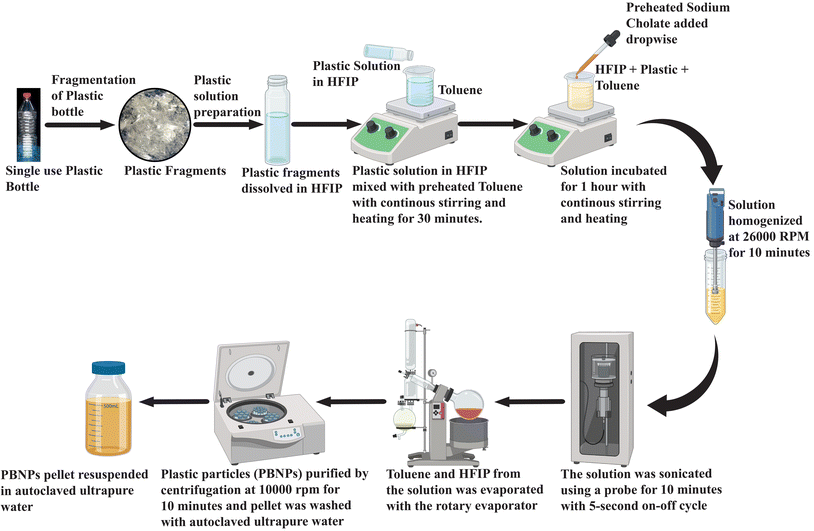
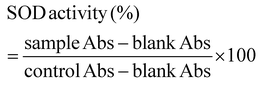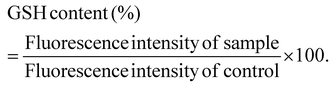Single-use polyethylene terephthalate bottle-derived nanoplastics propagate antibiotic resistance in bacteria via transformation and outer membrane vesicle secretion†
Prashant
Sharma‡
 a,
Abhinoy
Kishore‡
ab and
Manish
Singh
a,
Abhinoy
Kishore‡
ab and
Manish
Singh
 *a
*a
aChemical Biology Unit, Institute of Nano Science and Technology (INST), Knowledge City, Sector-81, Mohali, 140306, Punjab, India. E-mail: manish@inst.ac.in
bDepartment of Biotechnology, Chandigarh College of Technology, CGC, Landran, 140307, Punjab, India
First published on 30th October 2024
Abstract
Plastic pollution arising from single-use plastic bottles (SUPBs) and containers leads to the formation of micro/nanoplastics (NPs). These NPs raise concerns due to their potential toxicity and interactions with microorganisms. In various environments, including our digestive system, both microorganisms and plastics coexist. The interactions between these NPs and microorganisms can have far-reaching consequences, potentially affecting the ecosystems and human health. Therefore, understanding these interactions is crucial for addressing the challenges posed by plastic pollution. This study investigated the role of NPs in propagating antibiotic resistance (AR), specifically through outer membrane vesicles (OMVs), which is a mechanism that has not been fully explored to date in terms of NPs' effects. To explore this, NPs were synthesized using polyethylene terephthalate (PET) SUPBs, mimicking the natural chemical composition of environmental nanoplastics, unlike previous studies, which used pure PET, polystyrene (PS) or other pure plastic materials. The resulting PET bottle-derived nanoplastics (PBNPs), which exhibited diverse shapes and sizes (50–850 nm), were found to facilitate horizontal gene transfer (HGT) through transformation and outer membrane vesicles (OMVs), enabling the transport of plasmids among bacteria. In transformation, PBNPs physically carried plasmids across the bacterial membrane. In another scenario, PBNPs induced oxidative stress and bacterial surface damage, which led to the upregulation of stress response-associated genes and the escalation of OMV secretion in E. coli. This novel pathway highlights how PBNPs contribute to AR gene dissemination, potentially exacerbating the global antibiotic resistance crisis. Furthermore, PBNPs mediate cross-species gene transfer from E. coli to Lactobacillus acidophilus, underscoring their impact on diverse microorganisms, including those in the human gut. Our findings suggest that nanoplastics may be an unrecognized contributor to the rising tide of antibiotic resistance, with significant consequences for human health and the environment. Molecular analyses revealed the upregulated expression of genes associated with stress response and OMV secretion, offering deeper insights into the biological mechanisms affected by PBNPs. This study offers crucial insights into the interactions of NPs and microorganisms for developing strategies to address the ecological and health implications of nanoplastic contamination.
1. Introduction
Widespread issue of plastic pollution poses a significant global concern, with extensive ramifications for both ecosystems and human health. Omnipresence of plastic consumer products, especially single-use plastics, is unsustainable in current production and consumption patterns. In the last 50 years, plastic production has surged to over 9200 million metric tons (Mt), among which just 9% has been recycled, 12% has been incinerated, and the remaining 79% has accumulated in our natural ecosystems globally.1 Regarding the scenario in India, annual production of plastic materials for manufacturing industries is approximately 3.3 million tons.2 Plastic undergoes some form of degradation through various mechanisms, including UV radiation, chemical leaching, mechanical breakdown in air and water, and biological breakdown by microorganisms.1,3–5 Consequently, these processes result in the fragmentation of plastic into smaller particles, referred to as microplastics and nanoplastics (MPs and NPs), with sizes ranging from 1 μm to 5 mm for MPs and less than 1 μm for NPs.1,3–5 These small particles have infiltrated various ecosystems, ranging from oceans and rivers to soil, and have even been detected in polar region snowfall.6 Recently, research has revealed that microplastics were detectable in nearly all analysed samples of table salt.7,8 This widespread distribution of MPs/NPs increases the risk of human exposure through inhalation, ingestion, and dermal contact. Presently, various studies on animal models suggest that microplastics can accumulate in the gastrointestinal tract, and also migrate to organs including the liver, kidneys, brain, and testes.9,10 Long-term exposure to these MPs and NPs has been documented to induce a range of serious health issues, including respiratory infections and inflammation, gastrointestinal, liver, and kidney damage, cardiovascular complications, neurological disorders, cancer, and reproductive dysfunction.10 Thus, MPs and NPs are undoubtedly a cause of concern not only due to their various toxic effects but also their crucial interactions with microorganisms given that they are present both in proximity in our environment and the gut. Interestingly, these MPs/NPs have been reported to create a unique environment called the “plastisphere,” where tiny life forms, such as bacteria, can easily flourish.11,12 These studies highlight the ability of MPs/NPs to act as a substrate for various free bacteria in the environment and human gut to come in close proximity, and the possibility of exchanging their genetic content particularly antibiotic resistance genes (ARGs).13 The proliferation of ARGs on a global scale is a significant concern.14,15 The World Health Organization and European Commission have pointed out that the fast dissemination of antimicrobial resistance is a global health and environment problem. The misuse and overuse of antibiotics are the prime cause of the increasing antibiotic resistance, which has caused 1.27 million deaths globally in 2019.16–18 There are scientific predictions that antimicrobial resistance will lead to over 10 million deaths each year by 2050.19 This situation may become even worse if the rate of antibiotic resistant gene transfer increases due to certain anthropogenic activities, where NPs seem to be a potential one.Horizontal gene transfer (HGT) is a common environmental event and a major way for ARGs to disseminate in nature, which mainly occurs by three methods, viz. conjugation, transformation and transduction.20 Usually, conjugation, which involves direct cell-to-cell contact for the transfer of genetic elements such as plasmids, plays a substantial role in the transmission of antibiotic resistance. Transformation is when the exogenous genetic material is assimilated directly from the environment, whereas transduction involves bacteriophages for the same process.21 Besides these methods, another non-canonical method of HGT has recently been reported, where bacteria release genetic materials containing outer membrane vesicles (OMVs), which may be taken up by neighbouring bacteria, potentially facilitating the transfer of ARGs. However, the precise mechanisms of this process remain under investigation and require further research, particularly in the context of NP exposure. NPs are known to induce oxidative stress by increasing the bacterial reactive oxygen species (ROS) levels, which have been shown to enhance the secretion of OMVs, potentially amplifying this form of HGT.22 Therefore, we hypothesized that NPs can facilitate the horizontal transfer of ARGs. This may possibly occur by transformation, i.e. NPs directly augmenting bacterial permeability or serving as carriers to promote the internalization of ARGs, which generally lies on plasmids. Additionally, this process can be intensified by the NP-induced elevation of ROS levels, which stimulates bacteria to produce and release more OMVs. This heightened OMV secretion may increase the likelihood of antibiotic resistance propagation beyond normal levels.
Thus, to investigate this, an innovative oil-in-water nano-emulsion technique was used to synthesise single-use polyethylene terephthalate (PET) bottle-derived nanoplastics (PBNPs) similar to the plastics present in plastic-polluted natural environments. These PBNPs were further characterised using high-end characterization techniques including dynamic light scattering (DLS), field emission scanning electron microscopy (FESEM), transmission electron microscopy (TEM), atomic force microscopy (AFM), Raman spectroscopy and Fourier-transform infrared (FTIR) spectroscopy. Then, we specifically examined the capability of PBNPs to directly transfer ARGs to bacteria through the exchange of plasmids. Additionally, we evaluated the potential of outer membrane vesicles (OMVs) to transport and convey ARGs to probiotic bacteria such as Lactobacillus acidophilus. Furthermore, we focused on identifying the mechanisms responsible for the accelerated secretion of OMVs and gene transfer. These mechanisms involve alterations in the growth kinetics, intracellular ROS levels, cell morphology, and mRNA expression of genes associated with oxidative stress and OMV secretion. Thus, this study provides important insights into the interactions between NPs and microorganisms, which are essential for developing suitable strategies to mitigate the ecological and health impacts of nanoplastic contamination.
2. Methodology
2.1. Materials
All reagents and solvents were AR grade and sourced from SRL Chemicals. LB and MRS media were obtained from HiMedia Laboratories, while bacterial strains were acquired from MTCC, Chandigarh, India. Agarose low EEO grade (RC-149) was procured from the G-Biosciences. Primers were procured from Eurofins, Trizol reagent (TRI Reagent-T9424) from Sigma-Aldrich, and the cDNA synthesis kit (4368814) together with PCR master mix (K0171) from Thermo Fisher Scientific. Single-use PET plastic bottles were procured from a local market.2.2. Synthesis of nanoplastics from single-use plastic bottle
A polyethylene terephthalate single-use plastic bottle (PET-SUPB) was first cut into small fragments, and then meticulously cleaned using ethanol. Then, 100 mg of plastic was dissolved in 1 mL of 1,1,1,3,3,3-hexafluoroisopropanol (HFIP). This plastic solution was carefully added in droplets to 8 mL of toluene, which was placed on a magnetic stirrer set at 1100 rpm and heated to 100 °C. Continuous stirring was maintained for 30 min. Following this, a boiling solution of sodium cholate (0.27 mg mL−1) with a total volume of 80 mL was gradually introduced dropwise into the stirred mixture, while maintaining the same stirring speed and temperature of 100 °C. Subsequently, the mixture was allowed to stir for an additional hour, and then homogenized using a 125-watt CAT X120 hand-held homogenizer (CAT Scientific, Germany), running at 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min pulses over a total duration of 10 min. For further enhancement of the uniformity and reduction in size, the solution was subjected to probe sonication at a power level of 25 W, utilizing 5 s ON and OFF cycles for 10 min. The solution was promptly immersed in an ice bath after sonication. Then, the entire solution was carefully transferred to a 250 mL round-bottom flask and subjected to rotary evaporation under a vacuum of 80 psi, maintaining a temperature of 55 °C. The evaporated volume was gradually replenished with deionized water, and subsequently evaporated again. This process of rotary evaporation was repeated iteratively until the toluene was completely eliminated. Subsequently, the volume was gradually reduced to 20 mL. The suspended particles were purified through a series of centrifugation and resuspension steps. Each purification cycle involved centrifuging the suspension with an equal volume of 0.5 mg mL−1 BSA23 (pH 8.2) at 10
000 rpm for 2 min pulses over a total duration of 10 min. For further enhancement of the uniformity and reduction in size, the solution was subjected to probe sonication at a power level of 25 W, utilizing 5 s ON and OFF cycles for 10 min. The solution was promptly immersed in an ice bath after sonication. Then, the entire solution was carefully transferred to a 250 mL round-bottom flask and subjected to rotary evaporation under a vacuum of 80 psi, maintaining a temperature of 55 °C. The evaporated volume was gradually replenished with deionized water, and subsequently evaporated again. This process of rotary evaporation was repeated iteratively until the toluene was completely eliminated. Subsequently, the volume was gradually reduced to 20 mL. The suspended particles were purified through a series of centrifugation and resuspension steps. Each purification cycle involved centrifuging the suspension with an equal volume of 0.5 mg mL−1 BSA23 (pH 8.2) at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at room temperature. This washing step was repeated three times. Finally, the obtained pellet of nanoplastics was washed three times with autoclaved ultrapure water by inverting the tube. The nanoplastic pellet was resuspended in autoclaved ultrapure water, resulting in the desired purified suspended plastic particle (nanoplastic) solution. A schematic representation of the methodology for the synthesis of PBNPs is depicted in Fig. 1.
000 rpm for 10 min at room temperature. This washing step was repeated three times. Finally, the obtained pellet of nanoplastics was washed three times with autoclaved ultrapure water by inverting the tube. The nanoplastic pellet was resuspended in autoclaved ultrapure water, resulting in the desired purified suspended plastic particle (nanoplastic) solution. A schematic representation of the methodology for the synthesis of PBNPs is depicted in Fig. 1.
To quantify the NP concentration, a volume of 1 mL containing suspended NP particles was placed in 3 different pre-weighed microcentrifuge tubes. These tubes were placed in an oven at 40 °C and left to dry. Once fully dried, the Eppendorf tube was re-weighed. By subtracting the initial weight from the final weight, the concentration of synthesized NPs per mL was determined.23–25 The remainder of the stock suspension was stored at 4 °C, thereby avoiding prolonged exposure to any conditions that could cause its degradation.
2.3. Characterization of PET bottle-derived nanoplastics (PBNPs)
2.4. Investigation of PBNPs' potential to facilitate bacterial horizontal gene transfer
 | ||
| Fig. 2 Schematic illustrating the methodology used for investigation of PBNP-mediated horizontal gene transfer via outer membrane vesicles (OMVs). | ||
To quantify the binding efficiency of plasmid DNA to PBNPs, we employed a spectrophotometric method. Plasmid DNA (pEGFP-N1) was prepared at a concentration of 100 ng μL−1 in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0). PET nanoplastics were suspended at a concentration of 100 μg mL−1 in the same buffer. A mixture of 100 μL plasmid DNA solution (10 μg total DNA) and 900 μL nanoplastic suspension (90 μg total nanoplastic) was incubated at room temperature for 6 h with gentle shaking. Post-incubation, the samples were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min to pellet the nanoplastic-bound plasmid DNA. The supernatant, containing unbound plasmid DNA, was carefully collected and its absorbance at 260 nm was measured using an Infinite® 200 NanoQuant. The initial plasmid DNA concentration was determined by measuring the absorbance of the control sample containing only the plasmid DNA solution. The amount of plasmid DNA bound to the nanoplastics was determined by subtracting the unbound plasmid DNA from the initial plasmid DNA concentration.
000 rpm for 10 min to pellet the nanoplastic-bound plasmid DNA. The supernatant, containing unbound plasmid DNA, was carefully collected and its absorbance at 260 nm was measured using an Infinite® 200 NanoQuant. The initial plasmid DNA concentration was determined by measuring the absorbance of the control sample containing only the plasmid DNA solution. The amount of plasmid DNA bound to the nanoplastics was determined by subtracting the unbound plasmid DNA from the initial plasmid DNA concentration.
Further to assess the binding strength of the plasmids and PBNPs, we incubated both for 6 h at RT to sufficiently allow them to have proper interaction and adhesion with each other. Thereafter, they were washed with PBS two and four times by centrifugation. Also, the resulting pellets were resuspended in LB media and introduced into two separate E. coli DH5α cultures, which were incubated overnight under rotating condition at 120 rpm and 37 °C, and then screened against kanamycin-LB agar plates. Here, again only the E. coli DH5α cells that could form colonies had acquired the pEGFP-N1 plasmids, and hence resistance against kanamycin. To further confirm the presence of pEGFP-N1 plasmids, these E. coli DH5α colonies were scraped and cultured overnight, and then subjected to alkaline lysis and PCR.
2.4.2.1. Insertion of pEGFP-N1 in E. coli DH5α. Here, we required transformed E. coli already containing pEGFP-N1, a plasmid having the genes for kanamycin resistance. The insertion of pEGFP-N1 in E. coli was achieved using the standard heat shock method.27 Thereafter, the pEGFP-N1-containing E. coli was cultured on LB agar having kanamycin antibiotic (kanamycin-LB agar plate). Then, a single plasmid-transformed bacterial colony was selected and introduced into 5 mL of LB culture medium, allowing it to cultivate overnight at 120 rpm at 37 °C. This transformed kanamycin-resistant bacteria was used for further experiments.
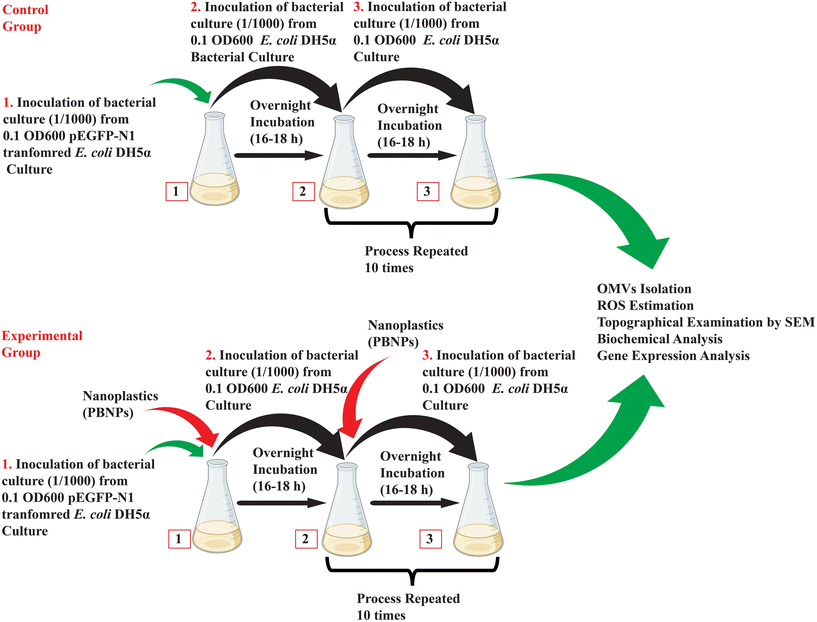
2.4.2.2. Exposure of E. coli DH5α to PBNPs. Considering the chance of multiple generational exposure of naturally existing bacteria and exponentially enhancing the pollution of nanoplastics, we tried to mimic this exposure experimentally, where kanamycin-resistant E. coli DH5α cells were cultured for a period of 10 days, during which it was subjected to repeated multiple generational exposure to 50 and 100 μg mL−1 concentrations of PBNPs. Initially, the bacterial culture was exposed to PBNPs, and the following day, inoculum from this treated culture was transferred to fresh LB medium together with the respective concentration of PBNPs. This process was repeated daily for 10 consecutive days, ensuring sustained exposure, and allowing the bacterial population to undergo multiple generations with PBNPs. The bacteria were considered the control group in the absence of any PBNP treatment. The illustration in Fig. 2 demonstrates the methodology of exposing PBNPs to bacteria.
2.4.2.3. Isolation of OMVs from the transformed E. coli DH5α using polyethylene glycol (PEG). As a result of multiple generational exposure of PBNPs, E. coli DH5α is supposed to secrete numerous OMVs, which may contain genetic information, i.e. pEGFP-N1 plasmid/kanamycin resistance. Thus, to test this, isolation of these OMVs was done by the precipitation method using PEG (MW 6000 Da).28 After the final exposure, and incubation, the culture was centrifuged at 500g for 5 min at 4 °C. The resulting supernatant was transferred to a separate tube and subjected to a second round of centrifugation at 2000g for 30 min at 4 °C. Post-centrifugation, the supernatant was carefully withdrawn and filtered using a 0.22 μm syringe filter. Polyethylene glycol (PEG) was introduced in the filtered supernatant to attain a final concentration of 16%. The PEG-supernatant mixture was placed on a rotary shaker and incubated overnight at 4 °C. Subsequent to the incubation period, the mixture was subjected centrifugation at 7000g for 1 h at 4 °C. During this centrifugation, the supernatant was gently decanted without disturbing the pellet. The resulting pellet containing OMVs was washed three times with phosphate-buffered saline (PBS), subsequently resuspended in PBS, and finally preserved at −80 °C for further experiments.
2.4.2.4. Confirmation and characterisation of OMVs. Confirmation and size estimation of OMVs was done by DLS and TEM, quantification was done by the Lowry method and the presence of pEGFP-N1 was confirmed using PCR. The detailed methods are presented below.
(I) Size estimation through DLS, FESEM and TEM: The OMVs' size distribution analysis was carried out using a Zetasizer (Nano ZS90 Malvern). The core size of OMVs was determined by FESEM and TEM. For the FESEM analysis, the OMV sample was treated with a 2.5% glutaraldehyde solution for 20 min at room temperature. Then, the fixed sample was drop-casted onto a cleaned glass slide and it was allowed to dry in a vacuum desiccator for a period of 4–5 days. Prior to mounting on the stage, the samples were subjected to gold coating using a sputtering machine. Subsequently, the images were acquired using a JEOL JSM-7610 Plus field emission scanning electron microscopy (FESEM).26 For TEM, the fixed sample was drop-casted onto a carbon-coated grid. Subsequently, a series of three PBS buffer washes was carried out, followed by staining with a 2% uranyl acetate solution for 10 min. Images were captured using HRTEM (JEOL JEM 2100) operating at an accelerating voltage of 120 kV.
(II) Quantification of OMVs based on protein content: Given that proteins make up a significant portion of the OMVs content, measuring the protein concentration provides an estimate of the OMVs concentration. It was performed by assessing the protein content of OMVs using the Lowry method, a well-established technique for measuring the total protein concentration. Bovine serum albumin was employed as a calibration standard. The scientifically validated and already reported protocol is given in the ESI (SI 2†).
(III) Validating the presence of pEGFP-N1 in OMVs: The presence of pEGFP-N1 plasmid in OMVs was confirmed using polymerase chain reaction (PCR). The primers (Table 1) were designed using the Gene Runner software. The OMVs were taken as the template and the annealing temperature was maintained at 60 °C for 35 cycles in a Thermocycler (Applied Biosystem). The PCR product was run on 1.2% agarose gel at 100 mV for 30 min in 1× TAE buffer.
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| gyrA | CAAGGTAATGCTCCAGGCATTGCT | CGAAGTTACCCTGACCGTCTAC |
| OxyR | TGATCTTGAGTACCTGGTGGCA | GGTGAACAACACTTTACGGCT |
| σE | ATGAGCGAGCAGTTAACGGAC | CCAGACTCGCCACTTTATGC |
| degP | GCTCTGAGTTTAGGTTTGGCG | CTGGAAATTACGCGGCATACG |
| 16s rRNA | AGTACGGCCGCAAGGTTAAA | CGCTTCTCTTTGTATGCGCC |
| tolB | GCAGGCATTACGAGTAGCATTTG | CGAGCGCGATCTAACGGA |
| pEGFP-N1 | CATGGTGAGCAAGGGCGAGGAG | CCGGCGGCGGTCACGAACTC |
2.4.2.5. Investigation of horizontal gene transfer capability of OMVs. To assess the HGT capability of OMVs between different bacterial species and within the same species, we conducted experiments using Lactobacillus acidophilus and E. coli MTCC 40 bacteria. The experimental procedures were as follows: OMVs obtained from E. coli DH5α bacteria carrying the pEGFP-N1 plasmids were incubated with Lactobacillus acidophilus (Gram-positive bacterium) and E. coli MTCC 40 (Gram-negative bacterium) separately overnight. For this, the bacterial cultures were exposed to two distinct growth conditions. These conditions involved exposing the bacterial culture to OMVs derived from different sources, includes transformed E. coli DH5α carrying the pEGFP-N1 plasmid, and transformed E. coli DH5α carrying pEGFP-N1 that had been exposed to PBNPs at a concentration of 100 μg mL−1 for multiple generations. Subsequently, both the Lactobacillus and E. coli MTCC 40 cultures were assessed for kanamycin resistance in MRS agar and LB agar media containing kanamycin (50 μg mL−1), respectively. Colony-forming units (CFU) were quantified, and images were captured using a gel doc system. Additionally, the presence of the pEGFP-N1 plasmid within the colonies was confirmed by isolation of the plasmids using alkaline lysis in ten distinctly selected colonies. The standard alkaline lysis protocol is given in the ESI (SI 3†).
2.5. Basic assessments investigating the effects of PBNPs on bacterial growth kinetics, morphology, ROS/antioxidant levels, and gene expressions linked to oxidative stress and OMV secretion
where sample Abs is the absorbance of the sample, blank Abs is the absorbance of the blank, and control Abs is the absorbance of the control.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio. A drop of this suspended bacterial solution was placed on a coverslip coated with poly-D-lysine, and the coverslip was allowed to incubate at room temperature for one hour. The coverslip was rinsed by applying PBS for a duration of 2 min, after which the excess PBS was carefully removed using filter paper. This rinsing process was repeated three times. Subsequently, the bacterial sample was fixed using a mixture containing 2.5% glutaraldehyde and 2% paraformaldehyde, and it was incubated at room temperature for 60 min. Following fixation, the sample was washed thrice for 2 min each with 0.1 M Na-cacodylate buffer. To prepare the sample for imaging, a gradual dehydration process was initiated using different ethanol concentrations, including 25%, 50%, 75%, 95%, and ultimately 100%, each step lasting for 5 min. The sample was further dehydrated using a mixture of HMDS (hexamethyldisilazane) and 100% ethanol in a 1
10 ratio. A drop of this suspended bacterial solution was placed on a coverslip coated with poly-D-lysine, and the coverslip was allowed to incubate at room temperature for one hour. The coverslip was rinsed by applying PBS for a duration of 2 min, after which the excess PBS was carefully removed using filter paper. This rinsing process was repeated three times. Subsequently, the bacterial sample was fixed using a mixture containing 2.5% glutaraldehyde and 2% paraformaldehyde, and it was incubated at room temperature for 60 min. Following fixation, the sample was washed thrice for 2 min each with 0.1 M Na-cacodylate buffer. To prepare the sample for imaging, a gradual dehydration process was initiated using different ethanol concentrations, including 25%, 50%, 75%, 95%, and ultimately 100%, each step lasting for 5 min. The sample was further dehydrated using a mixture of HMDS (hexamethyldisilazane) and 100% ethanol in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio for a duration of 15 min. Finally, complete dehydration was achieved through exposure to 100% HMDS for 15 min. Any excess solution was carefully removed, and the sample was placed in a vacuum jar to allow thorough drying. Subsequent imaging was performed using a JEOL JSM-7610 Plus electron microscope. The size distribution was determined by measuring the size of 200 bacteria, and a curve was generated using the Origin software.
1 ratio for a duration of 15 min. Finally, complete dehydration was achieved through exposure to 100% HMDS for 15 min. Any excess solution was carefully removed, and the sample was placed in a vacuum jar to allow thorough drying. Subsequent imaging was performed using a JEOL JSM-7610 Plus electron microscope. The size distribution was determined by measuring the size of 200 bacteria, and a curve was generated using the Origin software.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min at 4 °C, and the uppermost layer was carefully transferred to a fresh tube. To this solution, an equal volume of isopropanol was added, and the mixture was stored at −20 °C for 2 h before undergoing another centrifugation step at 15
000g for 15 min at 4 °C, and the uppermost layer was carefully transferred to a fresh tube. To this solution, an equal volume of isopropanol was added, and the mixture was stored at −20 °C for 2 h before undergoing another centrifugation step at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min at 4 °C. The supernatant was discarded, and the RNA pellet was washed twice with 75% chilled ethanol. Following these washes, the tube was left to dry for 20 min, and the RNA pellet was resuspended in nuclease-free water. The extracted RNA was then converted into cDNA using a kit obtained from Thermo Fisher Scientific, following the manufacturer's instructions. The resulting cDNA was quantified using a NanoQuant Plate Tecan Infinite 200 PRO instrument. Equal amounts of cDNA from both the control and treated groups (100 ng) were employed for semi-quantitative PCR to assess the expression levels of the target gene (Table 1). The semi-quantitative PCR protocol involved 28 cycles per reaction, and the primers for the target gene were designed using the NCBI database and Gene Runner software. In this study, we assessed the expression of gyrA (a housekeeping gene), oxyR, σE, degP, 16s rRNA, and tolB. The fold expression of each gene relative to the control was determined by dividing the band intensity of the treated group by the band intensity of the control.
000g for 15 min at 4 °C. The supernatant was discarded, and the RNA pellet was washed twice with 75% chilled ethanol. Following these washes, the tube was left to dry for 20 min, and the RNA pellet was resuspended in nuclease-free water. The extracted RNA was then converted into cDNA using a kit obtained from Thermo Fisher Scientific, following the manufacturer's instructions. The resulting cDNA was quantified using a NanoQuant Plate Tecan Infinite 200 PRO instrument. Equal amounts of cDNA from both the control and treated groups (100 ng) were employed for semi-quantitative PCR to assess the expression levels of the target gene (Table 1). The semi-quantitative PCR protocol involved 28 cycles per reaction, and the primers for the target gene were designed using the NCBI database and Gene Runner software. In this study, we assessed the expression of gyrA (a housekeeping gene), oxyR, σE, degP, 16s rRNA, and tolB. The fold expression of each gene relative to the control was determined by dividing the band intensity of the treated group by the band intensity of the control.
2.6. Statistical analysis
All experiments were performed in triplicate and repeated twice. The results are presented as mean ± SD, and the data were analysed by t-test or one-way ANOVA followed by Tukey's post hoc test. A p value < 0.05 was considered statistically significant.3. Results and discussion
3.1. Synthesis and characterization of PET bottle-derived nanoplastics (PBNPs)
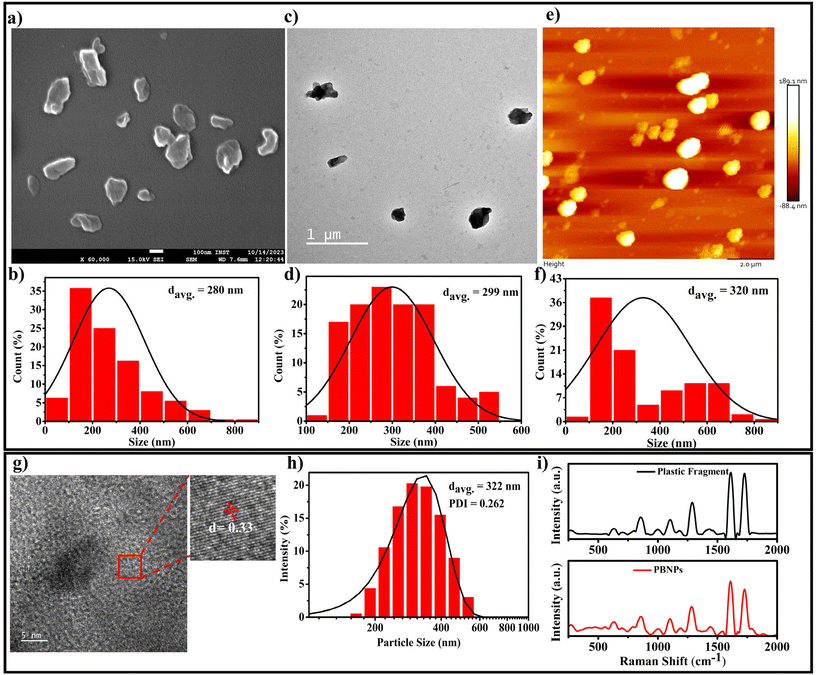
Considering that single-use plastic bottles and containers are the main source of MPs/NPs, causing the menace of plastic pollution, which has become a global crisis, we aimed to synthesise the NPs using these primary sources, i.e., plastic bottles composed of PET. Thus, the PBNPs used in this study were synthesized from fragments of these plastic bottles through the oil-in-water nano-emulsion technique, a method recently proposed for the production of polyethylene and polypropylene microplastic.33 The natural degradation of plastics into nanoplastics in the environment is an extremely slow process, taking years or even decades due to environmental factors such as UV radiation, mechanical abrasion, and microbial action. Various methods are available for the synthesis of nanoplastics, including mechanical grinding,34 ultrasonication,35 and laser or thermal degradation.34,36 However, these approaches often result in inconsistent particle sizes and contamination risks.35–38 In contrast, the oil-in-water nano-emulsion technique offers enhanced control of the particle size and lower energy consumption, while minimizing the contamination risks.36,38 Additionally, it provides a reliable and accelerated approach for the production of nanoplastics from PET fragments, closely mimicking that found in polluted ecosystems, while enabling timely and controlled laboratory experiments. In our procedure, we initially assessed the solubility of the ground plastic bottle fragments in various organic solvents, namely acetone, toluene and hexafluoroisopropanol (HFIP). Among them, HFIP proved to be the most effective in dissolving the plastic fragments. Therefore, we chose HFIP as the solvent for dissolving the plastic bottle fragments. To synthesize the PBNPs, we combined the solubilized plastic with toluene in a specific ratio. Then, this mixture was heated at 100 °C for 1 h until the plastic fragments were dissolved completely. Subsequently, a boiling solution of sodium cholate was added as a surfactant to stabilize the emulsion and disperse the plastic in the aqueous phase. To homogenize the two-phase system, we used a disperser (Ultra-Turrax T25, IKA) to create plastic droplets dispersed in the aqueous phase, which were subjected to ultrasonication to form nanoplastics (PBNPs). The mixture was rapidly cooled on an ice-water bath to induce the gelation of the plastic nanodroplets. Subsequently, toluene was removed via rotary evaporation, and the PBNPs were collected through centrifugation. To avoid the further aggregation of PBNPs during centrifugation and maintain a suitable particle dispersion, we utilized BSA protein solution as dispersant. Following this, the PBNP pellet was washed with ultrapure water, and subsequently resuspended in autoclaved ultrapure water for use in further experiments.The synthesis yield, calculated as the mean weight across three batches, was found to be 15% ± 2%. Thereafter, the as-synthesized PBNPs were subjected to characterization. The average hydrodynamic diameter of the synthesized PBNPs, as assessed by the dynamic light scattering (DLS) technique, was found to be 322 nm, with a polydispersity index (PDI) of 0.262 (Fig. 3h). The average hydrodynamic diameter signifies the effective size of these PBNPs when they are suspended in a liquid medium, with a size distribution ranging from 120 to 600 nm.39 Electron microscopy (SEM), transmission electron microscopy (TEM) and atomic force microscopy (AFM) were employed to further investigate the core sizes of the PBNPs. Notably, these particles exhibited an average size of 280 nm in the SEM image (Fig. 3a), with a size distribution in the range of 50 to 850 nm (Fig. 3b). In the TEM image, the average size was found to be nearly consistent with the SEM data, measuring 299 nm (Fig. 3c) and exhibiting a size distribution in the range of 100 to 550 nm (Fig. 3d). In the AFM image, the average size was 320 nm (Fig. 3e) with a size distribution in the range of 50 to 850 nm (Fig. 3f). It is important to note that the observations in SEM, TEM and AFM indicated that the PBNPs displayed non-uniform shapes. This wide range of sizes and variations in shape suggests that the synthesis process yielded a diverse population of PBNPs due to the differences in the nucleation and growth rates during synthesis. Additionally, we examined the lattice fringe spacing of the PBNPs using high-resolution transmission electron microscopy (HRTEM), which was measured to be 0.33 nm (Fig. 3g). This fringe spacing matches the lattice spacing observed in various crystalline materials,39 indicating that the PBNPs possess a crystalline nature. This finding is consistent with a prior study that reported an almost similar fringe spacing of 0.34 nm for plastic particles.40,41 Recent studies revealed the irregular shape of nanoplastic particles in various water bodies, such as oceans and river, freshwater, and even drinking waters.42–46 Therefore, in this study, the observed non-uniformity and size variations in the synthesized nanoplastics (PBNPs) closely resemble the naturally occurring nanoplastics found in the environment. These characteristics make our PBNPs well-suited for our further experiments.
Moreover, to determine the potential chemical modifications that may occur in PBNPs during their synthesis owing to the introduction of diverse chemicals and solvents, Raman spectroscopy and FTIR analyses were performed on both the original PET plastic bottle fragments and the synthesized PBNPs. The comparison of the Raman and FTIR spectra, as shown in Fig. 3i and ESI Fig. 1,† respectively, reveals no significant shifts in the characteristic peaks47–49 between the two samples, indicating that the chemical nature of the PBNPs remained largely unchanged. Both the Raman and FTIR spectra demonstrate that the chemical composition of the PBNPs closely mirrors that of the original SUPB fragments, with no major alterations induced during the synthesis process. This confirms that the production of PBNPs preserved the integrity of the original polymer, ensuring that the nanoplastics used in further experiments maintained their environmental relevance.
3.2. PBNPs enhance horizontal gene transfer via transformation in bacteria: facilitative role of PBNPs as permeability modulators and plasmid carriers
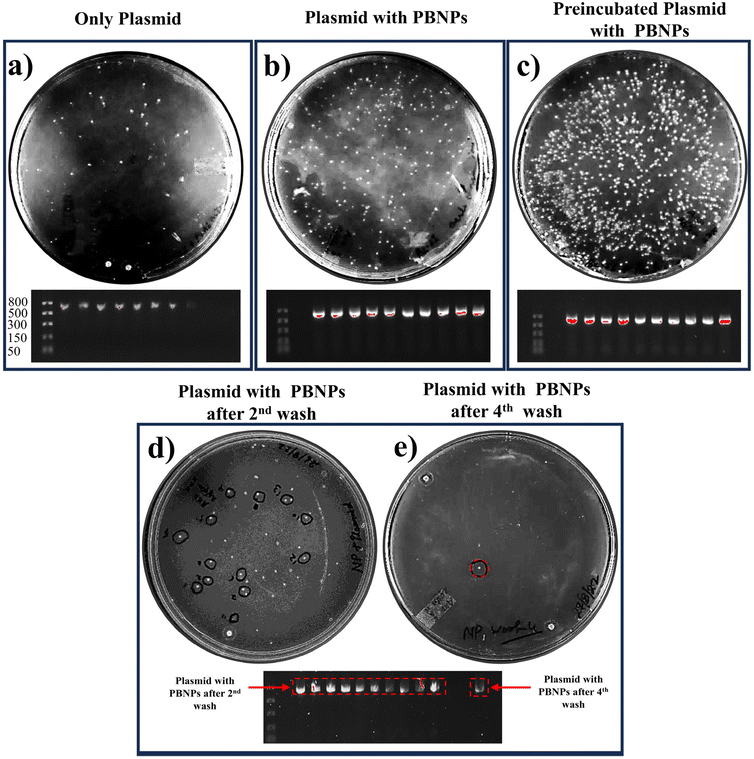
In our study, we aimed to unravel the possible involvement of nanoplastics (PBNPs) in the process of horizontal gene transfer (HGT) and its potential consequences on the propagation of AR in bacterial populations. The experimental paradigm of our study was devised to evaluate how the presence of PBNPs influenced the uptake and transfer of pEGFP-N1 plasmids within bacterial communities. Our findings yielded compelling insights into the interaction between PBNPs and plasmids in the context of HGT and antibiotic resistance. Notably, we observed a significant increase in the number of bacterial colonies, as shown in Fig. 4b, with a transformation efficiency of 2100 ± 180 CFU per μg (ESI Fig. 2†) in the presence of PBNPs (100 μg mL−1). In contrast, the cultures exposed only to plasmids had a transformation efficiency of 460 ± 80 CFU per μg. This indicates that PBNPs play a crucial role in facilitating the transfer of ARGs via plasmids. Notably, the group in which plasmids and PBNPs were pre-incubated for six hours showed the highest colony count (Fig. 4c) with a transformation efficiency of 7670 ± 610 CFU per μg (ESI Fig. 2†). This underscores the importance of prolonged interaction and strong binding between the plasmids and PBNPs, highlighting their role as permeability modulators and plasmid carriers, enhancing HGT via transformation in bacteria. The interpretation of these results raises several intriguing hypotheses. Firstly, the increased colony count in the presence of PBNPs can be attributed to the PBNPs inducing physical damage on the bacterial surface. This PBNP-induced alteration in the bacterial surface/wall was further elucidated and confirmed in the FESEM images, as depicted in Fig. 7. This damage could potentially make the cells more permeable, thereby facilitating the internalization of plasmids having ARGs, which is kanamycin resistance in our case. Secondly, the enhanced colony formation observed in the group with preincubated plasmids and PBNPs suggests that the PBNPs may have facilitated the transport of plasmids across the bacterial cell membranes. This notion is supported by the quantitative analysis of plasmid adsorption on the PBNPs. The amount of plasmid adsorbed onto the PBNPs was determined by incubating a known concentration of plasmid with the PBNPs, and then measuring the remaining plasmid concentration in the supernatant. The results showed that 12% ± 2% of the initial plasmid concentration was adsorbed onto the PBNPs. The significant adsorption of plasmid onto the PBNPs indicates that the nanoparticles can act as carriers, facilitating the transport of plasmids across the bacterial cell membrane. The prolonged interaction between the plasmid and PBNPs may allow stronger binding, promoting more efficient plasmid translocation. The strength of binding between PBNPs and plasmids was tested through multiple washing cycles. Remarkably, even after two rigorous washes, PBNPs retained residual plasmids, as evidenced by the continued growth of the bacterial colonies on the kanamycin-LB agar plates (Fig. 4d). However, after the fourth wash cycle, only one colony remained, indicating a gradually lost but still persisting plasmid attachment (Fig. 4e). These findings were further validated by the presence of the pEGFP-N1 plasmid in the PCR analysis of the corresponding colonies grown on the kanamycin-LB agar plates, as shown in Fig. 4. Thus, these findings suggest that nanoplastics may play a role in helping bacteria share genes, including ARGs.The 100 μg mL−1 dose used in this experiment was selected based on a prior dose–response screening (discussed later in Section 3.4). This preliminary screening indicated that 100 μg mL−1 elicited significant biological responses, including increased ROS levels and membrane damage, which are linked to enhanced bacterial transformation.50 Therefore, while we presented the HGT data first, where the decision to use 100 μg mL−1 was based on this screening.
Our results are in close agreement with another study demonstrating the greater ability of 100 nm polystyrene nanoparticles (PSNPs) in horizontal plasmid transfer. This study suggested that the small size of these nanoparticles might enhance the membrane permeability, facilitating plasmid penetration;51 however, the study was inconclusive about the significant plasmid adhesion to polystyrene nanoparticles and their binding strength. It is also worth noting that they utilized the readily available commercial monodispersed form of polystyrene nanoparticles, while we aimed to replicate the more natural process of nanoplastic formation from abandoned SUPB, which when introduced into aquatic environments, release various chemicals and plasticizers that create tiny cavities or openings on the plastic surface, could potentially serving as sites for plasmid adhesion and aiding their transport. Thus, our results have important implications for understanding how naturally occurring abandoned plastic bottles generate nanoplastics, which aid in the spread of AR in different environments and their potential impact on public health.
3.3. PBNPs enhance horizontal gene transfer via increased secretion of OMVs, a non-canonical way of propagating antibiotic resistance in bacteria
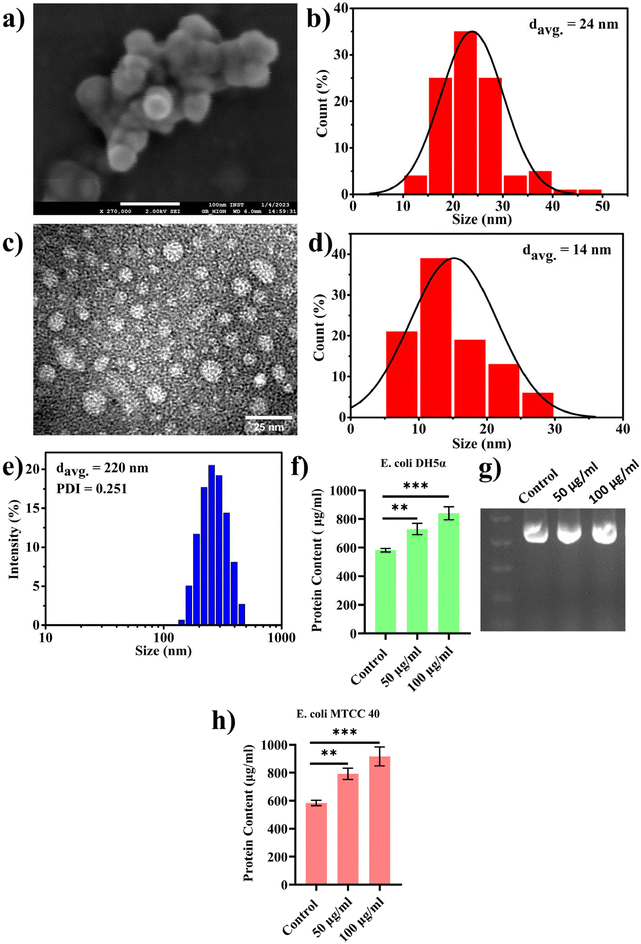
In the previous section, we presented our findings on the direct involvement of nanoplastics (PBNPs) in facilitating horizontal gene transfer (HGT), emphasizing their role in transferring ARGs among bacterial population via transformation. Together with this, we also explored the effects of PBNPs on a recently reported non-canonical method of HGT, where bacteria, particularly Gram-negative bacteria, release genetic materials containing outer membrane vesicles (OMVs), which potentially facilitate the transfer of ARGs. OMVs facilitate genetic exchange by encapsulating and transporting DNA fragments, thereby playing a pivotal role in enhancing the bacterial diversity and adaptation by aiding cell communication, delivery of virulence factors, formation of biofilms, response to stress, acquisition of nutrients, modulation of host–pathogen interactions, and facilitation of HGT.In the pursuit of exploring this non-canonical method of transferring antibiotic resistance in real-world situations in both the environment and human gut, where microorganisms may interact with NPs for longer periods spanning several generations, we cultured transformed E. coli DH5α cells inserted with pEGFP-N1 (a kanamycin-resistant plasmid) for multiple generations with exposure to PBNPs at concentrations of 50 and 100 μg mL−1 for 10 consecutive days. This approach ensured sustained exposure and allowed the bacterial population to undergo multiple generations in the presence of PBNPs. Subsequently, we isolated the OMVs using the PEG precipitation method and characterized them for their size and morphology by TEM, FESEM and DLS. The micrographs from TEM and FESEM (Fig. 5a and c) showed that these vesicles were intact nanosized structures with a spherical morphology, and the size of these nanovesicles ranged from 10 to 50 nm in diameter (Fig. 5b and d). The average hydrodynamic size determined by DLS was 220 nm with a polydispersity index of 0.251 (Fig. 5e). Further, to assess the impact of PBNP exposure on OMV secretions, OMVs were quantified based on their protein contents using the Lowry method to determine the effects of the test concentrations of PBNPs on the secretion of OMVs. Our results revealed a significant dose-dependent increase in the protein content in the OMVs isolated from the E. coli DH5α bacteria exposed to 50 and 100 μg mL−1 PBNPs compared with those isolated from the unexposed control (Fig. 5f). This indicates the increased secretion of OMVs from E. coli DH5α cells under the exposure of 50 and 100 μg mL−1 PBNPs. This finding strongly suggests that PBNPs have a stimulating effect on E. coli DH5α for OMV secretion. For the further validation of this PBNP-induced increase in OMV secretion, a similar experiment was conducted on another E. coli strain (i.e. MTCC 40) exposed to the same test concentrations of PBNPs (50 and 100 μg mL−1). Similar results of the Lowry's test further validated that the stimulating effect of PBNPs on OMV secretion is not limited to a single E. coli strain (Fig. 5h). Further, we validated the presence of the pEGFP-N1 plasmid in the secreted OMVs through PCR analysis, as shown in Fig. 5g. Our investigation conclusively detected the plasmid within the OMVs, providing compelling evidence that these vesicles were derived from the transformed E. coli DH5α carrying the pEGFP-N1 plasmid.
Our next aim was to assess the HGT capability of OMVs between different bacterial species and within the same species. For this, we tested the potential of OMVs isolated from transformed-E. coli DH5α in facilitating the transfer of ARG to Lactobacillus acidophilus, which is probiotic bacteria found in the human gut. To conduct this investigation, we isolated OMVs from the transformed E. coli DH5α carrying the pEGFP-N1 plasmid containing the gene responsible for ARGs. Subsequently, these OMVs were utilized to assess their effectiveness in transferring the kanamycin resistance to Lactobacillus acidophilus. For this, Lactobacillus acidophilus cultures were separately exposed to OMVs derived from the transformed E. coli carrying the pEGFP-N1 plasmid both in the absence and exposed to 100 μg mL−1 concentration of PBNPs for multiple generations. The 100 μg mL−1 dose was chosen based on a dose–response screening (discussed in Sections 3.3 and 3.4), which revealed that this concentration induced significant biological effects, including elevated ROS levels, membrane damage (Section 3.4), and enhanced OMV secretion (Section 3.3). Moreover, this dose was anticipated to produce more rapid and pronounced effects on the bacterial populations compared to lower concentrations.
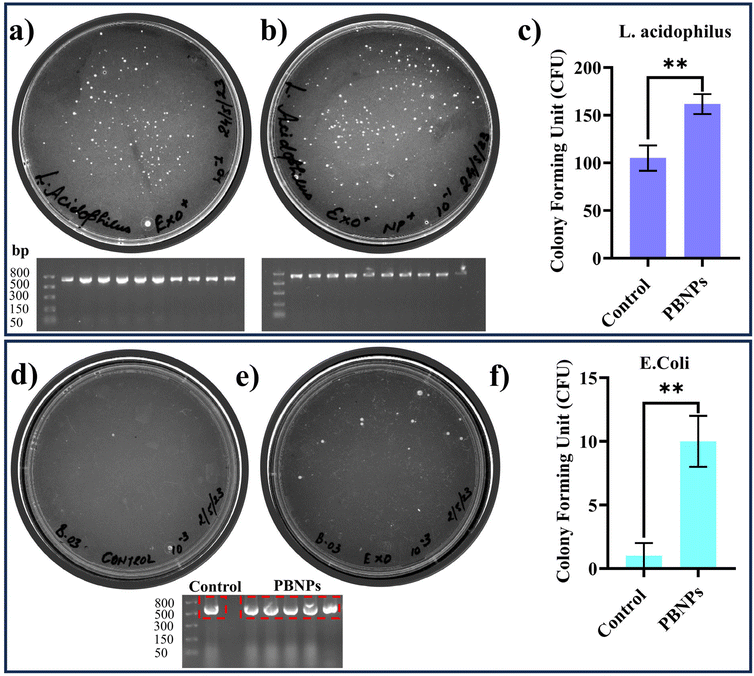
After overnight incubation, the cultures were plated on separate kanamycin-MRS agar plates, where the growth of the colonies confirmed the transfer of pEGFP-N1, i.e. kanamycin resistance from OMVs to L. acidophilus, while in the absence of transfer the bacteria would fail to grow on the kanamycin-MRS agar plates. The results, as depicted in Fig. 6a–c, demonstrated a significant increase in the number of kanamycin-resistant colonies of L. acidophilus incubated with OMVs isolated from the PBNP-exposed transformed E. coli carrying pEGFP-N1 plasmid (Fig. 6b and c) in comparison to the L. acidophilus exposed to OMVs isolated from the transformed E. coli carrying pEGFP-N1 (Fig. 6a and c). To further confirm the transfer of pEGFP-N1 plasmid to L. acidophilus, we conducted semi-quantitative PCR analysis of 10 randomly selected colonies from the kanamycin-MRS agar plate. The results showed the presence of the pEGFP-N1 plasmid in all 10 colonies, confirming the successful transfer of the pEGFP-N1 plasmids via OMVs (Fig. 6a and b). Moreover, a similar experiment was carried out on E. coli, consistently producing equivalent outcomes. The findings from this parallel investigation on E. coli, as depicted in Fig. 6d–f, mirrored that observed with L. acidophilus. Specifically, there was a visible increase in the number of kanamycin-resistant colonies when OMVs from 100 μg mL−1 PBNP-exposed transformed E. coli carrying the pEGFP-N1 plasmids were employed in contrast to OMVs from PBNP-unexposed transformed E. coli with pEGFP-N1. This concordance in results across different bacterial species further supports the efficacy and reproducibility of the observed HGT mediated by OMVs under the influence of PBNPs. These findings strongly suggest that PBNPs enhance the propagation of the ARGs via OMVs among the same and different species of bacteria. Additionally, the transfer of ARGs to Lactobacillus also raises concerns about human health given that these gut-dwelling bacteria can serve as reservoirs, transferring these genes to other harmful bacteria and making them resistant to respective antibiotics. This corroborates our hypothesis that the intervention of nanoplastics may more readily spread ARGs among gut bacteria, leading to the emergence of multi-drug resistant strains. This poses a direct threat to human health by compromising our primary defence against bacterial infections. As antibiotics lose their effectiveness, infections may persist or worsen, making individuals more susceptible to various diseases. Essentially, the accelerated spread of AR within the gut microbiota, facilitated by nanoplastics, may create a scenario where once-treatable common infections become challenging to manage. This can result in prolonged illnesses, increased healthcare costs, and heightened public health concerns. Thus, this study urges us to consider the broader implications of ARG transfer, not only in environmental settings but also within the human body. It underscores the need for responsible environmental practices and judicious antibiotic use to safeguard human health and the integrity of the gut microbiome together with the entire ecosystem.
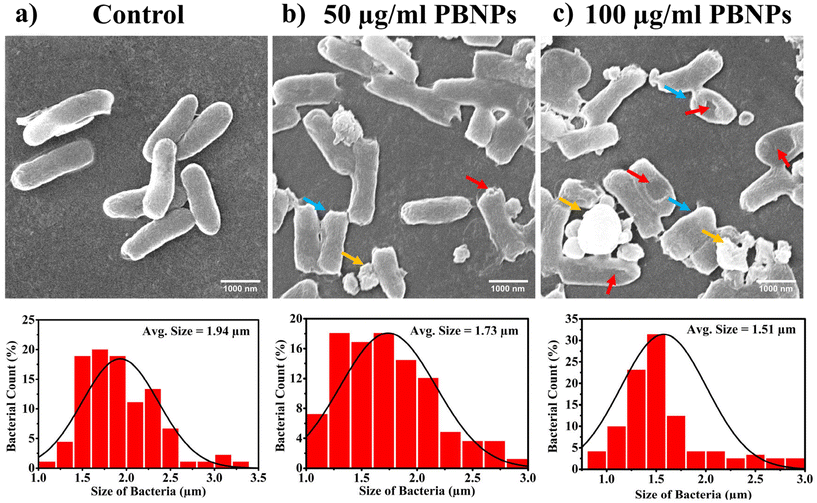
3.4. PBNPs exposure induces alterations in bacterial growth, morphology, ROS/antioxidant levels, and gene expressions linked to oxidative stress and OMV secretion
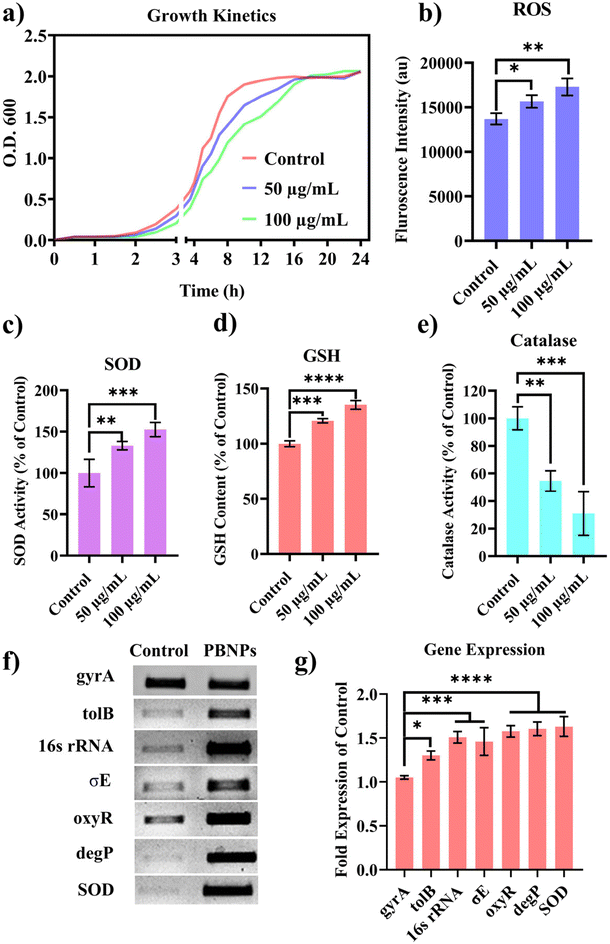
Following the confirmation of the impact of PBNPs on HGT via OMVs, we extended our investigation to their potential effects on bacterial growth, morphology, ROS/antioxidant levels, and the expression of genes involved in oxidative stress and OMV secretion. Firstly, we investigated the impact of the PBNP exposure on the growth kinetics of E. coli by monitoring the optical density (OD) of the bacterial cultures at defined intervals over a period of 24 h and plotted the recorded OD values against the respective time intervals of measurement. The resulting plot, as depicted in Fig. 7a, shows a significant delay in the log phase of bacteria exposed to PBNPs in a dose-dependent manner compared to the unexposed control group. This graph also shows that the duration of the lag phase spanned approximately 8 h (from 2 to 10 h) in the control group, whereas it increased to 12 h (from 2 to 14 h) and to 16 h (from 2.5 to 18.5 hours) with exposure to 50 and 100 μg mL−1 PBNPs, respectively. These findings suggest that PBNPs may induce alterations in the division cycle of the bacteria, consequently resulting in growth delays in the PBNP-exposed bacterial culture compared to the unexposed control. To further assess the impact of PBNP exposure on the size and morphology of the bacteria, the control and treated bacteria were examined under FESEM, which discerned notable morphological alterations, including diminished size appearance of wrinkles on the bacterial surface in the 50 μg mL−1 PBNP-exposed group (Fig. 8b), while a diminished size and the appearance of wrinkle and formation of pits were also observed on the bacterial surface in the 100 μg mL−1 PBNP-exposed group (Fig. 8c). In contrast, the morphology of the control bacteria remained unaffected, displaying integrity, as shown in Fig. 8a. The average length of the control E. coli was measured to be 1.94 μm (Fig. 8c), whereas the length of E. coli exposed to 50 and 100 μg mL−1 PBNPs was significantly reduced to 1.73 and 1.51 μm, respectively. Notably, in the presence of 100 μg mL−1 PBNPs, the size of 90% bacteria was observed to be smaller than the average size of the control bacteria. Though the most of studies done thus far considered plastic to be biochemically inert materials that do not interact with biomolecules,52 in our case, the smaller size and irregular morphology of the bacteria exposed PBNPs may be attributed to this. Due to the smaller size, PBNPs may penetrate the bacterial cell wall and disrupt its integrity, leading to cellular damage and potentially affecting the viability of the bacteria, resulting in a reduction in their size and delayed growth. Moreover, it is generally considered that particles having irregular shapes may possess edges that are more likely to cause damage to cellular surfaces,53,54 and thus the PBNPs having irregular shapes would have exerted mechanical force on the bacterial cells, contributing to the deformations in the bacteria. Moreover, the internal components of the bacterial cell, such as the cytoskeleton, could have been affected by this mechanical stress, potentially causing changes in the overall size of the bacteria. It has also been reported that mechanical stress caused due to exposure to particles having a small size and irregular shapes may induce ROS generation, and also alter the antioxidant defence mechanism in bacteria;55–57 therefore, the delayed growth and the altered morphology would have been induced by the increased levels of ROS or vice versa. Additionally, the involvement of ROS has also been reported in the secretion of OMVs.22,58,59 These deliberations prompted us to explore the influence of PBNPs on bacterial oxidative stress and the stress-busting processes.58 Our investigation began by assessing the levels of ROS together with the key molecules in the anti-oxidative process, such as superoxide dismutase (SOD), glutathione (GSH) and catalase. The examination of the ROS level serves as a crucial starting point, given that these species have been established as key players in the intricate network governing OMV secretion. After subjecting bacteria to multigenerational exposure of PBNP at concentrations of 50 and 100 μg mL−1, a noteworthy concentration-dependent increase in the ROS level was observed compared to the control (Fig. 7b). Furthermore, the SOD activity demonstrated a 34% and 53% elevation in the presence of 50 and 100 μg mL−1 concentrations of PBNPs, respectively, in comparison to the unexposed control (Fig. 7c). Similarly, the levels of glutathione (GSH) exhibited a 20% and 35% increase at 50 and 100 μg mL−1 of PBNP exposure, respectively (Fig. 7d). The increased SOD activity and elevated GSH levels likely signify adaptive responses to the increased ROS levels within the bacterial cells. It has been reported that in response to the surge in ROS, the cells activate their antioxidant defence mechanisms, leading to the upregulation of SOD and enhanced synthesis of GSH. However, the exposure of PBNPs at both 50 and 100 μg mL−1 concentrations notably decreased the catalase (CAT) activity to 46% and 71%, respectively, compared to the control (Fig. 7e). This reduction in catalase activity may be attributed to the potential induction of SOD activity by PBNPs. This induction may lead to the excessive production of hydrogen peroxide (H2O2), and consequently it may impede the CAT activity in this process. These findings are consistent with the previous studies by Liu et al.63 and Shang et al.,60 where a similar pattern of increased SOD activity inhibiting CAT activity was reported. To strengthen our findings, we studied how exposure to PBNPs (at a concentration of 100 μg mL−1) affects the expression of genes related to oxidative stress and the secretion of outer membrane vesicles (OMVs). Firstly, we investigated the oxyR gene, which is responsible for protecting cells from oxidative stress and regulating the production of antioxidant proteins. Our results showed a 1.5-fold increase in oxyR gene expression, confirming that exposure to PBNPs induces oxidative stress (Fig. 7f and g). Additionally, we examined the SOD gene, which aids in combating oxidative stress. In the presence of PBNPs, the expression of the SOD gene increased by 1.55-fold compared to the control (Fig. 7f and g). These findings are consistent with our biochemical assay data, which demonstrated an increase in ROS production and SOD activity (Fig. 7b and c), respectively. Moving forward, we explored the genes associated with OMV secretion, such as σE (sigma E) and tolB.61,62 The σE gene, also known as rpoE, regulates genes involved in OMV biogenesis and release, contributing to cell envelope stability. We observed a 1.55-fold increase in σE gene expression in the presence of 100 μg mL−1 PBNPs (Fig. 7f and g). Similarly, we noted an increase in the expression of the tolB gene (1.25-fold rise) when PBNPs were present. tolB is a key component of the Tol–Pal system in bacteria, which is crucial for the synthesis and secretion of OMVs (Fig. 7f and g). The increased expression of σE and tolB genes also confirmed the enhanced OMV secretion in the presence of PBNPs. In addition to these genes related to OMV secretion, we also investigated the expression of genes crucial for essential cellular functions such as protein synthesis and quality control, including 16S rRNA and degP.61,62 In response to oxidative stress during OMV release, bacteria upregulate the expression of components involved in protein synthesis, such as 16S rRNA. In the presence of PBNPs, we observed a significant 1.44-fold increase in 16S rRNA expression (Fig. 7f and g), indicating its significant role in protein synthesis during OMV release and oxidative stress. To cope with oxidative stress and protein misfolding during OMV secretion, bacteria enhance their ribosome production capacity. The degP gene, encoding the protease/chaperones degP responsible for proper protein folding, showed a 1.78-fold increase in expression compared to the control (Fig. 7f and g), respectively. This elevated expression of degP is likely a response to the increased oxidative stress induced by PBNPs, amplifying protein misfolding, and reflects the bacteria effort to manage this stress.Overall, the observed increase in the bacterial gene expressions, including oxyR, SOD, σE, tolB, 16S rRNA, and degP, can be directly linked to the exposure to PBNPs. The elevated expressions of the oxyR and SOD genes indicate that PBNPs induce oxidative stress within the E. coli cells. This oxidative stress triggers a cascade of responses, leading to an upregulation in the expression of the genes associated with OMV secretion, such as σE and tolB. The increased expression of 16S rRNA suggests the heightened involvement of ribosomes in protein synthesis during OMV release and oxidative stress, likely as a response to cope with the challenges posed by PBNP exposure. Furthermore, the heightened expression of degP can be justified as a mechanism to address the increased oxidative stress, which amplifies protein misfolding. The changes in the gene expression patterns provide valuable insights into the bacterial responses triggered by PBNP exposure, shedding light on the underlying molecular mechanisms for oxidative stress-driven OMV secretion.
4. Conclusion
This study revealed the multifaceted impact of nanoplastic particles derived from plastic bottles on bacterial cells, highlighting their potential environmental and public health consequences. Nanoplastic pollution, particularly from waste plastic bottles, is a significant global concern. This research provides insights into the environmental implications of nanoplastics, focusing on horizontal gene transfer (HGT) and antibiotic resistance (AR) propagation.By synthesizing nanoplastics (PBNPs) from waste PET bottles, this study closely mimicked naturally occurring aquatic nanoplastics. The characterization showed the formation of diverse, non-uniform, and crystalline PBNPs. The investigation found that PBNPs enhanced the transfer of antibiotic resistance genes (ARGs) via plasmids in bacteria, potentially by damaging the bacterial surfaces and facilitating plasmid translocation through transformation and outer membrane vesicle (OMV) secretion. Also, we explored OMV secretion, a lesser-known HGT method, revealing that PBNPs stimulated OMV secretion, leading to enhanced ARG transfer between bacteria. Further examination showed that PBNP exposure affected the bacterial growth, morphology, and oxidative stress, indicating a link to increased OMV secretion and HGT. The molecular analysis revealed changes in gene expressions related to oxidative stress, membrane integrity and protein synthesis, suggesting a cellular response to the PBNPs-induced stress. The broader implications of nanoplastic pollution include increased risk of ARG transfer in the human gut, emphasizing the need for discriminate usage of plastics and antibiotics. This study underscores the potential of nanoplastics to exacerbate antibiotic resistance by facilitating genetic material exchange among bacteria, posing threats to environmental stability and human health.
Future studies should focus on investigating the chronic impact of nanoplastics exposure on bacterial communities and human health, examining the impact of nanoplastics in diverse environments including soil and air, and delving deeper into the molecular mechanisms by which nanoplastics influence HGT and AR propagation. Additionally, developing methods to reduce nanoplastic pollution and its associated risks, as well as studying the potential effects of nanoplastics on the human gut-microbiota and the resulting health outcomes will be crucial. As plastic pollution increases, this research is essential for developing strategies to address the ecological and health impacts of nanoplastic contamination. This study enhances our understanding of how nanoplastics may intensify the challenges related to the antibiotic resistance, and paves the way for future investigations into their broader impacts on microbial communities and ecosystem stability.
Author contributions
PS and AK carried out all experiments and interpretations and prepared the manuscript draft. MS initially conceptualized the idea, supervised the experiments, analysed the data and then reviewed and finalized the manuscript. All the authors read and approved the final manuscript.Data availability
Data can be provided upon request.Conflicts of interest
Authors declare that they do not have any conflict of interest.Acknowledgements
Prashant Sharma is grateful to the University Grants Commission (UGC), India, for fellowship 1013/[CSIR-UGC NET, JUNE 2019]. Abhinoy Kishore is thankful to the Institute of Nanoscience and Technology for providing financial support. MS thankfully acknowledges INST Mohali for providing necessary infrastructure and lab facilities.References
- T. R. Walker and L. Fequet, TrAC, Trends Anal. Chem., 2023, 160, 116984 CrossRef CAS.
- C. Chinglenthoiba, B. Pukhrambam, K. T. Chanu, K. S. Devi, N. J. Meitei, Y. Devika and S. Valiyaveettil, Reg. Stud. Mar. Sci., 2023, 58, 102777 Search PubMed.
- K. L. Law and R. C. Thompson, Science, 2014, 345, 144–145 CrossRef CAS PubMed.
- J. M. M. Levy, C. G. Towers and A. Thorburn, Nat. Rev. Cancer, 2017, 17, 528–542 CrossRef CAS PubMed.
- J. R. Bermúdez and P. W. Swarzenski, MethodsX, 2021, 8, 101516 CrossRef PubMed.
- M. Bergmann, S. Mützel, S. Primpke, M. B. Tekman, J. Trachsel and G. Gerdts, Sci. Adv., 2019, 5, eaax1157 CrossRef CAS PubMed.
- H. Lee, A. Kunz, W. J. Shim and B. A. Walther, Sci. Rep., 2019, 9, 10145 CrossRef PubMed.
- K. Mallick, A. Sahu, N. K. Dubey and A. P. Das, J. Environ. Manage., 2023, 348, 119371 CrossRef CAS PubMed.
- X. Dong, X. Liu, Q. Hou and Z. Wang, Sci. Total Environ., 2023, 855, 158686 CrossRef CAS PubMed.
- R. P. Singh, S. Mishra and A. P. Das, Chemosphere, 2020, 257, 127199 CrossRef CAS PubMed.
- E. R. Zettler, T. J. Mincer and L. A. Amaral-Zettler, Environ. Sci. Technol., 2013, 47, 7137–7146 CrossRef CAS PubMed.
- S. Oberbeckmann, M. G. Loeder, G. Gerdts and A. M. Osborn, FEMS Microbiol. Ecol., 2014, 90, 478–492 CrossRef CAS PubMed.
- M. Arias-Andres, U. Klümper, K. Rojas-Jimenez and H.-P. Grossart, Environ. Pollut., 2018, 237, 253–261 CrossRef CAS PubMed.
- C. Smillie, M. P. Garcillán-Barcia, M. V. Francia, E. P. C. Rocha and F. De La Cruz, Microbiol. Mol. Biol. Rev., 2010, 74, 434–452 CrossRef CAS PubMed.
- N. K. Devanga Ragupathi, D. P. Muthuirulandi Sethuvel, R. Gajendran, S. Anandan, K. Walia and B. Veeraraghavan, Curr. Microbiol., 2019, 76, 666–672 CrossRef CAS PubMed.
- D. N. Pham, L. Clark and M. Li, J. Hazard. Mater. Lett., 2021, 2, 100014 CrossRef CAS.
- R. Ranjbar and M. Alam, Evidence-Based Nurs., 2023, 27(1), 16 CrossRef PubMed.
- World Bank, Drug-resistant infections: a threat to our economic future, 2007.
- J. O'Neill, Tackling drug-resistant infections globally: final report and recommendations, 2016.
- M. Juhas, Crit. Rev. Microbiol., 2015, 41, 101–108 CrossRef CAS PubMed.
- N. A. Lerminiaux and A. D. S. Cameron, Can. J. Microbiol., 2019, 65, 34–44 CrossRef CAS PubMed.
- I. A. MacDonald and M. J. Kuehn, J. Bacteriol., 2013, 195, 2971–2981 CrossRef CAS PubMed.
- L. M. Johnson, J. B. Mecham, S. A. Krovi, M. M. M. Caffaro, S. Aravamudhan, A. L. Kovach, T. R. Fennell and N. P. Mortensen, Nanoscale Adv., 2021, 3, 339–346 RSC.
- Y. Ji, C. Wang, Y. Wang, L. Fu, M. Man and L. Chen, Environ. Sci.: Nano, 2020, 7, 2313–2324 RSC.
- J. Austin, C. Minelli, D. Hamilton, M. Wywijas and H. J. Jones, J. Nanopart. Res., 2020, 22, 108 CrossRef CAS.
- S. Mishra, D. Dash and A. P. Das, Mar. Pollut. Bull., 2022, 185, 114254 CrossRef CAS PubMed.
- Bacterial Transformation, https://app.jove.com/t/5059/bacterial-transformation-the-heat-shock-method, accessed October 15, 2024.
- M. A. Rider, S. N. Hurwitz and D. G. Meckes, Sci. Rep., 2016, 6, 1–14 CrossRef PubMed.
- I. De, S. R, A. Kour, H. Wani, P. Sharma, J. J. Panda and M. Singh, Toxicol. Mech. Methods, 2021, 31, 159–168 CrossRef CAS PubMed.
- C. A. Garcia, E. S. Alcaraz, M. A. Franco and B. N. Passerini de Rossi, Front. Microbiol., 2015, 6, 926 Search PubMed.
- T. Roušar, O. Kučera, H. Lotková and Z. Červinková, Anal. Biochem., 2012, 423, 236–240 CrossRef PubMed.
- A. K. Sinha, Anal. Biochem., 1972, 47, 389–394 CrossRef CAS PubMed.
- D. Cassano, R. La Spina, J. Ponti, I. Bianchi and D. Gilliland, ACS Appl. Nano Mater., 2021, 4, 1551–1557 CrossRef CAS.
- L. A. Parker, E. M. Höppener, E. F. Van Amelrooij, S. Henke, I. M. Kooter, K. Grigoriadi, M. G. A. Nooijens, A. M. Brunner and A. Boersma, Microplast. Nanoplast., 2023, 3, 10 CrossRef.
- E. Von der Esch, M. Lanzinger, A. J. Kohles, C. Schwaferts, J. Weisser, T. Hofmann, K. Glas, M. Elsner and N. P. Ivleva, Front. Chem., 2020, 8, 169 CrossRef CAS PubMed.
- A. Galakhova, T. C. Meisel and G. Riess, Microplastics, 2024, 3, 433–448 CrossRef.
- P. Merdy, F. Delpy, A. Bonneau, S. Villain, L. Iordachescu, J. Vollertsen and Y. Lucas, Heliyon, 2023, 9(8), e18387 CrossRef CAS PubMed.
- C. J. McColley, J. A. Nason, B. J. Harper and S. L. Harper, Microplast. Nanoplast., 2023, 3, 20 CrossRef.
- M. K. Kumawat, M. Thakur, R. B. Gurung and R. Srivastava, Sci. Rep., 2017, 7, 15858 CrossRef PubMed.
- J. C. Aguilar-Guzmán, K. Bejtka, M. Fontana, E. Valsami-Jones, A. M. Villezcas, R. Vazquez-Duhalt and A. G. Rodríguez-Hernández, Microplast. Nanoplast., 2022, 2, 9 CrossRef.
- M. Ganguly and P. A. Ariya, ACS Earth Space Chem., 2019, 3, 1729–1739 CrossRef CAS.
- T. Luo, S. Moon, L. Martin, S. Kim, Q. Zhang and W. Xu, ChemRxiv., 2023 DOI:10.26434/chemrxiv-2023-375jp.
- N. Malygina, E. Mitrofanova, N. Kuryatnikova, R. Biryukov, D. Zolotov, D. Pershin and D. Chernykh, Water, 2021, 13, 2287 CrossRef.
- M. Piccardo, M. Renzi and A. Terlizzi, Mar. Pollut. Bull., 2020, 157, 111317 CrossRef CAS PubMed.
- O. S. Alimi, J. Farner Budarz, L. M. Hernandez and N. Tufenkji, Environ. Sci. Technol., 2018, 52, 1704–1724 CrossRef CAS PubMed.
- J. Zhang, M. Peng, E. Lian, L. Xia, A. G. Asimakopoulos, S. Luo and L. Wang, Environ. Sci. Technol., 2023, 57, 8365–8372 CrossRef CAS PubMed.
- M. J. Chinchillas-Chinchillas, V. M. Orozco-Carmona, C. G. Alvarado-Beltrán, J. L. Almaral-Sánchez, S. Sepulveda-Guzman, L. E. Jasso-Ramos and A. Castro-Beltrán, J. Polym. Environ., 2019, 27, 659–669 CrossRef CAS.
- F. Lionetto, M. G. Lionetto, C. Mele, C. E. Corcione, S. Bagheri, G. Udayan and A. Maffezzoli, Nanomaterials, 2022, 12, 1560 CrossRef CAS PubMed.
- H. H. Mohamed, A. A. Alsanea, N. A. Alomair, S. Akhtar and D. W. Bahnemann, Environ. Sci. Pollut. Res., 2019, 26, 12288–12301 CrossRef CAS PubMed.
- S. Zhang, Y. Wang, J. Lu, Z. Yu, H. Song, P. L. Bond and J. Guo, ISME J., 2021, 15, 2969–2985 CrossRef CAS PubMed.
- X. Hu, M. G. Waigi, B. Yang and Y. Gao, Environ. Sci. Technol., 2022, 56, 14948–14959 CrossRef CAS PubMed.
- E. L. Teuten, J. M. Saquing, D. R. Knappe, M. A. Barlaz, S. Jonsson, A. Bjorn, S. J. Rowland, R. C. Thompson, T. S. Galloway, R. Yamashita, D. Ochi, Y. Watanuki, C. Moore, P. H. Viet, T. S. Tana, M. Prudente, R. Boonyatumanond, M. P. Zakaria, K. Akkhavong, Y. Ogata, H. Hirai, S. Iwasa, K. Mizukawa, Y. Hagino, A. Imamura, M. Saha and H. Takada, Philos. Trans. R. Soc., B, 2009, 364, 2027–2045 CrossRef CAS PubMed.
- M. Österberg, M. H. Sipponen, B. D. Mattos and O. J. Rojas, Green Chem., 2020, 22, 2712–2733 RSC.
- X.-Y. Sun, J.-M. Ouyang and K. Yu, Sci. Rep., 2017, 7, 7250 CrossRef PubMed.
- M. Zhang, C. Zhang, X. Zhai, F. Luo, Y. Du and C. Yan, Sci. China Mater., 2019, 62, 1727–1739 CrossRef CAS.
- K. Krishnamoorthy, M. Veerapandian, L.-H. Zhang, K. Yun and S. J. Kim, J. Ind. Eng. Chem., 2014, 20, 3513–3517 CrossRef CAS.
- L. Peng, H. Zhu, H. Wang, Z. Guo, Q. Wu, C. Yang and H.-Y. Hu, Nat. Commun., 2023, 14, 5734 CrossRef CAS PubMed.
- B. van de Waterbeemd, G. Zomer, J. van den Ijssel, L. van Keulen, M. H. Eppink, P. van der Ley and L. A. van der Pol, PLoS One, 2013, 8, e54314 CrossRef CAS PubMed.
- S. Lekmeechai, Y.-C. Su, M. Brant, M. Alvarado-Kristensson, A. Vallström, I. Obi, A. Arnqvist and K. Riesbeck, Front. Microbiol., 2018, 9, 1837 CrossRef PubMed.
- A. H. Shang, J. Ye, D. H. Chen, X. X. Lu, H. D. Lu, C. N. Liu and L. M. Wang, J. Environ. Sci. Health, Part B, 2015, 50, 809–818 CrossRef CAS PubMed.
- A. J. McBroom and M. J. Kuehn, Mol. Microbiol., 2007, 63, 545–558 CrossRef CAS PubMed.
- A. Kulp and M. J. Kuehn, Annu. Rev. Microbiol., 2010, 64, 163–184 CrossRef CAS PubMed.
- F. Liu and S. J. Pang, J. Exp. Mar. Biol. Ecol., 2010, 382(2), 82–87 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr02613f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |