Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
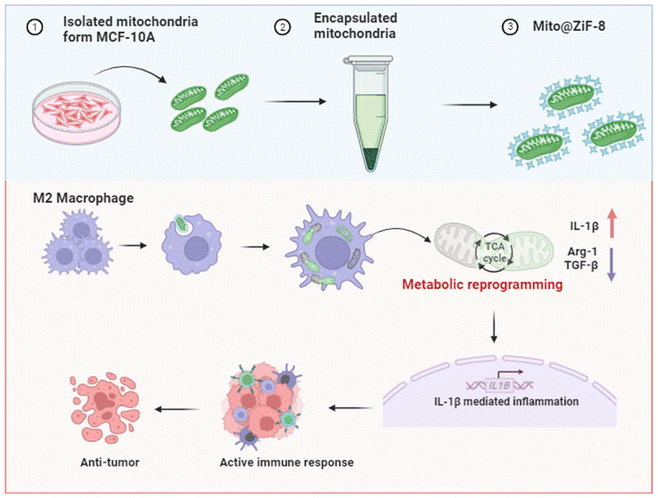
Encapsulated mitochondria to reprogram the metabolism of M2-type macrophages for anti-tumor therapy†
Yonghui
Wang
ab,
Chang
Liu
b,
Xiaodong
Ma
b,
Anne
Filppula
b,
Youbin
Cui
a,
Jiangbin
Ye
*c and
Hongbo
Zhang
 *abd
*abd
aDepartment of Thoracic Surgery, Organ Transplantation Center, The First Hospital of Jilin University, Changchun, China
bPharmaceutical Sciences Laboratory, Åbo Akademi University, Turku 20520, Finland. E-mail: hongbo.zhang@abo.fi
cDepartment of Radiation Oncology, Stanford University School of Medicine, Stanford, CA 94305, USA. E-mail: yej1@stanford.edu
dTurku Bioscience Centre, University of Turku and Åbo Akademi University, Turku 20520, Finland
First published on 23rd October 2024
Abstract
M2-type macrophages (M2Φ) play a pro-tumorigenic role and are closely associated with tumor development, where metabolic dysregulation exacerbates the immunosuppressive tumor microenvironment and fosters tumor growth. Mitochondria serve as the regulatory center of cellular metabolism, yet effective methods to modulate M2Φ mitochondria within the tumor microenvironment remain lacking. In this study, we developed a technique utilizing the bio-encapsulation of mitochondria in Zeolitic Imidazolate Framework-8 (ZiF-8), referred to as Mito@ZiF-8. Our findings demonstrated that this coating protects intact mitochondria and preserves their bioactivity over an extended period after isolation. We successfully delivered Mito@ZiF-8 into M2Φ, which inhibited the secretion of pro-inflammatory factors, promoted the release of anti-inflammatory factors, and reprogrammed M2Φ metabolism. This innovative approach has the potential to reduce breast cancer cell metastasis and enhance sensitivity to chemotherapy drugs such as 6-thioguanine, cisplatin, and doxorubicin (Dox). Mito@ZiF-8 aims to reprogram the M2Φ microenvironment to support anti-tumor therapies, offering a novel strategy for improving the effectiveness of breast cancer treatment.
1 Introduction
Metabolic dysregulation in immune cells may be a crucial factor in the occurrence and development of tumors.1 Under normal circumstances, the immune system exhibits high sensitivity and activity in detecting and clearing abnormal cells. However, when immune cell metabolism is disrupted, its ability to monitor and attack tumor cells may be compromised, creating a favorable environment for tumor initiation and progression. Metabolic reprogramming in immune cells can trigger immune escape, and the immunosuppressive strategies adopted by tumor cells, make it challenging for the immune system to clear tumor cells effectively.2,3In solid tumors, tumor-associated macrophages (TAMs) are a multifunctional and diverse group of immune cells, constituting up to 50% of the tumor mass.4 TAMs can differentiate into M1-type or M2-type macrophages, each characterized by specific gene expression patterns and metabolic regulatory pathways. In response to various signals within the tumor microenvironment, the metabolic reprogramming of TAMs changes, primarily polarizing into immunosuppressive M2Φ rather than anti-tumor M1Φ.5 M2Φ secrete anti-inflammatory factors such as interleukin 10 (IL-10) and transforming growth factor beta (TGF-β). IL-10 inhibits the activity of anti-tumor M1Φ and T cells, while TGF-β also suppresses the activity of anti-tumor M1Φ and T cells.6 TAMs upregulate immune checkpoint molecules such as PD-L1 (programmed death-ligand 1) in the tumor microenvironment. These molecules bind to PD-1 on the surface of T cells, inhibiting their activation and proliferation, thereby preventing them from attacking tumor cells. This mechanism is considered a crucial pathway by which tumor cells evade immune surveillance and is closely related to the efficacy of immune checkpoint inhibitor therapies.7 Therefore, studying TAMs, especially the metabolic changes in M2Φ within the tumor microenvironment, is of great importance for understanding tumor progression and developing new anti-cancer therapies.
As the center of cellular energy metabolism and biosynthesis, mitochondria produce ATP through oxidative phosphorylation to maintain cellular energy homeostasis, while metabolic intermediates play crucial roles in biosynthetic pathways.8 M2Φ rely on mitochondrial oxidative phosphorylation and fatty acid oxidation to participate in anti-inflammatory processes and tissue repair. In the tumor microenvironment, mitochondrial metabolism plays a key role in macrophage polarization and functional shifts, making it a potential target for anti-tumor immunotherapy.4 Therefore, therapies targeting mitochondrial functions, such as inhibiting oxidative phosphorylation or fatty acid oxidation, may improve the metabolic state of macrophages, suppress their immunosuppressive effects, and promote the development of cancer therapies. However, while some drugs or targeting systems are available to modify specific mitochondrial functions, they fall short of restoring the full mitochondrial function. Therefore, the effective medical approach to address abnormal metabolic disorders in tumors is to transfer intact mitochondria from healthy cells into cancerous cells, which is also known as “mitochondrial transplantation”.9,10
However, highly sensitive organelles, mitochondria pose numerous significant challenges and unsolved problems for mitochondrial transplantation therapy. First, the isolated mitochondria have a short survival time, which means short-lived bioactivity. With current technology, the bioactivity of isolated mitochondria typically lasts a maximum of six hours.11,12 Additionally, the cell capacity of taking up/absorbing isolated mitochondria is poor. Mitochondria have a negatively charged membrane, and they are typically 500–1000 nm in size; those properties have greatly limited their uptake efficiency in most cell types. This results in low treatment efficiency, as only a small fraction of dosed mitochondria can finally enter the metabolic abnormal cells in a bioactive form and take the therapeutic action. Hence, there's a pressing need to develop technology for the delivery of mitochondria to achieve a high therapeutic effect.
To address these challenges, we encapsulated mitochondria within a ZiF-8, a type of Metal–Organic Framework (MOF). MOFs represent a class of materials used for coating cells and other complex biological entities, such as viruses.13 Numerous MOFs remain stable in different solvents, including water, across broad temperature ranges and can be synthesized under physiological conditions. Among them, ZiF-8 is the most extensively studied. It can be fabricated in a few minutes in an aqueous solution without heating, organic solvents, or compatibilizers. More importantly, ZiF-8 has adjustable molecular-level pores that allow for basic ion and oxygen exchange while tightly stabilizing the structure of the encapsulated cargo and preventing the penetration of exogenous enzymes or other destructive factors, thereby protecting the cargo from biological, thermal, and chemical degradation.14 ZiF-8 has been used to encapsulate a variety of biomolecules, including globular proteins like albumin, enzymes such as horseradish peroxidase and urease, hormones, and oligonucleotides.15,16 Therefore, we developed a technology based on mitochondrial transplantation to deliver fully functional mitochondria into M2Φ. This technology regulates the abnormal metabolic microenvironment of M2Φ, in vitro results show that it can inhibit breast cancer cell metastasis and increase their sensitivity to chemotherapy drugs, potentially benefiting anti-breast cancer treatment (Scheme 1).
2 Materials and methods
2.1 Materials
Zinc acetate, 2-methylimidazole, hydrocortisone, 6-thioguanine, cisplatin, doxorubicin (Dox), and insulin solution were purchased from Sigma-Aldrich, (St Louis, Missouri, USA); glucose solution, phosphate buffered saline (PBS), Dulbecco's modified Eagle's medium (DMEM) cell culture medium, Dulbecco's modified Eagle's medium F12 (DMEM F12), sodium chloride, RNase-free, cholera toxin, fetal bovine serum (FBS), Mitochondria Isolation Kit for Cultured Cells (catalog number 89874), MitoTracker™ Red CMXRos, MitoTracker™ Green, CellTracker™ CM-DiI Dye, JC-1 Dye (Mitochondrial Membrane Potential Probe), Hoechst 33342, IL-4 protein, EGF protein, anti-CD44 APC, anti-CD24 PE, LysoTracker™ Green DND-26, and 0.25% trypsin–EDTA, and adenosine 5′-triphoshate (ATP) determination kit were purchased from Gibco, Thermo Fisher Scientific (Waltham, USA). The BCA protein assay kit from Bio-RAD (Waltham, USA). The WST-1 assay, Mouse IL-10 ELISA kit, Mouse IL-6 ELISA kit, Mouse TGF-β ELISA kit, Mouse CXCL10 ELISA kit, Mouse TNF-α ELISA, and Mouse IL-1β ELISA kit were from Abcam (Waltham, USA). The bovine serum albumin (BSA) from Amresco (Framingham, USA). Anti-HSP 90 antibody, anti-cytochrome C (anti-Cyto C) antibody, anti-CD206 antibody, anti-vimentin, anti-N-cadherin, anti-snail, anti-PD-L1, and anti-actin were from Novus Biologicals (R&D System, Minneapolis, USA). All seahorse-relevant regents were purchased from Agilent Technologies, Inc. (Santa Clara, California, USA).2.2 Methods
M2Φ were induced from RAW 264.7 with IL-4 at a final concentration of 40 ng ml−1.
Encapsulation: first, a 40 mM zinc acetate solution and a 160 mM dimethylimidazole solution were prepared using 0.9% sodium chloride. Next, the mitochondria from one of the tubes were resuspended in 100 μl of the zinc acetate solution, ensuring uniform dispersion without clumping. This suspension was then slowly added drop by drop into an EP tube containing 100 μl of dimethylimidazole under stirring conditions. After stirring for 3 minutes, the mixture was left undisturbed for 10 minutes, followed by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 minutes, and washed three times with 0.9% sodium chloride solution. Following the same encapsulation steps, the control group utilized a pure ZiF-8 system without mitochondria. Finally, according to the results of the previous BCA, unencapsulated mitochondria and encapsulated mitochondria were normalized with 0.9 sodium chloride solution, and the final volume was 200 μl and placed on ice for use. Pure ZiF-8 was also re-suspended with 200 μl 0.9% sodium chloride, while the blank group was treated with the same amount of 0.9% sodium chloride.
000 rpm for 3 minutes, and washed three times with 0.9% sodium chloride solution. Following the same encapsulation steps, the control group utilized a pure ZiF-8 system without mitochondria. Finally, according to the results of the previous BCA, unencapsulated mitochondria and encapsulated mitochondria were normalized with 0.9 sodium chloride solution, and the final volume was 200 μl and placed on ice for use. Pure ZiF-8 was also re-suspended with 200 μl 0.9% sodium chloride, while the blank group was treated with the same amount of 0.9% sodium chloride.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes for isolated mitochondria and at 2500 rpm for 5 minutes for cell samples, the supernatant was removed. Thereafter, 300 μl of a fixation solution, prepared by mixing 2 ml of S-collidine buffer and 1 ml of 25% glutaraldehyde, was carefully added, ensuring that the pellet remained undispersed during the process. The samples were then sent to the TEM sample preparation center at the University of Turku for preparation and observed using a JEM-1400 Plus (JEOL Ltd, Akishima, Tokyo, Japan).
000g for 10 minutes for isolated mitochondria and at 2500 rpm for 5 minutes for cell samples, the supernatant was removed. Thereafter, 300 μl of a fixation solution, prepared by mixing 2 ml of S-collidine buffer and 1 ml of 25% glutaraldehyde, was carefully added, ensuring that the pellet remained undispersed during the process. The samples were then sent to the TEM sample preparation center at the University of Turku for preparation and observed using a JEM-1400 Plus (JEOL Ltd, Akishima, Tokyo, Japan).
For the western blotting experiments involving cell samples, M2Φ cells were seeded at a density of 1 × 104 cells per well in a 6-well plate and cultured overnight. Then, mitochondria were isolated and encapsulated according to the procedure outlined in Section 2.2.2. Each well received 20 μl of the respective solution. After incubating for 5 days, the supernatant was removed, and the cells were washed three times with PBS before being collected. Subsequently, 200 μl of RIPA solution containing 1% protease inhibitor was added for complete lysis. After centrifugation to collect the supernatant, protein concentrations were determined using the BCA method, and samples were prepared. Finally, the experiments were conducted and analyzed following the protocol provided by Bio-Rad Company.
For the western blotting experiment of 4T-1 cells, cells collected were those harvested 48 hours post-treatment as described in Section 2.2.16, with all other procedures identical to those mentioned above.
The second method of detection involved isolating mitochondria first, followed by incubating them with CellTracker™ CM-DiI Dye at a final concentration of 5 μM at 4 °C for 5 minutes. After centrifugation and washing, encapsulation was performed. Then, 20 μl of this solution, along with M2Φ at a density of 2 × 104 cells, were mixed and added to confocal dishes. After 48 hours of continued incubation, the supernatant was removed, the cells were washed three times with PBS, fresh culture medium was added, and direct observation was carried out using a Zeiss LSM880 (Zeiss, Oberkochen, Germany) with Ex 553 nm/Em 570 nm.
At the designated time points, the supernatant was removed, and the cells were washed three times with PBS. Subsequently, 1 ml of LysoView Green dye, diluted with culture medium (final concentration 50 nM), was added, and the cells were incubated for an additional 2 hours. After discarding the staining solution, the cells were washed three times with PBS and then stained with Hoechst 33342 for 15 minutes. After removing the supernatant and washing three times with PBS, a fresh culture medium was added, and the results were observed using a Zeiss LSM880 (Zeiss, Oberkochen, Germany).
M2Φ were collected and mixed with 20 μl of each solution at a density of 2 × 104 cells per dish. The mixture was added to confocal dishes containing 1 ml of culture medium and incubated for two more days. Subsequently, the supernatant was removed, and the cells were washed three times with PBS. Hoechst 33342 was added for 15-minute staining, followed by the removal of the supernatant and three washes with PBS. Finally, a fresh culture medium was added, and the results were observed using a Zeiss LSM880 (Zeiss, Oberkochen, Germany).
Toxicity of 4T-1 to different chemo drugs. 4T1 cells were collected for each group and seeded in a 96-well plate at a density of 6 × 103 cells per well, followed by overnight incubation. The next day, different concentration gradients of 6-thioguanine (final concentrations: 0, 5, 10, 15, 20, 25, 30, 40, 50, 60 μM), cisplatin (final concentrations: 0, 1, 2, 4, 5, 8, 10, 20, 30, 50 μM), and doxorubicin (final concentrations: 0, 0.5, 1, 1.5, 2, 2.5, 3, 4, 5, 10 μM) were prepared. Then, 100 μl of drug-containing medium was added to each group, and the cells were incubated for an additional 48 hours. Afterward, the medium was removed, 100 μl of fresh medium was added, followed by 10 μl of WST-1. The cells were incubated for an additional 4 hours, and the absorbance was measured using a Varioskan FLASH (Thermo Fisher Scientific, Waltham, Massachusetts, United States).
Wound healing experiment. 4T1 cells were collected for each group and seeded in a 6-well plate at a density of 1 × 104 cells per well. The following day, a line was drawn in the middle of each 4T1 culture well using a 200 μl pipette tip, marking the 0-hour time point. Photographs were taken using an Evos XL Core optical microscope (Thermo Fisher Scientific, Waltham, Massachusetts, United States), and the cells were then cultured for an additional 48 hours before further photographs were taken. All images were uniformly analyzed using ImageJ.
Detection of tumor stem cell population. 4T1 cells were collected for each group, washed with PBS, and incubated with 100 μl of PBS containing CD44+ and CD24+ antibodies on ice for 15 minutes. After centrifugation (2500 rpm, 5 minutes) to remove the supernatant, 400 μl of PBS was added to resuspend the cells. Finally, detection and analysis were performed using the BD Fortessa Blues (BD Biosciences, Franklin Lakes, New Jersey, USA) at dual wavelengths of Ex 488 nm/Em 575 nm and Ex 635 nm/Em 660 nm.
3 Results
3.1 Synthesis and characterization of Mito@ZiF-8
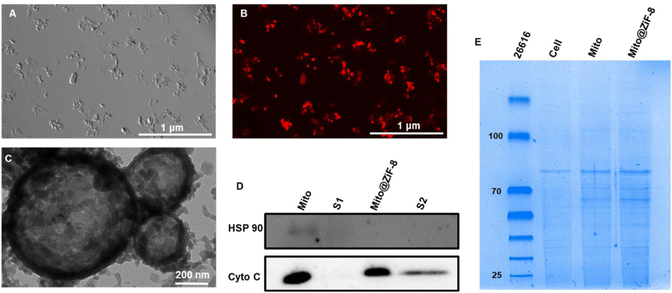
According to the results shown in Fig. S1A,† the outer mitochondrial membrane was smooth and intact after isolation, while the inner mitochondrial membrane was wrinkled inward, forming mitochondrial cristae. The cristae of the free mitochondria were visible under TEM, which rendered them favorable for respiration. In addition, we used the MitoTracker™ Red CMXRos, which can only label reactive mitochondria, to label mitochondria before isolation, and the free mitochondria were shown by brightfield and fluorescence microscopy (Fig. S1C and D†) that the mitochondria still process bioactivity after isolation. After encapsulation, while the morphology of the mitochondria changed slightly from the raindrop shape in Fig. S1C† to the powder-like structure in Fig. 1A, the mitochondria were still bioactive after encapsulation (Fig. 1B). Further morphological characterization was performed with TEM, and as can be seen from Fig. S1B,† the blank ZiF-8 carrier showed a polyhedral structure with a particle size of 50 nm or less, but after binding to the mitochondria, ZiF-8 formed a cage-like structure, generating a bionic membrane on the surface of the mitochondria, which protected the mitochondria therein. Cyto C as a mitochondria-associated protein located on the inner membrane and acts as an electron carrier in the mitochondrial respiratory chain, whereas the heat shock protein 90 (HSP90) is widely found in the cytoplasm and various organelles, as shown by the results of western blotting (Fig. 1D) experiments on isolated mitochondria pellet and its supernatant before and after encapsulation. While a small amount of non-mitochondria-associated proteins were present in the pellet of the Mito group, the majority obtained were indeed mitochondria. Notably, the high purity of the isolated mitochondria was evidenced by the presence of Cyto C proteins; moreover, in the process of encapsulation, due to the difference in the membrane potential of the different components, Zn2+ would preferentially bind to the mitochondria with a strong membrane potential, leading to a centrifugal collection of Mito@ZiF-8 with further improved purity. Further, the results of whole protein gel electrophoresis of mitochondria before and after encapsulation (Fig. 1E) showed that the protein electrophoretic bands of mitochondria before and after encapsulation were consistent, suggesting that our encapsulation method does not affect the synthesis and expression of mitochondria-associated proteins.3.2 In vitro validation of Mito@ZiF-8 function
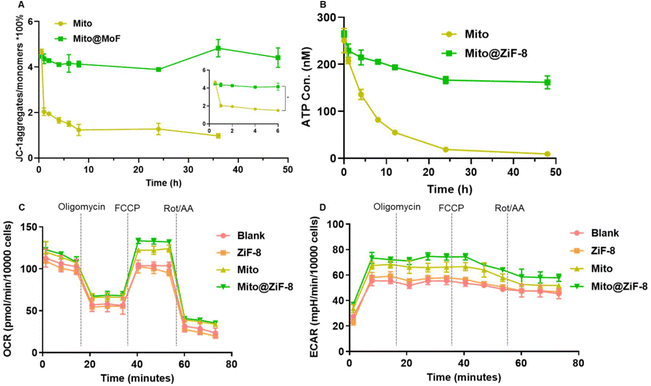
A stable mitochondrial membrane potential (MMP), often referred to as ΔΨm, is mitochondrial oxidative phosphorylation and is crucial to producing ATP, which is vital for maintaining normal cellular functions,17,18 so we used JC-1 dye to detect MMP. As shown in Fig. 2A, the membrane potential of the free Mito group rapidly decreased after isolation for 1 hour and continued to decline over the next 6 hours. In contrast, the Mito@ZiF-8 group maintained a relatively stable membrane potential for 48 hours. Furthermore, the results in Fig. 2B indicated that the free mitochondria retained the ability to produce ATP after isolation, while the Mito group could not sustain ATP production over an extended period. In contrast, Mito@ZiF-8 retained its ATP production capability even after 48 hours.Additionally, the Seahorse Energy Metabolism Analyzer was used to evaluate the energy metabolism of cells after mitochondrial transplantation by measuring its oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). As demonstrated in Fig. 2C and D, under the same cell count, ZiF-8 did not alter OCR and ECAR, while transferring mitochondria into cells enhanced both OCR and ECAR. Moreover, transferring Mito@ZiF-8 into cells further enhanced both OCR and ECAR. This confirmed that encapsulated mitochondria maintained their activity after entering the cells, demonstrating an advantage over uncoated mitochondria. In summary, ZiF-8 bioencapsulation not only preserved mitochondrial respiration but also extended their in vitro half-life and energy production due to its protective effect.
3.3 In vitro uptake of Mito@ZiF-8
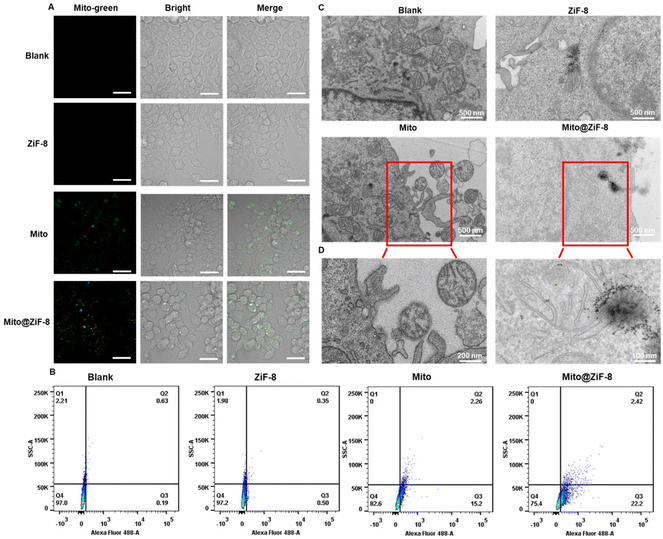
We assessed the cytotoxicity of free ZiF-8. As Fig. S3A† demonstrates the concentration of ZiF-8 added did not exhibit any cytotoxic effects on the cells. Next, we examined the efficiency of extracellular uptake of Mito@ZiF-8. Initially, we labeled mitochondria with CellTracker™ CM-DiI Dye after isolation. Fig. S3C† indicated that both uncoated and coated mitochondria were taken up by M2Φ. After encapsulation, the mitochondrial membrane charge was shielded and we can tailor the ζ potential to facilitate its uptake into the cells. More importantly, we labeled the mitochondria with a live mitochondrial dye MitoTracker™ Green before isolation and encapsulation, followed by transplantation into M2Φ. The results from Fig. 3A using confocal microscopy revealed that M2Φ exhibited a higher uptake of Mito@ZiF-8 than uncoated mitochondria. Furthermore, even after entering the cells, the mitochondria retained their respective activity. Flow cytometry results (Fig. 3B and Fig. S3B†) also indicated that ZiF-8-coating enhanced the uptake of mitochondria by M2Φ.Simultaneously, we employed TEM to observe the uptake process. As shown in Fig. 3C and D, most free mitochondria lingered outside the cells, whereas Mito@ZiF-8 was able to penetrate cells and co-localize with intracellular mitochondria. This may be attributed to the easy internalization of ZiF-8 and its accumulation around the mitochondria within the cells, as Zn2+ plays a role in regulating mitochondrial dynamics, apoptosis, and mitophagy.19 Furthermore, within mitochondria, regulatory proteins exist to maintain mitochondrial Zn2+ homeostasis. Fig. S3D† illustrates the entire process of Mito@ZiF-8 entering M2Φ. Together, these data demonstrated that M2Φ exhibits a capacity to uptake Mito@ZiF-8 to increase the concentration of mitochondria within the cells.
3.4 Regulation of M2Φ secretory functions by Mito@ZiF-8
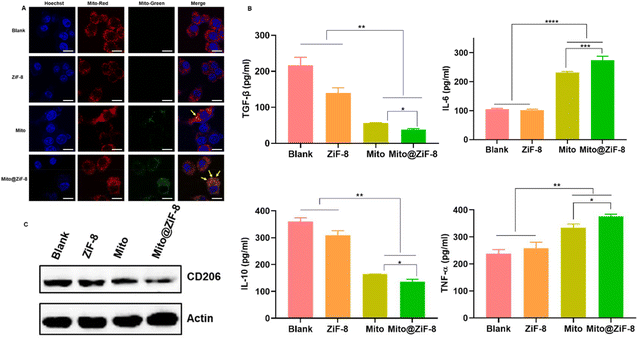
Based on the results shown in Fig. 3C, we observed that Mito@ZiF-8 had a higher uptake rate by M2Φ than uncoated mitochondria. To further validate this, the following experiments were conducted. Firstly, the nanocoating had to evade lysosomal degradation within the cells and successfully escape into the cytoplasm to allow the cargo to exert its intended functions. As determined by the lysosomal escape experiment in Fig. S4A,† after 24 hours of incubation of M2Φ with Mito@ZiF-8, the nanoparticles accumulate within the intracellular lysosomes. However, after 48 hours, a lysosomal rupture occurred, leading to the release of mitochondria. Further examination in Fig. 4A revealed that post-lysosomal escape, transplanted mitochondria could co-localize with intracellular mitochondria, potentially participating in normal mitochondrial fusion and fission cycles, thereby regulating cellular activities.M2Φ promotes tumor invasion, metastasis, and angiogenesis by producing chemokines and cytokines like TGF-β and IL-10, which activate tumor stem cells, whereas, M1Φ secretes pro-inflammatory cytokines such as IL-6, TNF-α, IL-1β, and CXCL10 to stimulate T helper cell responses to suppress tumor development.20–22 As illustrated in Fig. 4B after 5 days incubation, the transplantation of healthy mitochondria led to decreased secretion of anti-inflammatory cytokines TGF-β and IL-10. In contrast, the secretion of pro-inflammatory cytokines IL-6 and TNF-α increased. However, the impact on IL-1β and CXCL10 was minimal, especially as there were no significant differences in the effects on these two factors before and after mitochondrial encapsulation (Fig. S4B†). Overall, Mito@ZiF-8 showed a higher regulatory effect on pro-inflammatory and anti-inflammatory factors than the free Mito group. Additionally, western blotting experiments in Fig. 4C demonstrated that Mito@ZiF-8 can markedly inhibit the expression of CD206 protein, which is a specific marker for M2Φ and is closely associated with tumor cell proliferation and metastasis.20 In summary, these data suggested that transferring healthy mitochondria into M2Φ can effectively suppress their pro-tumor characteristics.
3.5 Mito@ZiF-8 regulates the metabolic environment of M2Φ
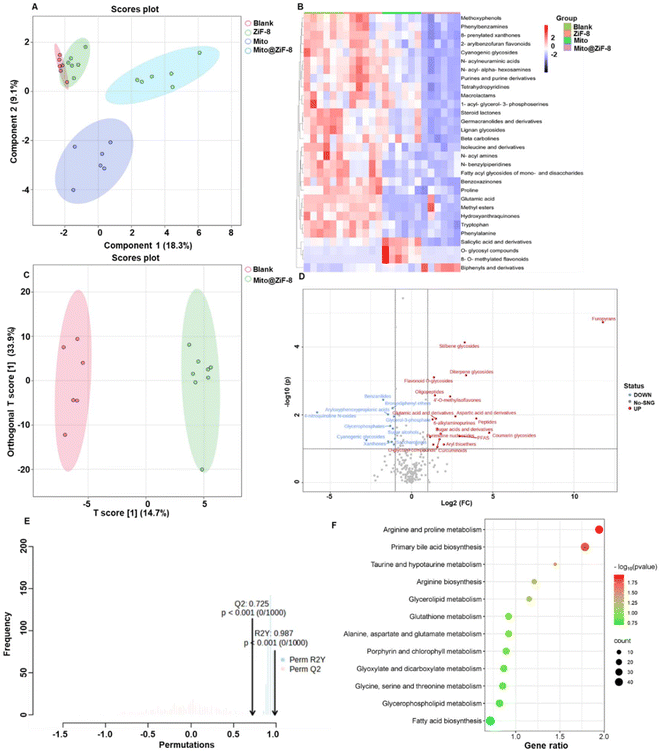
M1Φ tend to rely on glycolysis for rapid energy production and secrete large amounts of lactate.23,24 In contrast, M2 macrophages typically exhibit enhanced fatty acid oxidation and oxidative phosphorylation, generating large amounts of ATP as a key energy source.25–27 Therefore, we conducted specific investigations into the impact of mitochondrial transplantation on macrophage metabolism. Firstly, in Fig. 5A, the PLS-DA plot revealed the factors that can effectively differentiate between sample groups. It was evident that there were significant differences in metabolites between the blank group and the Mito group, as well as the Mito@ZiF-8 group, but there were no noticeable differences in metabolites between the ZiF-8 group and the blank group. Therefore, ZiF-8 did not appear to be regulating macrophage metabolism. Simultaneously, further analysis of the differences in metabolites among the four groups led to the identification of the top 30 metabolites with the most significant structural variations, as depicted in the heatmap in Fig. 5B. From Fig. 5A and B, it was apparent that the metabolism of M2Φ was altered upon treatment with Mito and Mito@ZiF-8. A more in-depth analysis of the regulatory effects of Mito and Mito@ZiF-8 on M2Φ metabolism was conducted. As evident from Fig. 5C and Fig. S5† OPLS-DA score plots, there were metabolic differences between the blank group and the Mito@ZiF-8 group. Notably, the differences were slightly more pronounced between the Mito and Mito@ZiF-8 groups. Furthermore, after model construction, model quality was assessed. In OPLS-DA evaluation, models were considered reliable when R2Y and Q2 were closer to 1, with Q2 > 0.5 indicating effectiveness. Therefore, Fig. 5E and Fig. S5C† indicated that the model was reliable.Additionally, through OPLS-DA analysis, each metabolite was assigned a Variable Importance in Projection (VIP) value. A higher VIP value suggested a greater contribution of that metabolite to differentiating between the two groups. Metabolites with a VIP greater than 1 were selected for further analysis. As shown in Fig. 5D, the delivery of Mito@ZiF-8 resulted in the significant upregulation of 19 metabolites, primarily involved in arginine and proline metabolism, primary bile acid biosynthesis, and more (Fig. 5F). Conversely, after delivering Mito, over 30 metabolites were upregulated (Fig. S5B†), mainly involved in unsaturated fatty acid biosynthesis, arginine synthesis, and other metabolic pathways (Fig. S5D†). These results collectively indicated that both Mito and Mito@ZiF-8 modulated the metabolic pathways of M2Φ. Moreover, Mito@ZiF-8 primarily regulated arginine metabolism and the primary bile acid synthesis pathway. However, the specific mechanisms of this regulation and whether ZiF-8 exerted a positive effect in conjunction with Mito would require further extensive validation in future studies.
3.6 Reversing the function of M2Φ can inhibit tumor progression
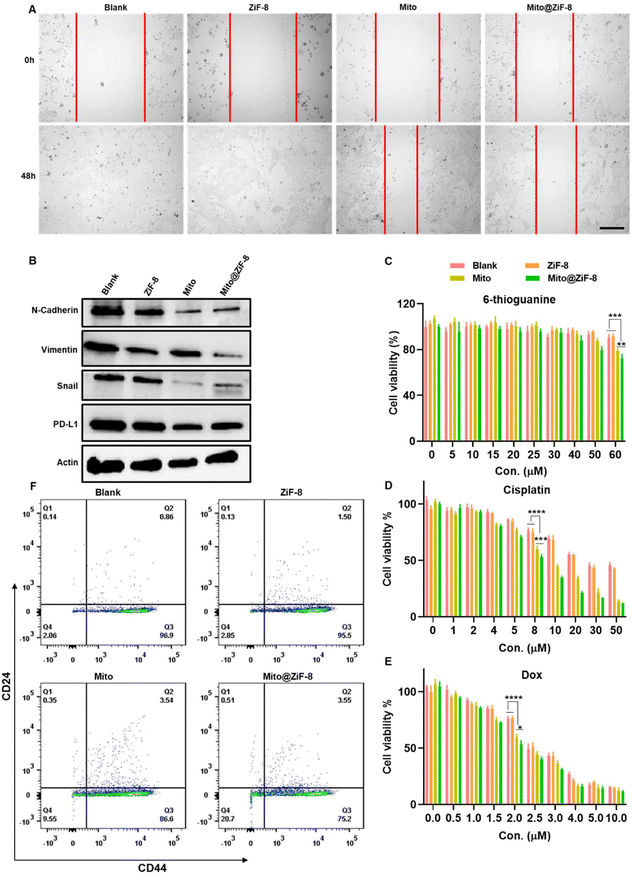
The tumor microenvironment and tumor cells themselves are interconnected. In regulating the microenvironment, there is an inevitable influence on the tumor cells. To explore the impact of metabolic reprogramming in M2Φ on tumor cells, the Transwell co-culture model was employed, as shown in Fig. S6A,† where M2Φ were seeded in the upper chamber and co-cultured with various groups, while 4T-1 cells, a metastatic mouse mammary carcinoma cell line, were placed in the lower chamber. Subsequently, 4T-1 cells were collected for the following assays after 5 days of co-incubation. First, as shown in Fig. 6A, 4T-1 cells exhibited strong metastatic ability, one of the main reasons for poor prognosis in breast cancer. However, we found that when M2Φ was reprogrammed with mitochondria transplantation, the migration of 4T-1 cells was suppressed. Epithelial-to-mesenchymal transition (EMT) activation is essential for cancer metastasis, where epithelial cells gain mesenchymal traits to enhance invasiveness by increasing expression of markers like N-cadherin, vimentin, and transcription factors (e.g., SNAI1, ZEB1).28,29 Western blotting results (Fig. 6B) showed downregulation of mesenchymal-like proteins such as N-cadherin, vimentin, and snail. Moreover, we also observed a downregulation of the PD-L1 protein, which is highly expressed in tumor cells, suggesting that mitochondria transplantation potentially benefits immunotherapy.4T-1 cells are resistant to 6-thioguanine, however, when 4T-1 cells were co-cultured with M2Φ after mitochondrial transplantation, they became more sensitive to high concentrations of 6-thioguanine (Fig. 6C). In contrast, for other chemotherapy drugs like cisplatin and doxorubicin (Dox), Fig. 6E showed that the IC50 values for the blank group and ZiF-8 group after cisplatin treatment were 29.28 μM and 25.98 μM, respectively. However, the mitochondrial transplantation group significantly enhanced the sensitivity of 4T-1 cells to cisplatin, with IC50 values of 10.98 μM (free Mito) and 8.22 μM (Mito@ZiF-8). For Dox, as shown in Fig. 6E, the IC50 values for the blank group and ZiF-8 group were 2.73 μM and 2.70 μM, while the IC50 values for the Mito group and Mito@ZiF-8 group were 2.32 μM and 2.16 μM, respectively. Overall, reversing the tumor microenvironment enhanced the sensitivity of breast cancer cells to chemotherapy drugs, allowing effective tumor suppression at lower chemotherapy concentrations, which is important for reducing the toxic side effects of chemotherapy. In addition, we also found that alterations in the functions of M2Φ can inhibit breast cancer stem-like cells. Breast cancer stem cells are highly correlated with the occurrence, development, recurrence, metastasis, and multidrug resistance of breast cancer.30,31 It was observed that Mito@ZiF-8 exhibited a higher efficacy in suppressing the stem cell population (CD24−CD44+) of breast cancer cells in comparison to Mito, as depicted in Fig. 6D and Fig. S6B.†
In summary, reprogramming M2Φ metabolism via bio-encapsulated mitochondrial transfer effectively inhibited breast cancer cell migration, reduced their stemness, and increased their sensitivity to chemotherapy drugs. However, further investigation through in vivo experiments will be necessary in the future to evaluate the efficacy of anti-tumor therapies.
4 Discussion
Mitochondrial transplantation, as an emerging therapeutic strategy, shows great potential in addressing a variety of diseases, including mitochondrial disorders, neurodegenerative conditions, cardiovascular diseases, and cancer. The primary aim of mitochondrial transplantation is to restore or enhance cellular function, thereby improving treatment outcomes. However, the viability of isolated mitochondria is limited in vitro, with current technologies only preserving mitochondrial bioactivity for up to six hours. Due to this short-lived bioactivity, transporting and storing freshly isolated mitochondria remains a significant challenge.32 We used MOFs for biological encapsulation to address this issue. This approach achieves efficient encapsulation under mild reaction conditions with short processing times. Our results demonstrate that, compared to free mitochondria, our encapsulation system prolongs mitochondrial bioactivity in vitro. Moreover, consistent with other studies, the stability of the ZiF-8 encapsulation system offers protection to biomolecules during storage and transportation, reducing their vulnerability to environmental factors such as oxidative stress and temperature fluctuations.33,34 The encapsulation also minimizes mechanical and chemical damage to mitochondria during handling and injection, preserving their function. The unique properties of ZiF-8 in bio-encapsulation contribute to the clinical translation of encapsulated mitochondria.Furthermore, the relatively large size of mitochondria, typically ranging from 500 nm to 1000 nm, makes cellular uptake challenging, leading to low therapeutic efficacy. The mechanism by which transplanted mitochondria are internalized remains unclear. Endocytosis is widely accepted as the main pathway through which exogenous mitochondria enter target cells, but the specific signaling interactions that trigger this process are still unknown.35,36 Our in vitro studies, however, indicate that ZiF-8 encapsulation enhances cellular uptake efficiency. Additionally, ZiF-8 can be functionalized by attaching specific ligands or binding proteins to anchor receptors on the cell membrane, further increasing the affinity between the mitochondria and the cell membrane, thereby improving uptake. We selected a zinc-based MOF for encapsulation, as Zn2+ plays multiple essential roles in cellular metabolism and function.37 Zn2+ are crucial cofactors for many enzymes, involved in numerous key biochemical reactions, with approximately 300 enzymes being zinc-dependent.38 Zn2+ also regulates gene transcription by influencing the structure and function of zinc finger proteins, which are transcription factors containing zinc ions that bind specific DNA sequences and regulate gene expression.39,40 However, the specific mechanisms of ZiF-8 in the Mito@ZiF-8 system and its therapeutic potential require further investigation.
In addition, whether the transplanted mitochondria can maintain long-term functionality in the new cellular microenvironment, including regulating mtDNA replication, expressing function-related proteins, and supporting physiological processes such as mitochondrial fission and fusion, is not yet fully understood. A deeper understanding of these mechanisms is crucial for advancing the theory of mitochondrial transplantation and refining treatment strategies for mitochondrial disorders. Our results indicate that encapsulated mitochondria can modulate the metabolic reprogramming of M2Φ, inhibiting their secretion of anti-inflammatory factors. Furthermore, changes in the metabolic microenvironment of M2Φ can suppress the metastasis of breast cancer cells, providing valuable insights for developing mitochondrial transplantation-based therapies.
However, successful clinical translation requires rigorous safety and efficacy evaluations, particularly in in vivo studies. The in vitro cellular environment differs from the in vivo tissue environment and further experimental results are needed to verify whether the delivery method and therapeutic efficacy remain as effective as in vitro studies. McCully's team found that direct mitochondrial perfusion during cardiac procedures can improve heart function within minutes.41 In future cancer therapies, direct peritumoral or intratumoral injection may enhance delivery efficiency compared to systemic administration (such as oral or intravenous routes), as it avoids dilution in other tissues and maintains a higher concentration of active mitochondria in the tumor, improving cytotoxic effects on cancer cells while reducing potential systemic toxicity.42 Additionally, studies have shown that inorganic/organic particles can induce endothelial leakage in vivo, enhancing the efficiency of nanoparticle delivery within tumors without affecting healthy endothelial vessels.43,44 These findings provide a reference for in vivo studies of Mito@ZIF-8 and are beneficial for further elucidating the mechanism of action of Mito@ZIF-8 in vivo. Another challenge to address is whether healthy donor-derived mitochondria could trigger an immune response. Some diseases are driven by mitochondrial genetic mutations, necessitating the transplantation of healthy mitochondria from donors.45 However, the immune system may recognize these transplanted mitochondria as foreign, leading to immune reactions and potential rejection. Whether our encapsulation technology can reduce immune rejection requires further investigation.
5 Conclusion
In conclusion, our study has further expanded the application of Mito@ZIF-8 in the biomedical field. Our results demonstrated that ZIF-8-coated mitochondria can maintain their biological activity in vitro, particularly by entering cells and regulating macrophage metabolic reprogramming, thereby inhibiting breast cancer cell metastasis and increasing sensitivity to chemotherapy drugs. Although our current research is still at the in vitro stage, it conceptually validates the feasibility of regulating cellular biological functions through mitochondrial transplantation. We aim to gradually optimize this technology to achieve practical applications of mitochondrial transplantation, providing new strategies and insights for its large-scale use in human diseases.Abbreviations
| HmIm | 2-Methylimidazole |
| ATP | Adenosine 5′-triphoshate |
| Cyto C | Cytochrome c |
| Dox | Doxorubicin |
| EP | Eppendorf |
| HSP90 | Heat shock protein 90 |
| IL-10 | Interleukin 10 |
| M1Φ | M1-type macrophages |
| M2Φ | M2-type macrophages |
| MoFs | Metal–organic frameworks |
| MMP | Mitochondrial membrane potential |
| PHD | Pyruvate dehydrogenase |
| ROS | Reactive oxygen species |
| TGF-β | Transforming growth factor beta |
| TEM | Transmission electron microscopy |
| TCA | Tricarboxylic acid cycle |
| TAMs | Tumor-associated macrophages |
| VIP | Variable importance in projection |
| ZiF-8 | Zeolitic imidazolate framework-8 |
Author contributions
Yonghui Wang led the data collection and organization and authored the manuscript. Chang Liu and Xiaodong Ma contributed to establishing and validating the methodology. Anne Filppula and Youbin Cui provided support in writing, reviewing, and editing. Jiangbin Ye and Hongbo Zhang oversaw project management and supervision, as well as writing, reviewing, and editing the manuscript.Data availability
They are included in the Results and Methods sections, and the relevant data is also available from the ESI† of the manuscript.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
We gratefully acknowledge the financial support provided by various sources, including the Academy of Finland through the Research Fellowship (Grant No. 328933), Research Project (Grant No. 347897), Health Profile Solutions (Grant No. 336355), and the InFLAMES Flagship (Grant No. 337531). We are also thankful for the funding from the Finland-China Food and Health International Pilot Project, supported by the Finnish Ministry of Education and Culture, as well as the China Scholarship Council. In addition, we express our sincere appreciation to the First Hospital of Jilin University for their collaboration and valuable contributions. We further extend our thanks to the Turku Bioscience Centre for facilitating access to advanced infrastructure, including Electron Microscopy, Confocal, and Flow Cytometry, which were essential for sample analysis, and to the Turku Metabolomics Centre for providing comprehensive sample analysis services.References
- W. X. Zong, J. D. Rabinowitz and E. White, Mitochondria and Cancer, Mol. Cell, 2016, 61, 667–676 CrossRef CAS
.
- P. E. Porporato, N. Filigheddu, J. M. B. Pedro, G. Kroemer and L. Galluzzi, Mitochondrial metabolism and cancer, Cell Res., 2018, 28, 265–280 CrossRef CAS PubMed
.
- M. P. King and G. Attardi, Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation, Science, 1989, 246, 500–503 CrossRef CAS PubMed
.
- K. K. Goswami, A. Bose and R. Baral, Macrophages in tumor: An inflammatory perspective, Clin. Immunol., 2021, 232, 108875 CrossRef CAS PubMed
.
- M. Bied, W. W. Ho, F. Ginhoux and C. Bleriot, Roles of macrophages in tumor development: a spatiotemporal perspective, Cell. Mol. Immunol., 2023, 20, 983–992 CrossRef CAS PubMed
.
- R. G. Jones and C. B. Thompson, Tumor suppressors and cell metabolism: a recipe for cancer growth, Genes Dev., 2009, 23, 537–548 CrossRef CAS
.
- H. Zhang, L. Liu and J. Liu,
et al., Roles of tumor-associated macrophages in anti-PD-1/PD-L1 immunotherapy for solid cancers, Mol. Cancer, 2023, 22, 58 CrossRef CAS PubMed
.
- T. Huang, T. Zhang and J. Gao, Targeted mitochondrial delivery: A therapeutic new era for disease treatment, J. Controlled Release, 2022, 343, 89–106 CrossRef CAS PubMed
.
- T. G. Zhang and C. Y. Miao, Mitochondrial transplantation as a promising therapy for mitochondrial diseases, Acta Pharm. Sin. B, 2023, 13, 1028–1035 CrossRef CAS
.
- D. C. Wallace, W. Fan and V. Procaccio, Mitochondrial energetics and therapeutics, Annu. Rev. Pathol.: Mech. Dis., 2010, 5, 297–348 Search PubMed
.
- M. Sun, W. Jiang, N. Mu, Z. Zhang, L. Yu and H. Ma, Mitochondrial transplantation as a novel therapeutic strategy for cardiovascular diseases, J. Transl. Med., 2023, 21, 347 CrossRef PubMed
.
- R. L. Elliott, X. P. Jiang and J. F. Head, Mitochondria organelle transplantation: introduction of normal epithelial mitochondria into human cancer cells inhibits proliferation and increases drug sensitivity, Breast Cancer Res. Treat., 2012, 136, 347–354 CrossRef CAS PubMed
.
- J. Cao, X. Li and H. Tian, Metal-Organic Framework (MOF)-Based Drug Delivery, Curr. Med. Chem., 2020, 27, 5949–5969 CrossRef CAS PubMed
.
- R. Ricco, W. Liang and S. Li,
et al., Metal-Organic Frameworks for Cell and Virus Biology: A Perspective, ACS Nano, 2018, 12, 13–23 CrossRef CAS
.
- K. Liang, R. Ricco and C. M. Doherty,
et al., Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules, Nat. Commun., 2015, 6, 7240 Search PubMed
.
- C. Doonan, R. Ricco, K. Liang, D. Bradshaw and P. Falcaro, Metal-Organic Frameworks at the Biointerface: Synthetic Strategies and Applications, Acc. Chem. Res., 2017, 50, 1423–1432 CrossRef CAS PubMed
.
- O. M. Russell, G. S. Gorman, R. N. Lightowlers and D. M. Turnbull, Mitochondrial Diseases: Hope for the Future, Cell, 2020, 181, 168–188 Search PubMed
.
- A. Suomalainen and B. J. Battersby, Mitochondrial diseases: the contribution of organelle stress responses to pathology, Nat. Rev. Mol. Cell Biol., 2018, 19, 77–92 CrossRef CAS PubMed
.
- H. Zhang, D. Liu and L. Wang,
et al., Microfluidic Encapsulation of Prickly Zinc-Doped Copper Oxide Nanoparticles with VD1142 Modified Spermine Acetalated Dextran for Efficient Cancer Therapy, Adv. Healthc. Mater., 2017, 6, 1601406 CrossRef
.
- N. R. Anderson, N. G. Minutolo, S. Gill and M. Klichinsky, Macrophage-Based Approaches for Cancer Immunotherapy, Cancer Res., 2021, 81, 1201–1208 CrossRef CAS PubMed
.
- J. Zhang, X. Zhou and H. Hao, Macrophage phenotype-switching in cancer, Eur. J. Pharmacol., 2022, 931, 175229 CrossRef CAS
.
- S. Mohapatra, C. Pioppini, B. Ozpolat and G. A. Calin, Non-coding RNAs regulation of macrophage polarization in cancer, Mol. Cancer, 2021, 20, 24 CrossRef CAS PubMed
.
- L. Zhu, Q. Zhao, T. Yang, W. Ding and Y. Zhao, Cellular metabolism and macrophage functional polarization, Int. Rev. Immunol., 2015, 34, 82–100 CrossRef CAS PubMed
.
- M. Li, Y. Yang, L. Xiong, P. Jiang, J. Wang and C. Li, Metabolism, metabolites, and macrophages in cancer, J. Hematol. Oncol., 2023, 16, 80 Search PubMed
.
- K. Mehla and P. K. Singh, Metabolic Regulation of Macrophage Polarization in Cancer, Trends Cancer, 2019, 5, 822–834 CrossRef CAS PubMed
.
- J. Zhang, J. Muri and G. Fitzgerald,
et al., Endothelial Lactate Controls Muscle Regeneration from Ischemia by Inducing M2-like Macrophage Polarization, Cell Metab., 2020, 31, 1136–1153 CrossRef CAS PubMed
.
- E. Vassiliou and R. Farias-Pereira, Impact of Lipid Metabolism on Macrophage Polarization: Implications for Inflammation and Tumor Immunity, Int. J. Mol. Sci., 2023, 24, 12032 CrossRef CAS
.
- B. Bakir, A. M. Chiarella, J. R. Pitarresi and A. K. Rustgi, EMT, MET, Plasticity, and Tumor Metastasis, Trends Cell Biol., 2020, 30, 764–776 CrossRef PubMed
.
- V. Ramesh, T. Brabletz and P. Ceppi, Targeting EMT in Cancer with Repurposed Metabolic Inhibitors, Trends Cancer, 2020, 6, 942–950 CrossRef CAS PubMed
.
- R. Camerlingo, G. A. Ferraro and F. De Francesco,
et al., The role of CD44+/CD24−/low biomarker for screening, diagnosis and monitoring of breast cancer, Oncol. Rep., 2014, 31, 1127–1132 CrossRef CAS
.
- J. C. Chang, H. S. Chang and Y. C. Wu,
et al., Mitochondrial transplantation regulates antitumour activity, chemoresistance and mitochondrial dynamics in breast cancer, J. Exp. Clin. Cancer Res., 2019, 38, 30 CrossRef
.
- G. B. Kubat, O. Ulger and S. Akin, Requirements for successful mitochondrial transplantation, J. Biochem. Mol. Toxicol., 2021, 35, e22898 CrossRef CAS
.
- J. H. Scott and R. Schekman, Lyticase: endoglucanase and protease activities that act together in yeast cell lysis, J. Bacteriol., 1980, 142, 414–423 CrossRef CAS
.
- S. H. Yang, S. M. Kang, K. B. Lee, T. D. Chung, H. Lee and I. S. Choi, Mussel-inspired encapsulation and functionalization of individual yeast cells, J. Am. Chem. Soc., 2011, 133, 2795–2797 CrossRef CAS
.
- C. Sun, X. Liu and B. Wang,
et al., Endocytosis-mediated mitochondrial transplantation: Transferring normal human astrocytic mitochondria into glioma cells rescues aerobic respiration and enhances radiosensitivity, Theranostics, 2019, 9, 3595–3607 CrossRef CAS PubMed
.
- R. Z. Lin, G. B. Im and A. C. Luo,
et al., Mitochondrial transfer mediates endothelial cell engraftment through mitophagy, Nature, 2024, 629, 660–668 CrossRef CAS PubMed
.
- X. Yang, H. Wang and C. Huang,
et al., Zinc enhances the cellular energy supply to improve cell motility and restore impaired energetic metabolism in a toxic environment induced by OTA, Sci. Rep., 2017, 7, 14669 CrossRef PubMed
.
- K. A. McCall, C. Huang and C. A. Fierke, Function and mechanism of zinc metalloenzymes, J. Nutr., 2000, 130, 1437S–1446S Search PubMed
.
- J. P. Liuzzi, L. Guo, C. Yoo and T. S. Stewart, Zinc and autophagy, BioMetals, 2014, 27, 1087–1096 CrossRef CAS
.
- M. Bendellaa, P. Lelievre, J. L. Coll, L. Sancey, A. Deniaud and B. Busser, Roles of zinc in cancers: From altered metabolism to therapeutic applications, Int. J. Cancer, 2024, 154, 7–20 CrossRef CAS
.
- A. Masuzawa, K. M. Black and C. A. Pacak,
et al., Transplantation of autologously derived mitochondria protects the heart from ischemia-reperfusion injury, Am. J. Physiol.: Heart Circ. Physiol., 2013, 304, H966–H982 CrossRef CAS
.
- L. Martinez-Ruiz, J. Florido and C. Rodriguez-Santana,
et al., Intratumoral injection of melatonin enhances tumor regression in cell line-derived and patient-derived xenografts of head and neck cancer by increasing mitochondrial oxidative stress, Biomed. Pharmacother., 2023, 167, 115518 CrossRef CAS
.
- M. I. Setyawati, Q. Wang and N. Ni,
et al., Engineering tumoral vascular leakiness with gold nanoparticles, Nat. Commun., 2023, 14, 4269 CrossRef CAS
.
- Y. Li, N. Ni and M. Lee,
et al., Endothelial leakiness elicited by amyloid protein aggregation, Nat. Commun., 2024, 15, 613 CrossRef CAS
.
- J. D. McCully, D. B. Cowan, S. M. Emani and P. J. Del Nido, Mitochondrial transplantation: From animal models to clinical use in humans, Mitochondrion, 2017, 34, 127–134 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr02471k |
| This journal is © The Royal Society of Chemistry 2024 |