Open Access Article
Open Access ArticleSurface-enhanced Raman spectroscopy for the characterization of bacterial pellets of Staphylococcus aureus infected by bacteriophage†
Nasir Mehmood‡
a,
Muhammad Waseem Akram‡a,
Muhammad Irfan Majeed *a,
Haq Nawaz
*a,
Haq Nawaz *a,
Muhammad Aamir Aslamb,
Abdul Namana,
Muhammad Wasima,
Usman Ghaffara,
Ali Kamrana,
Sana Nadeema,
Naeema Kanwala and
Muhammad Imran
*a,
Muhammad Aamir Aslamb,
Abdul Namana,
Muhammad Wasima,
Usman Ghaffara,
Ali Kamrana,
Sana Nadeema,
Naeema Kanwala and
Muhammad Imran c
c
aDepartment of Chemistry, University of Agriculture Faisalabad, Faisalabad (38000), Pakistan. E-mail: irfan.majeed@uaf.edu.pk; haqchemist@yahoo.com
bInstitute of Microbiology, Faculty of Veterinary, University of Agriculture Faisalabad, Faisalabad (38000), Pakistan
cDepartment of Chemistry, Faculty of Science, King Khalid University, P.O. Box 9004, Abha (61413), Saudi Arabia
First published on 12th February 2024
Abstract
Drug-resistant pathogenic bacteria are a major cause of infectious diseases in the world and they have become a major threat through the reduced efficacy of developed antibiotics. This issue can be addressed by using bacteriophages, which can kill lethal bacteria and prevent them from causing infections. Surface-enhanced Raman spectroscopy (SERS) is a promising technique for studying the degradation of infectious bacteria by the interaction of bacteriophages to break the vicious cycle of drug-resistant bacteria and help to develop chemotherapy-independent remedial strategies. The phage (viruses)-sensitive Staphylococcus aureus (S. aureus) bacteria are exposed to bacteriophages (Siphoviridae family) in the time frame from 0 min (control) to 50 minutes with intervals of 5 minutes and characterized by SERS using silver nanoparticles as SERS substrate. This allows us to explore the effects of the bacteriophages against lethal bacteria (S. aureus) at different time intervals. The differentiating SERS bands are observed at 575 (C–C skeletal mode), 620 (phenylalanine), 649 (tyrosine, guanine (ring breathing)), 657 (guanine (COO deformation)), 728–735 (adenine, glycosidic ring mode), 796 (tyrosine (C–N stretching)), 957 (C–N stretching (amide lipopolysaccharides)), 1096 (PO2 (nucleic acid)), 1113 (phenylalanine), 1249 (CH2 of amide III, N–H bending and C–O stretching (amide III)), 1273 (CH2, N–H, C–N, amide III), 1331 (C–N stretching mode of adenine), 1373 (in nucleic acids (ring breathing modes of the DNA/RNA bases)) and 1454 cm−1 (CH2 deformation of saturated lipids), indicating the degradation of bacteria and replication of bacteriophages. Multivariate data analysis was performed by employing principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) to study the biochemical differences in the S. aureus bacteria infected by the bacteriophage. The SERS spectral data sets were successfully differentiated by PLS-DA with 94.47% sensitivity, 98.61% specificity, 94.44% precision, 98.88% accuracy and 81.06% area under the curve (AUC), which shows that at 50 min interval S. aureus bacteria is degraded by the replicating bacteriophages.
1. Introduction
Infectious diseases caused by pathogenic bacteria that are resistant to antibiotics result in more than a quarter of deaths worldwide each year, after cardiovascular diseases.1,2 Despite food safety measurements, pathogenic foodborne infections are the major cause of infectious diseases in the world.3 Staphylococcus aureus (S. aureus) is a facultative Gram-positive coccus, displaying toxin-mediated virulence, and is capable of causing various infections, including sepsis, pneumonia, endocarditis, meningitis, food poisoning and postsurgical wound infections.4–6 The S. aureus concentration of 106 to 108 CFU per gram of food is enough to induce food poisoning;7,8 therefore, there is a legal requirement in many countries around the world to test for and ensure that food is free of S. aureus contamination. This bacterium is mostly found in the atmosphere, water, dust, warm-blooded animals, human skin and nasal cavity, which can contaminate food and cause nosocomial infections and community-acquired diseases.4,7Some strains of S. aureus are highly resistant to antibiotics, including penicillin and methicillin, despite the development of novel drugs to combat these pathogenic bacteria, and there is an urgent need to develop effective drugs to overcome these infections.9–11 Bacteriophages are a valuable therapeutic alternative to antibiotics to overcome infectious bacteria.12 Bacteriophages are viruses that infect and lyse bacteria and these phages are used as bioagents to treat pathogenic infections.1,13 The lytic phage cycle reduces the capability of bacteria to become phage-resistant by rapidly killing the target host.14 The clinical applications of phages are control of living bactericidal or natural mutants, the use of phage-encoded bacteriolytic cell wall lysin as trans glycosidase, phage proteins as metabolic inhibitors and phage display in proteins fused to antibodies.1 Staphylococcal phages have been extensively used in genetic study and phage typing of S. aureus; they have prophages of genomes that change the phenotype, resulting in lysogenic changes with virulence factors.15,16 The lysogenic action results in changes in the host macrorestriction maps due to the integration of prophages into bacterial chromosomes.15
S. aureus bacteria can be detected by culture-based method, which is time consuming (4–7 days), and some immunological techniques, such as immunoprecipitation, immunoblotting and enzyme-labeled immunosorbent assay (ELISA), in which S. aureus can be identified by the specific antigen and antibody binding.2,17 Time-saving and fast techniques have been introduced for the automated and quantitative identification of S. aureus in food, including mass spectrometry and polymerase chain reaction (PCR).7,18,19 However, these techniques need highly trained technicians with expensive instrumentation and pretreatment processes.20,21 SERS-based bacterial detection can be helpful for monitoring food safety as this technique is rapid, nondestructive and provides fingerprints with high sensitivity due to the surface enhancement from using nanoparticles as the substrate.6,22,23
SERS, with its high chemical specificity and nondestructive detection ability, has been used to analyze the chemical composition of biological samples using silver nanoparticles for enhancement in cancer screening, HCV detection and intraoperative surgical measurements to detect bacteria with valuable identification of biological compounds.24,25 Silver (Ag) or gold (Au) nanoparticles are extensively used as SERS substrates for the target recognition of biological molecules, playing a crucial role in SERS accuracy and specificity. Silver nanoparticles (AgNPs) have been used in various areas, including in food, medical fields and various industries, owing to their optical, electrical, thermal and specific chemical and physical properties.26,27 AgNPs have a high surface plasmon resonance due to their small surface area and large number of active sites.27
SERS has been used to characterize different pathogens, including Listeria and Salmonella, by the implementation of bacteriophage interaction.28,29 In this work, the temporal changes in pellets of phage-sensitive infectious S. aureus after interaction with bacteriophages are characterized by SERS using silver nanoparticles as the substrate to study the inactivation of infectious S. aureus. Chemometric tools including principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) are used to classify the SERS spectra of these bacterial samples on the basis of their characteristic SERS features representative of the temporal (0 to 50 minutes at intervals of 5 minutes) biochemical changes in phage-sensitive infectious S. aureus pellets as a result of lysis caused by the bacteriophages.
2. Materials and methods
2.1. Bacteriophage interaction with bacteria
The samples for the isolation of S. aureus strains were obtained from hospitalized patients suffering from burn wounds admitted to the burn ward of Allied Hospital, Faisalabad, Pakistan. For sampling from humans, ethical approval was obtained from the Institutional Biosafety Committee (IBC), University of Agriculture Faisalabad, Pakistan. Permit number 1292/ORIC was issued by the IBC. A lytic bacteriophage against S. aureus was isolated from sewage water. A time series analysis of the effect of the bacteriophage on the killing of the bacteria was conducted by mixing the phage and bacteria in nutrient broth at MOI 1, i.e., 108 CFU mL−1 of phage was mixed with 108 CFU mL−1 of bacterial strain. After mixing freshly grown overnight bacterial cultures with the phage, samples were taken for analysis at time intervals of 5 minutes from 0 to 50 minutes. The bacteriophage used in this study belongs to the Siphoviridae family, as concluded from electron microscopic studies. Samples were centrifuged and pellets of the phage-exposed bacteria were separated by centrifugation at 8000 rpm for 10 min.2.2. Synthesis of silver nanoparticles
Silver nanoparticles (Ag NPs) of monodispersed and intermediate size for use as the substrate for the SERS measurements were prepared by the chemical reduction method. Briefly, 51 mg of silver nitrate (AgNO3) was added to 300 mL of deionized water and heated with continuous stirring to obtain a temperature of 100 °C temperature. Then, 75 mg of tri-sodium citrate (Na3C6H5O7) was added as a reducing agent and capping ligand. The mixture was then heated for about 2 hours on a hot plate to obtain a gray-coloured suspension of AgNPs, which was centrifuged to obtain nanoparticles with size of around 45–65 nm.30,312.3. SERS spectral acquisition
For the SERS spectral measurements, 40 μl of S. aureus pellet, dispersed in deionized water, after interaction with bacteriophages, is mixed with 40 μl of silver nanoparticles in Eppendorf and left for 30 minutes incubation time to allow the effective interaction of the pellets with the silver nanoparticles. An Optosky Raman spectrometer (ATR8300BS) with a 785 nm diode laser as the excitation source delivering 50 mW laser through a 40× objective lens is used for the spectral data measurements for pellets of bacteriophage-attacked S. aureus samples. For this purpose, 50 μl of incubation mixture was transferred to an aluminum slide for the spectral measurements. In the spectral range of 300–1600 cm−1, 15 SERS spectra were collected with integration time of 15 seconds. These conditions were kept constant for all the phage-exposed S. aureus pellet samples taken at different times, including 0 (control), 5, 10, 15, 20, 25, 30, 35, 40, 45 and 50 minute intervals.2.4. Pre-processing of SERS spectral data
SERS spectral raw data was preprocessed using MATLAB 7.8.0.347 with in-house built chemometric codes to remove unwanted components like noise, baseline and substrate in order to obtain useful information and enhance the prominence of features.32 All the spectral data was imported in a single matrix in MATLAB and algorithms were applied for the substrate removal, baseline correction, data smoothing and vector normalization. Data smoothing was done using the Savitzky–Golay algorithm33 while for the baseline correction polynomial and rubber band algorithms were applied.2.5. Chemometric analysis of SERS spectral data
Chemometric statistical tools including principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) were applied for the analysis of multivariate SERS processed data. The differentiation of the SERS spectral data for the pellets of the bacteriophage-interacted S. aureus in the timeframe of control (0 min) to 50 min at intervals of 5 minutes was achieved using chemometric analysis. PCA is a qualitative analysis technique that is used to study the differentiation and variability of datasets in the form of PCA scatter plots and PCA loadings. In PCA, the variability remains unchanged by reducing the dimensionality as it converts the correlated variables to uncorrelated variables. Maximum variability can be calculated from the first principal component (PC-1) while the second principal component (PC-2) show less variability than PC-1, and so on.PLS-DA is a versatile modeling tool that can be used for the calibration and validation of processed SERS datasets by classifying the SERS spectra of the timeframe-based bacteriophage-infected bacterial strain (S. aureus) samples. This tool classifies the SERS spectra of timeframe-based samples on the basis of the information of x-variables against y-variables. Randomization is applied to spectral data in a matrix to mix up the data, which is split into 60% calibration and 40% validation. Monte Carlo cross-validation is used to get the latent variables. The receiver operating characteristic (ROC) curve is used to evaluate the binary classification algorithm to get information about the false positive, true positive, false negative and true negative results of classification of the SERS spectral groups.
3. Results and discussion
3.1. Silver nanoparticle analysis
SEM analysis is used to study the surface morphology along with the size of silver nanoparticles, as shown as Fig. S7(a),† which indicates that AgNPs are spherical in shape leading to mono-dispersity with the average size of 53 nm (Fig. S7(b)†). Aggregation is due to the refined form of AgNPs of intermediate size from 45–65 nm. The surface is smooth with well-defined crystalline structure. Elemental analysis through EDX, as shown in Fig. S7(c),† shows that the weight percentage of silver is 85.38% without any contaminants, making it suitable for the enhancement of the SERS signal.The antibacterial activity of silver nanoparticles depends upon their size as small size particles (5, 7 and 10 nm), due to their high surface area and high particle penetration, show higher antibacterial activity when incubated for 24 hours at 37 °C (ref. 34–36) as compared to intermediate size nanoparticles of 45–65 nm (average 53 nm), which are used in the current study. It is also reported in the literature that the minimum inhibitory concentration (MIC) of silver nanoparticles of 10, 50 and 63 nm size against S. aureus when incubated for 24 hours are 80, 130 and 160 mg L−1, respectively. Moreover, the minimum bactericidal concentration (MBC) values are found to be 100, 160 and 200 mg mL−1 for silver nanoparticles of 10, 50 and 63 nm, respectively, which indicates that the AgNPs of the size 50 and 63 nm show very little antibacterial activity after 24 hours of incubation.35 Notably, in the current work, silver nanoparticles with intermediate particle size (45–65 nm) were used for the enhancement of Raman signals. This helped to improve the Raman spectral signal while the incubation of samples with AgNPs for 30 minutes showed no antibacterial effect on the bacterial samples because the incubation time is very low for the NPs to interact with the bacteria.
3.2. SERS mean spectra
Fig. 1 shows the SERS spectra of bacteriophage-infected S. aureus pellets samples taken at 0 (control) 5, 25, 35 and 50 minutes of the degradation process of infectious phage-sensitive bacteria. The mean SERS spectra starting from 0 to 50 minutes with intervals of 5 minutes are shown in Fig. S1.† There are three sets of time intervals, 5 to 20 minutes, 25 and 30 minutes, 35 to 45 minutes, in which each set has differentiating SERS features indicating the biochemical changes taking place owing to the interaction of the phage with the host bacterial pellet samples, but there are no significant peak differences among the SERS spectra of the same set except for intensity-based differences, which are shown in Fig. S2–S4,† respectively. The differentiating SERS peaks are associated with the biochemical components, including carbohydrates, proteins, amino acids, DNA/RNA and lipids. The solid lines labeled in the spectra represent the differentiating SERS peaks while the dotted dash lines represent the intensity-based differences of SERS peaks. The literature revealed that when S. aureus bacteria is infected by bacteriophage, the phage replication is completed in approximately 50 minutes, which is the latent period (time interval between the absorption and the beginning of the first burst) after the infection.37,38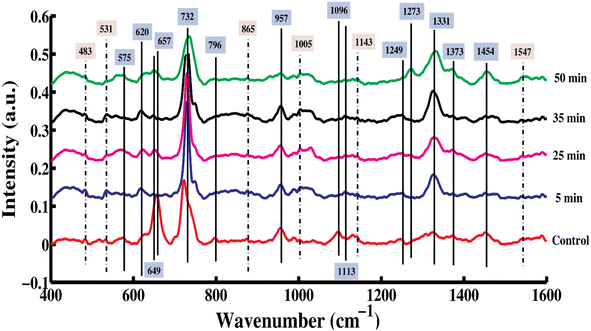 | ||
| Fig. 1 Mean SERS spectra of control and phage-exposed S. aureus bacterial samples including 5, 25, 35 and 50 minutes of exposure with the most differentiating time intervals of phage exposure. | ||
The differentiating SERS features include 575 (C–C skeletal mode), 620 (phenylalanine), 649 (tyrosine, guanine (ring breathing)), 657 (guanine (COO deformation)), 728–735 (adenine, glycosidic ring mode), 796 (tyrosine (C–N stretching)), 957 (C–N stretching (amide lipopolysaccharides)), 1096 (PO2 (nucleic acid)), 1113 (phenylalanine), 1249 (CH2 of amide III, N–H bending and C–O stretching (amide III)), 1273 (CH2, N–H, C–N, amide III), 1331 (C–N stretching mode of adenine), 1373 (in nucleic acids (ring breathing modes of the DNA/RNA bases)) and 1454 cm−1 (CH2 deformation of saturated lipids).
The intensity-based differences are denoted by dotted dash lines, which show that the biochemical features are present in all the bacteriophage-infected S. aureus pellet samples taken at 5 minute intervals from 0 to 50 minutes. These include 483 (C–C–C deformation), 531 (S–S stretching, C–O–C glycosidic ring deformation), 865 ((C–O–C) 1,4 glycosidic link), 1005 (C–C aromatic ring of phenylalanine and skeletal stretching of tryptophan, CCH stretching of carotenoids), 1143 (C–N amide stretching, C–C (stretching)) and 1547 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C asymmetric stretching).
C asymmetric stretching).
The SERS peaks observed at 483, 531, 575 and 865 cm−1 correspond to carbohydrates. The peak at 575 cm−1 is only observed in the control and disappeared in all other samples taken at 5–50 min, which means that the bacteriophage released the enzyme endolysins39 on the surface of the cell wall to destroy it, causing structural changes (S–S stretching, C–O–C glycosidic ring deformation) in peptidoglycan, which is associated with the carbohydrates. Peptidoglycan is the major component of the cell wall surrounding the cytoplasmic membrane; it is not vital for the cell wall to function, but acts as an Achilles' heel.40 Bands with differences in intensity were observed at 483 (C–C–C deformation), 531 (S–S stretching, C–O–C glycosidic ring deformation) and 865 cm−1 ((C–O–C) 1,4 glycosidic link). These bands all showed increasing intensity from the control sample to the one exposed to bacteriophages for 50 minutes. This suggests an increasing amount of carbohydrates with increasing exposure to bacteriophages, which is attributed to the presence of virion-associated active enzymes, specifically polysaccharide depolymerases, which break down polysaccharides.41
SERS bands linked to DNA are observed at 649 (tyrosine, guanine (ring breathing)), 657 (guanine (COO deformation)), 728–735 (adenine, glycosidic ring mode), 1096 (PO2 (nucleic acid)), 1331 (C–N stretching mode of adenine) and 1373 cm−1 (in nucleic acids (ring breathing modes of the DNA/RNA bases)) in which 657 (guanine (COO deformation)), 728 (adenine, glycosidic ring mode) and 1096 cm−1 (PO2 (nucleic acid)) are observed only in the control and not in samples from 5 to 50 minutes, which means that there is a high quantity of DNA in the S. aureus samples due to the presence of double stranded chromosomal plasmid DNA, which performs the replication and transformation of genes in the other host colonies.42,43 The plasmid of S. aureus, which produces the virulence factors and toxins and encodes the host survival elements, is resistant to antimicrobials, heavy metals and biocides, which enables S. aureus to adapt to different environments.44 The SERS spectral bands at 649 (tyrosine, guanine (ring breathing)), 735 (adenine, glycosidic ring mode) and 1331 cm−1 (C–N stretching mode of adenine) increase in intensity in the samples taken from 5 to 50 minutes, which corresponds to the increase of the DNA of the infected S. aureus, while the spectral peak at 1373 cm−1 (ring breathing modes of the DNA/RNA bases in nucleic acids) is only observed with higher intensity in the 50 minutes exposed bacterial sample, which means that the bacteriophage DNA is replicating and its contents are increasing at different time intervals.45,46
SERS peaks corresponding to proteins are observed at 620 (phenylalanine), 796 (tyrosine (C–N stretching)), 957 (C–N stretching (amide lipopolysaccharides)), 1005 (C–C aromatic ring of phenylalanine and skeletal stretching of tryptophan, CCH stretching of carotenoids), 1113 (phenylalanine), 1143 (C–N amide stretching, C–C (stretching)), 1249 (CH2 of amide III, N–H bending and C–O), 1273 (CH2, N–H, C–N, amide III) and 1547 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C asymmetric stretching) cm−1, of which the peaks at 620 (phenylalanine), 957 (C–N stretching (amide lipopolysaccharides)) and 1249 cm−1 (CH2 of amide III, N–H bending and C–O), are present only in the control and the samples from 5 minutes to 45 minutes of phage exposure time. Moreover, the peak at 796 cm−1 (tyrosine (C–N stretching)) is observed only in the control sample and the feature at 1113 cm−1 is observed in the samples with 5–50 minutes of exposure time and the peak at 1273 cm−1 appears in the 50 minutes phage-exposed samples. The SERS peaks observed at 1005, 1143 and 1547 cm−1 are present in all the phage-exposed samples with intensity-based differences. The S. aureus bacteria have virulence-associated cell wall anchor (CWA) proteins that are covalently attached to the peptidoglycan of the cell wall. These proteins are anchored to trans peptidases of the cell wall, which are known as sortases.47 The bacteriophage has a capsid protein that encases the viral genome and also a tail that is made of protein. These proteins help to recognize the receptor surface of the host bacteria.48,49 The SERS peak related to lipids at 1454 cm−1 (C
C asymmetric stretching) cm−1, of which the peaks at 620 (phenylalanine), 957 (C–N stretching (amide lipopolysaccharides)) and 1249 cm−1 (CH2 of amide III, N–H bending and C–O), are present only in the control and the samples from 5 minutes to 45 minutes of phage exposure time. Moreover, the peak at 796 cm−1 (tyrosine (C–N stretching)) is observed only in the control sample and the feature at 1113 cm−1 is observed in the samples with 5–50 minutes of exposure time and the peak at 1273 cm−1 appears in the 50 minutes phage-exposed samples. The SERS peaks observed at 1005, 1143 and 1547 cm−1 are present in all the phage-exposed samples with intensity-based differences. The S. aureus bacteria have virulence-associated cell wall anchor (CWA) proteins that are covalently attached to the peptidoglycan of the cell wall. These proteins are anchored to trans peptidases of the cell wall, which are known as sortases.47 The bacteriophage has a capsid protein that encases the viral genome and also a tail that is made of protein. These proteins help to recognize the receptor surface of the host bacteria.48,49 The SERS peak related to lipids at 1454 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C asymmetric stretching) only appeared in the 50 minutes phage-exposed bacterial sample and is probably associated with the S. aureus cytoplasmic membrane where the presence of lipids protects the membrane from rupturing.50 All peak assignment for the SERS spectra for the control and phage-exposed S. aureus bacterial samples have been given in Table 1.
C asymmetric stretching) only appeared in the 50 minutes phage-exposed bacterial sample and is probably associated with the S. aureus cytoplasmic membrane where the presence of lipids protects the membrane from rupturing.50 All peak assignment for the SERS spectra for the control and phage-exposed S. aureus bacterial samples have been given in Table 1.
| SERS peak (cm−1) | Assignment | Component | References |
|---|---|---|---|
| 483 | C–C–C deformation | Carbohydrates | 51 and 52 |
| 531 | S–S stretching, C–O–C glycosidic ring deformation | Carbohydrates | 53 and 54 |
| 575 | C–C skeletal mode | Carbohydrates | 54 |
| 620 | Phenylalanine | Protein | 53 and 54 |
| 649 | Tyrosine, guanine (ring breathing) | DNA | 6 and 51 |
| 657 | Guanine (COO deformation) | DNA | 55 and 56 |
| 728–732 | Adenine, glycosidic ring mode | DNA | 6, 57 and 58 |
| 743 | C–S stretching | Protein | 54 |
| 796 | Tyrosine (C–N stretching) | Protein | 54 |
| 865 | C–O–C 1,4-glycosidic link | Carbohydrates | 54 |
| 957 | C–N stretching (amide lipopolysaccharides) | Protein | 6, 57 and 59 |
| 1005 | C–C aromatic ring of phenylalanine and skeletal stretching of tryptophan, CCH stretching of carotenoids | Protein | 60 and 61 |
| 1096 | PO2 (nucleic acid) | DNA | 62 |
| 1113 | Phenylalanine | Protein | 63 |
| 1143 | C–N amide stretching, C–C (stretching) | Protein | 62 |
| 1249 | CH2 of amide III, N–H bending and C–O stretching (amide III) | Protein | 63 and 64 |
| 1273 | CH2, N–H, C–N, amide III | Protein | 59 and 65 |
| 1331 | C–N stretching mode of adenine | DNA | 55 and 56 |
| 1373 | In nucleic acids (ring breathing modes of the DNA/RNA bases) | DNA/RNA | 66 |
| 1454 | CH2 deformation of saturated lipids | Lipids | 55 |
| 1547 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C asymmetric stretching C asymmetric stretching |
Protein | 67 |
The SERS bands at 575 (C–C skeletal mode), 657 (guanine (COO deformation)), 728 (adenine, glycosidic ring mode), 743 (C–S stretching), 796 (tyrosine (C–N stretching)) and 1096 cm−1 (PO2 (nucleic acid)) were only found in the control samples, which show that the bacteriophage started inactivation of infectious S. aureus after the first minute of exposure, while the SERS bands at 620 (phenylalanine), 649 (tyrosine, guanine (ring breathing)), 1096 (PO2 (nucleic acid)), 1113 (phenylalanine), 1273 (CH2, N–H, C–N, amide III), 1331 (C–N stretching mode of adenine), 1373 (in nucleic acids (ring breathing modes of the DNA/RNA bases)) and 1454 cm−1 (CH2 deformation of saturated lipids) are only observed in the spectra of the phage-exposed samples, which confirms the replication of the viral genome to make further copies of bacteriophage.
Moreover, as described in the proposed schematic diagram of SERS characterization of bacteriophage-infected S. aureus bacteria (Fig. S8†), the identified SERS spectral features can be correlated with the possible known mechanism of interaction of the bacteriophages with S. aureus bacteria. The bacteriophage injects its viral DNA into S. aureus at 0 min and the reaction starts from 0 to 5 minutes. At 5 min, the disappearance of peaks at 657 and 1096 cm−1, assigned to DNA, indicates that the host DNA is degraded.68 Moreover, from 5 min onward, the biosynthesis of phage DNA and proteins takes place,68 which is characterized by the appearance of peaks at 620, 743, 1113 and 1273 cm−1 (proteins) and the shift of the SERS peak assigned to DNA at 728 cm−1 (control) to 732 cm−1 (different time intervals). In addition to this, the appearance of the SERS peaks at 1331 cm−1 (assigned to DNA) and 1454 cm−1 (lipids) indicates the bacteriophage replication in the host bacteria.
3.3. Principal component analysis (PCA)
PCA is a chemometric statistical tool that can be used to differentiate the SERS spectral data sets for the inactivation of S. aureus by the interaction of the bacteriophage in the time frame from 0 to 50 minutes. Fig. 2 shows the PCA scatter plot of the SERS spectra for the control sample and samples exposed to the bacteriophage for 5, 25, 35 and 50 minutes. The scatter plot for the samples from 0 to 50 minutes is shown in Fig. S5.† In Fig. 2 the red dots represent the control SERS spectra, the blue dots represent 5 min phage-exposed bacteria, the purple dots represent 25 min phage-exposed bacteria, the black dots are for the 35 min phage-exposed bacteria and the green dots represent the 50 min phage-exposed bacteria. The spectra for the 5 min, 25 min and 35 min samples are along the positive side of the first principal component (PC-1) while the SERS spectra of the control are clustered at the negative side of PC-1 with explained variability of 67.95%. The SERS spectra for the exposure time of 50 min and the control are differentiated along the second principal component (PC-2) with 25.20% explained variability.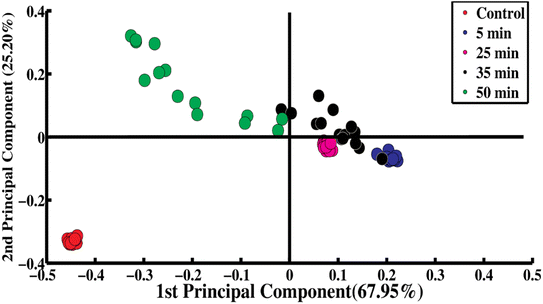 | ||
| Fig. 2 PCA scatter plot of SERS spectra of control and phage-exposed S. aureus bacterial samples including 5, 25, 35 and 50 minutes of exposure. | ||
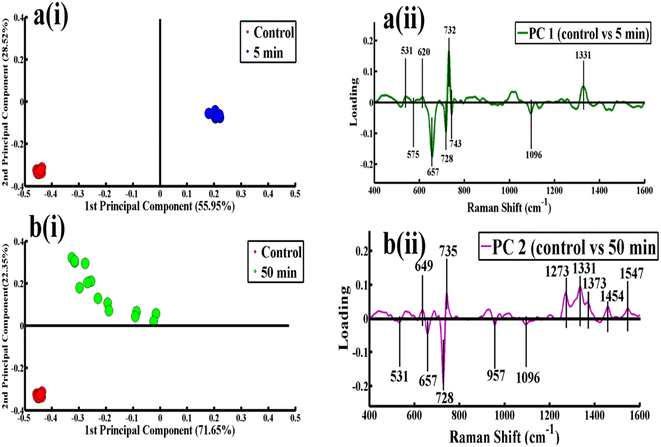
Fig. 3 shows the pairwise comparison of the SERS spectra for the control and the first (5 min) and last interval (50 min) of phage-exposed bacteria with their scatter plots and loadings. Fig. 3a(i) presents the comparison of the control with the first sample (5 min) where spectra are differentiated along PC-1 with 55.95% explained variability, in which the red dots on the negative side represent the control while the blue dots on the positive side represent the spectra of the 5 min phage-exposed bacteria. This variability shows that inactivation of infectious S. aureus started even within the first 5 min of exposure. Fig. 3a(ii) shows pairwise loadings of the control and first interval (5 min) phage-exposed bacteria, in which negative loadings represent the spectra of the control sample, which include peaks at 575 (C–C skeletal mode), 657 (guanine (COO deformation)), 728 (adenine, glycosidic ring mode), 743 (C–S stretching) and 1096 cm−1 (PO2 (nucleic acid)). The positive loadings represent the 5 min phage-exposed bacterial samples, which include 531 (S–S stretching, C–O–C glycosidic ring deformation), 620 (phenylalanine), 732 (adenine, glycosidic ring mode) and 1331 cm−1 (C–N stretching mode of adenine). Fig. 3b(i) shows a comparison of the SERS spectral data for the control sample with the sample taken after 50 min of phage exposure, which are differentiated along PC-2 with 22.35% explained variability, in which the red dots on the negative side represent the control while the green dots on the positive side represents the last interval (50 min). This variability shows that at the last interval of phage exposure replicated bacteriophage cells are produced and able to infect other infectious cells. Fig. 3b(ii) shows the pairwise loadings of the samples for the control and 50 min phage-exposed bacteria, in which negative loadings represent the spectra of the control, which includes 531 (S–S stretching, C–O–C glycosidic ring deformation), 657 (guanine (COO deformation)), 728 (adenine, glycosidic ring mode), 957 (C–N stretching (amide lipopolysaccharides)) and 1096 cm−1 (PO2 (nucleic acid)) while positive loadings represent the 50 min phage-exposed bacteria, which include 649 (tyrosine, guanine (ring breathing)), 732 (adenine, glycosidic ring mode), 1273 (CH2, N–H, C–N, amide III), 1331 (C–N stretching mode of adenine), 1373 (in nucleic acids (ring breathing modes of the DNA/RNA bases)), 1454 (CH2 deformation of saturated lipids) and 1547 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C asymmetric stretching).
C asymmetric stretching).
The SERS spectral loadings are taken by comparing the SERS spectra acquired at different intervals with the control; the samples taken at 5–45 min intervals are differentiated along PC-1, which has less variability because the S. aureus is in the process of degradation due to the replication of the bacteriophage. At the 50 min time interval, the S. aureus is completely degraded and bacteriophage replication has been completed as it is indicated by the SERS spectral features in the mean spectral plot (Fig. 1). This is why the SERS spectra acquired at 50 min are more differentiated along PC-2 from the control (S. aureus before attack of bacteriophage); hence the PCA loadings for the comparison of these groups of SERS spectra are very different from those acquired at other intervals.
3.4. Partial least squares-discriminant analysis (PLS-DA)
The chemometric tool PLS-DA was used to differentiate and discriminate the SERS spectra of bacteriophage-infected S. aureus at different time intervals. The most differentiating data sets for the time intervals of 0 (control), 5, 25, 35 and 50 minutes of exposure are used to build the PLS-DA model. To build the PLS-DA model, the five latent variables were selected by using Monte Carlo cross-validation by splitting the data sets into 60% calibration and 40% validation spectral data.Fig. 4 shows the scatter score of the most differentiating data sets of phage-exposed bacteria for the 5, 25, 35 and 50 minutes of exposure time. Red dots represent the spectra of the control sample, present on the negative side of the y-axis, blue dots present on the positive side of y-axis represent spectra for 5 min of exposure, black dots present on the positive side of the y-axis represent 25 min of exposure, purple dots present on the positive side of the y-axis represent 35 min of exposure and green dots present on the negative side of the y-axis represent 50 min of exposure. The scatter scores clearly differentiate the spectral data sets of the S. aureus samples exposed to the bacteriophage for different time intervals. It shows that bacteriophage attack started the inactivation of S. aureus even at 5 minutes of exposure as the SERS spectra for the control are clustered on the negative side of the y-axis while those for the 5 min exposure interval are clustered on the positive side of the y-axis. The spectra for the exposure times of 5–45 minutes are clustered on the positive side of the y-axis and representative dots are closer to the control (negative y-axis) with increasing exposure time, which shows the continuation of the inactivation process of S. aureus. Conversely, the spectra for the 50 minutes interval are on the negative side of the y-axis near to the control, which explains that at this interval the S. aureus is fully inactivated and the lytic or lysogenic cycle69,70 is completed, which means the bacteriophage has been replicated after the cell bursting process. The scatter score plot of the SERS spectra for these phage-exposed bacterial samples provides efficient discrimination and classification of SERS data sets with sensitivity of 94.47%, specificity of 98.61%, precision of 94.44% and accuracy of 98.88%.
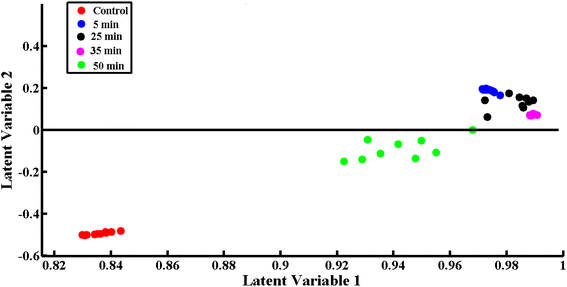 | ||
| Fig. 4 PLS-DA scatter score of SERS spectra of control (0 minutes) and 5, 25, 35 and 50 minutes phage-exposed S. aureus bacterial samples with most differentiating time intervals of phage exposure. | ||
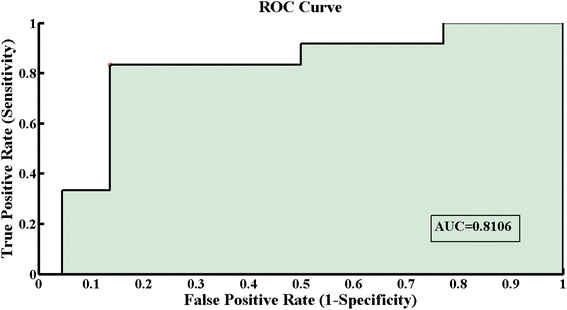
Fig. 5 shows the receiver operating curve (ROC) with an area under cure (AUC) value of 0.8106, which is near to 1, showing the best fitting model with high accuracy as an AUC near to zero indicates an inaccurate model. Table 2 presents the AUC, sensitivity, specificity, precision and accuracy values, indicating the excellent performance of this classification model. PLS-DA showed that the SERS data sets for the bacteriophage-infected S. aureus bacterial samples are well differentiated and provided a best fit model to discriminate the inactivation process of S. aureus bacteria.
| Parameters | Values |
|---|---|
| AUC | 0.8106 |
| Sensitivity | 94.47% |
| Specificity | 98.61% |
| Precision | 94.44% |
| Accuracy | 98.88% |
4. Conclusions
SERS was found to be a very effective technique for the temporal study of bacteriophage-exposed S. aureus bacterial samples. The characteristic SERS spectral features were identified to differentiate SERS spectra for the control and phage-exposed S. aureus bacterial samples. Chemometric statistical tools including PCA and PLS-DA were applied to differentiate and classify the SERS spectral data sets for the control and phage-exposed S. aureus bacterial samples. The SERS bands seen in the control sample at 575 (C–C skeletal mode), 657 (guanine (COO deformation)), 728 (adenine, glycosidic ring mode), 743 (C–S stretching) and 1096 cm−1 (PO2 (nucleic acid)) disappeared even in the first exposure interval (5 minutes), which shows that the bacteriophage started inactivation of infectious S. aureus in the first few minutes of exposure. The SERS bands observed at 620 (phenylalanine), 649 (tyrosine, guanine (ring breathing)), 1113 (phenylalanine), 1273 (CH2, N–H, C–N, amide III), 1331 (C–N stretching mode of adenine), 1373 (in nucleic acids (ring breathing modes of the DNA/RNA bases)) and 1454 cm−1 (CH2 deformation of saturated lipids) are only observed in the SERS spectra of phage-exposed S. aureus bacterial samples with exposure intervals of 5–50 minutes, which confirms the replication of the viral genome. PCA differentiated the exposure timeframe-based SERS spectral data sets with respect to the control, showing that the S. aureus bacteria are inactivated and the phage replicates and new copies of the viral genome are produced. PLS-DA is also an effective tool for differentiating uninfected and bacteriophage-infected S. aureus samples, which shows the continuous process of inactivation of this bacterium. PLS-DA discriminated and differentiated the SERS data sets with 94.47% sensitivity, 98.61% specificity, 94.44% precision, 98.88% accuracy and 81.06% AUC value, indicating the excellent performance of the classification model. Moreover, the bacteriophage only acts on specific species of bacteria due to its high specificity and narrow cleavage spectrum, which is a limitation of the current study as different Staphylococcus bacterial species may be affected by the bacteriophage to different extents and the mode of action may be different, hence leading to different spectral profiles. Thus, future studies should explore the application of SERS for the analysis of the interaction of bacteriophages with different bacterial species.Conflicts of interest
The authors have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
M. Imran expresses his appreciation to the Deanship of Scientific Research at King Khalid University, Saudi Arabia, for funding this work through the research group program under grant number R.G.P. 2/522/44.References
- S. Matsuzaki, M. Yasuda, H. Nishikawa, M. Kuroda, T. Ujihara, T. Shuin, Y. Shen, Z. Jin, S. Fujimoto and M. Nasimuzzaman, J. Infect. Dis., 2003, 187, 613–624 CrossRef CAS PubMed.
- X.-Y. Wang, J.-Y. Yang, Y.-T. Wang, H.-C. Zhang, M.-L. Chen, T. Yang and J.-H. Wang, Talanta, 2021, 221, 121668 CrossRef CAS PubMed.
- E. Abdollahzadeh, M. Rezaei and H. Hosseini, Food Control, 2014, 35, 177–183 CrossRef CAS.
- C. Liu, C. Shi, M. Li, M. Wang, C. Ma and Z. Wang, Front. Chem., 2019, 7, 124 CrossRef CAS PubMed.
- C. Catala, B. Mir-Simon, X. Feng, C. Cardozo, N. Pazos-Perez, E. Pazos, S. Gómez-de Pedro, L. Guerrini, A. Soriano and J. Vila, Adv. Mater. Technol., 2016, 1, 1600163 CrossRef.
- S. Wang, H. Dong, W. Shen, Y. Yang, Z. Li, Y. Liu, C. Wang, B. Gu and L. Zhang, RSC Adv., 2021, 11, 34425–34431 RSC.
- B. Alarcon, B. Vicedo and R. Aznar, J. Appl. Microbiol., 2006, 100, 352–364 CrossRef CAS PubMed.
- A. Wieneke, D. Roberts and R. Gilbert, Epidemiol. Infect., 1993, 110, 519–531 CrossRef CAS PubMed.
- S. Chang, D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp and W. J. Brown, N. Engl. J. Med., 2003, 348, 1342–1347 CrossRef PubMed.
- A. Liapikou and A. Torres, Expert Opin. Emerging Drugs, 2013, 18, 291–305 CrossRef CAS PubMed.
- J. Kurlenda and M. Grinholc, Acta Biochim. Pol., 2012, 59, 171–184 CrossRef CAS PubMed.
- B. Burrowes, D. R. Harper, J. Anderson, M. McConville and M. C. Enright, Expert Rev. Anti-Infect. Ther., 2011, 9, 775–785 CrossRef PubMed.
- R. Keary, O. McAuliffe, R. Ross, C. Hill, J. O'Mahony and A. Coffey, Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, ed. A. Méndez-Vilas, Formatex Research Center, Badajoz, Spain, 2013, pp. 1028–1040 Search PubMed.
- M. Skurnik, M. Pajunen and S. Kiljunen, Biotechnol. Lett., 2007, 29, 995–1003 CrossRef CAS PubMed.
- R. Pantůček, J. Doškař, V. Růžičková, P. Kašpárek, E. Oráčová, V. Kvardová and S. Rosypal, Arch. Virol., 2004, 149, 1689–1703 CrossRef PubMed.
- R. P. Novick, Molecular Biology of the Staphylococci, VCH Publishers, 1990 Search PubMed.
- M. Ruan, C.-G. Niu, G.-M. Zeng, P.-Z. Qin, X.-Y. Wang, D.-W. Huang, J. Huang and C.-Z. Fan, Microchim. Acta, 2011, 175, 105–112 CrossRef CAS.
- T. Trnčíková, V. Hrušková, K. Oravcová, D. Pangallo and E. Kaclíková, Food Anal. Methods, 2009, 2, 241–250 CrossRef.
- H. Li and J. Zhu, Anal. Chem., 2018, 90, 12108–12115 CrossRef CAS PubMed.
- N. Nicolaou, Y. Xu and R. Goodacre, Anal. Chem., 2011, 83, 5681–5687 CrossRef CAS PubMed.
- M. Yu, H. Wang, F. Fu, L. Li, J. Li, G. Li, Y. Song, M. T. Swihart and E. Song, Anal. Chem., 2017, 89, 4085–4090 CrossRef CAS PubMed.
- B. Yan, A. Thubagere, W. R. Premasiri, L. D. Ziegler, L. Dal Negro and B. M. Reinhard, ACS Nano, 2009, 3, 1190–1202 CrossRef CAS PubMed.
- X. Meng, H. Wang, N. Chen, P. Ding, H. Shi, X. Zhai, Y. Su and Y. He, Anal. Chem., 2018, 90, 5646–5653 CrossRef CAS PubMed.
- A. Sengupta, M. Mujacic and E. J. Davis, Anal. Bioanal. Chem., 2006, 386, 1379–1386 CrossRef CAS PubMed.
- H. Nawaz, N. Rashid, M. Saleem, M. Asif Hanif, M. Irfan Majeed, I. Amin, M. Iqbal, M. Rahman, O. Ibrahim and S. Baig, J. Raman Spectrosc., 2017, 48, 697–704 CrossRef CAS.
- P. Mukherjee, A. Ahmad, D. Mandal, S. Senapati, S. R. Sainkar, M. I. Khan, R. Parishcha, P. Ajaykumar, M. Alam and R. Kumar, Nano Lett., 2001, 1, 515–519 CrossRef CAS.
- X.-F. Zhang, Z.-G. Liu, W. Shen and S. Gurunathan, Int. J. Mol. Sci., 2016, 17, 1534 CrossRef PubMed.
- N. R. Stambach, S. A. Carr, C. R. Cox and K. J. Voorhees, Viruses, 2015, 7, 6631–6641 CrossRef CAS PubMed.
- L.-L. Tay, P.-J. Huang, J. Tanha, S. Ryan, X. Wu, J. Hulse and L.-K. Chau, Chem. Commun., 2012, 48, 1024–1026 RSC.
- H. Fang, X. Zhang, S. J. Zhang, L. Liu, Y. M. Zhao and H. J. Xu, Sens. Actuators, B, 2015, 213, 452–456 CrossRef CAS.
- S. Nasir, M. I. Majeed, H. Nawaz, N. Rashid, S. Ali, S. Farooq, M. Kashif, S. Rafiq, S. Bano and M. N. Ashraf, Photodiagn. Photodyn. Ther., 2021, 33, 102152 CrossRef CAS PubMed.
- H. Nawaz, F. Bonnier, P. Knief, O. Howe, F. M. Lyng, A. D. Meade and H. J. Byrne, Analyst, 2010, 135, 3070–3076 RSC.
- H. J. Butler, B. R. Smith, R. Fritzsch, P. Radhakrishnan, D. S. Palmer and M. J. Baker, Analyst, 2018, 143, 6121–6134 RSC.
- N. Durán, P. D. Marcato, R. D. Conti, O. L. Alves, F. Costa and M. Brocchi, J. Braz. Chem. Soc., 2010, 21, 949–959 CrossRef.
- S. Agnihotri, S. Mukherji and S. Mukherji, RSC Adv., 2014, 4, 3974–3983 RSC.
- S. Tang and J. Zheng, Adv. Healthcare Mater., 2018, 7, 1701503 CrossRef PubMed.
- L. Li and Z. Zhang, Mol. Biol. Rep., 2014, 41, 5829–5838 CrossRef CAS PubMed.
- T. Feng, S. Leptihn, K. Dong, B. Loh, Y. Zhang, M. I. Stefan, M. Li, X. Guo and Z. Cui, Front. Microbiol., 2021, 12, 602902 CrossRef PubMed.
- Ł. Grabowski, K. Łepek, M. Stasiłojć, K. Kosznik-Kwaśnicka, K. Zdrojewska, M. Maciąg-Dorszyńska, G. Węgrzyn and A. Węgrzyn, Microbiol. Res., 2021, 248, 126746 CrossRef PubMed.
- M. Wang, G. Buist and J. M. van Dijl, FEMS Microbiol. Rev., 2022, 46, fuac025 CrossRef PubMed.
- A. Latka, B. Maciejewska, G. Majkowska-Skrobek, Y. Briers and Z. Drulis-Kawa, Appl. Microbiol. Biotechnol., 2017, 101, 3103–3119 CrossRef CAS PubMed.
- T. Gu, S. Zhao, Y. Pi, W. Chen, C. Chen, Q. Liu, M. Li, D. Han and Q. Ji, Chem. Sci., 2018, 9, 3248–3253 RSC.
- Y. Li, K. Kurokawa, M. Matsuo, N. Fukuhara, K. Murakami and K. Sekimizu, Mol. Genet. Genomics, 2004, 271, 447–457 CrossRef CAS PubMed.
- L. Neyaz, N. Rajagopal, H. Wells and M. K. Fakhr, Front. Microbiol., 2020, 11, 223 CrossRef PubMed.
- C. Weigel and H. Seitz, FEMS Microbiol. Rev., 2006, 30, 321–381 CrossRef CAS PubMed.
- E. Noble, M. M. Spiering and S. J. Benkovic, Viruses, 2015, 7, 3186–3200 CrossRef CAS PubMed.
- K. A. Lacey, J. A. Geoghegan and R. M. McLoughlin, Pathogens, 2016, 5, 22 CrossRef PubMed.
- F. L. Nobrega, M. Vlot, P. A. de Jonge, L. L. Dreesens, H. J. Beaumont, R. Lavigne, B. E. Dutilh and S. J. Brouns, Nat. Rev. Microbiol., 2018, 16, 760–773 CrossRef CAS PubMed.
- M. Sanz-Gaitero, M. Seoane-Blanco and M. J. van Raaij, Bacteriophages: Biology, Technology, Therapy, 2021, pp. 19–91 Search PubMed.
- J. B. Parsons, J. Yao, M. W. Frank, P. Jackson and C. O. Rock, J. Bacteriol., 2012, 194, 5294–5304 CrossRef CAS PubMed.
- S. Adesoye and K. Dellinger, Sens. Bio-Sens. Res., 2022, 37, 100499 CrossRef.
- F. M. Lyng, E. Ó. Faoláin, J. Conroy, A. Meade, P. Knief, B. Duffy, M. Hunter, J. Byrne, P. Kelehan and H. Byrne, Exp. Mol. Pathol., 2007, 82, 121–129 CrossRef CAS PubMed.
- R. Z. A. Bari, H. Nawaz, M. I. Majeed, N. Rashid, M. Tahir, H. M. ul Hasan, S. Ishtiaq, N. Sadaf, A. Raza and A. Zulfiqar, Anal. Lett., 2023, 56, 1351–1365 CrossRef CAS.
- M. Saleem, H. Nawaz, M. I. Majeed, N. Rashid, F. Anjum, M. Tahir, R. Shahzad, A. Sehar, A. Sabir and N. Rafiq, Photodiagn. Photodyn. Ther., 2023, 41, 103278 CrossRef PubMed.
- F. U. Ciloglu, A. Caliskan, A. M. Saridag, I. H. Kilic, M. Tokmakci, M. Kahraman and O. Aydin, Sci. Rep., 2021, 11, 18444 CrossRef CAS PubMed.
- X. Chen, M. Tang, Y. Liu, J. Huang, Z. Liu, H. Tian, Y. Zheng, M. L. de La Chapelle, Y. Zhang and W. Fu, Microchim. Acta, 2019, 186, 1–8 CrossRef PubMed.
- Y. Liu, H. Zhou, Z. Hu, G. Yu, D. Yang and J. Zhao, Biosens. Bioelectron., 2017, 94, 131–140 CrossRef CAS PubMed.
- E. Akanny, A. Bonhommé, F. Bessueille, S. Bourgeois and C. Bordes, Appl. Spectrosc. Rev., 2021, 56, 380–422 CrossRef CAS.
- J. Li, C. Wang, L. Shi, L. Shao, P. Fu, K. Wang, R. Xiao, S. Wang and B. Gu, Microchim. Acta, 2019, 186, 1–12 CrossRef PubMed.
- W. E. Huang, M. Li, R. M. Jarvis, R. Goodacre and S. A. Banwart, Adv. Appl. Microbiol., 2010, 70, 153–186 CAS.
- J. Jehlička, H. G. Edwards and A. Oren, Appl. Environ. Microbiol., 2014, 80, 3286–3295 CrossRef PubMed.
- Y. Zhu, S. Liu, L. Zhao, N. Wang, M. Li, D. Liang, G. Zhao, W. Liu, L. Sun and L. Xu, PDMS Film SERS Substrate for Rapid Detection of Foodborne Pathogens in Beef, 2023 Search PubMed.
- K. Li, Y. Yang, C. Xu, Y. Ye, L. Huang, L. Sun, Y. Cai, W. Zhou, Y. Ge and Y. Li, Sens. Actuators, B, 2023, 380, 133381 CrossRef CAS.
- D. Kusić, B. Kampe, P. Rösch and J. Popp, Water Res., 2014, 48, 179–189 CrossRef PubMed.
- M. L. Laucks, A. Sengupta, K. Junge, E. J. Davis and B. D. Swanson, Appl. Spectrosc., 2005, 59, 1222–1228 CrossRef CAS PubMed.
- X. Lu, D. R. Samuelson, Y. Xu, H. Zhang, S. Wang, B. A. Rasco, J. Xu and M. E. Konkel, Anal. Chem., 2013, 85, 2320–2327 CrossRef CAS PubMed.
- C. Wei, M. Li and X. Zhao, Front. Microbiol., 2018, 9, 2857 CrossRef PubMed.
- S. M. Mandal, A. Roy, A. K. Ghosh, T. K. Hazra, A. Basak and O. L. Franco, Front. Pharmacol., 2014, 5, 105 Search PubMed.
- P. Sumby and M. K. Waldor, J. Bacteriol., 2003, 185, 6841–6851 CrossRef CAS PubMed.
- C. E. James, K. N. Stanley, H. E. Allison, H. J. Flint, C. S. Stewart, R. J. Sharp, J. R. Saunders and A. J. McCarthy, Appl. Environ. Microbiol., 2001, 67, 4335–4337 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra07575c |
| ‡ First two authors have equal contribution. |
| This journal is © The Royal Society of Chemistry 2024 |