Open Access Article
Open Access ArticleUnexpected in crystallo reactivity of the potential drug bis(maltolato)oxidovanadium(IV) with lysozyme†
Maddalena
Paolillo
a,
Giarita
Ferraro
a,
Irene
Cipollone
ab,
Eugenio
Garribba
 c,
Maria
Monti
*ab and
Antonello
Merlino
c,
Maria
Monti
*ab and
Antonello
Merlino
 *a
*a
aDepartment of Chemical Sciences, University of Naples Federico II, Complesso Universitario di Monte Sant'Angelo, Via Cintia, I-80126, Napoli, Italy. E-mail: montimar@unina.it; antonello.merlino@unina.it
bCEINGE Biotecnologie Avanzate ‘Franco Salvatore’, Via G. Salvatore, 486, 80131, Napoli
cDipartimento di Medicina, Chirurgia e Farmacia, Università di Sassari, Viale San Pietro, I-07100 Sassari, Italy
First published on 8th July 2024
Abstract
The high-resolution X-ray structure of the adduct formed upon the reaction of hen egg white lysozyme (HEWL) with the potential drug BMOV (bis(maltolato)oxidovanadium(IV) or [VIVO(malt)2], where malt = maltolato) from crystals grown in 1.1 M NaCl and 0.1 M sodium acetate at pH 4.0 reveals an unexpected reaction product, where the V compound is transformed, the maltolate ring is broken and a significant modification of the N-terminal lysine side chain is observed. The results, also supported by gel electrophoresis and mass spectrometry data, show an unusual reactivity of BMOV with HEWL in the solid state, prompting further research on the reactivity of vanadium compounds with proteins which could depend on the specific experimental conditions.
Introduction
Vanadium complexes (VCs) have a wide range of pharmacological properties, including antiviral, antiparasitic, and antituberculosis activities, although the most important potential application in medicine is the treatment of cancer and diabetes.1–10 VCs regulate multiple enzymes, including kinases and phosphatases, in ways that are still unknown, potentially bypassing non-functional components of the insulin signalling pathways.11–14 Some VCs normalize blood glucose and lipids in diabetic animals, and human therapeutic trials have shown a modest recovery of insulin sensitivity.11,15,16In vitro and in vivo investigations on animal models have confirmed that VCs can have various insulin-like or insulin-mimetic actions.17–20 Generally, VIV compounds are more effective at lowering blood sugar than their VIII and VV analogues11,21 and, based on this finding, various [VIVO(L)2] complexes, with L being a bidentate monoanionic organic ligand, were proposed for the possible treatment of diabetes. Among such complexes, bis(maltolato)oxidovanadium(IV) (BMOV or [VIVO(malt)2]) and bis(ethylmaltolato)oxidovanadium(IV) (BEOV or [VIVO(Etmalt)2]) (Fig. 1) have a prominent place and are considered as references for the development of new species with antidiabetic activity.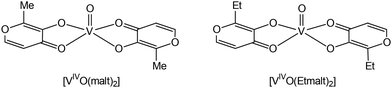 | ||
| Fig. 1 Structures of BMOV (bis(maltolato)oxidovanadium(IV)) and BEOV (bis(ethylmaltolato)oxidovanadium(IV)). | ||
In the 1990s it was demonstrated that BMOV and BEOV normalize both glucose and lipid values, are two–three times more effective than VIVOSO4, are better tolerated than inorganic V salts, and result in reliable glucose-lowering in all tested animal models of diabetes.19 Their activity was attributed to efficient absorption in the gastrointestinal tract, in turn related to the favorable hydro/lipophilic balance and neutral charge of the complexes which can be taken up by passive diffusion.22,23
Phases I and II of the clinical trials of BEOV have been completed, with the code AKP020,19,20 but the tests were interrupted due to the renal problems experienced by several patients, the financial problems of the company which performed the tests (Akesis Pharmaceuticals) and patent expiration.24,25 However, despite the trials as potential antidiabetic agents being stopped, the tests on BMOV were continued by CFM Pharma (with code CFM10 and commercial name Vanadis) and it entered the phase II clinical trials for the treatment of myocardial infarctions and tissue damage due to injuries on secondary tissues caused by fire or other accidents.26,27 In addition, it is worth mentioning that the development of new VIVO-pyrone complexes with antidiabetic effectiveness did not stop and a “second generation” of derivatives was proposed (for example, bis((5-hydroxy-4-oxo-4H-pyran-2-yl)-methylbenzoatato)oxidovanadium(IV) or BBOV has half the oral toxicity as BMOV and can be administered in a dose 1000 times lower than BMOV).28 Moreover, it has been shown that newly synthesised VCs, including those containing the decavanadate ion, might induce stronger insulin-mimetic effects than BMOV.29
In solution, VCs can undergo several chemical changes like hydrolysis, ligand replacement, redox reactions and formation of mixed species with low molecular mass bioligands (bL) and adducts with proteins, these transformations being particularly important at the low concentrations normally employed in biological assays.21,26,30 In most of the cases, VCs lose their identity and no longer exist in their initial form.30,31 This obviously influences significantly their biological activity. BMOV and BEOV can transform into octahedral cis-[VIVO(L)2(H2O)] and cis-[VIVO(L)2(OH)]− and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 [VIVO(L)(H2O)3]+ species; in addition, the formation of aqua ion [VIVO(H2O)5]2+ and hydroxido complexes plus oxidation to the corresponding VVO2 compounds are possible.26 Binding of BMOV to glutathione,32 citrate,33,34 oxalate,33,34 lactate,34 phosphate,33,34 ATP,32 human serum albumin (HSA),35–38 and apo- and holo-transferrin (apo-HTf and holo-HTf)36,37 has been studied. Depending on the pH and V concentration, in all the systems, VIVO–L–bL and VIVO–L–protein species were observed. Nuclear magnetic resonance (NMR), electron paramagnetic resonance (EPR) and circular dichroism (CD) measurements suggested the formation of an adduct with HSA, which seems suppressed in the presence of apo-transferrin.39 Using electrospray ionization-mass spectrometry (ESI-MS), EPR and computational methods it has been shown that BMOV reacts with myoglobin (Mb) and ubiquitin (Ub) forming adducts with the [VIVO]2+ ion, [VIVO(malt)]+ and [VIVO(malt)2] fragments.40,41 X-ray crystallography data on the adducts formed upon the reaction of BMOV with hen egg white lysozyme (HEWL) in 2.0 M sodium formate, 0.1 M Hepes at pH 7.5 or 20% ethylene glycol, 0.1 M sodium acetate at pH 4.0, and 0.6 M sodium nitrate showed non-covalent binding of cis-[VIVO(malt)2(H2O)] and [VIVO(malt)(H2O)3]+ and covalent binding of [VIVO(H2O)3–4]2+ and cis-[VIVO(malt)2] plus other V-containing fragments to the side chains of Glu35, Asp48, Asn65, Asp87, and Asp119 residues and to the C-terminal carboxylate.42
1 [VIVO(L)(H2O)3]+ species; in addition, the formation of aqua ion [VIVO(H2O)5]2+ and hydroxido complexes plus oxidation to the corresponding VVO2 compounds are possible.26 Binding of BMOV to glutathione,32 citrate,33,34 oxalate,33,34 lactate,34 phosphate,33,34 ATP,32 human serum albumin (HSA),35–38 and apo- and holo-transferrin (apo-HTf and holo-HTf)36,37 has been studied. Depending on the pH and V concentration, in all the systems, VIVO–L–bL and VIVO–L–protein species were observed. Nuclear magnetic resonance (NMR), electron paramagnetic resonance (EPR) and circular dichroism (CD) measurements suggested the formation of an adduct with HSA, which seems suppressed in the presence of apo-transferrin.39 Using electrospray ionization-mass spectrometry (ESI-MS), EPR and computational methods it has been shown that BMOV reacts with myoglobin (Mb) and ubiquitin (Ub) forming adducts with the [VIVO]2+ ion, [VIVO(malt)]+ and [VIVO(malt)2] fragments.40,41 X-ray crystallography data on the adducts formed upon the reaction of BMOV with hen egg white lysozyme (HEWL) in 2.0 M sodium formate, 0.1 M Hepes at pH 7.5 or 20% ethylene glycol, 0.1 M sodium acetate at pH 4.0, and 0.6 M sodium nitrate showed non-covalent binding of cis-[VIVO(malt)2(H2O)] and [VIVO(malt)(H2O)3]+ and covalent binding of [VIVO(H2O)3–4]2+ and cis-[VIVO(malt)2] plus other V-containing fragments to the side chains of Glu35, Asp48, Asn65, Asp87, and Asp119 residues and to the C-terminal carboxylate.42
We have recently shown that on changing the experimental conditions, different interactions between the same VC and the same protein are observed. Therefore, it is now clear that small variations of conditions, such as pH, ionic strength or buffer, result in a significant variation of the revealed adducts.43,44
This prompted us to extend the previous studies and to deepen our understanding of the binding between [VIVO(L)2] complexes and proteins. In this work the BMOV–HEWL system was studied under experimental conditions different from those previously examined,42 to evaluate if the change affects the number and structure of the formed adducts, the observed sites, and the type of binding – covalent or non-covalent – to the protein. In particular, crystals of HEWL grown in 1.1 M NaCl and 0.1 M sodium acetate at pH 4.0 were exposed to saturated solutions of BMOV for 21 days. The systems were characterized by a combined application of X-ray crystallography, gel electrophoresis, matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) and liquid chromatography coupled to electrospray ionization tandem mass spectrometry analysis (ESI-LC-MSMS). The change of conditions from those previously reported42 resulted in an unexpected process involving BMOV transformation, opening of the maltolate ring and a significant modification of the N-terminal lysine side chain.
Experimental section
Crystallization, data collection, structure resolution and refinement
To crystallize HEWL, 1 μL of the protein (13 mg mL−1) was mixed with an equal volume of a reservoir solution containing 1.1 M sodium chloride and 0.1 M sodium acetate at pH 4.0. The drop was then equilibrated against the reservoir solution (500 μL). Crystals were grown within 1–2 days at 20 °C. Metal-free HEWL crystals were exposed to a solution of the reservoir saturated with BMOV for a soaking time of 21 days. Exposure of the crystals to the solutions containing BMOV leads to a change in the crystal colour from colourless to yellow. As suggested by some authors,45 the colour change suggests a transformation of the VC that could be also due to a possible partial oxidation of the V centre. Crystals of the BMOV–HEWL system were cryoprotected using a solution of the reservoir with 25% glycerol. X-ray diffraction data collections were carried out on different crystals on Beamline XRD2 at Elettra synchrotron, Trieste, Italy, at 100 K, using λ = 1.00 Å. The crystals diffract X-rays at 1.09–1.65 Å resolution. Data processing and scaling were performed using Global Phasing autoPROC pipeline46 (see ESI for further details†). Molecular Replacement method (using the Phaser program from CCP4 suite47) was used to solve the structures with PDB 193L48 as template. Manual model building was carried out using Coot.49 Since the structures of the BMOV–HEWL system are basically identical to each other (root mean square deviation, rmsd, between 0.08 and 0.13 Å) and possess the same V-containing fragments bound to the same V-binding sites, only the structure at 1.09 Å resolution is here described. The final model refines to a working R-factor of 0.129 and Rfree of 0.152 (Table S1 of ESI†) with anisotropic restrained refinements carried out using Refmac5.0.50 Pymol (https://www.pymol.org) was used to generate figures and PDB validation server51 to validate the final model. The structure was deposited in the PDB under the accession code 9FMY.SDS-PAGE and mass spectrometry analyses
Dissolved crystals of the adduct obtained in the reaction of HEWL with BMOV were treated with Laemmli buffer (100 mM Tris–HCl pH = 6.8, 4% sodium dodecyl sulfate (SDS), 20% glycerol, bromophenol blue, and 100 mM dithiothreitol), boiled for 10 min and loaded on a 12% SDS PAGE gel for electrophoresis analysis. Metal-free HEWL was loaded as a negative control. The protein bands, stained with colloidal Coomassie, were cut and subjected to in situ hydrolysis with both Asp-N (V162A, Promega) and trypsin (T6567, Sigma-Aldrich) proteases. The mixtures were analyzed using both 4800 MALDI TOF/TOF Analyzer (Sciex) and nanoLC-MS/MS using a LTQ Orbitrap XL (Thermo Fisher Scientific) coupled to a nanoACQUITY UPLC system (Waters). Peptide mixtures were dissolved in 10 μL of 0.2% formic acid. MALDI-TOF analyses were performed by mixing 0.5 μL of each sample at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with an α-cyano-4-hydroxycinnamic acid matrix (10 mg mL−1 in 70% acetonitrile and 30% trifluoroacetic acid (TFA) 0.2%). The instrument setup was in positive reflectron mode; the MS acquiring range spanned 500 to 5600 m/z. External calibration with a standard peptide mixture was performed for each sample. For the LC-MS/MS analysis, 8 μL from the same mixtures were injected. The samples were firstly concentrated onto a C18 capillary reverse-phase pre-column (20 mm, 180 μm, 5 μm, Waters Bioseparation Technology) and then fractionated onto a C18 capillary reverse-phase analytical column (250 mm, 75 μm, 1.8 μm, Waters Bioseparation Technology) working at a flow rate of 300 nL min−1. A linear chromatographic gradient was used, consisting of a ramp of eluent B (0.2% formic acid in 95% acetonitrile) in eluent A [0.2% formic acid in 2% acetonitrile in liquid chromatography-mass spectrometry (LC-MS) grade (Merck)], ranging from 5 to 95% in 66 min. Tandem mass spectrometric (MS/MS) analyses were performed in Data-Dependent Acquisition (DDA) mode, by fragmenting the 10 most intense ions in the collision-induced dissociation (CID) method.
1 ratio with an α-cyano-4-hydroxycinnamic acid matrix (10 mg mL−1 in 70% acetonitrile and 30% trifluoroacetic acid (TFA) 0.2%). The instrument setup was in positive reflectron mode; the MS acquiring range spanned 500 to 5600 m/z. External calibration with a standard peptide mixture was performed for each sample. For the LC-MS/MS analysis, 8 μL from the same mixtures were injected. The samples were firstly concentrated onto a C18 capillary reverse-phase pre-column (20 mm, 180 μm, 5 μm, Waters Bioseparation Technology) and then fractionated onto a C18 capillary reverse-phase analytical column (250 mm, 75 μm, 1.8 μm, Waters Bioseparation Technology) working at a flow rate of 300 nL min−1. A linear chromatographic gradient was used, consisting of a ramp of eluent B (0.2% formic acid in 95% acetonitrile) in eluent A [0.2% formic acid in 2% acetonitrile in liquid chromatography-mass spectrometry (LC-MS) grade (Merck)], ranging from 5 to 95% in 66 min. Tandem mass spectrometric (MS/MS) analyses were performed in Data-Dependent Acquisition (DDA) mode, by fragmenting the 10 most intense ions in the collision-induced dissociation (CID) method.
Results and discussion
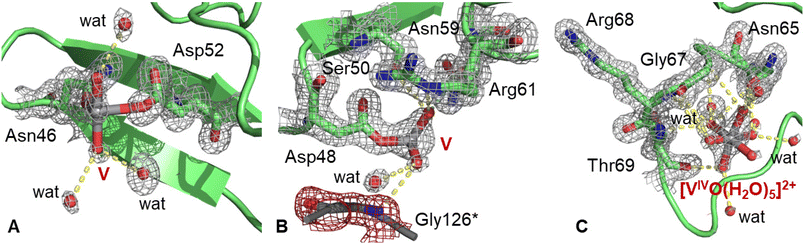
The best structure of the BMOV–HEWL system refines to an R-factor of 0.129 (Rfree = 0.152) against data at 1.09 Å resolution (Table S1 of ESI†) with anisotropic restrained refinements. In this structure, HEWL folding is not significantly affected by the V-compound binding. Cα root mean square deviations from the metal-free HEWL (PDB code 193L48) is 0.22 Å. Four distinct V binding sites were identified in the adduct. One V atom (occupancy = 0.30) was found close to the side chains of Asn46 and Asp52 (Fig. 2A). Such a site is involved in the binding of vanadium moieties like [VIVO(Cl)]+, [VIVO(phen)]2+ and [VIVO(bipy)]2+, where phen is 1,10-phenanthroline and bipy is 2,2′-bipyridine,52 but in our structure the V geometry is not well defined and it appears that at least two water molecules could be part of the metal coordination sphere. The V-containing fragment is solvent exposed and forms hydrogen bonds only with water molecules.An additional V atom (occupancy = 0.60) was observed close to the side chain of Asp48 (Fig. 2B), coordinating at least two oxygen atoms. This fragment forms hydrogen bonds with Ser50, Asn59 and Arg61 side chains and a water molecule and is in contact with the N atom of Gly126 of a symmetry-related molecule (Gly126*). Furthermore, a [VIVO(H2O)5]2+ ion (occupancy = 0.50) was also added to the model. This V-containing moiety is non-covalently bound to the protein and forms hydrogen bonds with the main chains of Gly67, Arg68 and Thr69, with the side chain of Asn65 and with water molecules (Fig. 2C). In this binding site, V adopts an octahedral geometry.
Finally, inspection of 2Fo–Fc and Fo–Fc electron density (e.d.) maps indicates the existence of a covalent modification of the side chain of the N-terminal lysine. The anomalous difference e.d. map corresponding to this region suggests the presence of two V atoms (Fig. S1 of the ESI†). The best interpretation of the e.d. map is reported in Fig. 3A. A schematic representation of the unusual structure modelled in the e.d. map is shown in Fig. 3B, compared to the structure of cis-[VIVO(malt)2(H22O)] reported in Fig. 3C. To build the (bis)V-containing fragment bound to Lys1, it was assumed that oxygen atoms were coordinated to the V centre. The interactions formed by the (bis)V-containing fragment with protein atoms are shown in Fig. S2 of the ESI:† it interacts with the side chains of Glu7, Arg14, water molecules, the main chains of Glu7, Arg14, Ser86, and Ile88, and the side chains of Arg14, His15, Thr89 and the (bis)V-containing fragment from a symmetry related molecule. The occupancy of the V centres in the structure is 0.60. In this respect, it should be noted that the distance between the NZ atom of Lys1 and the C atom of the (bis)V-containing fragment bound to the lysine side chain is 1.32 Å, thus suggesting the presence of a double bond N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C. To explain this result, a reaction between the amino group of the side chain of the N-terminal and the carbonyl group of the maltolato ligand could be guessed, with the nucleophilic addition generating a Schiff base. Moreover, a breakage of the maltolate ring has to be presumed, a process already observed in the literature in several studies,53–55 but never in protein-containing systems. It is known that pyrylium ions, once they are formed, can polymerize or react with O- and N-nucleophiles to produce pyridinium betains and isomaltol among other heterocyclic compounds (Fig. S3 of the ESI†).53 Furthermore, literature data support a quercetin-type oxygenative cleavage of malt−, with the formation of CO and 3-acetoxy-propenoic acid, the latter being chemically rather unstable and easily decomposing to acetate and 3-oxo-propanoic acid (Fig. S4 of the ESI†),54 and also the cleavage of malt− that produces 3-hydroxyacrylic acid and lactic acid (Fig. S5 of ESI†).55 As suggested by the inspection of Fig. 3, the (bis)V-containing fragment bound to the side chain of Lys1 is also coordinated to the side chain of Asp87* from a symmetry related molecule, suggesting the formation of a cross-linked HEWL dimer (Fig. S6 of the ESI†), mediated by one of the two V centres of the (bis)V-containing fragment. It must be observed that in the previous studies carried out in aqueous solution on the system BMOV–HEWL,1,56 there was no indication of these processes.
C. To explain this result, a reaction between the amino group of the side chain of the N-terminal and the carbonyl group of the maltolato ligand could be guessed, with the nucleophilic addition generating a Schiff base. Moreover, a breakage of the maltolate ring has to be presumed, a process already observed in the literature in several studies,53–55 but never in protein-containing systems. It is known that pyrylium ions, once they are formed, can polymerize or react with O- and N-nucleophiles to produce pyridinium betains and isomaltol among other heterocyclic compounds (Fig. S3 of the ESI†).53 Furthermore, literature data support a quercetin-type oxygenative cleavage of malt−, with the formation of CO and 3-acetoxy-propenoic acid, the latter being chemically rather unstable and easily decomposing to acetate and 3-oxo-propanoic acid (Fig. S4 of the ESI†),54 and also the cleavage of malt− that produces 3-hydroxyacrylic acid and lactic acid (Fig. S5 of ESI†).55 As suggested by the inspection of Fig. 3, the (bis)V-containing fragment bound to the side chain of Lys1 is also coordinated to the side chain of Asp87* from a symmetry related molecule, suggesting the formation of a cross-linked HEWL dimer (Fig. S6 of the ESI†), mediated by one of the two V centres of the (bis)V-containing fragment. It must be observed that in the previous studies carried out in aqueous solution on the system BMOV–HEWL,1,56 there was no indication of these processes.
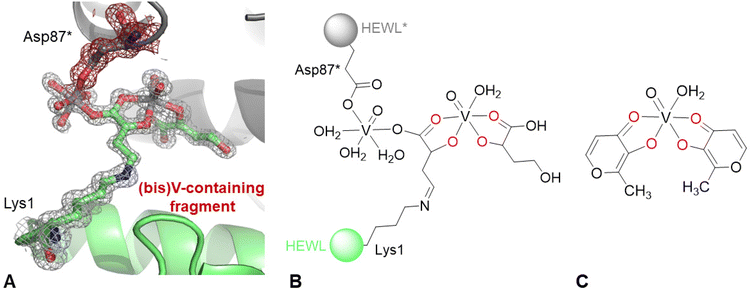 | ||
| Fig. 3 (A) Covalent modification of N-terminal lysine in the structure of the BMOV–HEWL system. 2Fo–Fc electron density map is reported at the 1.0 σ level in grey. Anomalous difference electron density map is reported in Fig. S1 of the ESI;† peak levels for V atoms are shown in Table S2 of the ESI.† Atoms from a symmetry related molecule are highlighted as * and coloured in grey; their density is shown in red. (B) Schematic representation of the covalent modification of N-terminal lysine. (C) Structure of cis-bis(maltolato)(aqua)oxidovanadium(IV), cis-[VIVO(malt)2(H2O)]. | ||
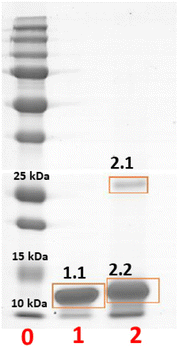
To further characterize this serendipitously found reaction product, crystals of HEWL were treated with BMOV, dissolved, and analysed by gel electrophoresis under denaturing conditions (Fig. 4). Crystals of the metal-free protein were used for comparison, as a negative control. The results of the experiment indicate the presence of an SDS-resistant HEWL dimer within the crystals of the protein treated with BMOV (band 2.1 in lane 2 of Fig. 4) in equilibrium with a metalated monomeric form of HEWL (band 2.2 in lane 2 of Fig. 4) that – in turn – has a molecular mass higher than that of metal-free HEWL (band 1.1 in lane 1 of Fig. 4). These findings are in perfect agreement with that expected on the basis of the crystal structure, which shows the existence of a cross-linked HEWL dimer with partial occupancy (0.60) and the binding of V-containing fragments to the protein.
The bands on the gel were then cut, hydrolysed with Asp-N protease and with trypsin, and investigated via MALDI-MS and ESI-LC-MSMS mass spectrometry methodologies. Peak assignments are reported in Table S3 of the ESI.† The inspection of the MS spectra derived from the enzymatic hydrolysis of the monomeric form of the V–protein adduct (band 2.2 in Fig. 4) was carried out in comparison with the results obtained for the band of the metal-free HEWL (band 1.1 in Fig. 4).
MALDI-MS and ESI-LC-MSMS fragmentation spectra reveal that band 2.2, derived from the crystal treated with BMOV and corresponding to metalated monomeric HEWL, generates many peptide ions absent in the analogous spectra of the metal-free HEWL band (band 1.1 in Fig. 4) in the 1110–1150 m/z and 2600–2800 m/z regions (Fig. S7 of the ESI,† panels A and B, respectively). In particular, in the 1110–1150 m/z region there are two signals, at m/z 1111.61 and 1127.60, assigned to the residues 62–68 carrying V + V![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and 2V
O and 2V![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively (Fig. 5A). Given that in the X-ray structure there is a [VIVO(H2O)5]2+ fragment non-covalently bound to the protein close to residues Asn65 and Asp66, it is possible to attribute these peaks to a covalent binding of V-containing fragments to HEWL, which could be induced by the operating conditions of the MS analysis.
O, respectively (Fig. 5A). Given that in the X-ray structure there is a [VIVO(H2O)5]2+ fragment non-covalently bound to the protein close to residues Asn65 and Asp66, it is possible to attribute these peaks to a covalent binding of V-containing fragments to HEWL, which could be induced by the operating conditions of the MS analysis.
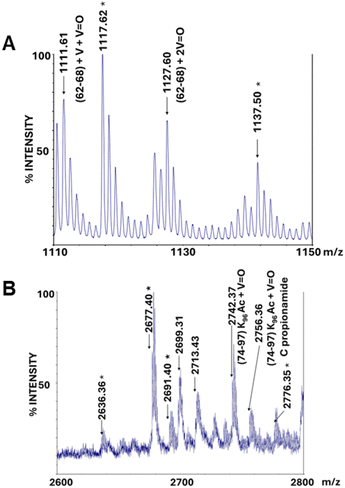 | ||
| Fig. 5 MALDI-MS spectra of the peptide mixture obtained from in situ hydrolysis of the metalated HEWL monomer (band 2.2 in Fig. 4): magnification of 1110–1150 (A) and 2600–2800 (B) m/z ranges. The signals also present in the reference (non-metalated HEWL, band 1.1 in Fig. 4) are marked with asterisks. | ||
Many signals in the 2600–2800 m/z region are present in the spectra of metalated monomeric HEWL and absent in those of metal-free protein. In particular, in this range, signals related to adducts of different nature deriving from the peptides 74–97 acetylated on Lys96 (m/z 2677.40) fall. Among all, we highlight signals at m/z 2742.37 and 2756.36; these peaks are attributed to the peptides 74–97, acetylated on Lys96 and carrying a V![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group, and its derivative containing a propionamidated cysteine formed during the migration of the protein on the polyacrylamide gel (Fig. 5B). These data agree with crystallographic findings which show the binding of a V-containing fragment close to the side chain of Asp87.
O group, and its derivative containing a propionamidated cysteine formed during the migration of the protein on the polyacrylamide gel (Fig. 5B). These data agree with crystallographic findings which show the binding of a V-containing fragment close to the side chain of Asp87.
The MS analysis of the band corresponding to the cross-linked protein dimer (band 2.1 in Fig. 4) was carried out in comparison with the band of the metalated monomeric HEWL (band 2.2 in Fig. 4). In the MALDI-MS spectra of the peptide mixture derived from the in situ hydrolysis of band 2.1, the peaks at m/z 1111 and 1127 are partially retained (Fig. S8 of ESI†), while the cluster included in the 2600–2800 m/z region is completely absent (Fig. 6A). Similarly, the peak at m/z 3007.42, attributable to the peptide encompassing residues 69–96, and thus containing Asp87, present in the spectra of the monomer (Fig. S9A of ESI†), disappears in the spectra of the cross-linked protein dimer (Fig. S9B of ESI†). Once again, these findings perfectly fit with crystallographic results, suggesting that signals associated with peptides containing the Asp87 residue disappear, since the latter might be involved in the cross-linking of the two protein chains of the HEWL dimer. Also, the canonical signal at m/z 2508.32, assigned to the peptides 76–96, in the spectra of the cross-linked protein dimer, is largely reduced in intensity (data not shown).
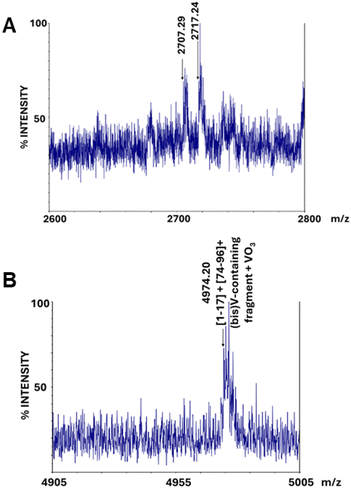 | ||
| Fig. 6 MALDI MS spectra of the peptide mixture obtained from in situ hydrolysis of the metalated HEWL dimer (band 2.1 in Fig. 4): magnification of 2600–2800 (A) and 4905–5005 (B) m/z ranges. | ||
Finally, the comparison between the spectra also reveals an additional relevant difference: in the spectra of the cross-linked HEWL dimer a peak at 4974.20 Da is observed (Fig. 6B). This peak could be attributed to the peptides formed by residues 1–17 and 74–96 plus the (bis)V-containing fragment and a VO3 group, in agreement with crystallographic data.
Conclusions
The study of the binding of vanadium compounds with proteins is essential to understand their biological activity and mechanism of action in the organism. More than 50 years of research has been carried out in this field and relevant papers have been published,57–60 but the understanding of the interaction of VCs with proteins is still in its infancy.14,26,61–64 Here, we have reported the X-ray structure and mass spectrometry data of the adduct formed upon the reaction of HEWL with BMOV in the solid state. The structure shows that the V compound transforms, and V-containing fragments bind covalently and non-covalently to protein atoms.Furthermore, an unexpected reaction product, which is highly reproducible in the solid state, is found: a (bis)V-containing fragment is bound at the interface between symmetry-related HEWL molecules. This unusual fragment is generated by BMOV transformation, re-assembly, and breakage of the maltolato ligand ring. It is involved in the formation of a cross-linked HEWL dimer; this dimer – in turn – is formed thanks to a modification of a Lys side chain and the binding of a V centre to the side chain of Asp87 from another protein molecule. The change in the structure of the Lys side chain is a sort of post-translational modification, which has never been reported before for a protein. The modification of Lys side chains by chemical methods could offer a valuable approach for enzyme functionalization.65
At this moment, we cannot exclude that the reaction that leads to the modification of Lys and opening of the maltolate ring can occur in solution. It is possible that the reaction is accelerated by the peculiar conditions that are formed within the porous protein crystals and by the high concentration of NaCl (1.1 M) and the low pH (4.0) used for protein crystallization. The NaCl concentration value is much larger than those in blood and cytosol, but the pH is close to that found in the gastric environment or in other acidic compartments within cells. Although there is no evidence that the observed process could occur in vivo, this possibility should be considered if special conditions are used in the biological assays and in vitro experiments.
Therefore, the possible formation of a modified lysine side chain when proteins are in the presence of BMOV in relation to the mechanism of action of the potential vanadium drugs formed by pyrones and pyridinones deserves deeper study. In any case, the data provide an indication of an unexpected reactivity of BMOV with proteins, which could be overlooked in other studies, suggesting that not everything is still understood about the reactivity of VCs with these biological macromolecules.
Similarly, we do not know if the formation of the unusual, cross-linked dimer is just a space filling artefact or if it could have a significance when BMOV binds to proteins. As a general comment, we highlight that it is known that intermolecular crystal contacts can provide insights into protein–protein interaction domains and can be biologically relevant.66
Overall, our results, once again,43,44 demonstrate that protein crystals are a useful vehicle to trap unusual structures and that, thanks to biological macromolecule X-ray crystallography, it is possible to unveil an unexpected reactivity of metal complexes with proteins. Data also underline that it is necessary to study in more detail the interaction between V drugs and proteins, and in general between metal-based drugs and proteins, specially under peculiar experimental conditions, since unexpected results may be obtained.
Author contributions
M. P., G. F. and A. M. performed crystallography experiments; I. C. performed SDS page and mass spectrometry analyses; E. G., M. M. and A. M. designed the concept and supervised the experiments; E. G., M. M. and A. M. wrote the manuscript. All authors have read and approved the final version of the manuscript.Data availability
Crystallographic data have been deposited at the PBD under the accession number 9FMY.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Elettra staff for their assistance during data collection. A. Merlino thanks MIUR PRIN 2022 – Cod. 2022JMFC3X, “Protein Metalation by Anticancer Metal-based Drugs” for financial support. E. G. thanks Fondazione di Sardegna (grant FdS2017Garribba) for the financial support.References
- V. Ugone, D. Sanna, G. Sciortino, D. C. Crans and E. Garribba, ESI-MS study of the interaction of potential oxidovanadium(IV) drugs and amavadin with model proteins, Inorg. Chem., 2020, 59, 9739–9755 CrossRef CAS.
- I. E. León, J. F. Cadavid-Vargas, A. Resasco, F. Maschi, M. A. Ayala, C. Carbone and S. B. Etcheverry, In vitro and in vivo antitumor effects of the VO-chrysin complex on a new three-dimensional osteosarcoma spheroids model and a xenograft tumor in mice, JBIC, J. Biol. Inorg. Chem., 2016, 21, 1009–1020 CrossRef PubMed.
- A. Banerjee, S. P. Dash, M. Mohanty, G. Sahu, G. Sciortino, E. Garribba, M. F. N. N. Carvalho, F. Marques, J. Costa Pessoa, W. Kaminsky, K. Brzezinski and R. Dinda, New VIV, VIVO, VVO, and VVO2 Systems: Exploring their Interconversion in Solution, Protein Interactions, and Cytotoxicity, Inorg. Chem., 2020, 59, 14042–14057 CrossRef CAS PubMed.
- J. Costa Pessoa, S. Etcheverry and D. Gambino, Vanadium compounds in medicine, Coord. Chem. Rev., 2015, 301–302, 24–48 CrossRef PubMed.
- E. Kioseoglou, S. Petanidis, C. Gabriel and A. Salifoglou, The chemistry and biology of vanadium compounds in cancer therapeutics, Coord. Chem. Rev., 2015, 301–302, 87–105 CrossRef CAS.
- D. Rehder, Perspectives for vanadium in health issues, Future Med. Chem., 2016, 8, 325–338 CrossRef CAS PubMed.
- Metallo-Drugs: Development and Action of Anticancer Agents, ed. A. Sigel, H. Sigel, E. Freisinger and R. K. O. Sigel, De Gruyter, 2018, vol. 18 Search PubMed.
- A. Ścibior, Ł. Pietrzyk, Z. Plewa and A. Skiba, Vanadium: Risks and possible benefits in the light of a comprehensive overview of its pharmacotoxicological mechanisms and multi-applications with a summary of further research trends, J. Trace Elem. Med. Biol., 2020, 61, 126508 CrossRef.
- M. Aureliano, N. I. Gumerova, G. Sciortino, E. Garribba, A. Rompel and D. C. Crans, Polyoxovanadates with emerging biomedical activities, Coord. Chem. Rev., 2021, 447, 214143 CrossRef CAS.
- A. L. De Sousa-Coelho, G. Fraqueza and M. Aureliano, Repurposing therapeutic drugs complexed to vanadium in cancer, Pharmaceuticals, 2024, 17, 12 CrossRef CAS.
- K. H. Thompson and C. Orvig, Boon and bane of metal ions in medicine, Science, 2003, 300, 936–939 CrossRef CAS.
- D. Rehder, The role of vanadium in biology, Metallomics, 2015, 7, 730–742 CrossRef CAS PubMed.
- E. Irving and A. Stoker, Vanadium compounds as PTP inhibitors, Molecules, 2017, 22, 2269 CrossRef PubMed.
- M. Aureliano, N. I. Gumerova, G. Sciortino, E. Garribba, C. C. McLauchlan, A. Rompel and D. C. Crans, Polyoxidovanadates’ interactions with proteins: An overview, Coord. Chem. Rev., 2022, 454, 214344 CrossRef CAS.
- S. Treviño, A. Díaz, E. Sánchez-Lara, B. L. Sanchez-Gaytan, J. M. Perez-Aguilar and E. González-Vergara, Vanadium in biological action: chemical, pharmacological aspects, and metabolic implications in diabetes mellitus, Biol. Trace Elem. Res., 2019, 188, 68–98 CrossRef.
- S. Treviño and A. Diaz, Vanadium and insulin: Partners in metabolic regulation, J. Inorg. Biochem., 2020, 208, 111094 CrossRef.
- C. Orvig, P. Caravan, L. Gelmini, N. Glover, F. G. Herring, H. Li, J. H. McNeill, S. J. Rettig and I. A. Setyawati, Reaction chemistry of BMOV, bis(maltolato)oxovanadium(IV), a potent insulin mimetic agent, J. Am. Chem. Soc., 1995, 117, 12759–12770 CrossRef CAS.
- X.-Q. Cong, M.-H. Piao, Y. Li, L. Xie and Y. Liu, Bis(maltolato)oxovanadium(IV) (BMOV) Attenuates Apoptosis in High Glucose-Treated Cardiac Cells and Diabetic Rat Hearts by Regulating the Unfolded Protein Responses (UPRs), Biol. Trace Elem. Res., 2016, 173, 390–398 CrossRef CAS PubMed.
- K. H. Thompson and C. Orvig, Vanadium in diabetes: 100 years from Phase 0 to Phase I, J. Inorg. Biochem., 2006, 100, 1925–1935 CrossRef CAS PubMed.
- K. H. Thompson, J. Lichter, C. LeBel, M. C. Scaife, J. H. McNeill and C. Orvig, Vanadium treatment of type 2 diabetes: a view to the future, J. Inorg. Biochem., 2009, 103, 554–558 CrossRef CAS PubMed.
- D. Rehder, J. Costa Pessoa, C. F. Geraldes, M. M. Castro, T. Kabanos, T. Kiss, B. Meier, G. Micera, L. Pettersson, M. Rangel, A. Salifoglou, I. Turel and D. Wang, In vitro study of the insulin-mimetic behaviour of vanadium(IV, V) coordination compounds, JBIC, J. Biol. Inorg. Chem., 2002, 7, 384–396 CrossRef CAS PubMed.
- V. Nischwitz, J. T. Davies, D. Marshall, M. González, J. L. Gómez Ariza and H. Goenaga-Infante, Speciation studies of vanadium in human liver (HepG2) cells after in vitro exposure to bis(maltolato)oxovanadium(IV) using HPLC online with elemental and molecular mass spectrometry, Metallomics, 2013, 5, 1685 CrossRef CAS PubMed.
- V. G. Yuen, C. Orvig and J. H. McNeill, Glucose-lowering effects of a new organic vanadium complex, bis(maltolato)oxovanadium(IV), Can. J. Physiol. Pharmacol., 1993, 71, 263–269 CrossRef CAS.
- J. Costa Pessoa and I. Tomaz, Transport of therapeutic vanadium and ruthenium complexes by blood plasma components, Curr. Med. Chem., 2010, 17, 3701–3738 CrossRef.
- K. D. Mjos and C. Orvig, Metallodrugs in medicinal inorganic chemistry, Chem. Rev., 2014, 114, 4540–4563 CrossRef CAS.
- D. Sanna and E. Garribba, Pharmacologically active vanadium species: Distribution in biological media and interaction with molecular targets, Curr. Med. Chem., 2021, 28, 7339–7384 CrossRef CAS PubMed.
- B. V. Rotterdam, CFM-Pharma https://cfmpharma.com/the-new-drug-vanadis.
- Y.-B. Wei and X.-D. Yang, Synthesis, characterization and anti-diabetic therapeutic potential of a new benzyl acid-derivatized kojic acid vanadyl complex, BioMetals, 2012, 25, 1261–1268 CrossRef CAS PubMed.
- M. J. Pereira, E. Carvalho, J. W. Eriksson, D. C. Crans and M. Aureliano, Effects of decavanadate and insulin enhancing vanadium compounds on glucose uptake in isolated rat adipocytes, J. Inorg. Biochem., 2009, 103, 1687–1692 CrossRef CAS.
- J. Costa Pessoa and I. Correia, Misinterpretations in evaluating interactions of vanadium complexes with proteins and other biological targets, Inorganics, 2021, 9, 17 CrossRef.
- P. Nunes, I. Correia, I. Cavaco, F. Marques, T. Pinheiro, F. Avecilla and J. Costa, Pessoa. Therapeutic potential of vanadium complexes with 1,10-phenanthroline ligands, quo vadis? Fate of complexes in cell media and cancer cells, J. Inorg. Biochem., 2021, 217, 111350 CrossRef CAS.
- Á. Dörnyei, S. Marcão, J. Costa Pessoa, T. Jakusch and T. Kiss, Interactions of Insulin–Mimetic Vanadium Complexes with the Cell Constituents ATP and Glutathione, Eur. J. Inorg. Chem., 2006, 2006, 3614–3621 CrossRef.
- T. Kiss, E. Kiss, G. Micera and D. Sanna, The formation of ternary complexes between VO(maltolate)2 and small bioligands, Inorg. Chim. Acta, 1998, 283, 202–210 CrossRef CAS.
- T. Kiss, E. Kiss, E. Garribba and H. Sakurai, Speciation of insulin-mimetic VO(IV)-containing drugs in blood serum, J. Inorg. Biochem., 2000, 80, 65–73 CrossRef CAS.
- B. D. Liboiron, K. H. Thompson, G. R. Hanson, E. Lam, N. Aebischer and C. Orvig, New insights into the interactions of serum proteins with bis(maltolato)oxovanadium(IV): transport and biotransformation of insulin-enhancing vanadium pharmaceuticals, J. Am. Chem. Soc., 2005, 127, 5104–5115 CrossRef CAS PubMed.
- D. Sanna, L. Bíró, P. Buglyó, G. Micera and E. Garribba, Biotransformation of BMOV in the presence of blood serum proteins, Metallomics, 2012, 4, 33–36 CAS.
- D. Sanna, G. Micera and E. Garribba, Interaction of insulin-enhancing vanadium compounds with human serum holo-transferrin, Inorg. Chem., 2013, 52, 11975–11985 CrossRef CAS PubMed.
- I. Correia, T. Jakusch, E. Cobbinna, S. Mehtab, I. Tomaz, N. V. Nagy, A. Rockenbauer, J. Costa Pessoa and T. Kiss, Evaluation of the binding of oxovanadium(IV) to human serum albumin, Dalton Trans., 2012, 41, 6477–6487 RSC.
- T. Kiss, T. Jakusch, D. Hollender, Á. Dörnyei, É. A. Enyedy, J. Costa Pessoa, H. Sakurai and A. Sanz-Medel, Biospeciation of antidiabetic VO(IV) complexes, Coord. Chem. Rev., 2008, 252, 1153–1162 CrossRef CAS.
- V. Ugone, D. Sanna, G. Sciortino, J.-D. Maréchal and E. Garribba, Interaction of vanadium(IV) species with Ubiquitin: a combined instrumental and computational approach, Inorg. Chem., 2019, 58, 8064–8078 CrossRef CAS.
- G. Sciortino, D. Sanna, V. Ugone, J.-D. Maréchal and E. Garribba, Integrated ESI-MS/EPR/computational characterization of the binding of metal species to proteins: vanadium drug–myoglobin application, Inorg. Chem. Front., 2019, 6, 1561–1578 RSC.
- G. Ferraro, M. Paolillo, G. Sciortino, E. Garribba and A. Merlino, Multiple and variable binding of pharmacologically active bis(maltolato)oxidovanadium(IV) to lysozyme, Inorg. Chem., 2022, 61, 16458–16467 CrossRef CAS PubMed.
- G. Ferraro, G. Tito, G. Sciortino, E. Garribba and A. Merlino, Stabilization and Binding of [V4O12]4− and Unprecedented [V20O54(NO3)]n− to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug VIVO(acetylacetonato)2, Angew. Chem., Int. Ed., 2023, 62, e202315577 CrossRef CAS.
- G. Tito, G. Ferraro, F. Pisanu, E. Garribba and A. Merlino, Non–Covalent and Covalent Binding of New Mixed–Valence Cage–like Polyoxidovanadate Clusters to Lysozyme, Angew. Chem., Int. Ed., 2024, 63, e202406669 CrossRef PubMed.
- M. Aureliano and D. C. Crans, Decavanadate (V10O286−) and oxovanadates: Oxometalates with many biological activities, J. Inorg. Biochem., 2009, 103, 536–546 CrossRef CAS.
- C. Vonrhein, C. Flensburg, P. Keller, A. Sharff, O. Smart, W. Paciorek, T. Womack and G. Bricogne, Data processing and analysis with the autoPROC, toolbox, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 293–302 CrossRef CAS.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, Phaser crystallographic software, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS.
- M. C. Vaney, S. Maignan, M. Riès-Kautt and A. Ducruix, High-Resolution Structure (1.33 Å) of a HEW Lysozyme Tetragonal Crystal Grown in the APCF Apparatus. Data and Structural Comparison with a Crystal Grown under Microgravity from SpaceHab-01 Mission, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1996, 52, 505–517 CrossRef CAS PubMed.
- P. Emsley and K. Cowtan, Coot: model-building tools for molecular graphics, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef.
- G. N. Murshudov, A. A. Vagin and E. J. Dodson, Refinement of macromolecular structures by the maximum-likelihood method, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1997, 53, 240–255 CrossRef CAS.
- H. Berman, K. Henrick and H. Nakamura, Announcing the worldwide Protein Data Bank, Nat. Struct. Mol. Biol., 2003, 10, 980–980 CrossRef CAS.
- M. F. A. Santos, G. Sciortino, I. Correia, A. C. P. Fernandes, T. Santos-Silva, F. Pisanu, E. Garribba and J. Costa Pessoa, Binding of VIVO2+, VIVOL, VIVOL2 and VVO2L Moieties to Proteins: X-ray/Theoretical Characterization and Biological Implications, Chem. – Eur. J., 2022, 28, e202200105 CrossRef CAS PubMed.
- V. A. Yaylayan and S. Mandeville, Stereochemical control of maltol formation in Maillard reaction, J. Agric. Food Chem., 1994, 42, 771–775 CrossRef CAS.
- C. M. L. Di Giuro, D. Buongiorno, E. Leitner and G. D. Straganz, Exploring the catalytic potential of the 3-His mononuclear nonheme Fe(II) center: discovery and characterization of an unprecedented maltol cleavage activity, J. Inorg. Biochem., 2011, 105, 1204–1211 CrossRef CAS PubMed.
- C. Kanzler, P. T. Haase, H. Schestkowa and L. W. Kroh, Antioxidant properties of heterocyclic intermediates of the Maillard reaction and structurally related compounds, J. Agric. Food Chem., 2016, 64, 7829–7837 CrossRef CAS PubMed.
- G. Sciortino, D. Sanna, V. Ugone, G. Micera, A. Lledós, J.-D. Maréchal and E. Garribba, Elucidation of binding site and chiral specificity of oxidovanadium drugs with lysozyme through theoretical calculations, Inorg. Chem., 2017, 56, 12938–12951 CrossRef CAS PubMed.
- M. Aureliano, Recent perspectives into biochemistry of decavanadate, World J. Biol. Chem., 2011, 2, 215–225 CrossRef PubMed.
- T. Tiago, M. Aureliano and C. Gutiérrez-Merino, Decavanadate binding to a high affinity site near the myosin catalytic centre inhibits F-actin-stimulated myosin ATPase activity, Biochemistry, 2004, 43, 5551–5561 CrossRef CAS.
- T. Tiago, P. Martel, C. Gutiérrez-Merino and M. Aureliano, Binding modes of decavanadate to myosin and inhibition of the actomyosin ATPase activity, Biochim. Biophys. Acta, Proteins Proteomics, 2007, 1774, 474–480 CrossRef CAS PubMed.
- M. Aureliano, A. L. De Sousa-Coelho, C. C. Dolan, D. A. Roess and D. C. Crans, Biological consequences of vanadium effects on formation of reactive oxygen species and lipid peroxidation, Int. J. Mol. Sci., 2023, 24, 5382 CrossRef CAS PubMed.
- J. Costa Pessoa, E. Garribba, M. F. A. Santos and T. Santos-Silva, Vanadium and proteins: Uptake, transport, structure, activity and function, Coord. Chem. Rev., 2015, 301–302, 49–86 CrossRef CAS.
- G. Sciortino, J.-D. Maréchal and E. Garribba, Integrated experimental/computational approaches to characterize the systems formed by vanadium with proteins and enzymes, Inorg. Chem. Front., 2021, 8, 1951–1974 RSC.
- J. Costa Pessoa, M. F. A. Santos, I. Correia, D. Sanna, G. Sciortino and E. Garribba, Binding of vanadium ions and complexes to proteins and enzymes in aqueous solution, Coord. Chem. Rev., 2021, 449, 214192 CrossRef.
- M. F. A. Santos and J. Costa Pessoa, Interaction of vanadium complexes with proteins: revisiting the reported structures in the protein data bank (PDB) since 2015, Molecules, 2023, 28, 6538 CrossRef CAS PubMed.
- A. D. Pagar, M. D. Patil, D. T. Flood, T. H. Yoo, P. E. Dawson and H. Yun, Recent advances in biocatalysis with chemical modification and expanded amino acid alphabet, Chem. Rev., 2021, 121, 6173–6245 CrossRef CAS.
- J. Janin and F. Rodier, Protein–protein interaction at crystal contacts, Proteins: Struct., Funct., Bioinf., 1995, 23, 580–587 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4qi01528b |
| This journal is © the Partner Organisations 2024 |