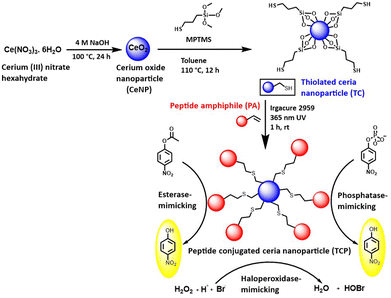
Ceria nanoparticles immobilized with self-assembling peptide for biocatalytic applications†‡
Moumita
Halder
,
Vatan
Chawla
and
Yashveer
Singh
 *
*
Department of Chemistry, Indian Institute of Technology Ropar, Rupnagar-140 001, Punjab, India. E-mail: 2018cyz0011@iitrpr.ac.in; vatan.19cyz0016@iitrpr.ac.in; yash@iitrpr.ac.in
First published on 16th August 2024
Abstract
Peptide-based artificial enzymes exhibit structure and catalytic mechanisms comparable to natural enzymes but they suffer from limited reusability due to their existence in homogenous solutions. Immobilization of self-assembling peptides on the surface of nanoparticles can be used to overcome limitations associated with artificial enzymes. A high, local density of peptides can be obtained on nanoparticles to exert cooperative or synergistic effects, resulting in an accelerated rate of reaction, distinct catalytic properties, and excellent biocompatibility. In this work, we have immobilized a branched, self-assembled, and nanofibrous catalytic peptide, (C12-SHD)2KK(Alloc)-NH2, onto thiolated ceria nanoparticles to generate a heterogeneous catalyst with an enhanced number of catalytic sites. This artificial enzyme mimics the activities of esterase, phosphatase, and haloperoxidase enzymes and the catalytic efficiency remains nearly unaltered when reused. The enzyme-mimicking property is investigated for pesticide detection, bone regeneration, and antibiofouling applications. Overall, this work presents a facile approach to develop a multifunctional heterogeneous biocatalyst that addresses the challenges associated with unstable peptide-based homogeneous catalysts and, thus, shows a strong potential for industrial applications.
Introduction
Biocatalysis refers to the use of natural catalysts, such as enzymes or whole cells, to perform chemical transformations.1 These catalysts accelerate the rate of chemical reactions by providing an alternative pathway with lower activation energy, allowing reactions to occur under milder conditions. Enzymes are proteins that act as highly efficient biocatalysts in living organisms.2 They possess specific active sites that can bind to substrates and facilitate their conversion into products. Enzymes are incredibly selective, often catalyzing specific reactions and producing specific products, which makes them valuable tools in industries.3 Biocatalysis offers several advantages over traditional chemical catalysis. Enzymes exhibit high selectivity, which minimizes the formation of by-products4 and they work under mild conditions, including lower temperatures and pressures, which can reduce energy consumption and production costs. Enzymes are also more environment friendly as they avoid the use of toxic or hazardous chemicals, and they find application in a wide range of industries, including pharmaceuticals, agriculture, food and beverage, biofuels, and manufacturing.5,6 Examples of biocatalytic reactions include the production of antibiotics, synthesis of fine chemicals, degradation of environmental pollutants, and conversion of biomass into biofuels. Overall, biocatalysis harnesses the power of biological catalysts to enable more sustainable and efficient chemical processes, with significant potential for advancing industries and reducing environmental impact.While biocatalysis offers numerous benefits, there are also several challenges associated with the use of biocatalysts. The lack of availability of these enzymes in large quantity is the major concern for the industrial use.7 Some enzymes may be derived from rare or difficult-to-cultivate organisms, making them less accessible, which significantly increases their costs. Many biocatalysts, especially enzymes, can be sensitive to environmental conditions, such as temperature and pH.8 They may undergo denaturation or loss of activity over time, which reduces their stability and shelf-life. There are issues related to the use of enzymes in the development of heterogeneous catalysts.9,10 The enzymes are attached to solid surfaces either through adsorption or covalent bonding. Adsorption-based immobilizations invariably result in a loss of adsorbed proteins during the course of subsequent cycles, whereas the covalent linkages generated through chemical processes may cause the connected enzyme to become denatured.11 As a result, it is increasingly difficult to reuse catalysts and the cost-effectiveness of process is also reduced. Artificial enzymes or compounds mimicking enzymes can be used to surmount these challenges.1,12,13 An enzyme's catalytic activity comes from supramolecular interactions that form a three-dimensional structure within the protein molecule. The correct spatial arrangement of amino acids inside these structures produces catalytic pockets needed to bind to a particular substrate and carry out chemical reactions.14 The minimalistic approach to develop artificial biocatalysts with same catalytic efficiency, stereoselectivity, and specificity as enzymes is to imitate their catalytic sites by incorporating amino acids found in catalytic pockets.
Self-assembling peptides can act as biocatalysts by organizing into well-defined structures or scaffolds that provide an environment for catalytic reactions to occur.15,16 These peptides have the unique ability to spontaneously form ordered structures through non-covalent interactions. The self-assembling peptides can be designed to incorporate catalytic residues or functional groups within their sequence, allowing them to perform specific enzymatic reactions. The catalytic activity of self-assembling peptides can be influenced by factors such as the peptide sequence, secondary structure, and the microenvironment created by the self-assembled structure.17 Proline-containing peptides, for instance, are known to catalyze Mannich, Michael, aldol, and acyl transfer processes.18,19 In a similar way, ester hydrolysis reactions are known to be catalyzed by histidine (His).20,21 For the regeneration of hydrolase and esterase activities, a variety of His-containing hydrogels and nanofibrils have been developed. Similarly, hydrogel-trapped hemin chloride has been found to have peroxidase-like activity.22 In this regard, hydrolytic enzymes, such as lipase and α-chymotrypsin, have drawn a lot of attention due to their prevalence in living things and growing economic significance.23 Therefore, much effort has been made to develop artificial esterase and phosphatase.24
Several design techniques have been used to generate peptide-based artificial enzymes. For instance, Korendovych et al. developed a set of heptapeptides that formed β-sheets, and by self-assembling these peptides, they obtained a Zn(II)-coordinated structure,25 which mimicked the catalytic center of natural carbonic anhydrase and demonstrated catalytic activity towards carbonic anhydrase substrates. Numata et al. developed an enzyme-like catalyst by incorporating the conventional serine-protease catalytic triad to peptides that can self-assemble into fibrils and resemble amyloids.26 The formation of β-sheet backbone was essential to imitate the protease binding site. Gulseren et al. reported esterase-mimicking catalytic nano-system with essential residues, Ser, His, and Asp in the peptide sequence.27 Mikolajczak et al. developed peptide-gold nanoparticle conjugates as sequential cascade catalyst for the hydrolysis followed by the hydrogenation reaction.28 Dowari et al. reported the immobilization of peptide amphiphile containing Asp(Ser)His triad on silica surface for hydrolase-mimicking properties.29 Reja et al. developed nanotubes from self-assembled lipidated short peptide (C10-FFVK) for aldolase-mimicking properties.30 Overall, self-assembling peptide provide a versatile platform for creating functional materials with catalytic capabilities and expanding the scope of biocatalysis beyond traditional enzymes.
There has been a lot of reports available in literature, where self-assembled peptides have been extensively explored to develop artificial enzymes. The major limitation associated with such system is that they are in homogenous solution with reaction mixture and cannot be removed once the reaction is over. Unbranched, linear peptides reported so far can only expose their single binding pocket to the substrate, which limits the rate of reaction. Additionally, there are very few reports on the use of artificial enzymes exhibiting multiple catalytic activities simultaneously. Developing multifunctional peptide catalysts that can perform diverse reactions or exhibit additive/synergistic effects has the potential to open new possibilities in biocatalysis.
The main aim of this work was to develop multifunctional, heterogenous catalyst with esterase, phosphatase, and haloperoxidase-mimicking activities and use them for pesticide detection (organophosphorus and carbamate), bone regeneration, and anti-biofouling applications. In this work, we have immobilized self-assembled, nanofibrous catalytic peptide on ceria nanoparticles and evaluated their potential to act as a biocatalyst. Ceria nanoparticles (CeNP) were fabricated using hydrothermal method and thiolated (TC) using (3-mercaptopropyl)trimethoxysilane, which was further conjugated to peptide amphiphile (PA) by “thiol–ene” reaction, thus, forming thioether bonds. The branched peptide amphiphile contains catalytic triad ‘Ser–His–Asp,’ which is also present in the active site of serine protease.31 These residues work together to perform various hydrolytic reactions by facilitating binding to the substrate and catalysis. This peptide was immobilized on thiolated ceria nanoparticle surface to generate a heterogenous catalyst (TCP) with a greater number of catalytic sites. CeNPs were selected due to their haloperoxidase-mimicking properties, which involves catalyzing the oxidation of halide ions by hydrogen peroxide.32 Peptide conjugated ceria nanoparticles were thoroughly characterized and they were found to mimic esterase, phosphatase, and haloperoxidase enzymes. This enzyme-mimicking activities were used to elucidate their role in pesticide detection, bone regeneration, and anti-biofouling material preparation.
Experimental
Materials and methods
All solvents and chemicals were of high analytical quality and utilized without further purification unless otherwise stated. Rink amide AM resins (0.80 mmol g−1 loading) was procured from Novabiochem. Fmoc-protected amino acids, such as Fmoc-Lys(Alloc)-OH, Fmoc-Lys(Fmoc)-OH, Fmoc-His(trt)-OH, Fmoc-Ser(trt)-OH, Fmoc-Asp(OtBu)-OH were purchased from BLD Pharm. The 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b] pyridinium 3-oxide hexafluorophosphate (HATU), trifluoroacetic acid (TFA), N,N-diisopropylethylamine (DIEA), triisopropyl silane (TIS), lauric acid, 5,5′-dithiobis-2-nitrobenzoic acid (DTNB), and 3-mercaptopropyl trimethoxysilane (MPTMS) were bought from TCI Chemicals. Dimethyl sulfoxide (DMSO) and HPLC grade solvents, such as acetonitrile, methanol, and isopropyl alcohol were purchased from Merck and used for reverse phase high-pressure liquid chromatography (RP-HPLC). Piperidine, N,N dimethyl formamide (DMF), and dichloromethane (DCM) were bought from Spectrochem and Rankem Laboratories. p-Nitrophenyl phosphate (pNPP) and p-nitrophenyl acetate (pNPA) were procured from Sisco Research Laboratories Pvt. Ltd (SRL). Cerium nitrate hexahydrate 99.9% AR and Irgacure 2959 were purchased from Loba Chemie and Sigma Aldrich. All pesticides (azamethiphos, profenofos, and carbendazim) were procured from Sigma-Aldrich. PolyPrep chromatography columns from Bio-Rad were used for solid-phase peptide synthesis (SPPS). Deionized water (DI, 18.2 MΩ cm) was obtained from a Milli-Q system and used in all experiments. MC3T3-E1 cells (CRL-2593, Subclone-4) were purchased from ATCC. Minimum Essential Medium alpha (MEM-α), 0.25% trypsin/EDTA, penstrep, SYBR™ Green Master Mix, and trizol were purchased from Thermo Fisher Scientific. The iScript cDNA synthesis kit was bought from Bio-Rad. The 3-(4,5-dimethylthiazol)-2,5-diphenyltetrazolium bromide (MTT reagent), ascorbic acid, and fetal bovine serum (FBS) were procured from HiMedia. E. coli (MTCC 1687) was purchased from CSIR- IMTECH, Chandigarh. For antibacterial studies, Luria broth (Himedia) was used as culture media.Synthesis of branched peptide amphiphile (PA) and characterization
The self-assembled, branched peptide amphiphile was synthesized using standard Fmoc-based SPPS method. Rink amide AM resin was employed as a solid support. The amino acids were coupled using a mixture of HATU (2.85 equiv.), and DIEA (5.7 equiv.) and 20% piperidine in DMF (3 mL) were used to deprotect Fmoc group. The synthesized peptide was cleaved from resin by using a cocktail comprising of 3 mL of 95% TFA, 2.5% TIS, and 2.5% water (v/v). Subsequently, the amide terminated peptide/PA (C12-SHD)2KK(Alloc)-NH2 was cleaved, and precipitated from 30 mL of cold diethyl ether, which was followed by centrifugation and drying under vacuum. The purity of these peptides was determined using a RP-HPLC equipped with Xbridge BEH C18 column (250 × 4.6 mm, 5 μm) and a gradient flow of acetonitrile/water for 35 min with 0.1% TFA as a mobile phase at a flow rate of 1 mL min−1. The peptides were further characterized by mass (XEVO G2-XS QTOF) and 1H NMR (JEOL JNM-ECS, 400 MHz) analysis.Secondary structure of peptide
The secondary structure of peptide was determined by circular dichroism (CD) and Thioflavin T (ThT) assay. CD analysis was performed on a CD spectrometer (JASCO J-1500) using a quartz cuvette with a 0.1 cm path length. In order to induce the formation of secondary structures, peptide solutions were prepared in DI water and incubated at room temperature for five days prior to investigation. Spectra of peptides were recorded at a concentration of 0.5 mM in the region of 195–350 nm at a continuous scanning rate of 200 nm min−1. Fluorescence spectra was used to analyze the binding of peptides with ThT according to a previously described method.33 The fluorescence was recorded using a Tecan multimode microplate reader at an excitation wavelength of 440 nm. ThT solution without peptides was used as a blank.Preparation of cerium oxide nanoparticles (CeNPs)
A conventional hydrothermal technique was used for the preparation of CeNPs.34 A 60 mL of 4 M NaOH was used to make a suspension of 0.4 g of cerium nitrate hexahydrate (1 mmol) followed by stirring for 30 min. The mixture was transferred to a 100 mL Teflon-lined stainless-steel autoclave and incubated for 24 h at 100 °C. After centrifuging, the CeNPs were washed with water and ethanol, and dried at 70 °C to get a distinctive dark yellow powder.Thiol functionalization of CeNPs (TC)
The surface of CeNP was activated by dehydrating the sample at 80 °C overnight. Around 75 mg of CeNPs were dispersed in 15 mL of toluene in a round bottom flask and ultrasonicated for 30 min.35 To this suspension, MPTMS (0.6 mL) was added, and the suspension was refluxed at 110 °C overnight followed by centrifugation. The resulting thiol functionalized ceria nanoparticles (TC) were washed with toluene and ethanol and air-dried at 50 °C overnight.Conjugation of peptide to TC (TCP)
The branched peptide amphiphile was conjugated to thiol functionalized ceria via thiol–ene reaction.36–38 The 20 mg of TC was dispersed in 1 mL of methanol and sonicated for 15 min, followed by the addition of 2 mM, 5 mL solution of PA. Radical initiator (Irgacure 2959, 0.2 equiv., 2.81 mg) was added to the mixture and suspension was exposed to UV light (365 nm) with stirring at room temperature for 1 h. The peptide-functionalized ceria nanoparticles (TCPs) were centrifuged at 6000 rpm, washed thoroughly with methanol, and finally dried overnight at 50 °C.Characterization
All materials (CeNPs, TCs, and TCPs) were characterized as follows:Thiol group quantification
Ellman's assay was used to quantify the functionalization of CeNPs with thiol group.39 Working solution of DTNB (Ellman's reagent) was prepared by dissolving 4 mg of DTNB and 20.5 g of sodium acetate in 5 mL of DI water. About 50 μL of this Ellman's reagent was mixed with 2.5 mL of tris buffer (1 M, pH: 8) to make blank solution. Samples were suspended in tris buffer and 250 μL of each sample (1 mg mL−1) was added to blank solution and incubated for 15 min at room temperature. The absorbance was measured at 412 nm, and the quantity of surface thiol group was determined by dividing absorbance by the extinction co-efficient of the reagent (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 M−1 cm−1).
600 M−1 cm−1).
Hydrolase-mimicking activity
The esterase-mimicking activity was measured using p-nitro phenyl acetate (pNPA) as a substrate.29 All nanomaterials (CeNP, TC, TCP, and PA) were dispersed in 20 mM phosphate buffer of pH 7 at a concentration of 1 mg mL−1. The suspension was thoroughly vortexed and sonicated for 10 min to get a homogeneous suspension. The catalytic suspension (100 μL) was added into the well of a 96-well plate, followed by the addition of 6 μL of pNPA solution (12.5 mM in ACN). The absorbance was measured at 405 nm, every 15 min for 150 min. Catalyst without pNPA was considered as the reference and its absorbance was recorded simultaneously. The absorbance value of reference was subtracted from that of sample to eliminate the contribution of catalyst. Absorbance of substrate without catalyst was observed to check their self-hydrolysis potential and these values were also subtracted to obtain the values exclusively from catalysis. The calibration curve of p-nitrophenol (pNP) was used to determine the amount of pNP formed and the rate of hydrolysis was calculated using the extinction coefficient of pNP, which is 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 014 M−1 cm−1. The phosphatase-mimicking activity was measured using p-nitro phenyl phosphate (pNPP) as a substrate and its working solution was prepared (12.5 mM) in 25 mM tris buffer of pH 8. The experiment was performed in a similar way as mentioned above. In order to calculate kinetic parameters, pNPA or pNPP concentrations between 0.5 and 10 mM were used and the data was fitted to Michaelis–Menten equation. Catalytic rate constant of biocatalyst (kcat) was obtained from linear Lineweaver–Burk plots.
014 M−1 cm−1. The phosphatase-mimicking activity was measured using p-nitro phenyl phosphate (pNPP) as a substrate and its working solution was prepared (12.5 mM) in 25 mM tris buffer of pH 8. The experiment was performed in a similar way as mentioned above. In order to calculate kinetic parameters, pNPA or pNPP concentrations between 0.5 and 10 mM were used and the data was fitted to Michaelis–Menten equation. Catalytic rate constant of biocatalyst (kcat) was obtained from linear Lineweaver–Burk plots.
Reusability of catalyst
We used a different method to test the reusability of catalyst because the 96-well plate was not suitable for it. In this instance, catalysts were suspended (1 mg mL−1) in 940 μL of 20 mM phosphate buffer, pH 7 using the same procedure as previously described. Substrate (60 μL of 12.5 mM solution of pNPA and pNPP in ACN or Tris buffer) was added to this suspension, and the sample was agitated on a mechanical shaker for 45 min. The sample was immediately centrifuged, supernatant was transferred to cuvette, and solution's absorbance was measured at 405 nm. The absorbances of catalyst (same concentrations as used for samples and similar treatment but without adding the substrate) or the substrate (same concentrations as used for samples) were recorded at 405 nm and subtracted from data obtained for samples in order to remove any absorbance that might come from catalyst or substrate. The entire process was carried out five times. PNP's calibration curve was used to quantify PNP formed in each case. The catalysts were cleaned five times with water and ACN, and vacuum-dried at 50 °C for 12 h after each cycle. The dried catalyst was used in additional cycles using the same procedure. In order to confirm the morphology and structure of recycled TCP, FT-IR and FESEM studies were performed after fifth cycle.Haloperoxidase-mimicking activity
The haloperoxidase-mimicking potential of catalyst was investigated by a protocol reported earlier with little modifications.40 Phenol red (0.6 mg, 50 μM) and ammonium bromide (0.1 mg, 25 μM) were dissolved in water, which was followed by the addition of H2O2 (1 μL, 300 μM). The pH of solution was maintained between 5 and 6 and each sample (1 mg mL−1) was incubated with reactant solution for 5 h at room temperature and absorbance was recorded in the range of 400–600 nm using a UV-Vis spectrophotometer.Acetylcholine esterase (AChE)-mimicking activity
Stock solutions of acetylthiocholine and DTNB (40 mg, 100 mM) were prepared in 1 mL DMSO. About 1 mg of each material (CeNP, TC, TCP, and PA) were suspended in 1 mL of PBS, and 10 μL of DTNB and acetylthiocholine were added to the suspension of catalysts. The absorbance data was collected at 405 nm, every 15 min for 150 min. The rate of reaction was obtained by using the molar extinction coefficient of 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 M−1 cm−1.
600 M−1 cm−1.
Detection of pesticides
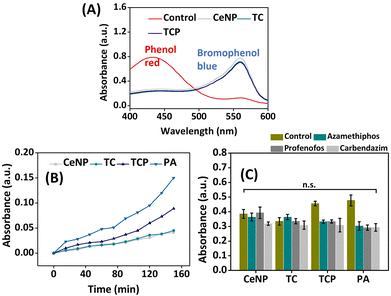
Pesticides, such as organophosphorus (OP) and carbamates, can be detected by nanocatalysts using colorimetric assay. Stock solution of acetylthiocholine and DTNB were prepared as mentioned above and 10 μL of both solutions were added to the suspension of CeNP, TC, TCP, and PA (1 mg mL−1). To this control solution, 10 mM (10 μL) of pesticide (azamethiphos, profenofos, or carbendazim) was added and incubated at 37 °C for 15 min followed by the measurement of absorbance at 405 nm.In vitro biomineralization
The capability of catalyst to imitate alkaline phosphatase (ALP) enzyme was assessed by Ca2+ mineralization study.24,41 Wells of a 24-well plate were coated with 1 mg mL−1 suspension of materials (CeNP, TC, TCP, and PA) and dried over night at 37 °C. An osteogenic solution was prepared containing 15 mM of β-glycerophosphate as a source of Pi and 20 mM of CaCl2 as a source of Ca2+ in Tris-HCl buffer (25 mM, pH 7.4). About 800 μL of this solution was added into each well and incubated at 37 °C for 7 days. Every day, osteogenic solution was replenished with fresh osteogenic solution. At 7th day, wells were washed after removing the solution with distilled water and allowed to air dry. The mineralized calcium developed due to the ALP-mimicking activity was further measured by using 0.1% alizarin red S staining. After staining, the excess dye was removed and wells were washed with DI water and 10% cetylpyridinium chloride was added to dissolve calcium-bound dye. Finally, absorbance was measured at 562 nm to quantify the mineralized calcium using a Tecan multimode microplate reader. Wells containing only osteogenic solution without any sample were treated as a control.Cell culture study
The MC3T3-E1, passage 5–6 were cultured in MEM-α, supplemented with 10% FBS and 1% Pen-Strep antibiotic solution at 37 °C with 5% CO2 in a humidified atmosphere. For experiments involving long term cell culture studies, medium was changed every 72 h and cells were harvested using a solution of trypsin-EDTA at around 70–80% confluency. The cells were further redispersed in complete media and seeded in dishes or cell culture plates to conduct various cell culture studies. Osteogenic media (OM) composed of complete MEM-α containing 100 nM dexamethasone, 50 μg mL−1 ascorbic acid, and 10 mM β-glycerophosphate disodium salt hydrate was considered as a positive control, while complete MEM-α without sample was treated as a negative control.Anti-biofouling activity
The anti-biofouling property of nanomaterials was evaluated using bacteria E. coli. Suspension (300 μL) of nanomaterials (1 mg mL−1) were casted on 24-well plates and air dried for 24 h.44,45 Meanwhile, E. coli was grown till mid-log phase in Luria broth (LB) media overnight and optical density was adjusted to 0.1 at 600 nm. Samples were incubated with 500 μL of bacterial suspension, 4.4 mM NH4Br, and 0.42 mM H2O2 for 24 h at 37 °C inside an incubator. Wells containing bacterial suspension, NH4Br, and H2O2 without samples were considered as controls. After 24 h of incubation, suspension was removed and wells were gently washed with distilled water to remove the weakly attached bacteria. Next, the biofilm was air-dried and stained with crystal violet (0.1%) for 15 min and washed again with DI water to remove excess dye. Finally, 33% acetic acid solution was added to dissolve crystal violet bound to biofilm and absorbance was measured at 590 nm to quantify biofilm formation.Statistical analysis
The studies were performed in triplicates and presented as average values and standard deviations from the mean value. The data were analyzed using Student's t-test. *p value ≤ 0.05 was considered significant, whereas ns represents non-significant difference.Results and discussion
The aim of this work was to develop multifunctional, artificial enzyme to mimic esterase, phosphatase, and haloperoxidase activity for pesticide detection, bone regeneration, and anti-biofouling material application. In order to achieve this goal, we have synthesized self-assembled, branched peptide amphiphile (PA) containing the catalytic triad (Ser: S; His: H; and Asp: D) present in serine protease enzyme. Histidine acts as a base to abstract proton from the side chain of serine and nucleophilic oxygen of serine attacks carbonyl carbon of a substrate to form a covalent bond during catalysis. The negatively charged Asp stabilizes the positive charge that forms on His residue through hydrogen bonding and helps in the orientation of substrate within the active site. These three amino acid residues together create a microenvironment within enzyme's active site that promotes specific chemical reactions, thus, making peptide-based artificial enzymes capable of catalyzing various biochemical transformations.27 Immobilization of peptide on the surface of thiol-modified ceria nanoparticles (TC) helped us in developing a recyclable, heterogenous biocatalyst. The self-assembled peptide rendered the hydrolase-mimicking activity, whereas CeNPs imparted the haloperoxidase-mimicking activity. Ceria nanoparticles undergo reversible oxidation and reduction reactions, and shuttle between Ce4+ and Ce3+ oxidation states. This redox cycling property allows ceria nanoparticles to act as catalysts for the oxidation of halide ions by hydrogen peroxide. The multifunctional, enzyme mimic was explored for pesticide detection, bone regeneration, and antibiofouling applications (Scheme 1).Design, synthesis, and characterization of PA
We have rationally designed the self-assembled, peptide amphiphile (PA) by incorporating a catalytic triad (S, H, and D) to mimic the catalytic site of serine protease, which belongs to the class of hydrolase enzyme (Fig. 1A). The branched peptide was synthesized in order to provide two catalytic sites in a single molecule. Based on literature review, we hypothesized that participating amino acids from nearby peptides will work together to increase the activity of peptide in its self-assembled state and local substrate concentration will be higher at the surface of aggregates. Therefore, we incorporated hydrophobic chain (C12) at the N-terminus of peptide to make it amphiphilic. As our aim was to conjugate the peptide to nanoparticle by thiol–ene reaction, we did not deprotect the Alloc-protecting group in peptide sequence.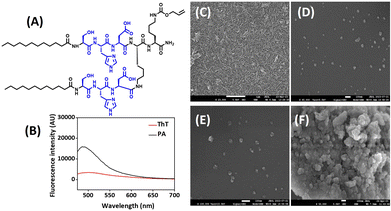 | ||
| Fig. 1 (A) Structure of self-assembled, branched peptide amphiphile (PA). (B) ThT assay. (C–F) FESEM images of: (C) PA, (D) CeNP, (E) TC, and (F) TCP. Scale bar: (C) 1 μm, and (D–F) 100 nm. | ||
Fmoc-based SPPS technique was used to synthesize peptide and rink amide resin was used as a solid support to generate carboxamide group at C-terminus. The peptide was characterized by RP-HPLC, 1H NMR, and mass spectrometry. The RP-HPLC showed that peptide is 90–95% pure and retention time was 4.6 min (Fig. S1‡). The MW derived from the mass spectrum matched to its theoretical value (Fig. S2‡). The 1H NMR spectrum of peptide along with assigned peaks is included in the ESI (Fig. S3‡). The secondary structure of self-assembled peptide was investigated by CD spectroscopy and ThT assay. Negative peaks around 200 and 215 nm indicated the formation of α-helical morphology of peptide (Fig. S4‡). ThT assay demonstrated the enhancement of fluorescence intensity in the range of 490–500 nm, thereby implying the binding of ThT to the hydrophobic region of peptide (Fig. 1B). Unbound ThT without any sample was considered as a control. FT-IR spectra further indicated the presence of α-helical structure as it showed characteristic peak at 1673 cm−1 (Fig. S5‡). The nanoscale architecture of self-assembled peptide was evaluated by FESEM investigations. The microscopic image suggested the formation of long nanofibers by peptide in its self-assembled form (Fig. 1C). The width of nanofibers was calculated by ImageJ software and was found to be below 20 nm, which can be attributed to long hydrophobic carbon chain attached at the N-terminus of peptide.
Preparation and functionalization of CeNPs
CeNPs were fabricated using hydrothermal method by hydrolyzing a ceria precursor, cerium nitrate hexahydrate, in a basic environment. The surface was modified post synthesis using MPTMS to obtain thiol modified ceria nanoparticles (TCs). The thiol grafting on surface was quantified by Ellman's assay, which suggested a 1306.88 μmol g−1 of thiol density. The nanoparticles were characterized by XRD, TGA, BET, elemental mapping, and XPS. The morphology and size of nanoparticles were confirmed by FESEM and DLS, as discussed below.Conjugation of peptide amphiphile to ceria
Our objective was to develop a multifunctional peptide-based catalyst in which the hydrolase-mimicking PA was immobilized on a ceria surface to produce a heterogeneous catalyst. Immobilization on solid surface is expected to maintain its catalytic efficiency. The assembly of catalytic peptides on a solid surface increases the peptide catalyst's robustness and reusability features in an integrated manner. With this aim, the PA was conjugated on the surface of thiolated ceria (TC) via thiol–ene click reaction. The successful grafting was confirmed by Ellman's assay, which showed that free thiol density on ceria surface has reduced after conjugation to the peptide.Characterization
All nanomaterials (CeNP, TC, and TCP) were characterized by PXRD, TGA, elemental mapping, FESEM, and DLS. FESEM images revealed that CeNPs have a spherical morphology and grafting of thiol groups and peptide on the surface of CeNPs did not change its morphology (Fig. 1D–F). It is interesting to note that the conjugation of branched PA on the surface of CeNPs reduced their aggregation, thus, increasing their stability. The size of nanoparticles increased a little after functionalization. The particle size distribution is evident from DLS study. The size of CeNPs, TC, and TCP were found to be 41.6, 49.7, and 54.3 nm (Fig. S6‡). Zeta potential of nanoparticles was also investigated to measure their surface charge and found to be 0.5, −1.5, and 5.9 mV for CeNP, TC, and TCP. It was observed that zeta potential becomes more negative after thiolation of CeNPs and it increased after conjugation to peptide. XRD patterns of materials were investigated to determine their crystal structure (Fig. 2A). CeNPs showed characteristic sharp peaks at 28.38, 32.82, 47.16, and 56.1° corresponding to 111, 200, 220, and 311 planes.40 This data suggested that CeNPs have a fluorite-type crystalline nature as per JCPDS file no. 34-0394.46 Neither surface modification with thiol groups nor the integration of peptide on it changed the PXRD patten or induced any new peak. However, the intensity of peaks was significantly reduced. This data suggests that crystallinity decreased after modification of ceria surface.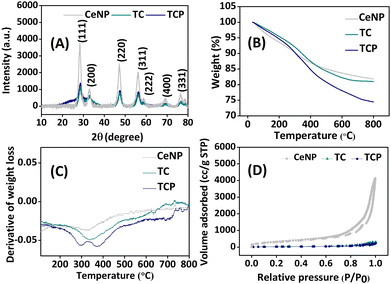 | ||
| Fig. 2 Characterization of nanoparticles: (A) PXRD patterns, (B) TGA study, (C) derivative of thermogravimetry (DTG) curve, and (D) N2 adsorption/desorption isotherm. | ||
Successful grafting of thiol group and PA on ceria surface was further confirmed by TGA. The percentage mass loss was observed from TGA curve (Fig. 2B), whereas derivative thermogravimetry curve (DTG) demonstrated the rate of material weight change upon heating (Fig. 2C). The weight loss at 100 °C signified the loss of adsorbed water molecules from nanomaterials. The weight loss from 250–400 °C in DTG curve was due to the loss of organic moiety from ceria surface.29 The NPs obtained at different stages were also investigated for pore size measurements and surface area using Brunauer–Emmett–Teller (BET) method. The pore size parameters and specific surface area are tabulated in Table S1 (ESI).‡ The BET surface area analysis revealed type IV isotherm, which indicated mesoporous structures for all NPs (Fig. 2D). The BET surface area of CeNP was found to be 1120 m2 g−1, which sharply decreased to 105 m2 g−1 and 76 m2 g−1 after thiol-functionalization (TC) and subsequent peptide conjugation (TCP). This considerable reduction in surface area implies that surface functions are blocking pores. All data confirm the functionalization of CeNP with thiol (–SH) and PA. EDX spectra and elemental mapping helped in understanding the elemental distribution of all NPs (Fig. 3A and S7‡). It was observed from the elemental mapping that CeNP surfaces were functionalized with thiol and PA.
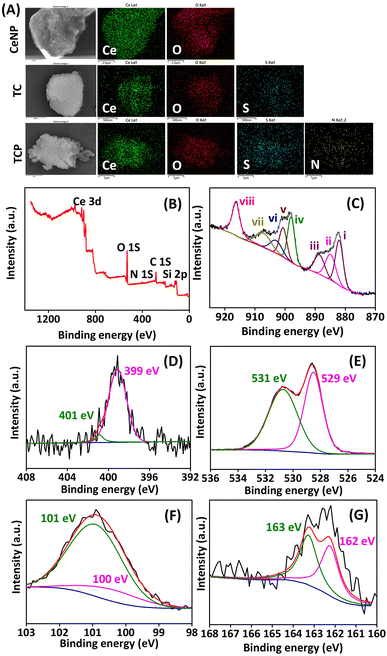 | ||
| Fig. 3 (A) Elemental mapping of CeNP, TC, and TCP. (B–G) XPS spectra of TCP: (B) surface survey spectrum, (C) Ce 3d, (D) N 1s, (E) O 1s, (F) Si 2p, and (G) S 2p. | ||
The oxidation state of components of nanocomposite was revealed by XPS spectra (Fig. 3B–G). High-resolution Ce 3D spectrum of TCP showed spin–orbit coupling peaks corresponding to Ce 3d3/2 and Ce 3d5/2. The peaks designated at II (884.9 eV), IV (898.0 eV), and VI (903.0 eV) corresponds to Ce3+ oxidation state, whereas peaks at I (882.1 eV), III (888.5 eV), V (900.6 eV), VII (907.0 eV), and VIII (916.3 eV) corresponds to Ce4+ oxidation state.40 The ratio of Ce3+ to Ce4+ was calculated as 0.57 (Table S2, ESI‡). The oxygen vacancy in material was assessed using the expression Ce3+/(Ce3+ + Ce4+) × 100, and found to be 37%. TCPs with 37% oxygen vacancy can demonstrate efficient catalytic activity, as this activity is enhanced by oxygen vacancy.
Catalytic activity
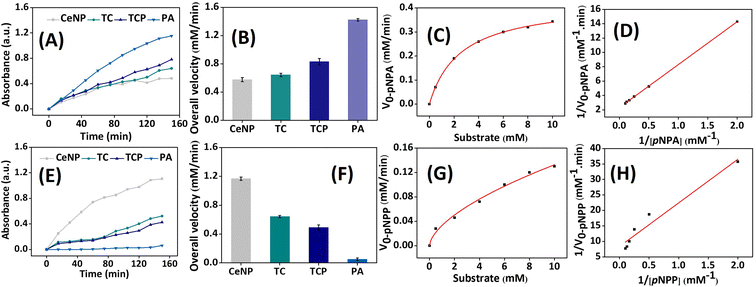
The individual and cooperative role of amino acids in catalytic pockets of enzymes are best understood using peptide nano catalysts, which are simple to synthesize and modify. His residues can be used to create PA nano catalysts with catalytic domains. Here, we developed branched, amphiphilic peptide with two catalytic domains, composed of Ser–His–Asp. The artificial enzyme can undergo self-assembly to mimic the folding mechanism of natural enzyme. Accordingly, we integrated a catalytic peptide on the surface of CeNPs to evaluate their multiple enzyme-mimicking properties. The esterase-mimicking activity was assessed using pNPA as a substrate, which upon hydrolysis generates yellow colored product, pNP (λmax = 405 nm). Autohydrolysis of pNPA was considered as a blank and its absorbance value was subtracted from that of final absorbance (Fig. 4A). The PA exhibited maximum absorbance, which signifies highest amount of p-NP production due to the hydrolysis of substrate by esterase-mimicking enzyme. The activity was reduced when PA was conjugated to ceria NP surface but it was better than CeNPs and TCs. The overall velocity was also determined and it was found to be 0.57, 0.64, 0.82, and 1.42 μM min−1 for CeNP, TC, TCP, and PA (Fig. 4B). The esterase-mimicking activity was comparable to catalyst reported by Gulseren et al.27The phosphatase-mimicking activity was determined using pNPP as a substrate. It was fascinating to note that CeNPs exhibited the highest rate of hydrolysis of phosphoester bond (Fig. 4E). This dephosphorylation by CeNPs was attributed to the Lewis acidity of metal, which facilitates the interaction with negatively charged phosphate moiety. It results in a series of phenomenon like, Lewis acid activation and nucleophilic attack, followed by the removal of phosphate group. In particular, Ce4+ tend to exhibit higher catalytic reaction than Ce3+ due to its oxygen vacancy and better efficacy in attracting electron.47–49 After the functionalization of CeNPs with thiol and PA, the phosphoesterase-mimicking activity was reduced. The overall velocity of TCP was better than PA (Fig. 4F). Herein, the catalytic activity of peptide conjugated ceria nanoparticles (TCP) toward pNPA and pNPP was assessed. The initial hydrolysis rates V0-pNPA for the substrate pNPA are shown as a function of pNPA concentration (0.5–10 mM) in Fig. 4C and V0-pNPP was measured at different concentrations (0.5–10 mM) of pNPP (Fig. 4G). The reactions followed conventional Michaelis–Menten model, confirming that TCP exhibited catalytic behavior. An enzyme's catalytic rate constant (kcat) represents the highest quantity of substrate molecules transformed into product for each active site in a unit of time. This value indicates the rate at which an enzyme–substrate complex proceeds with its forward process. The kcat was measured based on linear Lineweaver–Burk plot and it was calculated as 0.041 min−1 and 0.01 min−1 for TCP to catalyse pNPA and pNPP (Fig. 4D and H). A comprehensive parameter to assess an enzyme's capacity to catalyse a particular substrate is its catalytic efficiency (kcat/Km). The catalytic activity of TCP toward pNPA and pNPP was examined in this study and calculated as 0.0160 μM−1 min−1 and 0.0082 μM−1 min−1. This data validates the superior esterase-like activity of TCP compared to its phosphatase-mimicking property.
A catalyst's recyclability is one of main issues for the industrial use. We carried out a single batch of catalysis followed by the removal of catalyst by filtration, washing, and drying in order to determine whether the heterogenous catalyst (TCP) can be recycled after each usage. The catalyst was reused for subsequent batch after drying. The same procedure was carried out five times (Fig. S8‡), and no significant loss of activity was observed even during the fifth cycle. This demonstrates that catalyst can be recycled using a simple and affordable process. The catalyst retained its morphology and structure after fifth cycle, as evident from FESEM and FT-IR analysis (Fig. S9, ESI‡).
A class of peroxidase enzyme called haloperoxidase catalyzes the oxidation of halides.45 The material's haloperoxidase-like activity was assessed utilizing the phenol red bromination assay. In the presence of halide ions and peroxide, phenol red can be brominated to generate bromophenol blue using a catalyst that resembles haloperoxidase. A successful reaction is indicated by a drop in absorbance intensity of phenol red (λmax = 432 nm) and a corresponding increase in absorbance intensity of bromophenol blue (λmax = 590 nm). It can be observed from Fig. 5A that CeNPs exhibited highest haloperoxidase-mimicking activity with a shift of wavelength from 430 to 590 nm and absorbance was highest at 590 nm among other nanomaterials. TCP also exhibited significant potential in bromination of phenol red while showing haloperoxidase-mimicking activity.
Applications
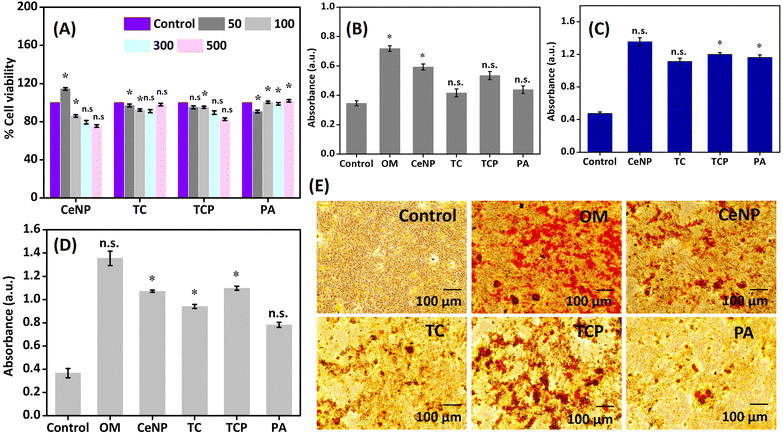
The heterogenous biocatalyst, developed in this work was explored for several applications.We evaluated the cell viability of mesenchymal stem cells (MC3T3-E1) at different concentrations of nanomaterials (50, 100, 300 and 500 μg mL−1). It was evident from the data shown in Fig. 6A that CeNPs exhibited toxicity at higher concentrations, whereas PA helped in the proliferation of cells at increased concentration. Surface modification of CeNPs with thiol and PA decreased its toxicity and enhanced the cell viability. The same trend was observed in long-term cell viability analysis, where toxicity increased with increase in the concentration of nanomaterials but it was masked after conjugating the peptide (Fig. S11‡). As far as ALP assay was concerned, CeNPs exhibited highest ALP activity on MSCs at 300 μg mL−1 concentration, which can be attributed to their inherent phosphatase-mimicking properties owing to their reversible Ce3+/Ce4+ redox pair and oxygen vacancy in the outer shell47 (Fig. 6B). Modification of their surface with thiol and peptide interferes with their oxidation state and deteriorate their ALP-mimicking potential.
Considering the phosphatase like catalytic behavior of nanomaterials, we anticipated that they will have a great potential in stimulating osteogenic activity of natural ALP. In order to verify this, 24-well plates were coated with nanomaterials and incubated with osteogenic solution containing β-glycerophosphate and CaCl2 for 7 days. Alizarin red S staining was carried out to quantify the amount of calcium deposition on the surface of well. As shown in Fig. 6C, the deposited calcium on the surface of well coated with nanomaterial was higher compared to the control (osteogenic media without any sample), thus, confirming the ALP-like catalytic property of nanomaterials, which accelerated the hydrolysis of β-glycerophosphate. The phosphatase-mimicking activity was again evaluated on MC3T3-E1 cells and similar trend was observed where TCP allowed a significant calcium deposition (Fig. 6D). Controls used were cells treated with complete media and those treated with osteogenic media (OM). Inverted microscopy revealed red-stained calcium nodules representing the extracellular calcium accumulation (Fig. 6E). The results are comparable to earlier reports by Wang and coworkers and Gulseren et al.41 Understanding the expression of genes that regulate bone tissue regeneration is essential for developing therapies and interventions to enhance bone healing in various clinical contexts, including fractures, bone defects, and osteoporosis. Therefore, we studied the expression of genes (ALP, OPN, OCN, and RUNX2) in the presence of CeNP, TC, TCP, and PA on MC3TC-E1 cell lines for 7 and 14 days by RTPCR. We observed that TCP was able to upregulate expressions of ALP, OPN, OCN, and RUNX2 by 5, 7, 8, and 16-fold (Fig. S12‡). The data was also compared to positive control (OM). The results suggest that this material holds significant potential for advancing bone regeneration therapies.
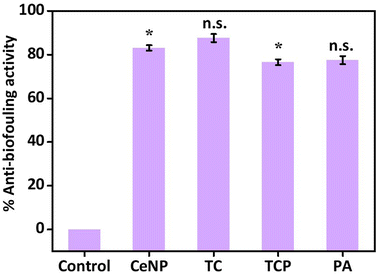
Finally, the haloperoxidase-like activity of nanomaterials was exploited for the development of anti-biofouling materials.53 The non-specific surface adherence of microorganisms is known as biofouling, which can harm materials’ performance and decrease its effectiveness by causing corrosion on metallic surfaces and bacterial infections on surgically implanted medical equipment.32,54 Haloperoxidase can catalyze the oxidation of halides with hydrogen peroxide to generate hypohalous acids, such as HOBr, which is known to disrupt the quorum sensing of microorganisms, thus inhibiting biofilm formation, the initial step for biofouling of a surface. We determined the inhibition of biofilm formation by crystal violet staining method (Fig. 7) and heterogenous catalyst (TCP) demonstrated around 80% anti-biofouling activity. Therefore, the material can be a good candidate for integration on the surface of implant or as a coating material to prevent the adhesion of bacteria.
Conclusions
In summary, we have developed self-assembling, branched peptide amphiphile containing catalytic triad, ‘Ser–His–Asp’, to mimic the active site of serine protease, which can effectively bind with esters and hydrolyze the bond. This hydrolase mimetic catalyst was immobilized on the surface of ceria nanoparticles using thiol–ene click reaction, which exhibits haloperoxidase-mimicking properties due to their reversible change between two oxidation states, Ce3+/Ce4+. The nanofibrous morphology of self-assembling peptide was not retained upon conjugation with ceria nanoparticles and the heterogenous catalyst adopted a spherical shape, with a diameter of 54.3 nm. Integration of peptide on nanoparticle surface minimized the aggregation of nanoparticles and made the catalyst recyclable without the loss of activity. The peptide-based nanomaterial showed multifunctional catalytic activity by mimicking the esterase, phosphatase, and haloperoxidase enzymes. The enzyme mimicking potential of biocatalyst was further explored for various applications. The acetylcholinesterase (AChE)-mimicking property was used for the detection of pesticides, like organophosphates and carbamates. Alkaline phosphatase (ALP)-like activity was exploited to develop material for bone regeneration and anti-biofouling applications. Thus, the catalyst created in this work can be quite valuable for industrial applications. Overall, this work showcases the benefits of developing multifunctional heterogenous biocatalyst to reduce the problem of instability associated with peptide-based homogenous catalyst. We also exploited the potential enzyme-mimicking role of catalyst in different fields of human welfare.Author contributions
The manuscript was written through contributions of all authors, and they have given their approval to the final version of the manuscript.Data availability
The data supporting this article have been included as part of the ESI.‡Conflicts of interest
Authors declare no competing financial interest.Acknowledgements
The authors acknowledge financial support from the DBT, India (grant # BT/PR40669/MED/32/761/2020) to YS and FIST (Level-I) grant of DST, India (SR/FST/CS-I/2018/55) to the chemistry department for the LC-MS facility. The authors are thankful to the Departments of Chemistry and Biomedical Engineering and Central Research Facilities at IIT Ropar for giving access to their research facilities. The authors also thank Prof. Asish Pal, Chemical Biology Unit, Institute of Nano Science and Technology, Mohali for helping with CD studies. M. H. is thankful to IIT Ropar for the institute fellowship and V. C. is thankful to Prime Minister Research Fellowship (PMRF) from the Ministry of Education, India (Application No. PMRF-192002-167).References
- W. Hamley, Biomacromolecules, 2021, 22, 1835–1855 CrossRef PubMed.
- D. Yi, T. Bayer, C. P. S. Badenhorst, S. Wu, M. Doerr, M. Höhne and U. T. Bornscheuer, Chem. Soc. Rev., 2021, 50, 8003–8049 RSC.
- E. L. Bell, W. Finnigan, S. P. France, A. P. Green, M. A. Hayes, L. J. Hepworth, S. L. Lovelock, H. Niikura, S. Osuna, E. Romero, K. S. Ryan, N. J. Turner and S. L. Flitsch, Nat. Rev. Methods Primers, 2021, 1, 46 CrossRef CAS.
- A. Schmid, J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts and B. Witholt, Nature, 2001, 409, 258–268 CrossRef CAS PubMed.
- Y. R. Maghraby, R. M. El-Shabasy, A. H. Ibrahim and H. M. E.-S. Azzazy, ACS Omega, 2023, 8, 5184–5196 CrossRef CAS PubMed.
- R. Singh, S. Langyan, B. Rohtagi, S. Darjee, A. Khandelwal, M. Shrivastava, R. Kothari, H. Mohan, S. Raina, J. Kaur and A. Singh, Mater. Sci. Energy Technol., 2022, 5, 294–310 CAS.
- F. Rudroff, M. D. Mihovilovic, H. Gröger, R. Snajdrova, H. Iding and U. T. Bornscheuer, Nat. Catal., 2018, 1, 12–22 CrossRef.
- G. Alvarez, T. Shahzad, L. Andanson, M. Bahn, M. D. Wallenstein and S. Fontaine, Global Change Biol., 2018, 24, 4238–4250 CrossRef PubMed.
- N. F. Mokhtar, R. N. A. Rahman, N. D. Md. Noor, F. Md. Shariff and M. S. Md. Ali, Catalysts, 2020, 10, 744 CrossRef CAS.
- A. Basso and S. Serban, Mol. Catal., 2019, 479, 110607 CrossRef CAS.
- S. Sabater, J. A. Mata and E. Peris, ACS Catal., 2014, 4, 2038–2047 CrossRef CAS.
- H. J. Federsel, T. S. Moody and S. J. C. Taylor, Molecules, 2021, 26, 2822 CrossRef CAS PubMed.
- A. M. Garcia, M. Kurbasic, S. Kralj, M. Melchionna and S. Marchesan, Chem. Commun., 2017, 53, 8110–8113 RSC.
- C. M. Rufo, Y. S. Moroz, O. V. Moroz, J. Stöhr, T. A. Smith, X. Hu, W. F. DeGrado and I. V. Korendovych, Nat. Chem., 2014, 6, 303–309 CrossRef CAS PubMed.
- M. O. Guler and S. I. Stupp, J. Am. Chem. Soc., 2007, 129, 12082–12083 CrossRef CAS PubMed.
- Y. Lou, B. Zhang, X. Ye and Z.-G. Wang, Mater. Today Nano, 2023, 21, 100302 CrossRef CAS.
- J. N. B. D. Pelin, B. B. Gerbelli, C. J. C. Edwards-Gayle, A. M. Aguilar, V. Castelletto, I. W. Hamley and W. A. Alves, Langmuir, 2020, 36, 2767–2774 CrossRef CAS PubMed.
- B. List, R. A. Lerner and C. F. Barbas, J. Am. Chem. Soc., 2000, 122, 2395–2396 CrossRef CAS.
- S. J. Miller, Acc. Chem. Res., 2004, 37, 601–610 CrossRef CAS PubMed.
- C. Zhang, X. Xue, Q. Luo, Y. Li, K. Yang, X. Zhuang, Y. Jiang, J. Zhang, J. Liu, G. Zou and X.-J. Liang, ACS Nano, 2014, 8, 11715–11723 CrossRef CAS PubMed.
- Z. Huang, S. Guan, Y. Wang, G. Shi, L. Cao, Y. Gao, Z. Dong, J. Xu, Q. Luo and J. Liu, J. Mater. Chem. B, 2013, 1, 2297 RSC.
- D. W. Watkins, J. M. X. Jenkins, K. J. Grayson, N. Wood, J. W. Steventon, K. K. Le Vay, M. I. Goodwin, A. S. Mullen, H. J. Bailey, M. P. Crump, F. MacMillan, A. J. Mulholland, G. Cameron, R. B. Sessions, S. Mann and J. L. R. Anderson, Nat. Commun., 2017, 8, 358 CrossRef PubMed.
- S. C. Patel, L. H. Bradley, S. P. Jinadasa and M. H. Hecht, Protein Sci., 2009, 18, 1388–1400 CrossRef CAS PubMed.
- Y. Wang, L. Yang, M. Wang, J. Zhang, W. Qi, R. Su and Z. He, ACS Catal., 2021, 11, 5839–5849 CrossRef CAS.
- O. Zozulia and I. V. Korendovych, Angew. Chem., Int. Ed., 2020, 59, 8108–8112 CrossRef CAS PubMed.
- Y.-M. Wong, H. Masunaga, J.-A. Chuah, K. Sudesh and K. Numata, Biomacromolecules, 2016, 17, 3375–3385 CrossRef CAS PubMed.
- G. Gulseren, M. A. Khalily, A. B. Tekinay and M. O. Guler, J. Mater. Chem. B, 2016, 4, 4605–4611 RSC.
- D. J. Mikolajczak and B. Koksch, ChemCatChem, 2018, 10, 4324–4328 CrossRef CAS.
- P. Dowari, M. K. Baroi, T. Das, B. K. Das, S. Das, S. Chowdhuri, A. Garg, A. Debnath and D. Das, J. Colloid Interface Sci., 2022, 618, 98–110 CrossRef CAS PubMed.
- A. Reja, S. P. Afrose and D. Das, Angew. Chem., Int. Ed., 2020, 59, 4329–4334 CrossRef CAS PubMed.
- L. Hedstrom, Chem. Rev., 2002, 102, 4501–4524 CrossRef CAS PubMed.
- K. Herget, P. Hubach, S. Pusch, P. Deglmann, H. Götz, T. E. Gorelik, I. A. Gural'skiy, F. Pfitzner, T. Link, S. Schenk, M. Panthöfer, V. Ksenofontov, U. Kolb, T. Opatz, R. André and W. Tremel, Adv. Mater., 2017, 29, 1603823 CrossRef PubMed.
- M. Halder, Y. Bhatia and Y. Singh, Biomater. Sci., 2022, 10, 2248–2262 RSC.
- I. Trenque, G. C. Magnano, M. A. Bolzinger, L. Roiban, F. Chaput, I. Pitault, S. Briançon, T. Devers, K. Masenelli-Varlot, M. Bugnet and D. Amans, Phys. Chem. Chem. Phys., 2019, 21, 5455–5465 RSC.
- N. Rasool, D. Negi and Y. Singh, ACS Biomater. Sci. Eng., 2023, 9, 3535–3545 CrossRef CAS PubMed.
- A. A. Aimetti, R. K. Shoemaker, C.-C. Lin and K. S. Anseth, Chem. Commun., 2010, 46, 4061 RSC.
- Y. Liu, W. Hou, H. Sun, C. Cui, L. Zhang, Y. Jiang, Y. Wu, Y. Wang, J. Li, B. S. Sumerlin, Q. Liu and W. Tan, Chem. Sci., 2017, 8, 6182–6187 RSC.
- Y. Li, M. Yang, Y. Huang, X. Song, L. Liu and P. R. Chen, Chem. Sci., 2012, 3, 2766 RSC.
- M. Kalantari, T. Ghosh, Y. Liu, J. Zhang, J. Zou, C. Lei and C. Yu, ACS Appl. Mater. Interfaces, 2019, 11, 13264–13272 CrossRef CAS PubMed.
- N. Wang, W. Li, Y. Ren, J. Duan, X. Zhai, F. Guan, L. Wang and B. Hou, Colloids Surf., A, 2021, 608, 125592 CrossRef CAS.
- G. Gulseren, I. C. Yasa, O. Ustahuseyin, E. D. Tekin, A. B. Tekinay and M. O. Guler, Biomacromolecules, 2015, 16, 2198–2208 CrossRef CAS PubMed.
- M. Halder, M. Narula and Y. Singh, Bioconjugate Chem., 2023, 34, 645–663 CAS.
- X. Cui, L. Xu, Y. Shan, J. Li, J. Ji, E. Wang, B. Zhang, X. Wen, Y. Bai, D. Luo, C. Chen and Z. Li, Sci. Bull., 2024, 69, 1895–1908 CrossRef CAS PubMed.
- H. Meng, S. Cheng, L. Wang, Y. L. Balachandran, W. Zhang and X. Jiang, Mater. Adv., 2022, 3, 8137–8140 RSC.
- M. Hu, K. Korschelt, M. Viel, N. Wiesmann, M. Kappl, J. Brieger, K. Landfester, H. Thérien-Aubin and W. Tremel, ACS Appl. Mater. Interfaces, 2018, 10, 44722–44730 CrossRef CAS PubMed.
- T. S. Sreeremya, K. M. Thulasi, A. Krishnan and S. Ghosh, Ind. Eng. Chem. Res., 2012, 51, 318–326 CrossRef CAS.
- A. Dhall, A. Burns, J. Dowding, S. Das, S. Seal and W. Self, Environ. Sci.: Nano, 2017, 4, 1742–1749 RSC.
- F. Tan, Y. Zhang, J. Wang, J. Wei, Y. Cai and X. Qian, J. Mass Spectrom., 2008, 43, 628–632 CrossRef CAS PubMed.
- Y. Bai, Y. Li, Y. Li and L. Tian, ACS Omega, 2024, 9, 8601–8614 CrossRef CAS PubMed.
- J. S. Sidhu, K. Rajendran, A. B. Mathew, T. Iqbal, D. K. Saini and D. Das, Anal. Chem., 2023, 95, 7594–7602 CrossRef CAS PubMed.
- S. Qian and H. Lin, Anal. Chem., 2015, 87, 5395–5400 CrossRef CAS PubMed.
- C. Uslu, S. Narin, Z. Demirsoy, H. B. Öksüz and G. Gülseren, Int. J. Biol. Macromol., 2023, 233, 123604 CrossRef CAS PubMed.
- E. Pütz, I. Tutzschky, H. Frerichs and W. Tremel, Nanoscale, 2023, 15, 5209–5218 RSC.
- H. Xiong, X. He, T. Lou and X. Bai, Biochem. Eng. J., 2023, 195, 108931 CrossRef CAS.
Footnotes |
| † This article is dedicated to Prof. Santanu Bhattacharya on his 65th birthday. We wish him many years of active research ahead and hope that he continues to inspire and motivate research groups around the world. YS carried out postdoctoral research under his mentorship. |
| ‡ Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr02672a |
| This journal is © The Royal Society of Chemistry 2024 |