2018.01326.
- R. R. Mizenko, T. Brostoff and T. Rojalin, et al., Tetraspanin immunocapture phenotypes extracellular vesicles according to biofluid source but may limit identification of multiplexed cancer biomarkers, bioRxiv, 2021:2021.03.02.433595. DOI:10.1101/2021.03.02.433595.
- W. Shen, K. Guo and G. B. Adkins, et al., A Single Extracellular Vesicle (EV) Flow Cytometry Approach to Reveal EV Heterogeneity, Angew. Chem., Int. Ed., 2018, 57(48), 15675–15680, DOI:10.1002/ANIE.201806901.
- A. Görgens, M. Bremer and R. Ferrer-Tur, et al., Optimisation of imaging flow cytometry for the analysis of single extracellular vesicles by using fluorescence-tagged vesicles as biological reference material, J. Extracell. Vesicles, 2019, 8(1), 1587567, DOI:10.1080/20013078.2019.1587567.
- B. Nastasijevic, B. R. Wright, J. Smestad, A. E. Warrington and M. Rodriguez, Remyelination Induced by a DNA Aptamer in a Mouse Model of Multiple Sclerosis, PLoS One, 2012, 7(6), 39595, DOI:10.1371/journal.pone.0039595.
- M. Fereidan-Esfahani, W. Y. Yue and B. Wilbanks, et al., Remyelination-promoting dna aptamer conjugate myaptavin-3064 binds to adult oligodendrocytes in vitro, Pharmaceuticals, 2020, 13(11), 1–11, DOI:10.3390/ph13110403.
- M. K. Perwein, J. A. Smestad and A. E. Warrington, et al., A comparison of human natural monoclonal antibodies and aptamer conjugates for promotion of CNS remyelination: where are we now and what comes next?, Expert Opin. Biol. Ther., 2018, 18(5), 545–560, DOI:10.1080/14712598.2018.1441284.
- P. Suárez-Meade and A. Ibarra, Protective Role of the Immune System in Spinal Cord Injury: Immunomodulation with Altered Peptide Ligands, in Recovery of Motor Function Following Spinal Cord Injury, InTech, 2016, ch. 3, DOI:10.5772/63221.
- A. D. Keefe, S. Pai and A. Ellington, Aptamers as therapeutics, Nat. Rev. Drug Discovery, 2010, 9(7), 537–550, DOI:10.1038/nrd3141.
- A. Trivedi, A. D. Olivas and L. J. Noble-Haeusslein, Inflammation and Spinal Cord Injury: Infiltrating Leukocytes as Determinants of Injury and Repair Processes, Clin. Neurosci. Res., 2006, 6(5), 283, DOI:10.1016/J.CNR.2006.09.007.
- K. Clark, S. Zhang and S. Barthe, et al., Placental Mesenchymal Stem Cell-Derived Extracellular Vesicles Promote Myelin Regeneration in an Animal Model of Multiple Sclerosis, Cells, 2019, 8(12), 1497, DOI:10.3390/cells8121497.
- M. Palviainen, H. Saari and O. Kärkkäinen, et al., Metabolic signature of extracellular vesicles depends on the cell culture conditions, J. Extracell. Vesicles, 2019, 8(1), 1596669, DOI:10.1080/20013078.2019.1596669.
- J. P. Mitchell, J. Court, M. D. Mason, Z. Tabi and A. Clayton, Increased exosome production from tumour cell cultures using the Integra CELLine Culture System, J. Immunol. Methods, 2008, 335(1–2), 98–105, DOI:10.1016/j.jim.2008.03.001.
- C. Théry, S. Amigorena, G. Raposo and A. Clayton, Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids, Curr. Protoc. Cell Biol., 2006, 30(1), 3.22.1–3.22.29, DOI:10.1002/0471143030.CB0322S30.
- T. Zeev-Ben-Mordehai, D. Vasishtan, C. A. Siebert, C. Whittle and K. Grü Newald, Extracellular Vesicles: A Platform for the Structure Determination of Membrane Proteins by Cryo-EM, Structure, 2014, 22(11), 1687–1692, DOI:10.1016/j.str.2014.09.005.
- P. Kumar, J. C. Becker and K. Gao, et al., Neuroprotective effect of placenta-derived mesenchymal stromal cells: role of exosomes, FASEB J., 2019, 33(5), 5836–5849, DOI:10.1096/fj.201800972R.
- J. A. Welsh, J. C. Jones and V. A. Tang, Fluorescence and Light Scatter Calibration Allow Comparisons of Small Particle Data in Standard Units across Different Flow Cytometry Platforms and Detector Settings, Cytometry, Part A, 2020, 97(6), 592–601, DOI:10.1002/CYTO.A.24029.
- J. A. Welsh, E. Van Der Pol and G. J. A. Arkesteijn, et al., MIFlowCyt-EV: a framework for standardized reporting of extracellular vesicle flow cytometry experiments, J. Extracell. Vesicles, 2020, 9(1), 1713526, DOI:10.1080/20013078.2020.1713526.
- C. Théry, K. W. Witwer and E. Aikawa, et al., Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines, J. Extracell. Vesicles, 2018, 7(1), 1535750, DOI:10.1080/20013078.2018.1535750.
- D. Bachurski, M. Schuldner and P. H. Nguyen, et al., Extracellular vesicle measurements with nanoparticle tracking analysis – An accuracy and repeatability comparison between NanoSight NS300 and ZetaView, J. Extracell. Vesicles, 2019, 8(1), 1596016, DOI:10.1080/20013078.2019.1596016.
- G. G. Daaboul, P. Gagni and L. Benussi, et al., Digital Detection of Exosomes by Interferometric Imaging, Sci. Rep., 2016, 6(1), 1–10, DOI:10.1038/srep37246.
- S. K. George, L. Lauková and R. Weiss, et al., Comparative analysis of platelet–derived extracellular vesicles using flow cytometry and nanoparticle tracking analysis, Int. J. Mol. Sci., 2021, 22(8), 3839, DOI:10.3390/IJMS22083839/S1.
- G. C. Brittain, Y. Q. Chen and E. Martinez, et al., A Novel Semiconductor-Based Flow Cytometer with Enhanced Light-Scatter Sensitivity for the Analysis of Biological Nanoparticles, Sci. Rep., 2019, 9(1), 1–13, DOI:10.1038/s41598-019-52366-4.
- M. A. Lopez-Verrilli, A. Caviedes, A. Cabrera, S. Sandoval, U. Wyneken and M. Khoury, Mesenchymal stem cell-derived exosomes from different sources selectively promote neuritic outgrowth, Neuroscience, 2016, 320, 129–139, DOI:10.1016/j.neuroscience.2016.01.061.
- M. Wu, Y. Ouyang and Z. Wang, et al., Isolation of exosomes from whole blood by integrating acoustics and microfluidics, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(40), 10584–10589, DOI:10.1073/pnas.1709210114.
- E. van der Pol, F. A. W. Coumans and A. E. Grootemaat, et al., Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing, J. Thromb. Haemost., 2014, 12(7), 1182–1192, DOI:10.1111/JTH.12602.
- S. Salunkhe, Dheeraj, M. Basak, D. Chitkara and A. Mittal, Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: Strategies and significance, J. Controlled Release, 2020, 326, 599–614, DOI:10.1016/j.jconrel.2020.07.042.
- K. Zhang, Y. Yue, S. Wu, W. Liu, J. Shi and Z. Zhang, Rapid Capture and Nondestructive Release of Extracellular Vesicles Using Aptamer-Based Magnetic Isolation, ACS Sens., 2019, 4(5), 1245–1251, DOI:10.1021/acssensors.9b00060.
- Y. Nojima, K. Iguchi, Y. Suzuki and A. Sato, The pH-Dependent Formation of PEGylated Bovine Lactoferrin by Branched Polyethylene Glycol (PEG)-N-Hydroxysuccinimide (NHS) Active Esters, Biol. Pharm. Bull., 2009, 32(3), 523–526, DOI:10.1248/BPB.32.523.
- H. J. Lee, G. M. Fernandes-Cunha, K. S. Na, S. M. Hull and D. Myung, Bio-Orthogonally Crosslinked, In Situ Forming Corneal Stromal Tissue Substitute, Adv. Healthc. Mater., 2018, 7(19), 1800560, DOI:10.1002/ADHM.201800560.
- L. Lu, V. T. Duong, A. O. Shalash, M. Skwarczynski and I. Toth, Chemical Conjugation Strategies for the Development of Protein-Based Subunit Nanovaccines, Vaccines, 2021, 9(6), 563, DOI:10.3390/VACCINES9060563.
- E. Trenkenschuh, M. Richter, E. Heinrich, M. Koch, G. Fuhrmann and W. Friess, Enhancing the Stabilization Potential of Lyophilization for Extracellular Vesicles, Adv. Healthc. Mater., 2022, 11(5), 2100538, DOI:10.1002/ADHM.202100538.
- H. Cheng, M. Byrska-Bishop and C. T. Zhang, et al., Stem cell membrane engineering for cell rolling using peptide conjugation and tuning of cell-selectin interaction kinetics, Biomaterials, 2012, 33(20), 5004–5012, DOI:10.1016/J.BIOMATERIALS.2012.03.065.
- H. Zhou, Z. Fan, P. K. Lemons and H. Cheng, A Facile Approach to Functionalize Cell Membrane-Coated Nanoparticles, Theranostics, 2016, 6(7), 1012–1022, DOI:10.7150/THNO.15095.
- J. Kowal, G. Arras and M. Colombo, et al., Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(8), E968–E977, DOI:10.1073/PNAS.1521230113/-/DCSUPPLEMENTAL/PNAS.201521230SI.PDF.
| This journal is © The Royal Society of Chemistry 2024 |