Open Access Article
Open Access ArticleBiocatalytic ZIF-8 surface-functionalized micromotors navigating in the cerebrospinal fluid: toward Alzheimer management†
J.
Bujalance-Fernández
 a,
E.
Carro
bc,
B.
Jurado-Sánchez
a,
E.
Carro
bc,
B.
Jurado-Sánchez
 *ad and
A.
Escarpa
*ad and
A.
Escarpa
 *ad
*ad
aDepartment of Analytical Chemistry, Physical Chemistry, and Chemical Engineering, Universidad de Alcala, Alcala de Henares, E-28802 Madrid, Spain. E-mail: beatriz.jurado@uah.es; alberto.escarpa@uah.es
bChronic Disease Programme, UFIEC, Carlos III Health Institute, Majadahonda, Madrid 28220, Spain
cCIBERNED, Madrid, Spain
dChemical Research Institute “Andres M. del Rio”, Universidad de Alcala, E-28802, Madrid, Spain
First published on 12th October 2024
Abstract
Alzheimer's disease (AD) is the major cause of irreversible dementia in the elderly population worldwide and one of the major causes of the decrease in the quality of life. Efficient diagnosis and monitoring would allow a fast treatment to delay the appearance of symptoms. Herein, zeolitic imidazole framework (ZIF-8)@Au@catalase micromotors are described for motion-based sensing of copper as a marker of AD. The synthesis design was based on enzyme covalent immobilization instead of encapsulation to maximize the contact with the sample at the microscale for the potential use of extremely low AD-diagnosed sample volumes. The micromotors are prepared by asymmetric modification of ZIF-8 with a gold layer for functionalization of catalase as a compatible biocatalyst. The micromotors can propel at speeds of up to 287 ± 41 μm s−1 in cerebrospinal fluid (CSF) samples of healthy volunteers. Yet, in the presence of copper, catalase poisoning results in a decrease in the speed that can be monitored for motion-based sensing detection, as illustrated in the analysis of CSF samples from AD patients from mild to severe stages (Braak III to Braak VI). The copper-mediated modulation of catalase activity proposed here as an indicator of progression states in AD disease possesses distinct advantages such as ultrafast analysis (less than 1 min) and requiring only 1 μL of sample, holding considerable promise as a supporting prescreening tool for fast diagnosis of AD and other neurodegenerative diseases.
1. Introduction
Metal–organic frameworks (MOFs) are fascinating materials with unique properties such as a high internal surface area (up to 6000 m2 g−1), porosity and biocompatibility.1 MOFs can be classified according to the metal node and organic linker used into zeolitic imidazole frameworks (ZIFs), materials Institute Lavoisier and University of Oslo (UiO) families.2 The remarkable properties of MOFs have attracted the attention of the micromotor community for the development of multifunctional entities with infinite possibilities in the environmental, biomedical or analytical fields.3–5Micromotors are self-propelled particles capable of autonomous movement in solution. This leads to a dynamic, on-the-move process, with enhanced performance as compared with the use of their static counterparts.6–9 The first MOF micromotor was proposed by Matsui et al.10 using a Cu2+ 1,4-benzenedicarboxylate–triethylenediamine MOFs encapsulating diphenylalanine peptides. Dissolution of MOFs and release of the peptide generate an autonomous Marangoni-based propulsion.10 Catalytic designs were later explored accounting for the enhanced fluid mixing and propulsion of such micromotors. Different catalytic metals such as cobalt in connection with ZIF-8/ZIF-67 MOFs11 or cobalt and manganese with Zr-based UiO-67 bipyridine-type MOFs were used.12 Directional micromotor propulsion at speeds of up to 1000 μm s−1 (10% H2O2) was achieved. On-demand braking and acceleration of micromotors was illustrated using chelating agents. The introduction of catalytic layers can also be achieved by asymmetric sputter deposition of a Pt layer, as illustrated with ZIF-8, UiO-66, and UiO-66-SH MOFs, reaching a speed of over 1500 μm s−1 (20% H2O2).13
Concerns about the toxicity of peroxide fuel in biomedical applications motivated the development of fuel-free MOF micromotors driven by electromagnetic fields, but this direction is still in early infancy, with few developments to date.14,15 Another interesting alternative towards biofriendly MOFs micromotors is the use of enzymes as catalytic elements. Catalase is the preferred choice to mimic the widely used Pt catalyst at a much lower cost and with efficient catalytic activity. Thus, mesoporous UiO-Zr MOFs were used for the encapsulation (via absorption) of such enzymes, achieving efficient propulsion at levels as low as 0.5% H2O2.16 Catalase-loaded ZIF-L MOFs containing the polymer poly(2-diisopropylamino)ethyl methacrylate exhibit a buoyancy motion triggered by the polymer for controlled drug delivery, as illustrated in MCF-7 cell media.17 In another interesting approach, the pair catalase–glucose oxidase has been encapsulated into ZIF-8 MOFs. The resulting micromotors were used for electrochemical monitoring of glucose. The principle behind the assay relies on the initiation of cascade reactions in the presence of glucose, generating hydrogen peroxide substrates for catalase, resulting in propulsion and collision with the electrode.18 Another example is this direction investigates the introduction of peroxidase nanozymes into MOFs to enhance the amplification of electrochemiluminescence (ECL) signals in immunoassays, thereby optimizing the ultrasensitive detection of disease markers for clinical diagnostics.19 Also, metal azole framework 6 can be used to immobilize esterases to improve yield in transesterification reactions.20 Another study investigated how the crystallization of MOFs affects the mobility and performance of encapsulated enzymes to optimize the design of robust platforms for biotechnological and catalytic applications.21 Such early reports indicate the potential of biocatalytic propelled MOFs micromotors for analytical sensing of clinical analytes, which yet remains unexplored.
Inspired by the promising potential of MOFs micromotors for analytical sensing and the biocompatibility features of binomial enzymes–MOFs, herein we describe the synthesis and application of ZIF-8/Au/catalase micromotors for motion-based sensing of copper as a modulation marker of AD. AD is the main cause of dementia of the elderly population worldwide. Early diagnosis for prompt treatment can increase the quality of life, greatly delaying the progression of degenerative symptoms. The diagnosis of AD combines a clinical and biological approach, supported by the detection of specific biomarkers to monitor the state of the disease. The target biomarkers are divided into 3 main categories, A (amyloid), T (phosphorylated tau), and N (neurodegeneration, measured by total tau).22 In this context, the disease is characterized by the formation of extracellular amyloid plaques (mainly 39–43 amyloid-β peptides), with an altered concentration of metal ions, mainly copper and zinc.23 There is a controversy in the literature about the role of the levels of such metals in the generation of amyloid plaques. Yet, it is well known that in severe AD states, elevated copper concentrations can be found in the brain, and subsequently in the CSF.24–26 As such, a fast and supporting tool to add to the battery of tests for AD diagnosis is the monitoring of the levels of Cu, as a new additional manner to classify the state of the disease.
Herein, we used a ZIF-8@Au@catalase strategy for motion-based sensing of copper in CSF samples from AD patients. Previous literature works have carried out quantitative studies of copper in neurodegenerative patients, using serum as a sample due to its less invasive nature.27,28 Usually these studies have been done in serum samples due to the accessibility and ease of extraction rather than CSF samples which require a more invasive extraction. Our work has the unique advantage of having a sample from a healthy volunteer, this fact allows us to have a control to compare the results with respect to a normal value. In the following sections, we first describe the synthesis and characterization of ZIF-8 micromotors. The asymmetric modification of the ZIF-8 unit with an Au layer and subsequent covalent functionalization with catalase result in biocompatible and efficient micromotors that can navigate in just 1 μL of CSF. Poisoning of the catalase later with the Cu metal results in a decrease in the concentration-dependent speed that can be used to monitor and classify the samples. We then also illustrate the feasibility of the approach as illustrated in the screened analysis of samples at different stages (Braak III to Braak VI).
2. Materials and methods
2.1. Reagents and materials
Copper (cat. 38996), sodium hydroxide (cat. 106498), methanol (cat. 34860), 1-methylimidazole (cat. M50834), 2-methylimidazole (cat. M50850), zinc nitrate hexahydrate (cat. 96482), catalase from Aspergillus niger (cat. C3515), 11-mercaptoundecanoid acid (cat. 450561), 6-mercapto-1-hexanol (cat. 451088), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) (cat. 800907), N-hydroxysuccinimide (NHS) (cat. 130672), N,N-dimethylformamide (cat. 543897), Triton X-100 solution (cat. 93443) and sodium dodecyl sulfate (SDS, cat. 822050) were purchased from Merck, Spain. Phosphate buffer saline (cat. 28372), 4-mercaptobenzoic acid (cat. 373300050), MES buffered saline (cat. 28390), and hydrogen peroxide 30% solution (cat. 10687022) were purchased from Thermo Fisher Scientific, Spain. Ethanol (cat. ET0002005P) was purchased from Scharlau, Spain. All reagents were used without further purification. Milli Q water was obtained using a Millipak Express filter (cat. MPGP04001) and a vent filter (cat. TANKMPK01) which were purchased from Merck Millipore.2.2. Equipment
An ultrasonic bath (Elmasonic S 30 H) was used to detach ZIF-8 from the gold sputtered glass slide. A 3D printer Original Prusa i3 MK3S+ was used to print the gold-sputtered glass slide support to detach the gold sputtered ZIF-8. An Eppendorf centrifuge 5430 attached with an FA-45-30-11 rotor was used to wash the ZIF-8 and micromotors. An inverted Nikon Eclipse Instrument Inc. Ti-S/L100 optical microscope, coupled with a Zyla sCMOS camera, was used to capture images and record videos. The speed of the micromotors was recorded and measured using NIS-5.41 elements software. X-ray diffraction (XRD) analysis was performed in the “X-ray center” of the University Complutense de Madrid. Scanning electron microscopy characterization of the micromotors was performed using a JSM 6335F (JEOL) coupled to an energy-ray dispersive (EDX) system (Xflash Detector 4010, Bruker). Transmission electron (TEM) microscopy characterization was performed with a Zeiss EM10C microscope. (3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) (MTT) assays were performed in the Cell Culture Centre of the Universidad de Alcala. The results were obtained from 3 replicates of HeLa cell incubation under different conditions.2.3. ZIF-8 micromotors synthesis
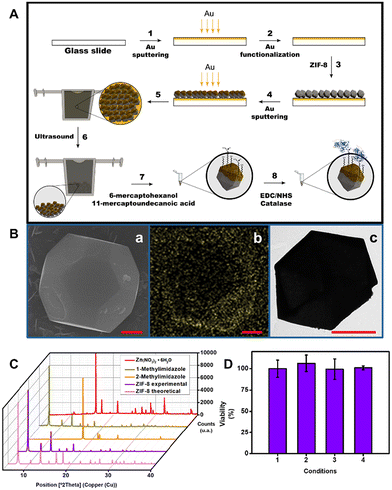
ZIF-8 MOFs were synthesized using a procedure adapted from the literature.29 To this end, a 50 mM solution of Zn(NO3)2·6H2O in methanol was mixed with 200 mM solutions of 1-methylimidazole and 2-methylimidazole. The mixture was then covered and incubated overnight at room temperature. A white precipitate was then obtained, which was separated from the solution by centrifugation, followed by resuspension and cleaning with methanol. The resulting solution was filtered and collected in a 0.2 μm membrane as dried powder under vacuum conditions.For micromotor synthesis, a ZIF-8 solution (1 mg mL−1) in dimethylformamide was drop-cast on a modified glass slide. For modification, the slide was sputtered with an ∼1000 Å gold layer, followed by incubation with a 0.1 mM 4-mercaptobenzoic acid solution in ethanol overnight. Next, the slide was washed with ethanol and incubated with a 0.1% (w/w) sodium hydroxide solution for 20 min. The slide was washed 3 times with water and dried prior to drop-casting of the ZIF-8 solution. After the formation of the ZIF-8 monolayer in the glass slide, this was dried in a heater at 60 °C. Later, it was sputtered with an ∼1500 Å gold layer for ZIF-8 modification. The glass slide was next inserted into a homemade designed 3D printed stand filled with MeOH to release the micromotors (Fig. S1†).
For functionalization, the solution containing the ZIF-8@Au particles was cleaned by centrifugation at 4000g for 4 min to eliminate the supernatant with small particles of gold that had been detached too. Next, the ZIF-8@Au particles were resuspended in an ethanolic solution containing 7.5 mM 6-mercapto-1-hexanol and 2.5 mM 11-mercaptoundecanoid acid and incubated overnight. The supernatant was then removed by centrifugation and the particles were washed with ethanol. After removing the ethanol, the ZIF-8@Au MOFs were resuspended and incubated in 200 μL of an MES solution (25 mM, pH 6.5) containing 0.4 M EDC, 0.1 M NHS and catalase (7000 U) for 6 h at 37 °C. The ZIF-8@Au@catalase micromotors were collected by centrifugation, shaken in 0.01 M PBS (pH 7.2) to wash them twice, and finally centrifuged and resuspended in water.
2.4. Micromotor counting
A concentration of 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 micromotors per mL was used for each experiment. To get this ratio, 1 μL of the sample was photographed at 4× objective and then four more times at 20× objective at different locations within the drop. To determine the average number of micromotors per area and, therefore, per μL, the area of the 1 μL drop (4× objective) was measured and the number of MMs per each 20× image (with a known area) was counted. This method was applied four times per batch of micromotors. Finally, they were resuspended in a volume determined to obtain the required concentration and were ready for the next experiments. In all experiments, monodispersion of the micromotors was assured prior counting (see the image in Fig. S2†).
000 micromotors per mL was used for each experiment. To get this ratio, 1 μL of the sample was photographed at 4× objective and then four more times at 20× objective at different locations within the drop. To determine the average number of micromotors per area and, therefore, per μL, the area of the 1 μL drop (4× objective) was measured and the number of MMs per each 20× image (with a known area) was counted. This method was applied four times per batch of micromotors. Finally, they were resuspended in a volume determined to obtain the required concentration and were ready for the next experiments. In all experiments, monodispersion of the micromotors was assured prior counting (see the image in Fig. S2†).
2.5. Sample collection
The Institutional Ethical Review Board of the Spanish Research Center for Neurological Diseases Foundation (CIEN), the Instituto de Salud Carlos III, and the University of Alcalá de Henares approved this study on marker analyses of AD (CEID2021/4/108).Subjects’ consent was obtained following the Declaration of Helsinki and the approval of the Research Ethics Committee of the institutions involved. Subjects were selected based on the post-mortem diagnosis of AD according to neurofibrillary tangle pathology and Aβ plaques.30 The absence of neuropathological change was reported in the control healthy participant (CSF0274) obtained from the 12 de Octubre University Hospital (Madrid, Spain) with the approval of its Research Ethic Committee (CEIm2018/459). AD biological samples at Braak stages III (BCPA167), IV (BCPA067) (intermediate AD), V (BCPA897) and VI (BCPA799) (advanced AD) were obtained from the CIEN Foundation's Tissue Bank (BT-CIEN) according to the brain bank's protocols. CSF samples were collected and processed according to standardized procedures by lumbar puncture in 15 mL sterile polypropylene tubes. The samples were then centrifuged at 3000 rpm at 4 °C for 10 min. Supernatant aliquots were stored at −80 °C into 0.5 mL polypropylene cryogenic tubes with protease inhibitor cocktails (Roche, Basel, Switzerland). All participants gave written informed consent for participation. The project was approved by the 12 de Octubre University Hospital (Madrid, Spain) Ethical Review Committee (CEIm2018/459).
3. Results and discussion
A schematic of the strategy used for copper detection is shown in Fig. 1A. ZIF-8 MOFs half-coated with gold were used as biocompatible templates. The micromotors possess a hexagonal morphology (see Fig. 1B) with unique features to generate directional and efficient propulsion. Thus, as illustrated in the time-lapse images of Fig. 1C, in the presence of hydrogen peroxide as a fuel, decomposition by catalase generates oxygen bubbles for highly efficient propulsion in the samples. Yet, in the presence of copper, strong inhibition of the catalase activity is observed.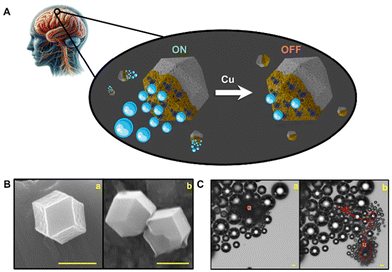 | ||
| Fig. 1 (A) Schematic of ZIF-8 composition and catalytic movement of the ZIF-8@Au@catalase micromotors through CSF. (B) SEM images showing the morphologies of ZIF-8 (a) and the ZIF-8@Au@catalase micromotors (b). Scale bars, 1 μm. (C) Time-lapse microscopy images (taken from Video S1†) of the ZIF-8@Au@catalase micromotors after 0 s (a) and 2 s (b) navigation in water. Conditions: 1% Triton X-100 and 10% H2O2 as the fuel. Scale bars, 10 μm. | ||
Catalase is a naturally occurring enzyme in aerobic organisms to regulate oxidative stress by hydrogen peroxide decomposition. Its structure is composed of four sub-units with ∼500 amino acids and a heme active site.31 Certain metals can inhibit enzyme activity by blocking such active sites. Some studies indicate that copper is the strongest inhibitor of catalase activity, followed by mercury, iron, chromium and cadmium.32 Wang et al. illustrated the inhibition of catalase-propelled micromotors by mercury, sodium azide, aminotriazole, and copper.33 The mechanism for inhibition can be explained by interactions with a His 74 residue close to the heme group unit, which hinders hydrogen peroxide accessibility, thus inducing enzyme inhibition.31 While the feasibility of tubular micromotors modified with an inner catalase layer for copper detection has been illustrated in water samples at relevant environmental levels (according to EC50), our goal here is to apply the concept for detection in clinically diagnosed samples such as CSF. Also, copper levels in AD disease samples can be much higher than those tested in the previous work.33 Thus, and considering the distinctive MOFs features, we opted for a micromotor design in which catalase is present at the outer micromotor layer, using MOFs as a template to increase the surface area, and thus the amount of the immobilized enzyme for efficient work at the microscale in ultra-low sample volumes with diagnosis purposes.
The ZIF-8 (for details on the MOFs synthesis, please see the Experimental section) micromotor functionalization is depicted in Fig. 2, along with further characterization studies.
To generate the asymmetric gold layer for catalase immobilization and directional propulsion, it is essential to generate a ZIF-8 monolayer on the glass slide used as a solid support. This demands its functionalization to deprotonate the carboxylic groups,13 as described in the Experimental section (Au functionalization, steps 1 and 2 in Fig. 2A). Next, a solution containing 1 mg mL−1 of ZIF-8 MOFs in dimethylformamide was dropped in the functionalized glass slide. After drying the slide at 60 °C, an asymmetric gold layer (≈1500 Å) for catalase immobilization was generated by sputter deposition. Next, for micromotor release, the slide was introduced in a homemade designed 3D-printed stand filled with ∼8 mL MeOH (for the design and magnification, see Fig. S1†) and sonicated in an ultrasonic bath for no more than 1 minute (more time could detach the gold layer of ZIF-8). Please note here the importance of the 3D printed stand, thanks to this homemade design, we can use the minimum volume of MeOH to release the micromotors, avoiding the use of any glass that needs to fill with large volumes to immerse the glass slide; with this design, we can adapt the shape to need the minimum volume. After release and cleaning by centrifugation, the MOFs were modified with catalase by immobilization via EDC/NHS chemistry. For more details, see the Experimental section. Morphological and element analyses of the ZIF-8@Au@catalase micromotors were next conducted. The SEM and corresponding EDX images, as well as the TEM images in Fig. 2B show the octahedral morphology of the micromotors and the presence of an asymmetric Au layer. The average micromotor size is approximately 4 μm in length. Additional XRD observation (Fig. 2C) reveals the characteristics peaks of ZIF-8, with the characteristic diffraction peaks at 2θ = 8°, 10°, 13°, 15°, 16°, 18°, 22°, 25°, 27° and 30°, which correspond to the (011), (002), (112), (022), (013), (222), (114), (233), (134) and (044) planes respectively.34,35 A good correlation was observed with the literature values, indicating the successful generation of ZIF-8@Au@catalase micromotors. To check the biocompatibility of our micromotors, MTT assays (see the Experimental section for more details) were performed, illustrating the almost 100% viability rates in all cases (see Fig. 2D). These data reveal the suitability of the micromotors for their application in biological and clinical samples.
After assuring the successful micromotor synthesis by morphological and elemental characterization, we studied the propulsion performance. To this end, we evaluated first the effect of different surfactants on oxygen bubble nucleation and enzyme stability. To this end, we dropped on a glass slide 1 μL of ZIF-8@Au@catalase micromotor solutions, 1 μL of SDS or Triton X-100 as a surfactant (to a final concentration of 1%) and 1 μL of H2O2 solution (to a final concentration of 3.3%). No movement is observed using SDS as a surfactant, probably due to denaturation or conformational changes in the enzyme catalase that prevents peroxide substrate's fuel decomposition.36 Efficient propulsion is observed using Triton X-100 as a surfactant, with speeds ranging from 72 ± 9 μm s−1 (3.3% H2O2) to 368 ± 130 μm s−1 (10% H2O2). To maximize the linear dynamic range for calibration, 10% H2O2 concentration was selected as the fuel. Under such conditions, flow dynamics simulations were performed with the CFD tool embedded in the Solidworks software. Video S2† used for simulations and the results are listed in Fig. 3. To this end, the ZIF-8@Au@catalase micromotors were modelled and the relevant flow dynamics parameters were obtained by applying the standard Launder and Spalding K-epsilon model. The corresponding boundary and initial conditions were applied, namely a linear velocity of 237.08 μm s−1 in the x-axis and an angular frequency (ω) of 0.35 Hz around the y-axis were considered according to the experimental observations in the steady state. Mean fluid velocity cut plots and flow lines were generated as an output and reported. As can be seen, the flow velocities in the Z and X sections were the highest, indicating good fuel accessibility, as also corroborated with the simulations of Y–X axis sections (please see also Video S2†). The flow-like propulsion associated with the catalytic mechanisms is reflected in the simulated trajectories of Fig. 3D and E, with a turbulent but directional flow.37 These results show why the flow generated by ZIF-8@Au@catalase allows excellent mixing in the drop, providing the encounter between catalase and free Cu, which can be translated into efficient measurement in small volumes of a sample as difficult to obtain as this one.
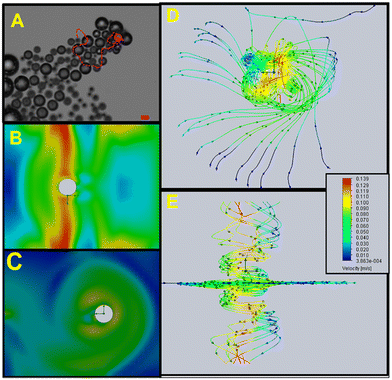 | ||
| Fig. 3 (A) Time-lapse image showing the propulsion of a ZIF-8@Au@catalase micromotor (taken from Video S2†) and (B) corresponding schematic of the flow dynamics simulations of the Z- and X-axis sections. (C) Y- and X-axis sections. (D) Bottom view and (E) back view trajectories. Conditions: 250 μg L−1 of Cu, 1% Triton X-100 and 10% H2O2 as the fuel. Scale bar, 20 μm. | ||
The applicability of the ZIF-8@Au@catalase micromotors to detect Cu ions in the CSF was next tested. While there is some controversy in the literature,38 some studies have illustrated a general increase in free Cu levels in the CSFs in AD and other degenerative illnesses.39–41 While other protein aggregates may interfere with this system, some clinical studies suggests the primary role of copper in the modulation of catalase activity. Indeed, amyloid β is able to decrease the catalase activity, resulting in an elevation of Cu levels.42 Additionally, Cu accumulates in the amyloid β plaque of patients with AD,43 and Cu levels are elevated in AD patients.44 Finally, Cu decreases the catalase activity.45,46 Far from contradicting our results, this complex system reinforced our hypothesis on copper-mediated modulation of catalase, without excluding the participation of other related molecules. Thus, to test the suitability of our micromotors as fast screening tools for the sample analysis of AD patients, we choose Cu as the target analyte, capable of catalase inhibition, for motion-based sensing performance. We performed a calibration plot by tracking the speed on the micromotors in the absence and the presence of increasing concentrations of copper. The results are listed in Fig. 4A and Video S3.† As can be seen, as the Cu concentration increases, there is a marked decrease in the speed of the micromotors. Speed data were plotted against copper concentration, obtaining a linear plot. The adjusted regression weighting equation is speed = −0.52 ± 0.03 [Cu, μg L−1] + 359 ± 6 (r = 0.990). Limits of detection and quantification were calculated as 3 and 10 times the error of the intercept divided by the slope of the calibration plot and are 35 and 115 μg L−1 of Cu, respectively.
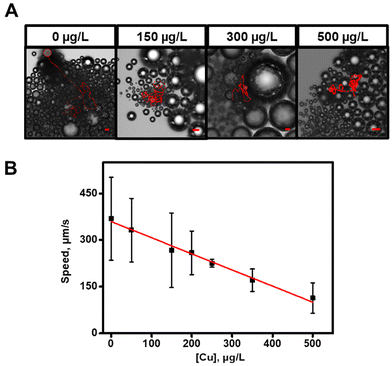 | ||
| Fig. 4 (A) Time-lapse microscopy images (taken from Video S3†) after 3 s of navigation of the ZIF-8@Au@catalase micromotors in solutions containing different free Cu concentrations (μg L−1) in water. (B) Graphical representation of a regression weighting curve relating the rate of ZIF-8@Au@catalase to the concentrations of free Cu (μg L−1) in water. Conditions: 1% Triton X-100 and 10% H2O2 as the fuel. Scale bars, 20 μm. Error bars represent the standard deviation of 5 measurements. | ||
Next, CSFs of healthy and AD patients at different Braak stages were analyzed with our micromotors. Details of sample collection are described in the Experimental section. We also tested the micromotor propulsion in samples from healthy volunteers. As can be seen in Fig. 5A and Video S4,† efficient propulsion at a speed of 287 ± 41 μm s−1 is observed, which is statistically like the speed of the micromotor in the absence of Cu, considering the slight differences in the viscosity of the media. Yet, in AD patients’ samples, a marked decrease in the speed was noted with a progression of the degenerative stages, from 255 ± 41 μm s−1 in Braak stage III to a total stop in the micromotor motion in severe stage VI. The analysis of a sample from a healthy volunteer allows the comparison of the values of different stages of the disease with normal values. Calculated Cu concentrations were 138 ± 79 μg L−1 (μg L−1) in the healthy volunteer and 199 ± 79, 295 ± 34, 323 ± 18 and >500 μg L−1 in Braak stages III to VI, respectively (see Fig. 5B). Braak stage VI is not shown in this graph because its copper concentration is unknown.
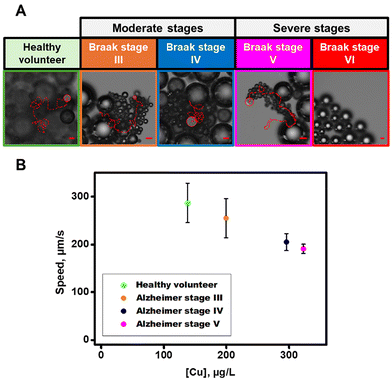 | ||
| Fig. 5 (A) Time-lapse microscopy images (taken from Video S4†) showing ZIF-8@Au@catalase propulsion in the CSF samples from different stages of AD. (B) Graphical representation of the relationship between ZIF-8@Au@catalase speed in different CSFs and its free Cu concentration calculated from the regression weighting curve. Conditions: 1% Triton X-100 and 10% H2O2 as the fuel. Scale bars, 20 μm. Error bars represent the standard deviation of 5 measurements. | ||
The results show that Cu-mediated modulation of catalase activity can be proposed as an indicator of different progression stages in AD. Indeed, in the brains of patients suffering from AD, both metals, Cu and zinc, are significantly elevated, in comparison with surrounding healthy tissues.47 The neurocytotoxicity of Cu and zinc, and the interaction of zinc and Cu with β-amyloid and tau protein are involved in the pathogenesis of AD. A protective role for zinc can be explained by its competition with Cu to interact with Aβ, since the coordination of zinc to Aβ changes its conformation and prevents it from interacting with copper, thus circumventing the formation of hydroxyl radicals via the Fenton reaction that is otherwise catalyzed by the Cu–Aβ complex.48 Indeed, zinc can exchange redox active metals, such as Cu and iron, in certain binding sites and attenuate cellular site-specific oxidative injury. It has been described that zinc increases the activation and levels of catalase.49,50 Thus, although zinc could be interacting in this cellular process, this interaction would be trying to compensate for the copper-mediated down regulation of catalase.
4. Conclusions
A ZIF-8@Au@catalase micromotor has successfully been synthesized and used for motion-based sensing of Cu as a marker of AD. The micromotor relies on catalase as a biocatalyst for highly efficient propulsion, which also acts as an active layer for sensing. Compared with previous MOF micromotor works,16,17 herein we designed and optimized the approach for covalent immobilization instead of encapsulation, maximizing contact with the sample with enhanced detection at the microscale using extremely low AD-diagnosed sample volumes. While the analysis of AD markers is highly complex, our initial results illustrated a valuable trend among the micromotor speed and progression in AD stages. The decrease in the motion can be attributed to Cu poisoning by catalase, assuming our hypotheses of elevated concentrations of such a metal as the illness progresses, and some cellular rest. Given that Zn has been shown to enhance catalytic activity, and considering the drastic decrease in the micromotor speed in cerebrospinal fluid samples as the stage of the disease increases (with three distinct groups clearly: healthy (287 ± 41 μm s−1) and Braak III (255 ± 41 μm s−1) [an early/moderate stage of the disease], Braak IV (205 ± 17 μm s−1) and V (191 ± 10 μm s−1) [a moderate/severe stage of the disease], and Braak VI (0 μm s−1) [as the more severe stage of the disease]), this decrease was attributed to Cu, whose influence was demonstrated, mainly in the advanced stages of the disease. As such, we propose Cu-mediated modulation of catalase activity as a supporting tool for fast AD pre-screened early diagnosis. The enhanced fluid mixing and capability for autonomous motion allow for ultrafast detection (1 min) in just 1 μL of the CSF sample, which is particularly difficult to obtain and is available in low quantities. The new approach holds considerable promise as an additional supporting tool for fast neurodegenerative disease diagnosis and classification.Author contributions
J. Bujalance-Fernández: conceptualization, data curation, formal analysis, investigation, visualization, and writing – review and editing. E. Carro: resources and writing – review and editing. B. Jurado-Sánchez: conceptualization, data curation, formal analysis, visualization, funding acquisition, project administration, resources, supervision, writing – original draft, and writing – review and editing. A. Escarpa: conceptualization, formal analysis, funding acquisition, project administration, resources, supervision, and writing – review and editing.Data availability
Data for the article are available upon reasonable request to the authors due to confidentiality requirements. Data collected from human participants are not available for confidentiality reasons.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by The Spanish Ministry of Science, Innovation and Universities [grant PID2020-118154GB-I00 funded by MCIN/AEI/10.13039/501100011033 (A. E., B. J. S.), grant TED2021-132720B-I00 funded by MCIN/AEI/10.13039/501100011033 and the European Union “NextGenerationEU”/PRTR (A. E., B. J. S.), and grant CNS2023-144653 funded by MCIN/AEI/10.13039/501100011033 and the European Union “NextGenerationEU”/PRTR (B. J. S.)]; the Community of Madrid [grant number CM/JIN/2021-012 and Y2020/NMT6312 (NEURO-CHIP-CM)] and the Universidad de Alcalá [FPI contract, Plan Propio UAH (J. B. F.) and Línea de Actuación Excelencia para el Profesorado Universitario de la UAH, EPU-INV-UAH/2022/003 (B. J. S.)]. We would like to thank Víctor de la Asunción-Nadal for his help in the development of the simulation for MOF movement, Guillermo Sánchez-Esteban for his help in the beginning of the design of ZIF-8@Au@catalase micromotor and Rodrigo Barderas for his valuable help and assistance with cerebrospinal fluid sample acquisition.Notes and references
- H. G. Zhou, J. R. Long and O. M. Yaghi, Chem. Rev., 2012, 112, 673–674 CrossRef.
- S. L. Griffin and N. R. Champness, Coord. Chem. Rev., 2020, 414, 213295 CrossRef.
- A. Terzopoulou, J. D. Nicholas, X.-Z. Chen, B. J. Nelson, S. Pané and J. Puigmartí-Luis, Chem. Rev., 2020, 120, 11175–11193 CrossRef PubMed.
- B. Khezri and M. Pumera, Adv. Mater., 2019, 31, 1806530 CrossRef.
- J. Bujalance-Fernández, B. Jurado-Sánchez and A. Escarpa, Chem. Commun., 2023, 59, 10464–10475 RSC.
- G. A. Ozin, I. Manners, S. Fournier-Bidoz and A. Arsenault, Adv. Mater., 2005, 17, 3011–3018 CrossRef.
- W. Gao, S. Sattayasamitsathit, J. Orozco and J. Wang, J. Am. Chem. Soc., 2011, 133, 11862–11864 CrossRef.
- Y. Mei, A. A. Solovev, S. Sanchez and O. G. Schmidt, Chem. Soc. Rev., 2011, 40, 2109–2119 RSC.
- J. Wang, Nanomachines: Fundamentals and Applications, Wiley, 2013 Search PubMed.
- Y. Ikezoe, G. Washino, T. Uemura, S. Kitagawa and H. Matsui, Nat. Mater., 2012, 11, 1081–1085 CrossRef CAS.
- T. T. Y. Tan, J. T. M. Cham, M. R. Reithofer, T. S. Andy Hor and J. M. Chin, Chem. Commun., 2014, 50, 15175–15178 RSC.
- J. Li, X. Yu, M. Xu, W. Liu, E. Sandraz, H. Lan, J. Wang and S. M. Cohen, J. Am. Chem. Soc., 2017, 139, 611–614 CrossRef PubMed.
- A. Ayala, C. Carbonell, I. Imaz and D. Maspoch, Chem. Commun., 2016, 52, 5096–5099 RSC.
- L. Dekanovsky, Y. Ying, J. Zelenka, J. Plutnar, S. M. Beladi-Mousavi, I. Křížová, F. Novotný, T. Ruml and M. Pumera, Adv. Funct. Mater., 2022, 32, 2205062 CrossRef.
- X. Wang, X.-Z. Chen, C. C. J. Alcântara, S. Sevim, M. Hoop, A. Terzopoulou, C. de Marco, C. Hu, A. J. de Mello, P. Falcaro, S. Furukawa, B. J. Nelson, J. Puigmartí-Luis and S. Pané, Adv. Mater., 2019, 31, 1901592 CrossRef.
- Y. Yang, X. Arqué, T. Patiño, V. Guillerm, P.-R. Blersch, J. Pérez-Carvajal, I. Imaz, D. Maspoch and S. Sánchez, J. Am. Chem. Soc., 2020, 142, 20962–20967 CrossRef PubMed.
- Z. Guo, T. Wang, A. Rawal, J. Hou, Z. Cao, H. Zhang, J. Xu, Z. Gu, V. Chen and K. Liang, Mater. Today, 2019, 28, 10–16 CrossRef.
- Z. Guo, Y. Wu, Z. Xie, J. Shao, J. Liu, Y. Yao, J. Wang, Y. Shen, J. J. Gooding and K. Liang, Angew. Chem., Int. Ed., 2022, 61, e202209747 CrossRef CAS.
- C. Li, T. Hang and Y. Jin, Exploration, 2023, 3, 20220151 CrossRef CAS.
- W. Liang, K. Flint, Y. Yao, J. Wu, L. Wang, C. Doonan and J. Huang, J. Am. Chem. Soc., 2023, 145, 20365–20374 CrossRef CAS.
- Z. Armstrong, A. MacRae, M. Lenertz, Q. Li, K. Johnson, A. Scheiwiller, P. Shen, L. Feng, M. Quadir and Z. Yang, ACS Appl. Mater. Interfaces, 2023, 15, 38124–38131 CrossRef CAS.
- P. Scheltens, B. De Strooper, M. Kivipelto, H. Holstege, G. Chételat, C. E. Teunissen, J. Cummings and W. M. van der Flier, Lancet, 2021, 397, 1577–1590 CrossRef CAS.
- C. J. Sarell, S. R. Wilkinson and J. H. Viles, J. Biol. Chem., 2010, 285, 41533–41540 CrossRef CAS PubMed.
- M. W. Bourassa, A. C. Leskovjan, R. V. Tappero, E. R. Farquhar, C. A. Colton, W. E. Van Nostrand and L. M. Miller, Biomed. Spectrosc. Imaging, 2013, 2, 129–139 CAS.
- K. L. Summers, K. M. Schilling, G. Roseman, K. A. Markham, N. V. Dolgova, T. Kroll, D. Sokaras, G. L. Millhauser, I. J. Pickering and G. N. George, Inorg. Chem., 2019, 58, 6294–6311 CrossRef CAS.
- E. Andrási, É. Farkas, H. Scheibler, A. Réffy and L. Bezúr, Arch. Gerontol. Geriatr., 1995, 21, 89–97 CrossRef PubMed.
- R. Squitti, M. Siotto, E. Cassetta, I. G. E. Idrissi and N. A. Colabufo, Clin. Chem. Lab. Med., 2017, 55, 1360–1367 CAS.
- A. Miglione, M. Spinelli, A. Amoresano and S. Cinti, ACS Meas. Sci. Au, 2022, 2, 177–184 CrossRef CAS.
- J. Cravillon, R. Nayuk, S. Springer, A. Feldhoff, K. Huber and M. Wiebcke, Chem. Mater., 2011, 23, 2130–2141 CrossRef CAS.
- B. T. Hyman, C. H. Phelps, T. G. Beach, E. H. Bigio, N. J. Cairns, M. C. Carrillo, D. W. Dickson, C. Duyckaerts, M. P. Frosch, E. Masliah, S. S. Mirra, P. T. Nelson, J. A. Schneider, D. R. Thal, B. Thies, J. Q. Trojanowski, H. V. Vinters and T. J. Montine, Alzheimer's Dementia, 2012, 8, 1–13 CrossRef PubMed.
- F. Hao, M. Jing, X. Zhao and R. Liu, J. Photochem. Photobiol., B, 2015, 143, 100–106 CrossRef.
- S. M. Singh and P. M. Sivalingam, J. Fish Biol., 1982, 20, 683–688 CrossRef.
- J. Orozco, V. García-Gradilla, M. D'Agostino, W. Gao, A. Cortés and J. Wang, ACS Nano, 2013, 7, 818–824 CrossRef.
- Y. Pan, Y. Liu, G. Zeng, L. Zhao and Z. Lai, Chem. Commun., 2011, 47, 2071–2073 RSC.
- K. S. Park, Z. Ni, A. P. Côté, J. Y. Choi, R. Huang, F. J. Uribe-Romo, H. K. Chae, M. O'Keeffe and O. M. Yaghi, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10186–10191 CrossRef PubMed.
- A. Takeda, A. Hachimori, K. Sato and T. Samejima, J. Biochem., 1975, 78, 911–924 CrossRef PubMed.
- P. Wrede, M. Medina-Sánchez, V. M. Fomin and O. G. Schmidt, Small, 2021, 17, 2006449 CrossRef PubMed.
- S. Bucossi, M. Ventriglia, V. Panetta, C. Salustri, P. Pasqualetti, S. Mariani, M. Siotto, P. M. Rossini and R. Squitti, J. Alzheimer's Dis., 2011, 24, 175–185 Search PubMed.
- P. M. Roos, O. Vesterberg, T. Syversen, T. P. Flaten and M. Nordberg, Biol. Trace Elem. Res., 2013, 151, 159–170 CrossRef PubMed.
- G. J. Brewer, Chem. Res. Toxicol., 2010, 23, 319–326 Search PubMed.
- Z. Zhou, S. Chen, Y. Huang, B. Gu, J. Li, C. Wu, P. Yin, Y. Zhang and H. Li, Biosens. Bioelectron., 2022, 198, 113858 CrossRef PubMed.
- Y. G. Kaminsky and E. A. Kosenko, Free Radical Res., 2008, 42, 564–573 CrossRef PubMed.
- R. Squitti, J. Trace Elem. Med. Biol., 2014, 28, 482–485 CrossRef PubMed.
- R. Squitti, I. Simonelli, M. Ventriglia, M. Siotto, P. Pasqualetti, A. Rembach, J. Doecke and A. I. Bush, J. Alzheimer's Dis., 2014, 38, 809–822 Search PubMed.
- M. Lamtai, O. Zghari, S. Ouakki, I. Marmouzi, A. Mesfioui, A. El Hessni and A. Ouichou, Toxicol. Res., 2020, 36, 359–366 CrossRef PubMed.
- J. Arowoogun, O. O. Akanni, A. O. Adefisan, S. E. Owumi, A. S. Tijani and O. A. Adaramoye, J. Biochem. Mol. Toxicol., 2021, 35, e22623 CrossRef.
- R. Rajendran, R. Minqin, M. D. Ynsa, G. Casadesus, M. A. Smith, G. Perry, B. Halliwell and F. Watt, Biochem. Biophys. Res. Commun., 2009, 382, 91–95 CrossRef PubMed.
- L. Rink and H. Kirchner, J. Nutr., 2000, 130, 1407S–1411S CrossRef PubMed.
- M. Jarosz, M. Olbert, G. Wyszogrodzka, K. Młyniec and T. Librowski, Inflammopharmacology, 2017, 25, 11–24 CrossRef PubMed.
- Y. Qi, X. Li, S. Guo, F. He and R. Liu, J. Mol. Liq., 2024, 394, 123760 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Supporting figures and videos. See DOI: https://doi.org/10.1039/d4nr02044h |
| This journal is © The Royal Society of Chemistry 2024 |