Folic acid-targeted redox responsive polylactic acid-based nanoparticles co-delivering pirarubicin and salinomycin suppress breast cancer tumor growth in vivo†
Priya
Gupta
,
Ankushi
Bansal
 *,
Harshdeep
Kaur
,
Mohd
Anees
,
Neetu
Singh
and
Harpal
Singh
*,
Harshdeep
Kaur
,
Mohd
Anees
,
Neetu
Singh
and
Harpal
Singh
 *
*
Centre for Biomedical Engineering, Indian Institute of Technology Delhi, New Delhi, India. E-mail: harpal2000@yahoo.com; bansalankushi21@gmail.com
First published on 18th October 2024
Abstract
Targeted cancer therapy using nanocarriers has emerged as a promising solution to the majority of drawbacks associated with conventional chemotherapy. The present research work describes the development of folic acid (FA)-targeted redox responsive [S-(PLA-b-PEG-CONH)]2 polymeric nanoparticles for the co-delivery of pirarubicin (Pira) and salinomycin (Sal). The nanoparticles’ redox responsiveness arises from embedded disulfide bonds within the polymer, which gradually break in response to high GSH levels in tumors, enabling sustained drug release. The nanoparticles exhibited a hydrodynamic size of ∼104 nm and a surface charge density of −15.5 mV with low PDI values. Blank nanoparticles (w/o drug) showed negligible toxicity towards both non-malignant human and murine cells and exhibited excellent stability under different environmental conditions for up to 3 weeks. A cellular internalization study conducted using Rho B/C6 dual-dye-encapsulated nanoparticles showed efficient uptake of the nanoparticles after just 1 hour of incubation with SUM-149 2D adherent cells and 3D spheroids. The release of Pira and Sal from Pira/Sal dual-loaded nanoparticles increased significantly in a reducing environment. The % cumulative release of Pira increased from 20.5% ± 1.0 in PBS (pH 7.4) to 40.1% ± 0.4 in dithiothreitol (DTT) after 20 days; similarly, the % cumulative release of Sal increased from 36.2% ± 1.7 in PBS (pH 7.4) to 51.5% ± 1.7 in DTT. The cytotoxicity studies of FA-targeted Pira/Sal dual-loaded nanoparticles with varying Pira![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sal ratios (1
Sal ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) revealed that the nanoparticles displayed 8–10 fold lower IC50 values than the respective free drug formulations across multiple breast cancer cell lines including SUM-149, MDA-MB-231 and EAC as well as in 3D mammospheres. Balb/c syngeneic mice bearing EAC tumors experienced ∼100% tumor regression upon treatment with FA-targeted Pira/Sal (3
3) revealed that the nanoparticles displayed 8–10 fold lower IC50 values than the respective free drug formulations across multiple breast cancer cell lines including SUM-149, MDA-MB-231 and EAC as well as in 3D mammospheres. Balb/c syngeneic mice bearing EAC tumors experienced ∼100% tumor regression upon treatment with FA-targeted Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) dual-loaded nanoparticles, indicating synergistic anti-tumor potency. In vivo survival and histopathological studies indicated no significant toxicity in vital organs of the body as compared to free drugs. Based on the performance, the currently investigated FA-targeted Pira/Sal dual-loaded nano-formulation is recommended to be further explored in other cancer types as well as in higher animals for cancer therapy.
1) dual-loaded nanoparticles, indicating synergistic anti-tumor potency. In vivo survival and histopathological studies indicated no significant toxicity in vital organs of the body as compared to free drugs. Based on the performance, the currently investigated FA-targeted Pira/Sal dual-loaded nano-formulation is recommended to be further explored in other cancer types as well as in higher animals for cancer therapy.
1. Introduction
Targeted drug delivery has emerged as an effective and promising strategy in cancer therapy.1,2 It provides the opportunity to improve treatment efficacy by delivering drugs directly to tumor cells while minimizing systemic toxicity and safeguarding normal cells. It utilizes various mechanisms to specifically target cancer cells within the tumor microenvironment.3 Key disparities such as hypoxia,4 overexpressed antigens or receptors,5 acidic pH,6 elevated ROS levels,7 and an increased number of tumor associated macrophages and fibroblasts8 serve as targets for selective drug delivery for cancer therapy.9,10Ligand-mediated targeting, particularly using folate receptors overexpressed in various cancer types, has attracted significant attention due to its ability to achieve selective drug delivery.11–13 For instance, Monteiro et al.14 developed a dual-targeted (folate and pH) liposome-based formulation for the effective delivery of paclitaxel to breast cancer cells. They demonstrated improved cellular uptake and enhanced antitumor efficacy compared to non-targeted liposomes. Similarly, Tonbul et al. utilized folic acid-conjugated mesoporous silica nanoparticles to deliver doxorubicin selectively to folate receptor-positive breast cancer cells, resulting in enhanced cytotoxicity and reduced systemic toxicity.15 This targeted approach harnessing the overexpression of folate receptors showcases the potential of folate in advancing cancer therapeutics.
Nanoparticles have further revolutionized targeted therapy by serving as effective delivery vehicles for anticancer agents.16,17 Their multifunctionality allows for tailored drug delivery approaches that can be fine-tuned to the specific characteristics of the tumor microenvironment.18 For instance, the integration of redox-responsive chemical linkers into nanocarriers provides the benefit of controlled release due to degradation in the presence of high glutathione concentration in tumor cells. Koul et al. developed redox-sensitive polymersomes for the delivery of doxorubicin (DOX) and in vivo results demonstrated that polymersomes achieved a 7.16-fold greater reduction in tumor volume compared to the control and showed 5.39 times better tumor inhibition than free DOX in EAC tumor bearing Swiss albino mice.19 In another study, redox-responsive Dox-conjugated poly-L-glutathione oxidized nanomedicine (Dox-Poly GSSG) was prepared and it was observed that the nanomedicine exhibited better anticancer properties in terms of off-target toxicity, maintaining mice's body weight and extending the survival and apoptosis of tumor cells in tumor tissues as compared to free DOX.20 Thus, redox-responsive nanoparticles offer a promising strategy for more precise and safer cancer therapies.
Pirarubicin (Pira), an anthracycline developed by Umeza in 1979, is widely gaining popularity as a potent substitute of doxorubicin due to its reportedly lower off-target cytotoxicity and higher anti-tumour efficacy and cellular uptake.21 However, its long-term usage shows the dose-limiting effect and chronic cardiac toxicity. Encapsulating pirarubicin in a nanoparticulate formulation is expected to reduce its off-target cytotoxicity and improve its anti-tumor efficacy. Despite the great tumor inhibiting potential of chemotherapeutic drugs, the elimination of cancer stem cells is still a challenging task. Salinomycin (Sal) is an ionophore antibiotic noted for its exceptional efficiency in targeting cancer stem cells, addressing the challenge of preventing cancer relapse by eliminating these cells.22,23
In the present work, we have developed folic acid-conjugated redox-responsive polylactic acid (PLA)-based biodegradable polymeric nanoparticles for co-delivery of pirarubicin and salinomycin for synergistic therapeutic action. To enhance the stability and stealth properties of the nanoparticles, polyethylene glycol (PEG) was incorporated with PLA via esterification.
The prepared nanoparticles were thoroughly characterized for their physiochemical properties and evaluated in vitro and in vivo for their biocompatibility, drug release kinetics, cellular uptake, and synergistic anti-tumor efficacy. To the best of our knowledge, folic acid-guided redox-responsive polymeric nanoparticles for co-delivery of Pira and Sal have not been previously reported. Hence, this innovative approach presents a promising avenue for further investigation as a potential candidate for cancer treatment.
2. Materials and methods
2.1 Materials
L(−)-Lactide, polyethylene glycol (PEG, 4 kDa), 2-hydroxyethyl disulfide (2-HEDS), folic acid (FA), succinic anhydride, 1,3-dicyclohexylcarbodiimide (DCC), 4-(dimethyl amino) pyridine (DMAP), dithiothreitol (DTT), 4′,6-diamino-2-phenylindole (DAPI), and N-hydroxysuccinimide (NHS) were procured from Sigma-Aldrich (USA). Both drugs, pirarubicin (Pira) and salinomycin (Sal), were obtained from MedChem Express (USA). Mammalian cell culture media, Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and antibiotic penicillin streptomycin solution were purchased from Gibco (MA, USA). A 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide MTT cell assay kit was bought from HiMedia Laboratories. All the solvents including dichloromethane (DCM), methanol (MeOH), hydrochloric acid (HCl), dimethyl formamide (DMF), dimethyl sulphoxide (DMSO), acetonitrile (ACN) and tetrahydrofuran (THF) were obtained from Merck & Co. India. Amicon ultra centrifugal filters (3 kDa) were obtained from Merck Millipore (Billerica, MA, USA).2.2 Synthesis and characterization of the FA-conjugated [S-(PLA-b-PEG-CONH)]2 block copolymer
The synthesis of the FA-conjugated [S-(PLA-b-PEG-CONH)]2 block copolymer was carried out by following previously reported literature and is detailed in the ESI.†24,25 In brief, the ring-opening polymerization of L-lactide was conducted using bifunctional 2-HEDS as the initiator and tin(II) 2-ethylhexanoate as the catalyst at 170 °C for 3 hours under an N2 flow to obtain PLA with a disulfide core (HO-PLA-s-s-PLA-OH, or [S-(PLA-OH)]2). Subsequently, the terminal –OH groups of the [S-(PLA-OH)]2 polymer were reacted with succinic anhydride to obtain a –COOH terminated polymer, i.e. HOOC-PLA-s-s-PLA-COOH, or [S-(PLA-COOH)]2. In the subsequent step, PEG was incorporated into the polymer via Steglich esterification, which was performed between the –OH group of PEG-diol (4 kDa) and the –COOH group of the PLA polymer. DCC and DMAP were utilized for PLA/PEG esterification at room temperature. Following the coupling reaction, the product was recovered by precipitation in cold diethyl ether and MeOH. It was further modified with succinic anhydride, as previously described, to obtain a –COOH-terminated block copolymer with a disulfide core, (HOOC-PEG-b-PLA-s-s-PLA-b-PEG-COOH, or [S-(PLA-b-PEG-COOH)]2). In the final step, the conjugation of FA to the diblock copolymer was achieved using DCC-NHS chemistry, where the –COOH group from the block copolymer was coupled with the –NH2 group of FA to obtain the desired polymer (FA-HNOC-PEG-b-PLA-s-s-PLA-b-PEG-CONH-FA, or FA-[S-(PLA-b-PEG-CONH)]2).The structures of the polymers at every stage were determined by proton nuclear magnetic resonance (1H NMR) spectroscopy (Bruker AC 400 MHz, USA). The samples were prepared at a concentration of 10 mg ml−1 in deuterated chloroform (CDCl3) and data were recorded at room temperature. The molecular weights and polydispersity indices (PDIs) of the polymers were also determined by gel permeation chromatography (GPC) using a GPCmax Viscotek equipped with RI, RALS, and LALS detectors and polystyrene (Mol. Wt. 105 kDa) was used as the internal standard for instrument calibration [mobile phase: THF solvent, flow rate: 1 ml min−1, ejection volume: 10 μl, and 25 °C].
2.3 Preparation of FA-conjugated [S-(PLA-b-PEG-CONH)]2 block copolymeric nanoparticles
Polymeric nanoparticles loaded with pirarubicin/salinomycin were prepared by using the nanoprecipitation technique. Concisely, 10 mg of FA-conjugated [S-(PLA-b-PEG-CONH)]2 polymer was dissolved in 500 μl of ACN with mild heating. Separately, 0.5 mg of Pira or Sal were dissolved in a minimum amount of DMSO or MeOH, respectively. Thereafter, the polymer solution in ACN and the drug solution in DMSO or MeOH were mixed via sonication. On the other hand, 10 mg of emulsifier, Pluronic® F-127, was dissolved in 2 ml of Milli-Q® water via stirring. Afterwards, the drug–polymer mixture was introduced dropwise into the emulsifier solution using a 1 ml syringe (Dispovan, 26 G × 1.5 inches) with continuous stirring. The nanoparticles were stirred at 600 rpm and R.T. for 12 hours to allow ACN evaporation and nanoparticle maturation. After 12 hours, the nanoparticles were filtered using an Amicon filter (3 kDa; Millipore, Sigma) at 4000 rpm for 40 minutes, followed by water washing to remove the unencapsulated drug from the nanoparticles. The purified nanoparticles were lyophilized using 5 mg of glucose as a cryoprotectant. The supernatant collected was used to quantify the free drug content indirectly and to calculate the drug encapsulation efficiency of the nanoparticles. For the quantification of pirarubicin, the lyophilized supernatant was dissolved in mobile phase solvent and analyzed using HPLC (Agilent 1260 Infinity, USA) [mobile phase: 0.1% TFA in ACN, flow rate: 1 ml min−1, ejection volume: 10 μl, 25 °C and λexcitation = 480 nm and λemmision = 580 nm]. Free Pira (concentration range 0.5 mg ml−1 to 0.0078 mg ml−1) was used to plot the calibration curve using a similar HPLC protocol. Salinomycin was quantified by using the vanillin assay, where the lyophilized supernatant was dissolved in 1 ml of MeOH via sonication. This supernatant solution was incubated with vanillin working solution (40 mg of vanillin in 2 ml of MeOH with 0.01% concentrated HCl) in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 v/v under dark conditions for 10 min, followed by measuring absorbance at λmax = 526 nm using a UV-Vis spectrometer.26 Free Sal (concentration range 1 mg ml−1 to 0.03125 mg ml−1 MeOH) was used to plot the calibration curve. The % encapsulation efficiency and the drug loading content of the nanoparticles were calculated using the formulas mentioned below.
9 v/v under dark conditions for 10 min, followed by measuring absorbance at λmax = 526 nm using a UV-Vis spectrometer.26 Free Sal (concentration range 1 mg ml−1 to 0.03125 mg ml−1 MeOH) was used to plot the calibration curve. The % encapsulation efficiency and the drug loading content of the nanoparticles were calculated using the formulas mentioned below. | (i) |
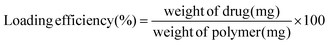 | (ii) |
2.4 Characterization of FA-conjugated Pira/Sal-loaded [S-(PLA-b-PEG-CONH)]2 block copolymeric nanoparticles
Pira/Sal-loaded FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles were investigated for their size, surface charge density and poly dispersity index (PDI) using dynamic light scattering (DLS, Anton Paar, Austria). For measuring their size and PDI, 20 μl of nanoparticles (10 mg ml−1) were diluted to 1 ml solution with Milli-Q® water and the same dilution was used for Zetasizer measurement. Moreover, the shape, size and morphology of the nanoparticles were also confirmed by using HR-TEM analysis (Tecnai™ G2 20). For sample preparation, diluted nanoparticles (1/10 of 10 mg ml−1) were drop-cast on a carbon-coated copper TEM grid, followed by staining with uranyl acetate.2.5 Stability evaluation of FA-conjugated [S-(PLA-b-PEG-CONH)]2 polymeric nanoparticles
The stability of the FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles was examined at three different temperatures: 37 °C, 4 °C and 25 °C ± 3.0 (R.T.) for a period of 30 days. Nanoparticles with a polymer concentration of 10 mg ml−1 were maintained at all three temperatures and their size variation was evaluated using DLS every four days. Origin 2020b software was used to plot and analyze the collective data of the study.2.6 Biocompatibility assessment of FA-conjugated [S-(PLA-b-PEG-CONH)]2 polymeric nanoparticles
To check whether the prepared nanoparticles are suitable for biological applications, their cellular cytocompatibility and hemocompatibility were evaluated. The cytocompatibility study was carried out on two different non-malignant cell lines: HEK 293 (human cells) and NIH 3T3 (mice fibroblast cells). The cells were seeded in a flat bottom 96-well plate with a cell density of 5000 cells per well, cultured in DMEM and left for 24 hours in a CO2 incubator. Meanwhile, lyophilized FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles were reconstituted at a concentration of 10 mg ml−1 and serially diluted up to a polymer concentration of 0.001 mg ml−1. Following 24 hours, the cells were incubated with a range of different concentrations of polymer solutions and analyzed after 72 hours of further incubation via the MTT assay protocol.For hemocompatibility evaluation, blood was collected from a healthy female Balb/c mouse using the retro-orbital technique. The blood sample was then processed via centrifugation at 1500 rpm for 10 min to separate RBCs, followed by washing and dilution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) with PBS pH 7.4 and then these RBCs were incubated with serially diluted different concentrations of lyophilized FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles in a volume ratio 1
10) with PBS pH 7.4 and then these RBCs were incubated with serially diluted different concentrations of lyophilized FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles in a volume ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 1 hour in a shaker incubator at 37 °C. After 1 hour, the samples were centrifuged at 1500 rpm for 10 min to allow RBCs to settle down and the supernatant was aspirated in a micro-well plate, which was used to measure absorbance at λmax = 540 nm using an ELISA microplate reader (BioTek Epoch, Agilent Technologies, USA). 1% solutions of Triton X 100 and PBS culture were used as the positive control and negative control, respectively. % Hemolysis was calculated using the equation given below.
1 for 1 hour in a shaker incubator at 37 °C. After 1 hour, the samples were centrifuged at 1500 rpm for 10 min to allow RBCs to settle down and the supernatant was aspirated in a micro-well plate, which was used to measure absorbance at λmax = 540 nm using an ELISA microplate reader (BioTek Epoch, Agilent Technologies, USA). 1% solutions of Triton X 100 and PBS culture were used as the positive control and negative control, respectively. % Hemolysis was calculated using the equation given below.
 | (iii) |
2.7 Drug release profile in vitro
To analyze the drug release rates of single- and dual-loaded Pira/Sal FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles, the nanoparticles were subjected to two different buffer environments for 20 days: neutral physiological conditions (PBS buffer, pH 7.4) and a disulfide reducing environment (10 mM DTT in PBS buffer, pH 7.4). In brief, lyophilized nanoparticles were reconstituted at a concentration of 10 mg ml−1 Milli-Q® water and dispersed in 10 ml of buffer medium in each case. To ensure a controlled drug release environment, the nanoparticles were kept under dark conditions in a shaker incubator at 150 rpm and 37 °C. At predetermined intervals, the nanoparticles were Amicon filtered (3 kDa; Millipore, Sigma) and washed twice with the respective buffer medium and again kept in fresh media for further release. The collected supernatant was lyophilized and the concentration of free Pira was analyzed using HPLC (Agilent 1260 Infinity, USA) [mobile phase: 0.1% TFA in ACN, flow rate: 1 ml min−1, ejection volume: 10 μl, 25 °C and λexcitation = 480 nm and λemmision = 580 nm]. For the quantification of Sal, the vanillin assay protocol was followed as mentioned above. For the quantification of Pira and Sal in the dual-loaded nanoparticles, the supernatant was divided into halves and processed individually for each drug. The study was performed in triplicate for every sample.2.8 Cellular uptake in 2D adherent cells and 3D mammospheres
The cellular internalization ability of the prepared nanoparticles was examined qualitatively using confocal microscopy (CLSM; Fluoview FV1000 Olympus), followed by quantification via FACS (BD FACSAria III). For the experiment, SUM-149 breast cancer cells were used. Briefly, a cell density of 5 × 105 cells was seeded in a 6-well adherent culture plate containing a gelatin-coated coverslip per well and left in incubation under a 5% CO2 humidified atmosphere at 37 °C. After ∼70% cellular confluency, old media were replaced with fresh media containing 10 μM concentration of Rho B/C6 dual-dye loaded FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles. Following a 4-hour incubation period, DAPI (2–3 μl per well) was added to stain the nucleus of the cells. After 15 minutes of staining, followed by 2–3 washes with PBS, the cover slips were elicited and imaged under a confocal microscope. A similar protocol was followed for preparing samples for FACS, except for omitting the coverslips, and standard 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were scanned for every sample in FACS.
000 events were scanned for every sample in FACS.
In addition, the cellular uptake was investigated in SUM-149-derived 3D mammospheres, grown in an ultra-low attachment 6-well culture plate in F-12 DMEM supplemented with 2 mM glutamine and other growth factors (EGF, bFGF and B-27). 5 × 103 cells were seeded per well and left in a CO2 incubator undisturbed for 7–8 days to allow the growth of 3D spheres until they attained a size >50 μm. Afterwards, FA-conjugated rhodamine B/coumarin 6 dual-dye-loaded nanoparticles were prepared and incubated with 3D spheres for 1 hour at a final concentration of 10 μM. Thereafter, the spheres were washed with PBS and imaged under a confocal microscope.
2.9 2D cell proliferation inhibition studies of free drug Pira/Sal vs. FA-conjugated Pira/Sal dual-loaded nanoparticles
The MTT assay protocol was used to investigate the cell proliferation inhibition activity of FA-targeted Pira/Sal single vs. dual-loaded nanoparticles and free Pira/Sal drug formulations on different cancer cell lines, including SUM-149, MDA-MB-231 and EAC. Briefly, cells with a cell density of 5 × 103 were seeded into a flat bottom 96-well tissue culture plate containing DMEM growth medium. After 24 hours, the cells were treated with serially diluted concentrations (ranging from 100 μM to 1 nM) of the prepared nanoparticles vs. free drugs, followed by incubation for 72 hours. Subsequently, the cells were washed with PBS and treated with MTT solution (5 mg ml−1 in PBS culture, pH 7.4) to achieve a final concentration of 10% of the total medium volume. Thereafter, the cells were then incubated in the dark for another 4 hours. After incubation, all the media were syringed out and 100 μl of DMSO was added to each well, followed by a 15-minute incubation on a shaker under dark conditions. Finally, the absorbance of the entire plate was recorded using an ELISA microplate spectrophotometer at λmax 570 nm. GraphPad Prism version 9.0 was used to plot dose–response curves using non-linear regression analysis. All the experiments were performed in triplicate to minimize the chances of manual errors.2.10 In vitro cytotoxicity on 3D mammospheres
SUM-149-derived 3D mammospheres were grown in an ultra-low attachment 24-well tissue culture plate using the protocol described earlier. As the size reached >50 μm, the mammospheres were treated with FA-targeted Pira, Sal and Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) nanoparticles for 72 hours. After this incubation period, the mammospheres were collected and centrifuged at 2000 rpm for 5 min. The supernatant was then discarded, the pellet was washed with PBS and the mammospheres were incubated with 50 μl of CCK-8 reagent for another 6 hours. Thereafter, the spheres were again centrifuged and 100 μl of supernatant was aspirated from each sample to record absorbance using an ELISA spectrometer (λmax = 450 nm). Control samples, which did not receive treatment, were included for comparison.
1) nanoparticles for 72 hours. After this incubation period, the mammospheres were collected and centrifuged at 2000 rpm for 5 min. The supernatant was then discarded, the pellet was washed with PBS and the mammospheres were incubated with 50 μl of CCK-8 reagent for another 6 hours. Thereafter, the spheres were again centrifuged and 100 μl of supernatant was aspirated from each sample to record absorbance using an ELISA spectrometer (λmax = 450 nm). Control samples, which did not receive treatment, were included for comparison.
2.11 Combination index and synergy evaluation
For the quantitative simulation of the synergism of Pira/Sal drug combination at different ratios of Pira/Sal dual-loaded nanoparticles, the combination index (CI values) was calculated over a range of affected cellular fractions (Fa) using the automated algorithms in CompuSyn software, where CI < 1 indicates synergism, CI = 1 is the additive effect and CI > 1 is the antagonism.2.12 In vivo tumor regression, toxicity, and survival studies
The studies were carried out in AIIMS Delhi with the ethical clearance no. 122/IAEC/2019 issued under the guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA). To study the therapeutic effect of the prepared nanoparticles in vivo, BALB/c (22–25 g) strain syngeneic mice were used with an EAC cell tumor model. A total of 108 EAC cells per 100 μl PBS were injected subcutaneously into the left flank of mice to generate a solid tumor. To measure the dimensions of the tumor, a Vernier caliper was used, and the tumor volume was calculated using the following formula: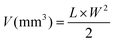 | (iv) |
For histopathological analysis, the harvested organs were embedded in paraffin wax, sectioned into slides, and stained with H&E staining and finally examined under a CLSM for tissue damage and toxicity. GraphPad Prism 9.0 was used to plot non-linear tumor regression curves and Kaplan–Meier survival plots.
2.13 Statistical analysis
All the studies were carried out in at least triplicate, and the data are reported as mean ± SD. One-way ANOVA with the Bonferroni test for multiple comparisons using statistical hypothesis was used to carry out significance value measurements. P values <0.0001(****), 0.0002(***), 0.0021(**) and 0.0332(*) were considered significant, whereas 0.1234 (ns) was considered non-significant.3. Results and discussion
3.1 Characterization of the FA-conjugated [S-(PLA-b-PEG-CONH)]2 polymer
The folic acid-conjugated redox-responsive PLA-based block copolymer, FA-conjugated [S-(PLA-b-PEG-CONH)]2, was successfully synthesized, as illustrated in Scheme 1, following a previously reported protocol with slight modifications.24,25 The synthesized polymer at various stages was characterized using 1H NMR (Fig. S(1–3)†) to assess its molecular structure. The 1H NMR spectra of the synthesized [S-(PLA-OH)]2 (Fig. S1†) polymer exhibited characteristic peaks at ∼1.6 ppm and ∼5.2 ppm corresponding to the –CH3 and –OCH protons of PLA. In addition, the appearance of small triplet peaks at 2.9 ppm and 3.8 ppm of –SCH2 and –SCH2CH2 protons of 2-HEDS, respectively, further confirmed the successful polymerization of the monomer L-lactide using 2-HEDS as an initiator to form [S-(PLA-OH)]2. The 1H NMR spectra of the –COOH-terminated polymer ([S-(PLA-COOH)]2, Fig. S2†) showed the appearance of new triplet peaks at 2.37 and 2.76 ppm, which correspond to the –CH2 groups of succinic anhydride, along with the previous peaks of PLA, further verifying the successful reaction of succinic anhydride with [S-(PLA-OH)]2 to yield a –COOH-terminated PLA polymer, [S-(PLA-COOH)]2. Next, the appearance of an additional new peak at 3.61 ppm corresponding to –OCH2 groups from PEG in 1H NMR (Fig. S3†) validated the successful conjugation of PEG–diol with [S-(PLA-COOH)]2 to yield the desired [S-(PLA-b-PEG-OH)]2 block copolymer. It is further modified with succinic anhydride to obtain a –COOH-terminated block copolymer with a disulfide core, [S-(PLA-b-PEG-COOH)]2. In the last step, the conjugation of FA to the [S-(PLA-b-PEG-OH)]2 block copolymer was confirmed by the appearance of a peak at ∼8.3 ppm due to the pteridine proton of folate, a peak at 7.95 ppm due to the para-aminobenzoic acid part of folate and a peak at 10.5 ppm due to the –COOH protons of FA in the 1H NMR [Fig. 1(A)].27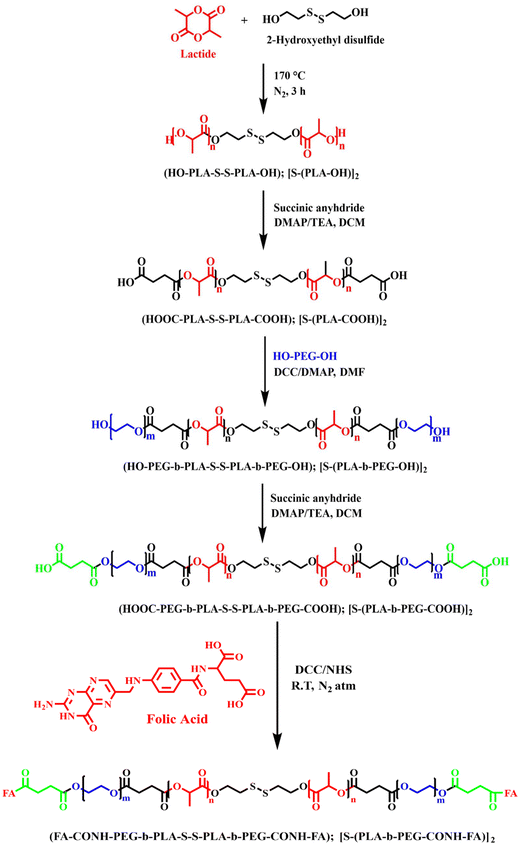 | ||
| Scheme 1 Reaction scheme of the synthesis of the FA-conjugated [S-(PLA-b-PEG-CONH)]2 block copolymeric polymer. | ||
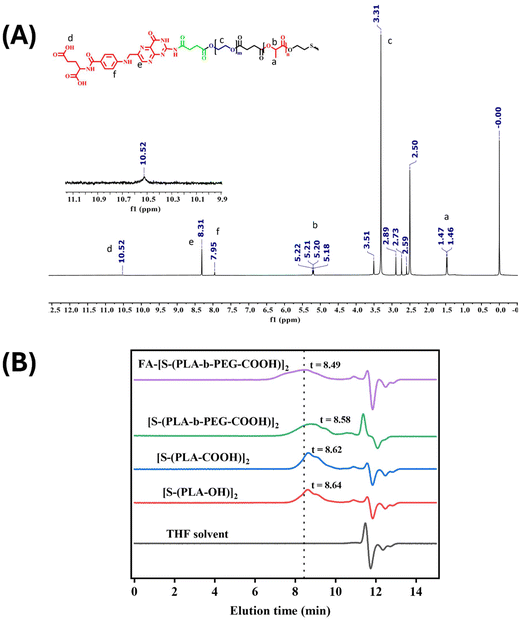 | ||
| Fig. 1 (A) 1H NMR spectra and (B) gel permeation chromatographic analysis of the FA-conjugated [S-(PLA-b-PEG-CONH)]2 polymer. | ||
According to GPC results, the number average molecular weight (Mn) and weight average molecular weight (Mw) of the [S-(PLA-OH)]2 polymer with a disulfide core were found to be 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 and 22
800 and 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 g mol−1, respectively, with a polydispersity index (Mw/Mn) of 1.1. After the conjugation of PEG and folic acid, the column elution time decreased to 8.49 min from 8.64 min due to an increase in the molecular weight of the polymer and the resulting final FA-conjugated [S-(PLA-b-PEG-CONH)]2 polymer showed an Mn and Mw of 27
400 g mol−1, respectively, with a polydispersity index (Mw/Mn) of 1.1. After the conjugation of PEG and folic acid, the column elution time decreased to 8.49 min from 8.64 min due to an increase in the molecular weight of the polymer and the resulting final FA-conjugated [S-(PLA-b-PEG-CONH)]2 polymer showed an Mn and Mw of 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 and 38
700 and 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 g mol−1, respectively, with a polydispersity index of 1.4 [Fig. 1(B)].
400 g mol−1, respectively, with a polydispersity index of 1.4 [Fig. 1(B)].
3.2 Preparation and characterization of FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles
The nanoprecipitation technique was employed to prepare blank, Pira/Sal single- or dual-loaded FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles, keeping the polymer-to-drug ratio constant at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
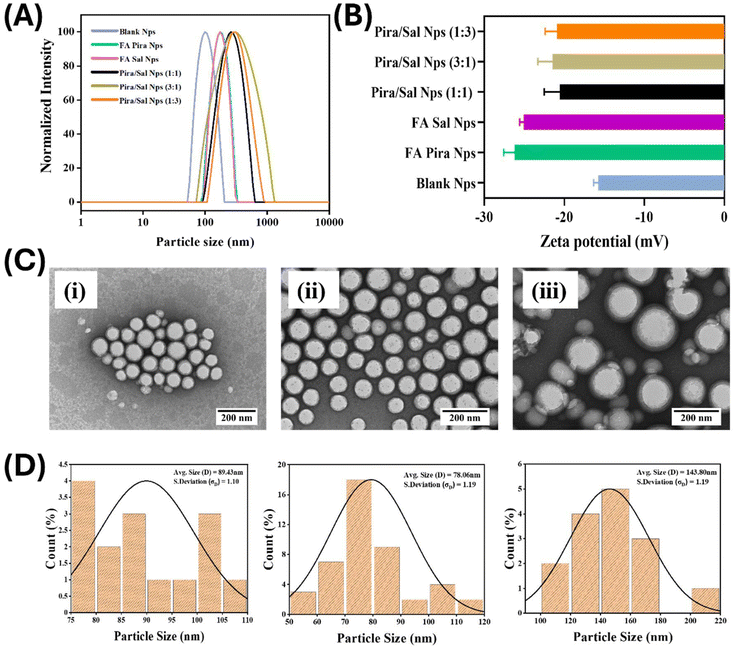
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The hydrodynamic sizes of the prepared Pira or Sal loaded nanoparticles were recorded as 185.5 ± 4.7 nm and 184.76 ± 4.1 nm, respectively, which are larger in size compared to blank nanoparticles, i.e. 104.47 ± 0.5 nm (Table 1). In the case of Pira/Sal dual-loaded nanoparticles, the hydrodynamic size increased by more than 100 nm compared to the single loaded nanoparticles due to a higher drug loading [Fig. 2(A)]. All the prepared nanoparticles displayed a narrow polydispersity index (PDI), confirming the homogeneous size distribution of the nanoparticles in the aqueous medium.28 In addition, the zeta potential of all nanoparticles, whether blank, single-, or dual-loaded, ranged between −15.8 and −25.0 mV [Fig. 2(B)]. The higher negative zeta potential values are attributed to the surface –COOH groups on folic acid. The absolute particle size and surface morphology of the nanoparticles were determined using TEM, which showed discrete core–shell like structures with fairly separated boundaries and low polydispersity indices in both the cases of Pira/Sal single- and dual-drug loaded nanoparticles [Fig. 2(C)]. The absolute size analysis using TEM particle size distribution histograms showed a similar trend to hydrodynamic size via DLS, validating the consistency of the prepared nanoparticles [Fig. 2(D)].
1. The hydrodynamic sizes of the prepared Pira or Sal loaded nanoparticles were recorded as 185.5 ± 4.7 nm and 184.76 ± 4.1 nm, respectively, which are larger in size compared to blank nanoparticles, i.e. 104.47 ± 0.5 nm (Table 1). In the case of Pira/Sal dual-loaded nanoparticles, the hydrodynamic size increased by more than 100 nm compared to the single loaded nanoparticles due to a higher drug loading [Fig. 2(A)]. All the prepared nanoparticles displayed a narrow polydispersity index (PDI), confirming the homogeneous size distribution of the nanoparticles in the aqueous medium.28 In addition, the zeta potential of all nanoparticles, whether blank, single-, or dual-loaded, ranged between −15.8 and −25.0 mV [Fig. 2(B)]. The higher negative zeta potential values are attributed to the surface –COOH groups on folic acid. The absolute particle size and surface morphology of the nanoparticles were determined using TEM, which showed discrete core–shell like structures with fairly separated boundaries and low polydispersity indices in both the cases of Pira/Sal single- and dual-drug loaded nanoparticles [Fig. 2(C)]. The absolute size analysis using TEM particle size distribution histograms showed a similar trend to hydrodynamic size via DLS, validating the consistency of the prepared nanoparticles [Fig. 2(D)].
| S. no. | Sample | Size (nm) | Zeta (mV) | PDI |
|---|---|---|---|---|
| 1. | Blank | 104.5 ± 5.3 | −15.8 ± 1.9 | 0.14 ± 0.04 |
| 2. | Pira-loaded | 185.5 ± 7.7 | −25.0 ± 3.2 | 0.18 ± 0.03 |
| 3. | Sal-loaded | 184.8 ± 4.0 | −24.1 ± 2.0 | 0.15 ± 0.01 |
| 4. | Pira/Sal (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
283.8 ± 4.2 | −22.7 ± 1.8 | 0.24 ± 0.04 |
| 5. | Pira/Sal (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
324.5 ± 2.8 | −23.4 ± 4.7 | 0.24 ± 0.02 |
| 6. | Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
327.2 ± 7.0 | −20.7 ± 2.8 | 0.18 ± 0.02 |
The FA-conjugated polymeric nanoparticles showed excellent encapsulation efficiency for both drugs. The encapsulation efficiency of Pira (∼95%) was found to be significantly higher as compared to that of Sal (∼80%) in the Pira- or Sal-loaded nanoparticles, indicating the better entrapment of Pira in the core of the nanoparticles due to its comparatively higher hydrophobicity. However, the encapsulation efficiency of Sal increased to 90% in the case of Pira/Sal dual-loaded nanoparticles due to the intermolecular interactions between the two drugs. The drug loading content in the nanoparticles was calculated using equation (ii) and the results are tabulated in Table 2.
3.3 Stability examination of FA-conjugated [S-(PLA-b-PEG-CONH)]2 block copolymeric nanoparticles
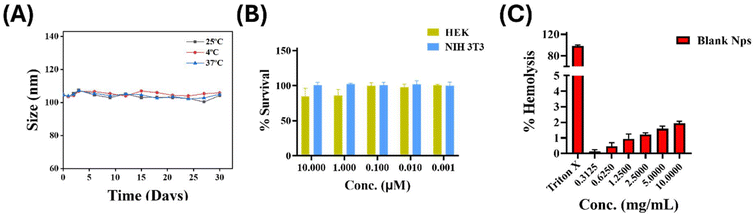
The stability of nanoparticles is important for those meant for the intravenous route (IV) of administration as agglomerated nanoparticles can obstruct blood capillaries or cause other complications. Therefore, the stability of the FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles was determined by measuring their hydrodynamic size under three different conditions – at low temperature (4 °C), room temperature (25 °C ± 3.0) and physiological temperature (37 °C) over 30 days [Fig. 3(A)]. As depicted in [Fig. 3(A)], the nanoparticles did not show any significant change in size until the end of the study period, thus indicating robust stability of the nanoparticles.3.4 Biocompatibility assessment of FA-conjugated [S-(PLA-b-PEG-COOH)]2 block copolymeric nanoparticles
Nano-formulations envisioned for intravenous administration must be hemocompatible and cytocompatible. For cytocompatibility, the formulation should exhibit selective cytotoxicity towards cancer cells, sparing healthy cells to avoid side effects. Therefore, the cytocompatibility of the prepared FA-conjugated [S-(PLA-b-PEG-CONH)]2 blank nanoparticles was evaluated on two healthy cell lines HEK 293 (human) and NIH 3T3 (murine) [Fig. 3(B)]. The results confirmed that even at the highest concentration of nanoparticles (10 mg ml−1), no significant cytotoxicity was seen in any of the healthy cells with nearly 100% cell survival after 72 hours of treatment. Since RBCs are the first point of interaction for any intravenous dose circulating in blood, a potential tendency to cause hemolysis may lead to anemia, compromised immunity and other pathological diseases in the subject. When tested for hemolysis, the results indicated that 10.0 mg ml−1 and 5.0 mg ml−1 nanoparticle concentrations showed only 1.94% and 1.59% hemolysis, respectively, as compared to 100% hemolysis observed in the positive control, Triton X 100. Overall, the % hemolysis was not observed >2% at any concentration of nanoparticles used [Fig. 3(C)], thereby confirming the suitability of the prepared nanoparticles for intravenous administration in in vivo applications.3.5 Drug release profile in vitro
The quantification of Sal was performed using the vanillin colorimetric assay. In this assay, the aldehydic group of the vanillin reagent reacts with the ketonic group of Sal in the presence of concentrated HCl via aldol condensation, producing a violet-pink colored product with UV-Vis absorption at λmax = 526 nm. The intensity of the violet-pink color produced is directly proportional to the amount of Sal present in the supernatant. For the quantification of Sal in the case of Pira/Sal dual-loaded nanoparticles, the lyophilized supernatant was first treated with diethyl ether to precipitate Pira and the resulting supernatant was then processed with the vanillin working solution to avoid any hindrance with the fluorescence emission spectra of Pira (λexcitation = 480 nm and λemmision = 580 nm). The % cumulative release of Pira from Pira-loaded nanoparticles was 43.7% ± 1.0 in PBS (pH 7.4) and 75.1% ± 0.4 in DTT, while the % cumulative release of Sal from the Sal-loaded nanoparticles was 50.1% ± 1.7 in PBS (pH 7.4) and 81.7% ± 1.7 in DTT, respectively [Fig. 5(A) and (B)]. The higher amount of drug release in DTT may be attributed to the presence of disulfide linkages in the carrier, for which DTT acts as a stimulus for faster degradation. Also, since Pira is more hydrophobic than Sal, it exhibited a relatively slower release than Sal. Similarly, in the case of the co-release of Pira and Sal from the Pira/Sal dual-loaded nanoparticles, the % cumulative release of Pira increased from 20.5% ± 1.0 in PBS (pH 7.4) to 40.1% ± 0.4 in DTT after 20 days; similarly, the % cumulative release of Sal increased from 36.2% ± 1.7 in PBS (pH 7.4) to 51.5% ± 1.7 in DTT (Fig. 5A and B), respectively. Notably, the % drug release observed in the Pira/Sal dual-loaded nanoparticles was relatively lower than that observed for the single Pira- or Sal-loaded nanoparticles. This decrease may be due to the presence of intermolecular forces of attraction between the two drugs, Pira and Sal, causing them to hold together in the nano-core for a longer period.3.6 Cellular uptake study on 2D adherent cells and 3D mammospheres
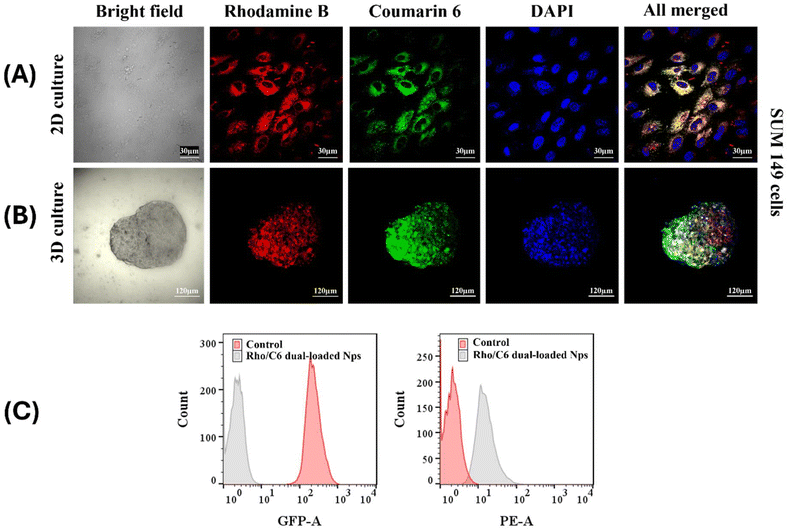
The therapeutic efficacy of nano-formulations largely depends on their ability to achieve cellular uptake and retention in abnormal cells as compared to free drugs. Therefore, nano-carriers that demonstrate good uptake in vitro provide preliminary evidence of eventual in vivo efficiency. In addition, 3D spheroids are known to mimic the realistic solid tumor more closely than their 2D counterparts; therefore, observing internalization in 3D spheroids is more valuable. Consequently, RhoB/C6 dual dye-loaded FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles were used to evaluate their cellular internalization ability on 2D adhered cells, SUM-149 [Fig. 4(A)] and 3D mammospheres [Fig. 4(B)]. As per CLSM images, in both 2D and 3D cases, rhodamine B (red) and coumarin 6 (green) fluorescence were found concentrated together in the cell cytosol around the DAPI (blue)-stained nucleus, just after 1 hour incubation of the cells with the nanoparticles. In the FACS analysis also, the RhoB/C6 dual dye-loaded nanoparticles showed significantly higher mean fluorescence intensity (%) as compared to the control after uptake in 2D adherent SUM-149 cells after 1 h [Fig. 4(C)]. The results clearly demonstrate that the prepared nanoparticles possess excellent cellular uptake in both 2D adherent cells and 3D spheroids.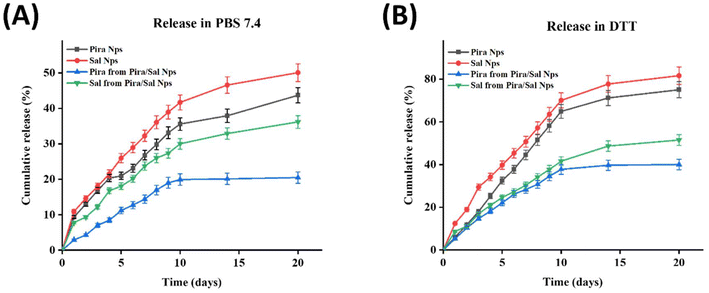 | ||
| Fig. 4 In vitro release study of the Pira/Sal single or dual-loaded FA-conjugated [S-(PLA-b-PEG-CONH)]2 nanoparticles in (A) PBS and (B) DTT medium at pH 7.4. | ||
3.7 2D cell proliferation inhibition studies of FA-conjugated Pira/Sal single or dual-loaded nanoparticles
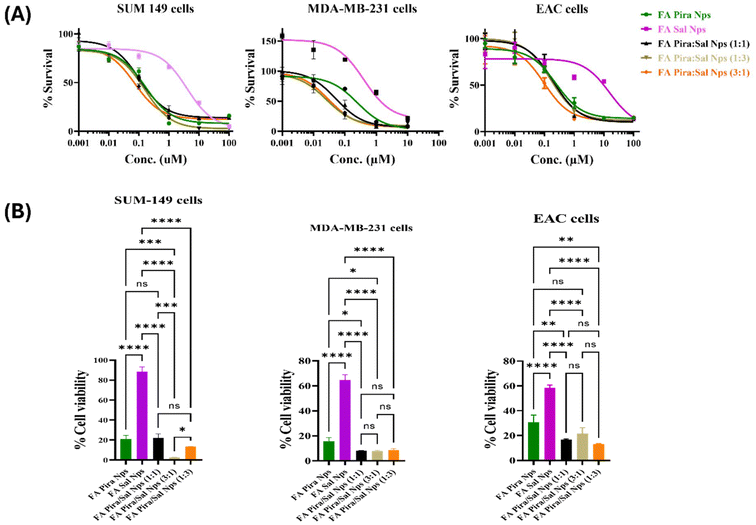
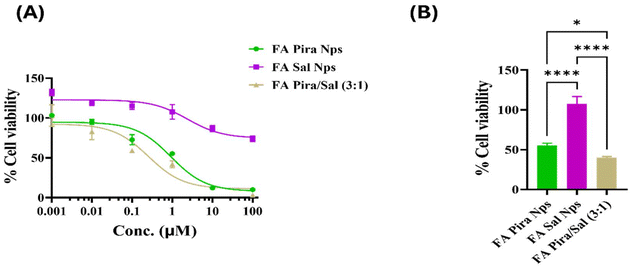
Cell proliferation inhibition studies were performed on different folate overexpressing breast cancer cells such as SUM-149, MDA-MB-231 and EAC via the MTT assay. The effectiveness of the FA-conjugated Pira/Sal dual-loaded nanoparticles was checked by observing the change in their IC50 values as compared to the Pira/Sal single-loaded nanoparticles and respective free drugs after 72 hours of treatment of the cells [Fig. 6(A)]. The FA-conjugated Pira-loaded nanoparticles showed a significant 8- to 10-fold decrease in IC50 values when used in combination with Sal in all three cell types used. Sal was observed to visibly complement Pira as a drug partner, as the Pira/Sal dual-loaded nanoparticles showed relatively lower IC50 values than individual drug-loaded nanoparticles in all Pira/Sal combination ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). In the case of SUM-149 cells, when the cells were treated with 1 μM concentration of each formulation, the % cell viability of the FA-conjugated Pira-loaded nanoparticles decreased from 23.15 ± 3.32 to 2.70 ± 1.96 in the FA-conjugated Pira/Sal (3
3). In the case of SUM-149 cells, when the cells were treated with 1 μM concentration of each formulation, the % cell viability of the FA-conjugated Pira-loaded nanoparticles decreased from 23.15 ± 3.32 to 2.70 ± 1.96 in the FA-conjugated Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) nanoparticles. A similar trend was observed in two other cell types: MDA-MB-231 and EAC, indicating the synergistic activity potential of the Pira and Sal combination [Fig. 6(B)]. Notably, the FA-conjugated Pira/Sal (3
1) nanoparticles. A similar trend was observed in two other cell types: MDA-MB-231 and EAC, indicating the synergistic activity potential of the Pira and Sal combination [Fig. 6(B)]. Notably, the FA-conjugated Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) dual-loaded nanoparticles showed the highest anti-cancer efficacy among all three types of breast cancer cells. In addition, the IC50 values of individual drug-loaded nanoparticles (Table 3) were found to be comparable to their respective free drug formulations (Table S1†), indicating that encapsulating the drugs into a polymeric nano-core has not compromised their therapeutic efficacy.
1) dual-loaded nanoparticles showed the highest anti-cancer efficacy among all three types of breast cancer cells. In addition, the IC50 values of individual drug-loaded nanoparticles (Table 3) were found to be comparable to their respective free drug formulations (Table S1†), indicating that encapsulating the drugs into a polymeric nano-core has not compromised their therapeutic efficacy.
| Sample | SUM-149 | MDA-MB-231 | EAC | |||
|---|---|---|---|---|---|---|
| Pira | Sal | Pira | Sal | Pira | Sal | |
| Pira NPs | 0.249 | 0.160 | 0.243 | |||
| Sal NPs | 1.536 | 3.955 | 15.63 | |||
Pira/Sal (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) NPs 1) NPs |
0.052 | 0.362 | 0.105 | 0.073 | 0.198 | 0.139 |
Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) NPs 1) NPs |
0.026 | 0.097 | 0.072 | 0.059 | 0.097 | 0.061 |
Pira/Sal (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) NPs 3) NPs |
0.030 | 0.478 | 0.156 | 0.114 | 0.161 | 0.154 |
3.8 In vitro cytotoxicity on 3D mammospheres
The IC50 values of the FA-conjugated Pira, FA-conjugated Sal, and FA-conjugated Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) nanoparticles were found to be 0.922 μM, 2.357 μM and 0.253 μM, respectively [Fig. 7(A)]. Since 3D cultured cancer cells are known to be more resistant to anti-cancer drugs than 2D culture cells, the IC50 values obtained in the case of 3D spheres were significantly higher than those in 2D adherent cells, as expected.29 Pira/Sal (3
1) nanoparticles were found to be 0.922 μM, 2.357 μM and 0.253 μM, respectively [Fig. 7(A)]. Since 3D cultured cancer cells are known to be more resistant to anti-cancer drugs than 2D culture cells, the IC50 values obtained in the case of 3D spheres were significantly higher than those in 2D adherent cells, as expected.29 Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) dual-loaded nanoparticles showed the highest % killing in comparison with Pira and Sal alone, due to the combined effect of Pira killing the primary cancer cells and Sal eliminating the core cancer stem cells [Fig. 7(B)].
1) dual-loaded nanoparticles showed the highest % killing in comparison with Pira and Sal alone, due to the combined effect of Pira killing the primary cancer cells and Sal eliminating the core cancer stem cells [Fig. 7(B)].
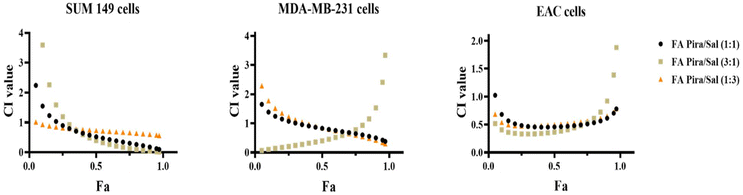
3.9 Combination index and synergy calculation
The synergistic efficacy of the Pira and Sal combination both in free drug formulation (Table S2†) and in nanoparticles was evaluated in three different breast cancer cells: SUM-149, MDA-MB-231 and EAC. The combination index values are tabulated in Table 4. The drug combination was found to be synergistic in all types of cancer across all three drug ratios used (Pira![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sal = 1
Sal = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). The FA-conjugated Pira/Sal (3
3). The FA-conjugated Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) dual-loaded nanoparticles were found to be most effective against breast cancer cells among all the formulations [Fig. 8] and thus selected for further in vivo therapeutic efficacy testing.
1) dual-loaded nanoparticles were found to be most effective against breast cancer cells among all the formulations [Fig. 8] and thus selected for further in vivo therapeutic efficacy testing.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sal – 1
Sal – 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) on breast cancer cells: SUM-149, MDA-MB-231 and EAC at Fa = 0.5
3) on breast cancer cells: SUM-149, MDA-MB-231 and EAC at Fa = 0.5
| Sample | SUM-149 | MDA-MB-231 | EAC |
|---|---|---|---|
Pira/Sal (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) NPs 1) NPs |
0.523 | 0.827 | 0.456 |
Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) NPs 1) NPs |
0.395 | 0.401 | 0.365 |
Pira/Sal (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) NPs 3) NPs |
0.728 | 0.847 | 0.497 |
3.10 In vivo tumor regression, toxicity, and survival study
The in vivo therapeutic potential of the prepared nanoparticles was investigated on a syngeneic breast cancer tumour model, EAC. All six groups, including free Pira, free Sal, FA-conjugated Pira nanoparticles, Sal nanoparticles, Pira + Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
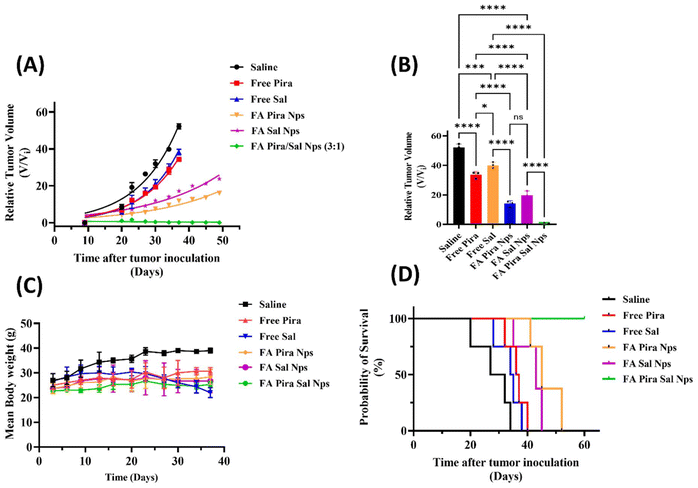
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) nanoparticles and saline as the control, were given an intravenous (IV) treatment dose of 2 mg kg−1 biweekly for 3 weeks via the lateral tail vein of mice. A relative tumour volume regression of 35.7% and 23.4% was observed in the case of free Pira and free Sal, respectively, whereas 69.3% and 54.5% were observed in the case of FA-conjugated Pira and FA-conjugated Sal nanoparticles, respectively, on day 37 after tumour inoculation [Fig. 9(A) and 9(B)].
1) nanoparticles and saline as the control, were given an intravenous (IV) treatment dose of 2 mg kg−1 biweekly for 3 weeks via the lateral tail vein of mice. A relative tumour volume regression of 35.7% and 23.4% was observed in the case of free Pira and free Sal, respectively, whereas 69.3% and 54.5% were observed in the case of FA-conjugated Pira and FA-conjugated Sal nanoparticles, respectively, on day 37 after tumour inoculation [Fig. 9(A) and 9(B)].
The nanoparticles showed better tumor growth inhibition than their respective free drug formulation, potentially due to the combined effects of folic acid targeting, GSH-responsive drug release and the EPR effect. Treatment with the FA-conjugated Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) dual-loaded nanoparticles resulted in ∼100% tumor regression as compared to the control group, indicating their synergistic anti-tumor activity. To identify any dose-related toxicity, the average body weight of each mouse was recorded at various time intervals until the end of the study [Fig. 9(C)]. The results showed no significant change in the body weight in any case except for the free Sal and saline treatment groups. Survival studies were carried out for 60 days and the results were analyzed using Kaplan–Meier plots [Fig. 9(D)]. The analysis showed that the mouse group treated with the FA-conjugated Pira nanoparticles experienced tumor relapse after 48 days of the study and died within 3 days, whereas those treated with the FA-conjugated Pira/Sal (3
1) dual-loaded nanoparticles resulted in ∼100% tumor regression as compared to the control group, indicating their synergistic anti-tumor activity. To identify any dose-related toxicity, the average body weight of each mouse was recorded at various time intervals until the end of the study [Fig. 9(C)]. The results showed no significant change in the body weight in any case except for the free Sal and saline treatment groups. Survival studies were carried out for 60 days and the results were analyzed using Kaplan–Meier plots [Fig. 9(D)]. The analysis showed that the mouse group treated with the FA-conjugated Pira nanoparticles experienced tumor relapse after 48 days of the study and died within 3 days, whereas those treated with the FA-conjugated Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) dual-loaded nanoparticles showed no signs of tumor relapse and survived until the end of the study. This manifests the role of Sal in combination with primary chemotherapeutic drugs in killing cancer stem cells to prevent cancer recurrence.
1) dual-loaded nanoparticles showed no signs of tumor relapse and survived until the end of the study. This manifests the role of Sal in combination with primary chemotherapeutic drugs in killing cancer stem cells to prevent cancer recurrence.
3.11 Histopathological analysis
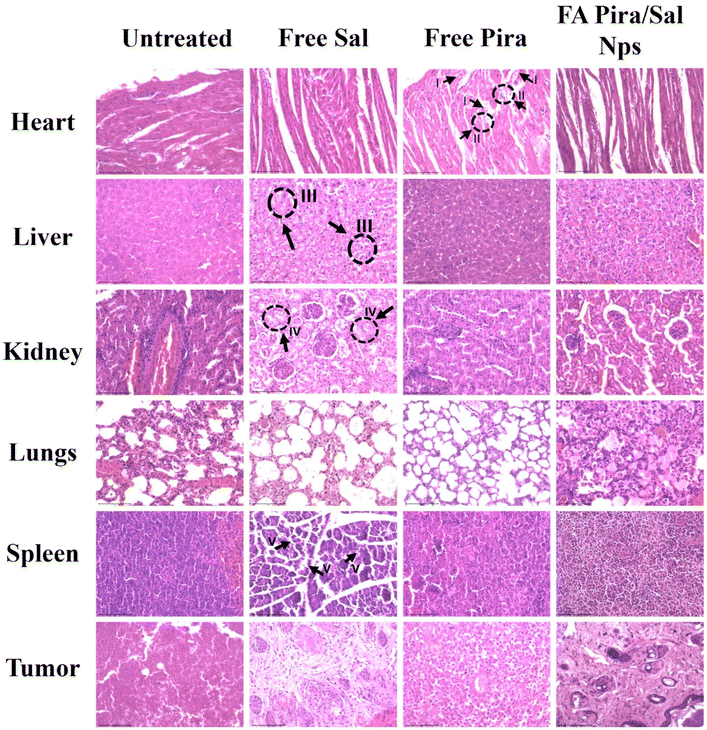
To gain better insight into the in vivo toxicity of the nanoparticles, histopathological analysis of vital organs like the heart, liver, kidneys, lungs, and spleen was performed along with tumor tissue [Fig. 10]. Free Pira showed inflammation in cardiomyocytes, fiber degeneration and cytoplasmic vacuolization in heart tissue. Abnormal hepatocytes and erratic necrosis in the liver were observed in the free Sal treated group.30 Furthermore, free Sal was found to cause significant tubular destruction in the kidneys and lead to spleen toxicity. In contrast, the FA-conjugated Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) dual-loaded nanoparticles displayed normal hepatic cells and sinusoids in the liver and showed mild cardiotoxicity as compared to free drugs. The presence of regular renal cortex and glomerulus indicated healthy kidney conditions. All these results suggested that the FA-conjugated Pira/Sal dual-loaded nanoparticles are efficient in tumor targeting with minimal off-target cytotoxicity.
1) dual-loaded nanoparticles displayed normal hepatic cells and sinusoids in the liver and showed mild cardiotoxicity as compared to free drugs. The presence of regular renal cortex and glomerulus indicated healthy kidney conditions. All these results suggested that the FA-conjugated Pira/Sal dual-loaded nanoparticles are efficient in tumor targeting with minimal off-target cytotoxicity.
4. Conclusion
An FA-conjugated redox responsive [S-(PLA-b-PEG-CONH)]2 block copolymer was successfully synthesized and characterized. The synthesized polymer served as an excellent nano-carrier, encapsulating anti-cancer drugs, pirarubicin and salinomycin, to form uniformly sized nanoparticles with good encapsulation efficiency, high surface charge density and low PDI. The designed nanoparticles serve a dual purpose: first, they selectively target tumor cells through folate receptors, enhancing drug delivery precisely to cancerous tissues while minimizing off-target effects. Second, their redox-responsive nature allows for controlled drug release in response to the high levels of glutathione within the tumor microenvironment, ensuring sustained and effective therapeutic action. In vitro experimental results confirmed that the FA-targeted Pira/Sal (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) dual-loaded nanoparticles possess remarkable cellular internalization ability and prolonged and sustained drug release, whereas in vivo experiments demonstrated excellent anti-tumor activity with ∼100% tumor regression and without any major organ toxicity or tumor relapse. The co-encapsulation of Sal along with Pira further enhanced the therapeutic efficacy of this combination due to the ability of Sal to attack cancer stem cells, which are mainly responsible for cancer reoccurrence. The outcomes of this work conclude that the prepared dual-targeted and dual-drug loaded systemic nano-formulation provides a promising approach for breast cancer therapy.
1) dual-loaded nanoparticles possess remarkable cellular internalization ability and prolonged and sustained drug release, whereas in vivo experiments demonstrated excellent anti-tumor activity with ∼100% tumor regression and without any major organ toxicity or tumor relapse. The co-encapsulation of Sal along with Pira further enhanced the therapeutic efficacy of this combination due to the ability of Sal to attack cancer stem cells, which are mainly responsible for cancer reoccurrence. The outcomes of this work conclude that the prepared dual-targeted and dual-drug loaded systemic nano-formulation provides a promising approach for breast cancer therapy.
Author contributions
Priya Gupta and Ankushi Bansal – conceptualization, methodology, investigation, formal analysis, data curation and writing – original draft. Harshdeep Kaur and Mohammad Anees – writing – review and editing. Neetu Singh – supervision, validation, and funding acquisition. Harpal Singh – supervision, resources, project administration and funding acquisition.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the Council of Scientific and Industrial Research (CSIR) for providing Ph.D. fellowship support, and the Department of Science & Technology (DST), India for financial assistance to the project under Grant ID - DST/WoS-B/NH-8/2021. All the authors also extend their appreciation to the the Central Research Facility (CRF), IIT Delhi, for access to characterization instruments. Additionaly, we would also like to express our gratitude to Mr Anil Pandey for helping with animal experiments.References
- J. Li, Q. Wang, G. Xia, N. Adilijiang, Y. Li, Z. Hou, Z. Fan and J. Li, Recent Advances in Targeted Drug Delivery Strategy for Enhancing Oncotherapy, Pharmaceutics, 2023, 15(9), 2233, DOI:10.3390/pharmaceutics15092233.
- J. Nel, K. Elkhoury, É. Velot, A. Bianchi, S. Acherar, G. Francius, A. Tamayol, S. Grandemange and E. Arab-Tehrany, Functionalized Liposomes for Targeted Breast Cancer Drug Delivery, Bioact. Mater., 2023, 24, 401–437, DOI:10.1016/j.bioactmat.2022.12.027.
- H. Cao, S. Gao, R. Jogani and R. Sugimura, The Tumor Microenvironment Reprograms Immune Cells, Cell. Reprogram., 2022, 24(6), 343–352, DOI:10.1089/cell.2022.0047.
- R. Kumari, D. Sunil and R. S. Ningthoujam, Hypoxia-Responsive Nanoparticle Based Drug Delivery Systems in Cancer Therapy: An up-to-Date Review, J. Controlled Release, 2020, 319, 135–156, DOI:10.1016/j.jconrel.2019.12.041.
- Z. Chen, R. K. Kankala, Z. Yang, W. Li, S. Xie, H. Li, A.-Z. Chen and L. Zou, Antibody-Based Drug Delivery Systems for Cancer Therapy: Mechanisms, Challenges, and Prospects, Theranostics, 2022, 12(8), 3719 CrossRef.
- H. Ding, P. Tan, S. Fu, X. Tian, H. Zhang, X. Ma, Z. Gu and K. Luo, Preparation and Application of PH-Responsive Drug Delivery Systems, J. Controlled Release, 2022, 348, 206–238, DOI:10.1016/j.jconrel.2022.05.056.
- M. Su, S. Xiao, M. Shu, Y. Lu, Q. Zeng, J. Xie, Z. Jiang and J. Liu, Enzymatic Multifunctional Biodegradable Polymers for PH- and ROS-Responsive Anticancer Drug Delivery, Colloids Surf., B, 2020, 193, 111067, DOI:10.1016/j.colsurfb.2020.111067.
- V. Dymicka-Piekarska, O. M. Koper-Lenkiewicz, J. Zińczuk, E. Kratz and J. Kamińska, Inflammatory Cell-Associated Tumors. Not Only Macrophages (TAMs), Fibroblasts (TAFs) and Neutrophils (TANs) Can Infiltrate the Tumor Microenvironment. The Unique Role of Tumor Associated Platelets (TAPs), Cancer Immunol. Immunother., 2021, 70(6), 1497–1510, DOI:10.1007/s00262-020-02758-7.
- S. Thakkar, D. Sharma, K. Kalia and R. K. Tekade, Tumor Microenvironment Targeted Nanotherapeutics for Cancer Therapy and Diagnosis: A Review, Acta Biomater., 2020, 101, 43–68, DOI:10.1016/j.actbio.2019.09.009.
- H. Sadeghi Rad, J. Monkman, M. E. Warkiani, R. Ladwa, K. O'Byrne, N. Rezaei and A. Kulasinghe, Understanding the Tumor Microenvironment for Effective Immunotherapy, Med. Res. Rev., 2021, 41(3), 1474–1498, DOI:10.1002/med.21765.
- S. Yan, J. Na, X. Liu and P. Wu, Different Targeting Ligands-Mediated Drug Delivery Systems for Tumor Therapy, Pharmaceutics, 2024, 16(2), 248, DOI:10.3390/pharmaceutics16020248.
- S. Kunjiappan, P. Pavadai, S. Vellaichamy, S. Ram Kumar Pandian, V. Ravishankar, P. Palanisamy, S. Govindaraj, G. Srinivasan, A. Premanand, M. Sankaranarayanan and P. Theivendren, Surface Receptor-Mediated Targeted Drug Delivery Systems for Enhanced Cancer Treatment: A State-of-the-Art Review, Drug Dev. Res., 2021, 82(3), 309–340, DOI:10.1002/ddr.21758.
- P. Tagde, G. T. Kulkarni, D. K. Mishra and P. Kesharwani, Recent Advances in Folic Acid Engineered Nanocarriers for Treatment of Breast Cancer, J. Drug Delivery Sci. Technol., 2020, 56, 101613, DOI:10.1016/j.jddst.2020.101613.
- L. O. F. Monteiro, R. S. Fernandes, L. Castro, D. Reis, G. D. Cassali, F. Evangelista, C. Loures, A. P. Sabino, V. Cardoso, M. C. Oliveira, A. Branco de Barros and E. A. Leite, Paclitaxel-Loaded Folate-Coated PH-Sensitive Liposomes Enhance Cellular Uptake and Antitumor Activity, Mol. Pharm., 2019, 16(8), 3477–3488, DOI:10.1021/acs.molpharmaceut.9b00329.
- H. Tonbul, A. Sahin, E. Tavukcuoglu, G. Ultav, S. Akbas, Y. Aktas, G. Esendaglı and Y. Capan, Folic Acid Decoration of Mesoporous Silica Nanoparticles to Increase Cellular Uptake and Cytotoxic Activity of Doxorubicin in Human Breast Cancer Cells, J. Drug Delivery Sci. Technol., 2021, 63, 102535, DOI:10.1016/j.jddst.2021.102535.
- Y. Yao, Y. Zhou, L. Liu, Y. Xu, Q. Chen, Y. Wang, S. Wu, Y. Deng, J. Zhang and A. Shao, Nanoparticle-Based Drug Delivery in Cancer Therapy and Its Role in Overcoming Drug Resistance, Front. Mol. Biosci., 2020, 7, 193 CrossRef PubMed.
- S. Tiwari, S. Liu, M. Anees, N. Mehrotra, A. Thakur, G. J. Tawa, G. Grewal, R. Stone, S. Kharbanda and H. Singh, QuatramerTM Encapsulation of Dual-Targeted PI3-Kδ/HDAC6 Inhibitor, HSB-510, Suppresses Growth of Breast Cancer, Bioeng. Transl. Med., 2023, 8(5), e10541, DOI:10.1002/btm2.10541.
- K. Elumalai, S. Srinivasan and A. Shanmugam, Review of the Efficacy of Nanoparticle-Based Drug Delivery Systems for Cancer Treatment, Biomed. Technol., 2024, 5, 109–122, DOI:10.1016/j.bmt.2023.09.001.
- C. Nehate, A. Nayal and V. Koul, Redox Responsive Polymersomes for Enhanced Doxorubicin Delivery, ACS Biomater. Sci. Eng., 2019, 5(1), 70–80, DOI:10.1021/acsbiomaterials.8b00238.
- X. Duan, Q. Wang, W. Che, T. Li, K. Zhang, L. Han, L. Song and W. Guo, Redox-Responsive Nanomedicine of Doxorubicin-Conjugated Poly-L-Glutathione Oxidized for Cancer Therapy, J. Taiwan Inst. Chem. Eng., 2024, 159, 105456, DOI:10.1016/j.jtice.2024.105456.
- R. Mattioli, A. Ilari, B. Colotti, L. Mosca, F. Fazi and G. Colotti, Doxorubicin and Other Anthracyclines in Cancers: Activity, Chemoresistance and Its Overcoming, Mol. Aspects Med., 2023, 93, 101205, DOI:10.1016/j.mam.2023.101205.
- D. Qi, Y. Liu, J. Li, J. H. Huang, X. Hu and E. Wu, Salinomycin as a Potent Anticancer Stem Cell Agent: State of the Art and Future Directions, Med. Res. Rev., 2022, 42(3), 1037–1063 CrossRef PubMed.
- M. Anees, N. Mehrotra, S. Tiwari, D. Kumar, S. Kharbanda and H. Singh, Polylactic Acid Based Biodegradable Hybrid Block Copolymeric Nanoparticle Mediated Co-Delivery of Salinomycin and Doxorubicin for Cancer Therapy, Int. J. Pharm., 2023, 635, 122779, DOI:10.1016/j.ijpharm.2023.122779.
- R. F. Pagels, N. M. Pinkerton, A. W. York and R. K. Prud'homme, Synthesis of Heterobifunctional Thiol-Poly(Lactic Acid)-b-Poly(Ethylene Glycol)-Hydroxyl for Nanoparticle Drug Delivery Applications, Macromol. Chem. Phys., 2020, 221(2), 1900396, DOI:10.1002/macp.201900396.
- S. V. Lale, A. Kumar, S. Prasad, A. C. Bharti and V. Koul, Folic Acid and Trastuzumab Functionalized Redox Responsive Polymersomes for Intracellular Doxorubicin Delivery in Breast Cancer, Biomacromolecules, 2015, 16(6), 1736–1752 CrossRef PubMed.
- J. Zhao, H. Xia, T. Yu, L. Jin, X. Li, Y. Zhang, L. Shu, L. Zeng and Z. He, A Colorimetric Assay for Vanillin Detection by Determination of the Luminescence of O-Toluidine Condensates, PLoS One, 2018, 13(4), e0194010 CrossRef.
- E. B. Kang, S. M. Sharker, I. In and S. Y. Park, Pluronic Mimicking Fluorescent Carbon Nanoparticles Conjugated with Doxorubicin via Acid-Cleavable Linkage for Tumor-Targeted Drug Delivery and Bioimaging, J. Ind. Eng. Chem., 2016, 43, 150–157, DOI:10.1016/j.jiec.2016.08.001.
- T. Z. Khan, S. M. Newaj, A. Rahman, R. Tabassum, K. N. Tasnim, H. M. Reza, M. S. Reza, S. Hong and S. M. Sharker, NIR-Light-Triggered Delivery of Doxorubicin-Loaded PLGA Nanoparticles for Synergistic Cancer Therapy on DMBA/TPA Induced Tumor-Bearing Mice, Mater. Adv., 2023, 4(21), 5175–5183, 10.1039/D3MA00375B.
- M. Muguruma, S. Teraoka, K. Miyahara, A. Ueda, M. Asaoka, M. Okazaki, T. Kawate, M. Kuroda, Y. Miyagi and T. Ishikawa, Differences in Drug Sensitivity between Two-Dimensional and Three-Dimensional Culture Systems in Triple-Negative Breast Cancer Cell Lines, Biochem. Biophys. Res. Commun., 2020, 533(3), 268–274, DOI:10.1016/j.bbrc.2020.08.075.
- İ. B Ekinci, A. Chłodowska and M. Olejnik, Ionophore Toxicity in Animals: A Review of Clinical and Molecular Aspects, Int. J. Mol. Sci., 2023, 24(2), 1696, DOI:10.3390/ijms24021696.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr02365j |
| This journal is © The Royal Society of Chemistry 2024 |