 Open Access Article
Open Access ArticleA simple and reliable QSPR model for prediction of chromatography retention indices of volatile organic compounds in peppers†
Shahin Ahmadi *a,
Shahram Lotfi
*a,
Shahram Lotfi b,
Hamideh Hamzehalic and
Parvin Kumar
b,
Hamideh Hamzehalic and
Parvin Kumar d
d
aDepartment of Pharmaceutical Chemistry, Tehran Medical Sciences, Islamic Azad University, Tehran, Iran. E-mail: ahmadi.chemometrics@gmail.com
bDepartment of Chemistry, Payame Noor University (PNU), 19395-4697 Tehran, Iran
cDepartment of Chemistry, Islamic Azad University, East Tehran Branch, Tehran, Iran
dDepartment of Chemistry, Kurukshetra University, Kurukshetra, Haryana 136119, India
First published on 19th January 2024
Abstract
Worldwide, various types of pepper are used in food as an additive due to their unique pungency, aroma, taste, and color. This spice is valued for its pungency contributed by the alkaloid piperine and aroma attributed to volatile essential oils. The essential oils are composed of volatile organic compounds (VOCs) in different concentrations and ratios. In chromatography, the identification of compounds is done by comparing obtained peaks with a reference standard. However, there are cases where reference standards are either unavailable or the chemical information of VOCs is not documented in reference libraries. To overcome these limitations, theoretical methodologies are applied to estimate the retention indices (RIs) of new VOCs. The aim of the present work is to develop a reliable QSPR model for the RIs of 273 identified VOCs of different types of pepper. Experimental retention indices were measured using comprehensive two-dimensional gas chromatography coupled to quadrupole mass spectrometry (GC × GC/qMS) using a coupled BPX5 and BP20 column system. The inbuilt Monte Carlo algorithm of CORAL software is used to generate QSPR models using the hybrid optimal descriptor extracted from a combination of SMILES and HFG (hydrogen-filled graph). The whole dataset of 273 VOCs is used to make ten splits, each of which is further divided into four sets: active training, passive training, calibration, and validation. The balance of correlation method with four target functions i.e. TF0 (WIIC = WCII = 0), TF1 (WIIC = 0.5 & WCII = 0), TF2 (WIIC = 0 & WCII = 0.3) and TF3 (WIIC = 0.5 & WCII = 0.3) is used. The results of the statistical parameters of each target function are compared with each other. The simultaneous application of the index of ideality of correlation (IIC) and correlation intensity index (CII) improves the predictive potential of the model. The best model is judged on the basis of the numerical value of R2 of the validation set. The statistical result of the best model for the validation set of split 6 computed with TF3 (WIIC = 0.5 & WCII = 0.3) is R2 = 0.9308, CCC = 0.9588, IIC = 0.7704, CII = 0.9549, Q2 = 0.9281 and RMSE = 0.544. The promoters of increase/decrease for RI are also extracted using the best model (split 6). Moreover, the proposed model was used for an external validation set.
1. Introduction
Peppers are among the most ancient spices known to man and are extensively harvested all over the entire globe. Pepper fruits contain a high quantity of constituents advantageous to human health, such as antioxidants, minerals, vitamins (mainly A, C, and E), polyphenols and carotene. All types of pepper are eaten fresh or dried and are used in the food industry as additives (coloring and flavoring agents) because of their unique pungency, color, flavor, and aroma.1–3 The piperine alkaloid (as the (E,E)-isomer), which is responsible for pungency, and the volatile essential oils that provide flavour and aroma are primarily accountable for the quality of peppers as rated by humans.4,5 However, other compounds have also been identified in peppers, such as terpenes, flavonoids, steroids, unsaturated fatty acids, and polysaccharides. Furthermore, the essential oil derived from the distillation of pepper contains various taste and flavouring components: e.g. oxygenated monoterpenoid compounds, monoterpene hydrocarbons and oxygenated compounds, sesquiterpene hydrocarbons and oxygenated compounds, oxygenated sesquiterpenes, and phenolic compounds. These compounds are designated as volatile organic compounds (VOCs).4,6,7 The essential oils of peppers can also be employed as antioxidant, antiseptic, antibacterial, antimycotic, anti-epileptic, anti-inflammatory, diuretic, antipyretic, anthelminthic, and carminative agents.8,9Numerous reports have been published for the identification and characterization of diverse VOCs of peppers.10–13 Gas chromatography (GC) and gas chromatography-mass spectrometry (GC-MS) techniques are generally employed for the quantitative determination of volatile compounds in peppers.14,15 Other techniques such as proton-transfer-reaction time-of-flight mass spectrometry (PTR-ToF-MS), two-dimensional gas chromatography with flame ionization detection (GC × GC-FID), quadrupole mass spectrometry (GC × GC-qMS) and time-of-flight mass spectrometry (GC × GC/TOFMS) are also applied to identify VOCs.5,10,16
In chromatography, the chemical structure of compounds is identified by comparing obtained peaks with a reference standard. However, in some cases, reference standards may be unavailable or the chemical information about VOCs may not be registered in reference libraries. To reduce these limitations, theoretical techniques for estimating the retention index (RI) of new VOCs are employed. Hence, the quantitative structure–property/activity relationship (QSPR/QSAR) is employed to predict the retention index (RI).17 QSPR/QSAR is a significant theoretical technique used to establish mathematical models that predict the properties/activities or endpoints of compounds, which have been newly designed or are undeveloped.18,19
A literature survey revealed that CORAL (CORrelation And Logic software available at http://www.insilico.eu/coral) software can be implemented for the development of predictive QSPR/QSAR models. CORAL is freeware software designed to calculate one-variable QSPR/QSAR models between an endpoint and descriptors using the Monte Carlo algorithm. In this software, the optimal descriptor of correlation weight (DCW) is calculated using the SMILES (Simplified Molecular-Input Line-Entry System) notation of the molecular structure.20–22 According to a literature report, the index of ideality of correlation (IIC) and correlation intensity index (CII) are applied as new criteria for judging the predictive potential of the QSPR model. It is often mentioned in the literature that the numerical value of the coefficient of determination (R2) for the validation and calibration set is improved by the IIC, whereas the CII improves the numerical value of the coefficient of determination (R2) for all four sets: i.e. active training, passive training, calibration and validation.23–31
The objective of this study is to construct a predictive QSPR model using the Monte Carlo technique of the CORAL software for the retention index property of 273 VOCs recognized in peppers. Ten random splits are made and each split is divided into four subsets. The IIC and CII statistical parameters are employed to predict a better model. The balance of correlation method with four target functions, i.e. TF0 (WIIC = WCII = 0), TF1 (WIIC = 0.5 & WCII = 0), TF2 (WIIC = 0 & WCII = 0.3) and TF3 (WIIC = 0.5 & WCII = 0.3), is used to examine the robustness and accuracy of the constructed QSPR model.
2. Data and method
2.1. Data
The retention index (RI) data for 273 VOCs identified in 13 peppers were obtained from the literature by Rojas et al.32 Polydimethylsiloxane/divinylbenzene (PDMS/DVB) fiber was used for extraction of the VOCs. The peaks on the two-dimensional GC with a quadrupole mass spectrometric detection (GC × GC-qMS) chromatogram were identified by the column set comprising a non-polar molecule (5% phenyl polysilphenylenesiloxane) as the primary column and a polar molecule (polyethylene glycol) in the second column. Experimental retention indices were obtained using the van den Dool and Kratz equation and Adams' retention indices. Data preprocessing details were reported in the literature by Rojas et al.32 The range of values for the retention index (RI) went from 930 to 1790. The IDs of the compound, SMILES codes, and corresponding experimental and predicted RI are provided in Table S1.† Ten splits were prepared and each split was further split randomly into four subsets: i.e. an active training set (≈26%), a passive training set (≈20%), a calibration set (≈20%), and a validation set (≈34%). The role of each set was fixed and is well explained in the literature.33–362.2. Method
The methodology for obtaining the results from the CORAL software can be summarized as a group by the following steps:(1) Data preparation involves converting the structure to SMILES and preparing the Total set file.
(2) The process of data splitting in CORAL software is carried out using random splitting. This can be done using the classical scheme or balance of correlation. In the balance of correlation the data is divided into four sets: active training, passive training, calibration, and validation sets. However, if the amount of data is small, the classical scheme is used and the data set includes training, calibration, and validation sets.
(3) The selection of descriptors is based on either SMILES or a graph, or a combination of both. The descriptors are chosen accordingly.
(4) The target function selection involves computing correlation weights using the Monte Carlo method and maximizing one of the target functions: namely TF0, TF1, TF2, or TF3. The formulas of these target functions are described in the corresponding section.
(5) Model building consists of two phases. In Phase 1, the preferable threshold and number of epochs are searched for using Monte Carlo optimization based on statistical results from the calibration set. In Phase II, the preferable model is constructed after optimization of the threshold and number of epochs.
(6) External validation is performed on the test sets after model building.
(7) Model interpretation is carried out in this step.
(8) New molecules can be designed based on the model interpretation.
2.3. Hybrid optimal descriptor
As previously mentioned in the preceding section, in CORAL software three types of optimal descriptors can be calculated: i.e., SMILES-based, graph-based, and hybrid descriptors (obtained by combining SMILES and graph-based). The graph-based descriptor can be computed by using a hydrogen-filled graph (HFG), a hydrogen-suppressed molecular graph (HSG) or a graph of atomic orbitals (GAO).37–39 A literature survey shows that QSPR models designed using the hybrid optimal descriptor provide robust models with higher statistical quality.40 Here, the QSPR model for the prediction of RI is designed by utilizing a hybrid optimal descriptor based on the correlation weights of SMILES attributes and vertex degrees in the hydrogen-filled graph (HFG).The hybrid optimal descriptor of the correlation weights (DCW) is computed using the following equation:
| DHybridCW(T*,N*) = DCWSMILES(T*,N*) + DCWHFG(T*,N*) | (1) |
The DCW of HFG and SMILES are calculated via mathematical eqn (2) and (3).
| DCWSMILES(T*,N*) = ∑CW(Sk) + ∑CW(SSk) + ∑CW(SSSk) + CW(BOND) + CW(MFCs) | (2) |
| DCWHFG(T*,N*) = ∑CW(EC0k) + ∑CW(EC1k) + ∑CW(pt2k) + ∑CW(VS2k) + ∑CW(nnk) + ∑CW(C5) + ∑(C6) | (3) |
In eqn (2), the structural attributes Sk, SSk, and SSSk are single SMILES symbols (e.g., Cl or S), two SMILES symbols and a combination of three SMILES symbols, respectively. The BOND code demonstrates the existence or absence of double (=), triple (#), or stereochemical bonds (@ or @@). Here, the molecular feature contributions (MFCs) are the total number of oxygen atoms (O), the number of double bonds (=), and the number of triple bonds (#). Therefore, in eqn (3), the attributes EC0 and EC1 are the number of neighbors of a vertex degree and Morgan's connectivity of first order; pt2k is the number of paths of length 2; VS2 is the valence shells of radius 2 in the HFG; the nn symbol implies nearest neighbors; C5 and C6 are descriptors that represent the five- and six-membered rings in the molecular structure, respectively. T is the threshold to separate SMILES attributes into noise or active. The active SMILES are applied to construct the model. The noise SMILES are not involved in constructing the model. T* and N* are the optimum threshold and number of epochs of the Monte Carlo optimization method. T* and N* provide the maximum statistical quality for the calibration set. The numerical values for CWs are acquired from the Monte Carlo optimization and the optimal descriptor is computed with the optimal CWs. Then the calculated CWs are employed to design a predictive model of the RI according to the following equation:
| RI = C0 + C1 × DCW(T*,N*) | (4) |
2.4. The Monte Carlo optimization
Here to design robust QSPR models, four different kinds of target functions, TF0, TF1, TF2 and TF3, are used. Then, the outcomes of the statistical results are compared with each other.The mathematical equations for each target function can be demonstrated follows:
| TF0 = RATRN + RPTRN − |RATRN − RPTRN| × drweight | (5) |
| TF1 = TF0 + IICCAL × weight for IIC (IICweight) | (6) |
| TF2 = TF0 + CIICAL × weight for CII (CIIweight) | (7) |
| TF3 = TF0 + IICCAL × IICweight + CIICAL × CIIweight | (8) |
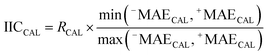 | (9) |
RCAL is the correlation coefficient between experimental values and calculated values of RI for the calibration set. The negative and positive mean absolute errors are indicated by −MAE and +MAE, which are computed as follows:
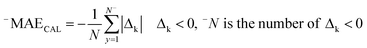 | (10) |
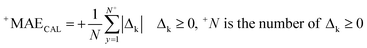 | (11) |
| Δk = observedk − calculatedk | (12) |
The ‘k’ is the index (1, 2, …, N) and the observedk and calculatedk are related to the endpoint.
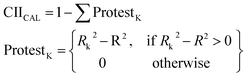 | (13) |
R2 is the correlation coefficient for a set that contains n substances. Rk2 is the correlation coefficient for n − 1 substances of a set after removing the kth substance. Hence, if (Rk2 − R2) is greater than zero, the kth substance is an “oppositionist” for the correlation between experimental and predicted values of the set. A small sum of “protests” means a more “intensive” correlation.
2.5. Applicability domain
In QSPR/QSAR models, the applicability domain (AD) is used to specify whether the designed model interpolates (correct predictions) or extrapolates (incorrect predictions). In the CORAL software, the distribution of SMILES attributes in the active training, passive training, and calibration sets is used to calculate the AD. Therefore, the AD for the model acquired as a result of Monte Carlo optimization varies depending on the distribution of the datasets in the training and calibration sets. In the QSPR/QSAR models designed by CORAL software, the statistical defects of SMILES are employed to define the AD. The “statistical defect,” d(A) is computed according to the following mathematical equation:
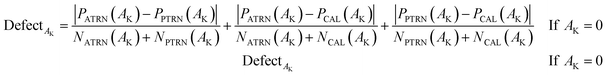 | (14) |
PATRN(Ak), PPTRN(Ak) and PCAL(Ak) are the probability of attributes in the active training set, passive training set, and calibration set, respectively; NATRN(Ak), NPTRN(Ak), and NCAL(Ak) are frequencies of attributes in the active training, passive training and calibration sets, respectively.
The SMILES-statistical defect (D) can be calculated as the sum of statistical defects of all attributes:
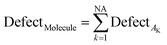 | (15) |
NA is the number of active SMILES attributes for the given compounds.
In CORAL, a SMILES is an outlier if:
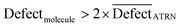 | (16) |
 D is an average of statistical defects for the dataset of the active training set.
D is an average of statistical defects for the dataset of the active training set.
3. Results and discussion
3.1. QSPR modelling for RI
Based on ten initial QSPR models, three compounds (compounds 49, 205, and 265) were identified as outliers. Therefore, these compounds were excluded from the data set before further processing. Herein, to achieve consistent statistical performance, ten different QSPR models were built for each type of target function (TF0, TF1, TF2, and TF3) employing hybrid optimal descriptors. The summary of statistical results for all QSPR models is summarized in Table S2.† The numerical value of R2 calculated with TF3 for the validation set of all splits is higher than the R2 calculated with the other target functions (TF0, TF1 and TF2); thus the TF3 calculated with eqn (8) was selected as the best target function. A comparison of the determination coefficients of the validation set for all splits computed via four target functions is represented in Fig. 1.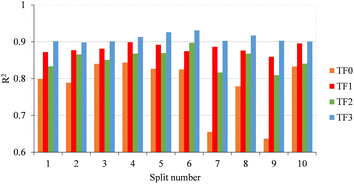 | ||
| Fig. 1 Comparison of determination coefficients computed with TF0, TF1, TF2 and TF3 of all ten splits. | ||
The QSPR models for the ten splits formulated with TF3 for prediction of the RI of the VOCs are given below:
Split 1
| RI = 73.8025(±7.0844) + 24.5924(±0.1499) × DCW(1,15) | (17) |
Split 2
| RI = 121.5622(±6.8196) + 26.4093(±0.1507) × DCW(1,15) | (18) |
Split 3
| RI = 315.0484(±6.5146) + 20.0374(±0.1270) × DCW(1,15) | (19) |
Split 4
| RI = 133.8434(±9.0780) + 16.6644(±0.1272) × DCW(1,15) | (20) |
Split 5
| RI = 120.8001(±5.6911) + 22.7349(±0.1122) × DCW(1,15) | (21) |
Split 6
| RI = 265.7739(±6.3219) + 23.9520(±0.1477) × DCW(1,15) | (22) |
Split 7
| RI = 107.8894(±7.4311) + 23.2382(±0.1417) × DCW(1,15) | (23) |
Split 8
| RI = 40.7742(±6.0698) + 25.9964(±0.1255) × DCW(1,20) | (24) |
Split 9
| RI = 25.2594(±6.5441) + 25.5860(±0.1587) × DCW(1,15) | (25) |
Split 10
| RI = 110.5639(±6.0444) + 21.8635(±0.1077) × DCW(1,15) | (26) |
3.2. Model validation
In this study, the RI of the VOCs was predicted using QSPR models based on Monte Carlo optimization employing four target functions TF0 (WIIC = WCII = 0), TF1 (WIIC = 0.5 & WCII = 0), TF2 (WIIC = 0 & WCII = 0.3) and TF3 (WIIC = 0.5 &WCII = 0.3), and each target function was checked with ten random splits. The balance of correlation method was applied to generate QSPR models. The statistical results presented in Table S2† indicate that all designed QSPR models are within the standard range in terms of statistical criteria and have robust predictability. It can also be seen from Table S2† that simultaneously adding the weight IIC and CII to the target function increases its ability to predict RI as well as improving the statistical results. The numerical value of R2 for the validation set of split 6 (R2 = 0.9308, eqn (22)) was found to be higher than the numerical value of R2 for the other models created with TF3, so it was identified as the best model. Fig. 2 displays the plot between observed and calculated data of the RI for the QSPR models computed with TF3. A good correlation between observed RI and calculated RI, as well as a uniform distribution of RI for active training, passive training, calibration and validation sets can be seen in Fig. 2. Finally, the validation metrics for each model are calculated using three strategies: (i) internal validation or cross-validation with the training set data; (ii) external validation with the test set data; and (iii) Y-scrambling or data randomization. If CR2p > 0.5 for the created model in a Y-randomization test, the model is free of chance correlation. For all constructed QSPR models the numerical value of CR2p was more than 0.5, indicating the robustness of the developed models.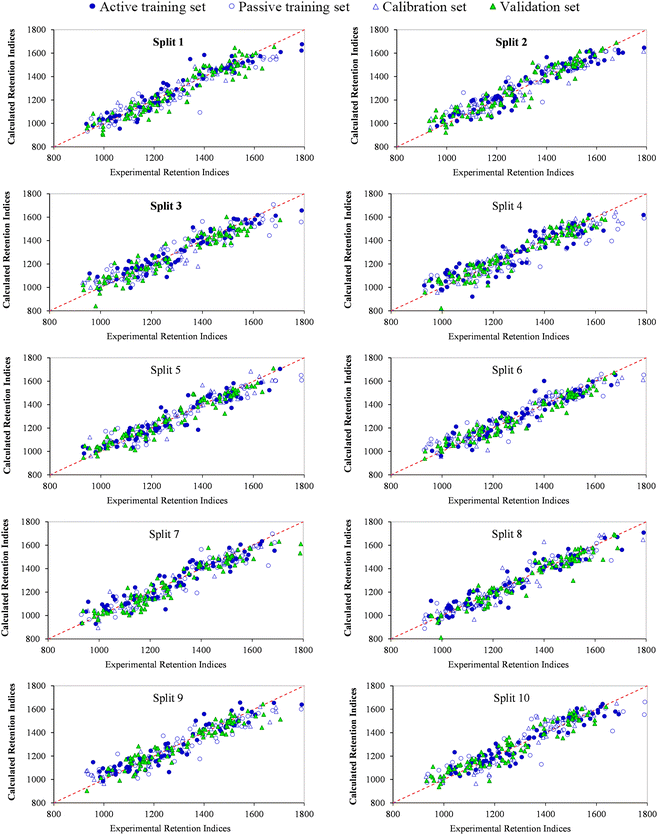 | ||
| Fig. 2 Experimental versus predicted retention indices of split 1 to 10 for VOCs detected in peppers by the Monte Carlo method based on target function TF3. | ||
3.3. Interpretation of the QSPR model
In the QSPR model developed by the CORAL software, mechanistic interpretation is defined as the description of structural attributes acquired from SMILES or hydrogen-filled graphs which are responsible for the increase or decrease of an endpoint. If the numerical value of correlation weights of these structural attributes is negative in three or more runs of the optimization, then these structural features are defined as a promoter of endpoint decrease. On the other hand, if the numerical value of correlation weights of these structural attributes is positive in three or more runs of the optimization, then these structural features are defined as a promoter of endpoint increase. However the structural attribute is undefined if the correlation weight of the structural descriptors has both positive and negative numerical values.The promoters for endpoint RI increase or decrease were computed from the best model (split 6) and are displayed in Table 1. Morgan extended connectivity of zero-order for hydrogen atom as 1 (ec0-h…1…), Morgan extended connectivity of first-order for hydrogen atom as 4 (ec1-h…4…), Morgan extended connectivity of first-order for carbon atom as 7 (ec1-c…7…), Morgan extended connectivity of zero-order for carbon atom as 3 (ec0-c…3…), Morgan extended connectivity of first-order for carbon atom as 4 (ec1-c…10…), the number of paths of length 2 which started from a hydrogen atom is equal to 3 (pt2-h…3…), the number of paths of length 2 which started from a carbon atom is equal to 5 (pt2-c…5…), the number of paths of length 2 which started from a hydrogen atom is equal to 2 (pt2-h…2…), two sp3 hybridized carbon joined by branching (c…(…c…), the presence of two consecutive aliphatic carbons (c…c…) etc. were some significant promoters of endpoint increase. The nearest neighbours code for carbon equal to 413 (nnc-c…413), the nearest neighbours code for carbon equal to 440 (nnc-c…440), a combination of the carbon atom, oxygen and branching (c…o…(…), and 28 as a sum of vertex degrees which take place at a topological distance of 2 relatively to carbon vertex (vs. 2-c…28) etc. were some significant promoters of endpoint decrease.
| No. | Structural attributes | CWs Probe 1 | CWs Probe 2 | CWs Probe 3 | NSs | NSc | NSv | Defect [SAk] | Description |
|---|---|---|---|---|---|---|---|---|---|
| The promoters of RI increase | |||||||||
| 1 | EC0-H…1… | 0.0509 | 0.16064 | 0.34295 | 68 | 58 | 54 | 0 | Morgan extended connectivity of zero-order for hydrogen atom as 1 |
| 2 | EC1-H…4… | 0.18025 | 0.22099 | 0.18401 | 68 | 57 | 54 | 0 | Morgan extended connectivity of first-order for hydrogen atom as 4 |
| 3 | PT2-H…3… | 0.16527 | 0.2584 | 0.3257 | 68 | 57 | 54 | 0 | The number of paths of length 2 which started from a hydrogen atom is equal to 3 |
| 4 | C⋯C…… | 0.44726 | 0.01345 | 0.30905 | 65 | 55 | 50 | 0.0003 | The presence of two consecutive aliphatic carbons |
| 5 | EC1-C…7… | 0.31326 | 0.46929 | 0.21017 | 63 | 56 | 50 | 0 | Morgan extended connectivity of first-order for carbon atom as 7 |
| 6 | VS2–H…6… | 0.10836 | 0.05545 | 0.26739 | 63 | 56 | 50 | 0 | 6 as a sum of vertex degrees which take place at a topological distance of 2 relatively to hydrogen vertex |
| 7 | EC0-C…3… | 0.56731 | 0.20537 | 0.33853 | 62 | 47 | 46 | 0.0006 | Morgan extended connectivity of zero-order for carbon atom as 3 |
| 8 | EC1-C…10 | 0.22378 | 0.25831 | 0.04221 | 61 | 52 | 47 | 0.0002 | Morgan extended connectivity of first-order for carbon atom as 4 |
| 9 | NNC-C…422 | 0.20503 | 0.01758 | 0.05181 | 60 | 53 | 48 | 0.0001 | The nearest neighbours codes for carbon equal to 422 |
| 10 | C…(…C… | 0.29947 | 0.13954 | 0.33177 | 59 | 49 | 49 | 0.0004 | Two sp3 hybridized carbon joined by branching |
| 11 | =…… | 0.15273 | 0.45098 | 0.39027 | 56 | 43 | 44 | 0.0001 | Presence of double covalent bond |
| 12 | 1…… | 0.26393 | 0.42999 | 0.64276 | 53 | 46 | 42 | 0 | Presence of at least one ring |
| 13 | NNC-C…321 | 0.44352 | 0.72343 | 0.10398 | 53 | 39 | 38 | 0.0008 | The nearest neighbours codes for carbon equal to 321 |
| 14 | PT2-H…2… | 0.06983 | 0.57052 | 0.40344 | 53 | 42 | 38 | 0.0008 | The number of paths of length 2 which started from a hydrogen atom is equal to 2 |
| 15 | PT2-C…5… | 0.21029 | 0.22211 | 0.19676 | 52 | 42 | 39 | 0.0005 | The number of paths of length 2 which started from a carbon atom is equal to 5 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| The promoters of RI decrease | |||||||||
| 1 | NNC-C…413 | −0.08801 | −0.07644 | −0.57505 | 67 | 57 | 52 | 0.0002 | The nearest neighbours codes for carbon equal to 413 |
| 2 | C…1…(… | −0.30101 | −0.02505 | −0.28091 | 26 | 27 | 20 | 0.0003 | Combination of aliphatic carbon, one ring and branching |
| 3 | NNC-C…440 | −0.3105 | −0.03567 | −0.80092 | 26 | 27 | 20 | 0.0003 | The nearest neighbours codes for carbon equal to 440 |
| 4 | C⋯O…(… | −0.39833 | −1.30355 | −0.7525 | 8 | 1 | 4 | 0.0036 | Combination of the carbon atom, oxygen and branching |
| 5 | O…(…(… | −0.15072 | −1.42791 | −2.29626 | 5 | 1 | 5 | 0.0019 | Oxygen atom with two branching |
| 6 | VS2–C…28 | −0.05713 | −0.5657 | −0.85747 | 5 | 8 | 4 | 0.0001 | 28 as a sum of vertex degrees which take place at a topological distance of 2 relatively to carbon vertex |
| 7 | 3⋯C…1… | −1.16603 | −0.21466 | −2.69557 | 1 | 6 | 1 | 0.0019 | |
3.4. A comparison of various QSPR models based on RI
A survey of the literature indicates that Rojas et al. (2019) reported only one QSPR model for retention index: the QSPR model for 273 VOCs of pepper.32 The molecular descriptors and molecular fingerprints were calculated using Dragon and PaDEL-Descriptor software. To create balanced subsets, the dataset was divided into training, validation, and test sets of molecules using the Balanced Subsets Method (BSM). Afterward, the Wootton, Sergent, and Phan-Tan-Luu (WSP) unsupervised variable reduction method was employed to reduce the presence of multicollinearity, redundancy, and noise among the initial pool of 4336 molecular descriptors and fingerprints. By implementing this method, a reduced pool consisting of 1664 descriptors was subjected to supervised selection through replacement method (RM) variable subset selection in order to establish a four-descriptor model. The efficacy of the model was assessed by evaluating the coefficient of determination and the root-mean-square deviation in fitting. Specifically, the values obtained for R2 and RMSD for training were 0.879 and 72.1, respectively. Similarly, R2 and RMSD were found to be 0.832 and 91.7 in the validation set, while R2 and RMSD were 0.915 and 55.4 in the test set. The minimal discrepancies observed among these parameters across the three sets indicate the stability and predictability of the QSPR model.Table 2 displays a comparison of the statistical results of the present QSPR model with the reported QSPR model. The previously reported model was implemented with only one split, but in the present QSPR models, 10 splits were used to design 40 QSRR models employing four target functions (TF0, TF1, TF2 and TF3). Two significant criteria, the index of ideality correlation (IIC) and correlation intensity index (CII), are also addressed in this work, which were not studied in earlier work. In the present QSPR models, only one descriptor, DCW, was used to construct the QSPR models but in the previously reported model, four descriptors were applied. The numerical value of the determination coefficient (Rval2) of the QSPR model generated with TF3 for split 6 is 0.9308, which is much better than the value for the reported model. Thus, the presented QSPR models are more robust and predictable.
| No. | Set | n | Descriptor generator | Regression method | R2 train | RMSD | IIC | CII | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Training | 92 | Dragon and PaDEL | MLR | 0.879 | 72.1 | — | — | 32 |
| Validation | 91 | 0.832 | 91.7 | — | — | ||||
| Test | 90 | 0.915 | 55.4 | — | — | ||||
| 2 | ActivTRN | 68 | CORAL package | LR | 0.885 | 62.7 | 0.658 | 0.928 | Present work |
| PassTRN | 58 | 0.900 | 70.8 | 0.677 | 0.943 | ||||
| Calib | 54 | 0.904 | 51.6 | 0.951 | 0.944 | ||||
| Valid | 90 | 0.931 | 54.4 | 0.770 | 0.955 |
3.5. External validation of the proposed models
An external dataset of 115 VOCs reported by Rojas et al.32 was used to predict the RI of molecules outside the dataset for modeling. The RI properties of these compounds were predicted by ten models based on TF3 and average values were compared with external predictions by Rojas et al. Table 3 shows the average predicted RI of ten models, the prediction by Rojas et al.32 for the BPX5 and BP20 column coupled system, and experimental retention indices from the literature.| No. | Name | CAS number | RIpredicted (this study) | RIpredicted (Rojas et al.) | Experimental RIliterature | Ref. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DB-5 column | HP-5MS stationary phase | HP-5 column | DB-Wax stationary phase | BPX5 column | RTX-Wax stationary phase | HP-20M column | DB-5MS stationary phase | BP20 column | ||||||
| a Not available. | ||||||||||||||
| 1 | Myrcenol | 543-39-5 | 1164.719 | 1159.6 | NA | NAa | NA | NA | NA | NA | NA | NA | NA | 41 |
| 2 | (E,E)-α-Farnesene | 502-61-4 | 1483.631 | 1496.2 | 1508 | NA | NA | NA | NA | NA | NA | NA | NA | 15, 42 and 43 |
| α-Farnesene | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | 41 and 44 | |||
| 3 | Elemicin | 487-11-6 | 1501.785 | 1565 | 1540 | NA | NA | NA | NA | NA | NA | NA | NA | 43 and 45 |
| 1554 | ||||||||||||||
| 1556 | ||||||||||||||
| 4 | Myristicin | 607-91-0 | 1391.435 | 1546.5 | 1520 | NA | NA | NA | NA | NA | NA | NA | NA | 43 |
| 5 | Apiole | 523-80-8 | 1541.468 | 1721.7 | 1679 | NA | NA | NA | NA | NA | NA | NA | NA | 43 |
| 1680 | ||||||||||||||
| 1685 | ||||||||||||||
| 6 | Dillapiole | 484-31-1 | 1565.236 | 1709.1 | 1622 | 1622 | NA | NA | NA | NA | NA | NA | NA | 14,43 |
| 1644 | ||||||||||||||
| 7 | Eugenol acetate | 93-28-7 | 1568.772 | 1577.7 | 1524 | NA | NA | NA | NA | NA | NA | NA | NA | 45 |
| 8 | Carvone oxide | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 383-49-8 383-49-8 |
1239.442 | 1367.3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41, 42 and 46–49 |
| trans-Carvone oxide | 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 204-74-9 204-74-9 |
─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 | |
| 9 | α-Bulnesene | 3691-11-0 | 1484.066 | 1508.7 | 1493 | 1505 | NA | NA | NA | NA | NA | NA | NA | 41 |
| 1503 | ||||||||||||||
| 1505 | ||||||||||||||
| δ-Guaiene | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 42 and 43 | ||
| 10 | Dihydrocarveol | 619-01-2 | 1164.505 | 1184.8 | 1192 | NA | 1195 | 1941 | NA | NA | NA | NA | NA | 41, 42, 44 and 46–49 |
| 1195 | ||||||||||||||
| p-Menth-8-en-2-ol | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 42 | ||
| trans-p-Menth-8-en-2-ol | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 47–49 | ||
| Neo-dihydrocarveol | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 675-34-8 675-34-8 |
─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 | |
| 11 | cis-β-Terpineol | 138-87-4 | 1196.492 | 1181 | 1159 | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 12 | β-Bisabolol | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 352-77-9 352-77-9 |
1647.933 | 1604.1 | NA | NA | 1668 | NA | NA | NA | 2021 | NA | NA | 41 and 42 |
| 13 | α-Bisabolol | 515-69-5 | 1644.237 | 1594.9 | 1683 | NA | NA | NA | NA | NA | NA | NA | NA | 14 and 44 |
| 1701 | ||||||||||||||
| 14 | Squalene | 111-02-4 | 2742.641 | 2726 | 2790 | NA | NA | NA | NA | NA | NA | NA | NA | 6 |
| 15 | δ-Terpinyl acetate | 93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 836-50-1 836-50-1 |
1382.135 | 1356 | 1313 | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 16 | 1,4-Cineole | 470-67-7 | 1139.433 | 1117.1 | 1016 | 1016 | NA | 1171 | NA | NA | NA | NA | NA | 41 |
| 17 | Cadina,1,4-dien-3-ol | 1608.026 | 1636.1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 | |
| 18 | trans-Piperitone oxide | 4713-38-6 | 1194.878 | 1303.8 | 1258 | NA | NA | NA | NA | NA | NA | NA | NA | 50 |
| 19 | 3-Buten-2-ol | 598-32-3 | 642.9435 | 758.6 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 20 | Vomifoliol | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 526-45-6 526-45-6 |
1573.99 | 1735.6 | NA | 1796 | NA | 3167 | 1814 | NA | NA | NA | NA | 41 |
| 3175 | ||||||||||||||
| 21 | Neryl isovalerate | 3915-83-1 | 1578.39 | 1593 | NA | NA | NA | NA | NA | NA | NA | NA | 1872 | 43 |
| 22 | Retrofractamide B | 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 794-74-0 794-74-0 |
2393.783 | 2624 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 23 | Guineensine | 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 038-30-7 038-30-7 |
2568.907 | 2783.1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 24 | Caryophyllene alcohol | 472-97-9 | 1586.876 | 1548.9 | 1568 | 1560 1564 | NA | NA | NA | NA | NA | NA | NA | 42 |
| Caryophyllenol | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41, 44 and 46–49 | ||
| 25 | Clovene | 469-92-1 | 1440.777 | 1460 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 42 |
| 26 | Piperitol | 491-04-3 | 1170.929 | 1143.6 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 43 |
| 27 | Humulene oxide II | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 888-34-7 888-34-7 |
1609.486 | 1589.6 | 1606 | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 1607 | ||||||||||||||
| 28 | α-Cedrene | 469-61-4 | 1447.516 | 1451.9 | 1409 | 1409 | NA | NA | NA | 1562 | NA | NA | NA | 43 |
| 1410 | ||||||||||||||
| 29 | Hedycaryol | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 657-90-9 657-90-9 |
1627.43 | 1612.5 | 1530 | NA | NA | NA | NA | NA | NA | NA | NA | 44 |
| 30 | Germacrene D-4-ol | 198![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 991-79-6 991-79-6 |
1594.602 | 1606.5 | 1511 | 1567 | NA | NA | NA | NA | NA | NA | NA | 43 |
| 1574 | ||||||||||||||
| 31 | α-Eudesmol | 473-16-5 | 1592.197 | 1595.8 | 1652 | NA | NA | NA | NA | NA | 2230 | NA | NA | 41 and 43 |
| 32 | Furanodiene | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 912-61-9 912-61-9 |
1586.736 | 1626.1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 43 |
| 33 | cis-p-Menth-8-en-2-ol | 1180.423 | 1193.1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 47–49 | |
| 34 | Isopulegol | 89-79-2 | 1200.764 | 1141.3 | 1145 | 1146 | NA | 1879 | NA | NA | NA | NA | NA | 43 |
| 1879 | ||||||||||||||
| 35 | Menthol | 89-78-1 | 1190.85 | 1113.6 | 1173 | NA | NA | 1626 | NA | NA | 1646 | NA | NA | 41 |
| 2103 | ||||||||||||||
| 36 | cis-Sabinene hydrate | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 537-55-0 537-55-0 |
1144.655 | 975 | 1069 | 1101 | NA | 1465 | NA | NA | NA | NA | NA | 14 and 43 |
| 1070 | ||||||||||||||
| 1097 | ||||||||||||||
| 37 | Cedrol | 77-53-2 | 1583.856 | 1544.5 | 1596 | NA | NA | NA | NA | NA | NA | NA | NA | 15, 41, 42 and 46–49 |
| 1604 | ||||||||||||||
| 38 | (Z)-Isosafrole | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 627-76-8 627-76-8 |
1271.586 | 1391.8 | 1308 | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 1336 | ||||||||||||||
| 39 | Nona-trans,cis-2,6-dienal | 557-48-2 | 1102.993 | 1130.7 | NA | NA | NA | 1597 1605 | NA | NA | NA | NA | NA | 51 |
| 40 | Howeveranediol | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 265-75-2 265-75-2 |
761.5132 | 733.4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 6 |
| 41 | Ethyl-2-hexenol | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 639-00-4 639-00-4 |
993.1246 | 1003.4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 42 | Hept-trans-3-en-2-one | 1119-44-4 | 940.3034 | 900.1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 51 |
| 43 | Nona-trans,trans-2,5- dien-4-one | 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 759-51-1 759-51-1 |
1101.811 | 1085.6 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 51 |
| 44 | Hex-trans-2-enal | 6728-26-3 | 838.5927 | 850.3 | 854 | NA | 857 | 1201 | NA | NA | 1209 | 848 | NA | 51 |
| (E)-2-Hexenal | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 | ||
| 45 | 1-Hepten-3-ol | 4938-52-7 | 902.912 | 925.4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 46 | (E)-3-Octenol | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 185-81-4 185-81-4 |
1063.265 | 1043.7 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 47 | 2-Heptanone | 110-43-0 | 900.1747 | 971.3 | 888 | NA | 882 | 1160 | 889 | NA | NA | NA | NA | 51 |
| 889 | ||||||||||||||
| 48 | Non-trans-2-en-4-one | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 064-72-5 064-72-5 |
1115.155 | 1156.7 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 51 |
| 49 | Non-1-en-4-one | 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 168-10-3 168-10-3 |
1084.331 | 1154.4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 51 |
| 50 | α-Ethyl hexanoate | 123-66-0 | 1021.078 | 1118 | 996 | 1001 | 997 | 1224 | NA | NA | NA | NA | NA | 6 |
| 997 | 1229 | |||||||||||||
| 998 | 1244 | |||||||||||||
| 1001 | 1270 | |||||||||||||
| 51 | 1-Octen-3-ol | 3391-86-4 | 990.474 | 1013.3 | 942 | 975 | 977 | 980 | NA | NA | 1423 | NA | NA | 41 |
| 978 | 991 | 1438 | ||||||||||||
| 980 | 1465 | |||||||||||||
| 52 | 2,4-Decadienoic acid | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 997-42-2 997-42-2 |
1235.616 | 1736.5 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| Piperidide | NA | NA | NA | NA | NA | NA | NA | NA | NA | |||||
| 53 | Pellitorin | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 836-52-7 836-52-7 |
1519.16 | 1624.7 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 54 | Deca-trans,cis-2,4-dienal | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 152-83-4 152-83-4 |
1189.996 | 1212.5 | 1291 | NA | NA | 1758 | NA | NA | NA | NA | NA | 51 |
| 1297 | ||||||||||||||
| Deca-trans,trans-2,4-dienal | 2363-88-4 | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 51 | |
| Deca-2,4-dienal | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 152-84-5 152-84-5 |
─ | ─ | 1311 | NA | NA | 1820 1832 | NA | NA | NA | NA | NA | 51 | |
| 1314 | ||||||||||||||
| 1319 | ||||||||||||||
| 55 | (E)-2-Octenal | 2363-89-5 | 1010.999 | 1023.6 | NA | 1060 | NA | NA | NA | NA | NA | NA | NA | 41 |
| 56 | N-Isobutyl-(2E,4E,12E)-octadecatrienamide | 943![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546-17-6 546-17-6 |
2222.115 | 2290 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 57 | N-Isobutyl-(2E,4E,14Z)-eicosatrienamide | — | 2397.239 | 2448.7 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 58 | Hexanal | 66-25-1 | 836.3585 | 921.5 | 784 | 805 | 800 | 1067 | NA | NA | 1075 | NA | NA | 16 and 51 |
| 797 | 803 | 1093 | ||||||||||||
| 799 | ||||||||||||||
| 800 | ||||||||||||||
| 819 | ||||||||||||||
| 59 | 2-Octanol | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 339-16-6 339-16-6 |
1019.923 | 981.9 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 60 | Nonane | 111-84-2 | 964.4392 | 965.9 | 899 | 900 | NA | NA | NA | NA | 900 | NA | NA | 41 |
| 61 | (E)-2-Tridecenal | 7774-82-5 | 1448.809 | 1429.9 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 43 |
| 62 | Tetradecane | 629-59-4 | 1402.249 | 1372.2 | 1116 | NA | NA | 1399 | 1400 | NA | 1400 | NA | NA | 14 |
| 1399 | ||||||||||||||
| 63 | N-Isobutyl-(2E,4E)-octadecadienamide | 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 794-70-6 794-70-6 |
2219.656 | 2266 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 64 | Pentadecanal | 2765-11-9 | 1624.416 | 1660.3 | 1513 | 1711 | NA | NA | NA | NA | NA | NA | NA | 50 |
| 1687 | ||||||||||||||
| 1710 | ||||||||||||||
| 65 | Hexadecane | 544-76-3 | 1577.373 | 1531.4 | 1600 | NA | NA | NA | NA | NA | 1600 | NA | NA | 14 |
| 66 | Palmitic acid | 57-10-3 | 1755.653 | 1811.2 | 1984 | NA | NA | NA | NA | NA | 2860 | NA | NA | 14 |
| Palmitic acid glyceride | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 6 | ||
| 67 | Heptadecane | 629-78-7 | 1664.935 | 1610.7 | 1700 | NA | NA | NA | NA | NA | 1700 | NA | NA | 14 |
| 68 | 1-Octadecene | 112-88-9 | 1759.766 | 1711.6 | 1793 | NA | NA | NA | 1774 | NA | NA | NA | NA | 14 |
| 1794 | ||||||||||||||
| 69 | Octadecane | 593-45-3 | 1752.497 | 1689.9 | 1800 | NA | NA | 1805 | NA | NA | 1800 | NA | NA | 14 |
| 70 | Nonadecane | 629-92-5 | 1840.059 | 1768.9 | 1900 | NA | NA | NA | NA | NA | 1900 | NA | NA | 14 |
| 71 | 1-Eicosene | 3452-07-1 | 1934.889 | 1869.5 | 1990 | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 1994 | ||||||||||||||
| 72 | Eicosane | 112-95-8 | 1927.621 | 1847.8 | 2000 | NA | NA | NA | NA | NA | 2000 | NA | NA | 14 |
| 73 | Heneicosane | 629-94-7 | 2015.183 | 1926.7 | 2100 | NA | NA | NA | NA | NA | 2100 | NA | NA | 14 |
| 74 | Docosane | 629-97-0 | 2102.745 | 2005.4 | 2200 | NA | NA | NA | NA | NA | 2200 | NA | NA | 14 |
| 75 | Nonadecanol | 1454-84-8 | 2015.34 | 1901.6 | NA | 2156 | NA | NA | NA | NA | NA | NA | NA | 50 |
| 76 | Hexanol | 111-27-3 | 877.0338 | 846.4 | 867 | NA | 865 | 1351 | NA | NA | 1325 | 869 | NA | 41 |
| 884 | 1354 | |||||||||||||
| 1360 | ||||||||||||||
| 1379 | ||||||||||||||
| 1392 | ||||||||||||||
| 77 | Amyl alcohol | 71-41-0 | 789.4719 | 751.3 | 766 | NA | 766 | 1244 | NA | NA | NA | NA | NA | 41 |
| 768 | ||||||||||||||
| 78 | Benzenepropanoic acid, ethyl ester | 2021-28-5 | 1339.896 | 1411.3 | 1390 | NA | NA | 1897 | NA | NA | NA | NA | NA | 43 |
| 1905 | ||||||||||||||
| 79 | Methyl salicylate | 119-36-8 | 1160.501 | 1248.3 | 1190 | 1190 | NA | NA | NA | NA | NA | NA | NA | 51 |
| 1191 | ||||||||||||||
| 1206 | ||||||||||||||
| 80 | Guaiacol | 90-05-1 | 1001.29 | 1087.9 | 1086 | NA | NA | 1872 | NA | NA | NA | NA | NA | 41 |
| 1091 | 1875 | |||||||||||||
| 1883 | ||||||||||||||
| 81 | 2-Methoxy-3-isobutylpyrazine | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 683-00-9 683-00-9 |
1055.625 | 1291 | 1135 | NA | NA | 1540 | NA | NA | NA | NA | NA | 51 |
| 1171 | ||||||||||||||
| 82 | Thymol | 89-83-8 | 1189.021 | 1203.6 | 1290 | 1290 | 1308 | NA | NA | NA | NA | NA | NA | 41 and 43 |
| 83 | 2-Methylnaphthalene | 91-57-6 | 1210.977 | 1379.3 | 1281 | NA | 1295 | NA | NA | NA | NA | NA | NA | 51 |
| 84 | 1-Methylnaphthalene | 90-12-0 | 1163.889 | 1355.6 | 1298 | NA | 1312 | NA | NA | NA | NA | NA | NA | 51 |
| 85 | Piperamide C 9:1 (8E) | 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510-52-5 510-52-5 |
2232.683 | 2552.6 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 86 | 4,5-Dihydropiperettine | 583-34-6 | 2105.33 | 2419.8 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 87 | Dehydropipernonaline | 107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 584-38-3 584-38-3 |
2279.967 | 2579.6 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 88 | Piperine | 94-62-2 | 1928.943 | 2235.5 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 89 | Piperanine | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 512-46-1 512-46-1 |
1932.372 | 2211.5 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 90 | 1-Cinnamoyl piperidine | 5422-81-1 | 1597.08 | 1759.2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 91 | Piperolein B | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 505-89-6 505-89-6 |
2301.005 | 2626.8 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 14 |
| 92 | Geranial | 5392-40-5 | 1182.447 | 1151.7 | 1240 | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| Neral | 106-26-3 | ─ | ─ | 1235 | NA | NA | 1630 | NA | NA | 1658 | NA | NA | 41 | |
| 1240 | 1690 | |||||||||||||
| 1294 | 1695 | |||||||||||||
| 93 | Citronella | 106-23-0 | 1155.695 | 1216.9 | 1153 | NA | NA | 1425 | NA | NA | NA | NA | NA | 41, 42 and 46–49 |
| 1159 | 1485 | |||||||||||||
| 1161 | 1488 | |||||||||||||
| 94 | Phenylacetaldehyde | 122-78-1 | 1046.275 | 1112.2 | 1043 | NA | NA | 1609 | NA | NA | 1615 | NA | NA | 51 |
| 1049 | 1671 | |||||||||||||
| 95 | Furfural | 98-01-1 | 753.1899 | 858.1 | 830 | NA | NA | 1458 | 832 | NA | NA | NA | NA | 51 |
| 1474 | ||||||||||||||
| 1485 | ||||||||||||||
| 96 | Oxalic acid | 144-62-7 | 740.7731 | 852.3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 6 |
| 97 | α-Hydroxypropionic acid | 50-21-5 | 699.8166 | 862.4 | NA | NA | NA | NA | 1058 | NA | NA | NA | NA | 6 |
| 98 | Howeverenedioic acid | 110-16-7 | 888.9371 | 943.8 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 6 |
| 99 | Dihydrolimonen-10-al | 3269-90-7 | 1223.518 | 1236 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 100 | Gluconate anion | 608-59-3 | 1350.775 | 1570.7 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 6 |
| 101 | Erythritol | 149-32-6 | 943.4734 | 1041.8 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 6 |
| 102 | Glycerin | 56-81-5 | 758.5946 | 864.6 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 6 |
| 103 | Myrtenol | 515-00-4 | 1159.949 | 1118.5 | 1194 | NA | NA | NA | NA | NA | NA | NA | NA | 42, 44 and 47–49 |
| 1196 | ||||||||||||||
| 1202 | ||||||||||||||
| 1214 | ||||||||||||||
| 104 | cis-p-Menth-2-en-7-oI | 1185.016 | 1202.4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 | |
| 105 | Glucopyranose | 492-62-6 | 1317.003 | 1440.1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 6 |
| 106 | Geraniol | 106-24-1 | 1175.015 | 1171.8 | 1255 | NA | 1240 | 1788 | NA | NA | 1814 | NA | NA | 41 |
| 1276 | 1850 | |||||||||||||
| 1862 | ||||||||||||||
| Nerol | 106-25-2 | ─ | ─ | 1228 | NA | NA | 1753 | NA | NA | 1770 | NA | NA | 41 and 50 | |
| 107 | (2E,6E)-Farnesol | 106-28-5 | 1606.869 | 1597.8 | 1706 | NA | NA | 2371 | NA | NA | NA | NA | NA | 43 |
| 1722 | ||||||||||||||
| (E,E)-Farnesol | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 | ||
| α-Farnesol | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 44 | ||
| (Z,Z)-Farnesol | 4602-84-0 | ─ | ─ | 1689 1713 | 1713 | 1713 | NA | NA | NA | NA | NA | NA | 41 | |
| 1718 | ||||||||||||||
| (2Z,6Z)-Farnesol | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 43 | ||
| (2E,6Z)-Farnesol | 3879-60-5 | ─ | ─ | 1734 | 1742 | NA | NA | NA | NA | NA | NA | NA | 43 | |
| 1742 | 1743 | |||||||||||||
| 1748 | ||||||||||||||
| (E,Z)-Farnesol | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 | ||
| (Z,E)-Farnesol | 3790-71-4 | ─ | ─ | 1697 | NA | NA | NA | NA | NA | NA | NA | NA | 41 | |
| 108 | Phytol | 150-86-7 | 1941.848 | 1928.7 | 1949 | NA | NA | NA | NA | NA | 2571 | NA | NA | 43 |
| 109 | (E)-Cinnamyl alcohol | 4407-36-7 | 1155.272 | 1238.7 | 1305 | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 110 | 2-Methyl cinnamyl alcohol | 1504-55-8 | 1212.211 | 1285.1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 |
| 111 | Isoamyl alcohol | 123-51-3 | 764.7509 | 679.6 | 734 | NA | 737 | 1169 | NA | NA | 1182 | NA | NA | 41 |
| 735 | 1206 | |||||||||||||
| 1230 | ||||||||||||||
| 112 | Hex-cis-3-enol | 928-96-1 | 880.2674 | 870.4 | 857 | NA | NA | 1378 | NA | NA | 1357 | 849 | NA | 51 |
| 1401 | ||||||||||||||
| 1407 | ||||||||||||||
| 3-Hexenol | 544-12-7 | ─ | ─ | NA | NA | NA | NA | NA | NA | NA | NA | NA | 41 | |
| 113 | Cumin alcohol | 536-60-7 | 1204.248 | 1262.4 | 1287 | NA | NA | 2099 | NA | NA | NA | NA | NA | 41 |
| 114 | Biphenyl | 92-52-4 | 1283.923 | 1486.7 | 1381 | NA | 1385 | NA | NA | NA | NA | NA | NA | 51 |
| 115 | Naphthalene | 91-20-3 | 1131.721 | 1316 | 1179 | NA | NA | NA | NA | NA | 1718 | NA | NA | 51 |
Fig. 3 shows a plot of the RI of the external set predicted by CORAL software versus the RI predicted by Rojas et al..32 There is good agreement between the external predictions by the two methods.
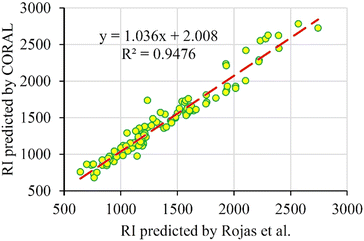 | ||
| Fig. 3 Plot of the RI of external set predicted by CORAL software versus the RI predicted by Rojas et al. | ||
4. Conclusions
In the present study, 40 QSPR models for the prediction of RI of 273 VOCs were developed from 10 random splits. The balance of correlation algorithm was used to design QSRR models. Four target functions, i.e. TF0 (without IIC or CII), TF1 (with IIC alone), TF2 (with CII alone) and TF3 (with IIC and CII, simultaneously) were employed to verify the significance of the present statistical method of QSPR model generation. The simultaneous use of IIC and CII (TF3) improves the predictive potential of the QSPR model. All suggested models render satisfactory predictive QSPR models for the RI of the VOCs, but the best predictive potential was computed withTF3 for split 6; thus it is specified as the best model. To evaluate the reliability and prediction ability of all created models, various statistical parameters, such as R2, IIC, CII, CCC, Q2, QF12, QF22, QF32, s, MAE, F, RMSE,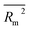 ,
, 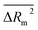 , CR2p and Y-test were utilized. A comparison of some statistical parameters of the present study, as analyzed by the QSPR model developed by Rojas et al.32 reveals that the R2 value for the training set has shown an improvement, increasing from 0.879 to 0.900. Similarly, the R2 value for the test set has also demonstrated an enhancement, rising from 0.915 to 0.931. In addition, the RMSD has exhibited a reduction, decreasing from 72.1 to 62.7 for the training set and from 55.4 to 54.4 for the test set. The applicability domain (AD) was studied based on “statistical defect” d(A). The structural attributes based on graph invariants and SMILES notation were also extracted from the split 6 (best model) and employed to recognize the promoters of RI increase and decrease. Morgan extended connectivity of zero-order for hydrogen atom as 1 (ec0-h…1…), Morgan extended connectivity of first-order for hydrogen atom as 4 (ec1-h…4…), Morgan extended connectivity of first-order for carbon atom as 7 (ec1-c…7…), Morgan extended connectivity of zero-order for carbon atom as 3 (ec0-c…3…), Morgan extended connectivity of first-order for carbon atom as 4 (ec1-c…10…) etc were some significant promoters of endpoint increase. The nearest neighbours codes for carbon equal to 413 (nnc-c…413), the nearest neighbours codes for carbon equal to 440 (nnc-c…440), a combination of the carbon atom, oxygen and branching (c…o…(…), and 28 as a sum of vertex degrees which take place at a topological distance of 2 relatively to carbon vertex (vs. 2-c…28) etc are some significant promoters of endpoint decrease.
, CR2p and Y-test were utilized. A comparison of some statistical parameters of the present study, as analyzed by the QSPR model developed by Rojas et al.32 reveals that the R2 value for the training set has shown an improvement, increasing from 0.879 to 0.900. Similarly, the R2 value for the test set has also demonstrated an enhancement, rising from 0.915 to 0.931. In addition, the RMSD has exhibited a reduction, decreasing from 72.1 to 62.7 for the training set and from 55.4 to 54.4 for the test set. The applicability domain (AD) was studied based on “statistical defect” d(A). The structural attributes based on graph invariants and SMILES notation were also extracted from the split 6 (best model) and employed to recognize the promoters of RI increase and decrease. Morgan extended connectivity of zero-order for hydrogen atom as 1 (ec0-h…1…), Morgan extended connectivity of first-order for hydrogen atom as 4 (ec1-h…4…), Morgan extended connectivity of first-order for carbon atom as 7 (ec1-c…7…), Morgan extended connectivity of zero-order for carbon atom as 3 (ec0-c…3…), Morgan extended connectivity of first-order for carbon atom as 4 (ec1-c…10…) etc were some significant promoters of endpoint increase. The nearest neighbours codes for carbon equal to 413 (nnc-c…413), the nearest neighbours codes for carbon equal to 440 (nnc-c…440), a combination of the carbon atom, oxygen and branching (c…o…(…), and 28 as a sum of vertex degrees which take place at a topological distance of 2 relatively to carbon vertex (vs. 2-c…28) etc are some significant promoters of endpoint decrease.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.Conflicts of interest
There are no conflicts to declare.References
- R. Sharma, V. K. Joshi and M. Kaushal, Effect of pre-treatments and drying methods on quality attributes of sweet bell-pepper (Capsicum annum) powder, J. Food Sci. Technol., 2015, 52(6), 3433–3439 CAS.
- G. Guclu, D. Keser, H. Kelebek, M. Keskin, Y. E. Sekerli, Y. Thereforeysal and S. Selli, Impact of production and drying methods on the volatile and phenolic characteristics of fresh and powdered sweet red peppers, Food Chem., 2021, 338, 128129 CrossRef CAS PubMed.
- L. Gorgani, M. Mohammadi, G. D. Najafpour and M. Nikzad, Piperine—the bioactive compound of black pepper: from isolation to medicinal formulations, Compr. Rev. Food Sci. Food Saf., 2017, 16(1), 124–140 CrossRef CAS PubMed.
- A. Musenga, R. Mandrioli, A. Ferranti, G. D'Orazio, S. Fanali and M. A. Raggi, Analysis of aromatic and terpenic constituents of pepper extracts by capillary electrochromatography, J. Separ. Sci., 2007, 30(4), 612–619 CrossRef CAS PubMed.
- C. Taiti, C. Costa, C. A. Migliori, D. Comparini, S. Figorilli and S. Mancuso, Correlation between volatile compounds and spiciness in domesticated and wild fresh chili peppers, Food Bioprocess Technol., 2019, 12(8), 1366–1380 CrossRef CAS.
- Q. Huo, Z. Wang, S. Xiong and R. Shi, Determination of volatile organic compounds in pepper seeds by GC/MS, Asian J. Chem., 2013, 25(16), 8909–8912 CrossRef CAS.
- S. K. Malhotra and K. Peter, Handbook of Herbs and Spices, Woodhead Publishing Series in Food Science, Technology and Nutrition, 2012, vol. 2 Search PubMed.
- G. Singh, P. Marimuthu, C. Catalan and M. DeLampasona, Chemical, antioxidant and antifungal activities of volatile oil of black pepper and its acetone extract, J. Sci. Food Agric., 2004, 84(14), 1878–1884 CrossRef CAS.
- K. L. Driffield, L. Mooney and K. G. Kerr, Temperature-dependent Changes in Susceptibility of Stenotrophomonas maltophilia. To the Essential Oils of Sweet Basil (Ocimum basilicum.) and Black Pepper (Piper nigrum.), Pharmaceut. Biol., 2006, 44(2), 113–115 CrossRef CAS.
- C. Taiti, C. Costa, P. Menesatti, D. Comparini, N. Bazihizina, E. Azzarello, E. Masi and S. Mancuso, Class-modeling approach to PTR-TOFMS data: a peppers case study, J. Sci. Food Agric., 2015, 95(8), 1757–1763 CrossRef CAS PubMed.
- M. Plessi, D. Bertelli and F. Miglietta, Effect of microwaves on volatile compounds in white and black pepper, LWT--Food Sci. Technol., 2002, 35(3), 260–264 CrossRef CAS.
- Y. Murakami, H. Iwabuchi, Y. Ohba and H. Fukami, Analysis of volatile compounds from chili peppers and characterization of Habanero (Capsicum chinense) volatiles, J. Oleo Sci., 2019, 68(12), 1251–1260 CrossRef CAS PubMed.
- Y. Murakami, H. Iwabuchi, M. Horikawa, S. Mori, Y. Ohba and H. Fukami, Isolation and Identification of a Volatile Compound in Habanero Pepper (Capsicum chinense), J. Oleo Sci., 2018, 67(10), 1219–1225 CrossRef CAS PubMed.
- I. Kapoor, B. Singh, G. Singh, C. S. De Heluani, M. De Lampasona and C. A. Catalan, Chemistry and in vitro antioxidant activity of volatile oil and oleoresins of black pepper (Piper nigrum), J. Agric. Food Chem., 2009, 57(12), 5358–5364 CrossRef CAS PubMed.
- B. Mamatha, M. Prakash, S. Nagarajan and K. Bhat, Evaluation of the flavor quality of pepper (Piper nigrum L.) cultivars by GC-MS, electronic nose and sensory analysis techniques, J. Sensory Stud., 2008, 23(4), 498–513 CrossRef.
- Z. d. L. Cardeal, M. Gomes da Silva and P. Marriott, Comprehensive two-dimensional gas chromatography/mass spectrometric analysis of pepper volatiles, Rapid Commun. Mass Spectrom., 2006, 20(19), 2823–2836 CrossRef CAS PubMed.
- S. Ahmadi, A. Abdolmaleki and M. Jebeli Javan, in Silico Study of Natural Antioxidants, Vitamins and Hormones, Academic Press, 2022 Search PubMed.
- A. Kumar and P. Kumar, Cytotoxicity of quantum dots: use of quasiSMILES in development of reliable models with index of ideality of correlation and the consensus modelling, J. Hazard. Mater., 2021, 402, 123777 CrossRef CAS.
- P. Kumar, A. Kumar, S. Lal, D. Singh, S. Lotfi and S. Ahmadi, CORAL: Quantitative Structure Retention Relationship (QSRR) of flavors and fragrances compounds studied on the stationary phase methyl silicone OV-101 column in gas chromatography using correlation intensity index and consensus modelling, J. Mol. Struct., 2022, 1265, 133437 CrossRef CAS.
- S. Lotfi, S. Ahmadi and P. Kumar, The Monte Carlo approach to model and predict the melting point of imidazolium ionic liquids using hybrid optimal descriptors, RSC Adv., 2021, 11(54), 33849–33857 RSC.
- A. Toropov, A. Toropova, G. Selvestrel and E. Benfenati, Idealization of correlations between optimal simplified molecular input-line entry system-based descriptors and skin sensitization, SAR QSAR Environ. Res., 2019, 30(6), 447–455 CrossRef CAS PubMed.
- A. P. Toropova, A. A. Toropov, R. Rallo, D. Leszczynska and J. Leszczynski, Optimal descriptor as a translator of eclectic data into prediction of cytotoxicity for metal oxide nanoparticles under different conditions, Ecotoxicol. Environ. Saf., 2015, 112, 39–45 CrossRef CAS PubMed.
- A. A. Toropov and A. P. Toropova, The Correlation Contradictions Index (CCI): building up reliable models of mutagenic potential of silver nanoparticles under different conditions using quasi-SMILES, Sci. Total Environ., 2019, 681, 102–109 CrossRef CAS PubMed.
- P. Kumar and A. Kumar, CORAL: QSAR models of CB1 cannabinoid receptor inhibitors based on local and global SMILES attributes with the index of ideality of correlation and the correlation contradiction index, Chemom. Intell. Lab. Syst., 2020, 200, 103982 CrossRef CAS.
- A. P. Toropova, A. A. Toropov, E. Carnesecchi, E. Benfenati and J. L. Dorne, The using of the Index of Ideality of Correlation (IIC) to improve predictive potential of models of water solubility for pesticides, Environ. Sci. Pollut. Res., 2020, 27(12), 13339–13347 CrossRef CAS PubMed.
- A. A. Toropov, R. Carbó-Dorca and A. P. Toropova, Index of Ideality of Correlation: new possibilities to validate QSAR: a case study, Struct. Chem., 2018, 29(1), 33–38 CrossRef CAS.
- S. Lotfi, S. Ahmadi and P. Kumar, A hybrid descriptor based QSPR model to predict the thermal decomposition temperature of imidazolium ionic liquids using Monte Carlo approach, J. Mol. Liq., 2021, 338, 116465 CrossRef CAS.
- P. Achary, A. Toropova and A. Toropov, Combinations of graph invariants and attributes of simplified molecular input-line entry system (SMILES) to build up models for sweetness, Food Res. Int., 2019, 122, 40–46 CrossRef CAS PubMed.
- T. Ghiasi, S. Ahmadi, E. Ahmadi, M. Talei Bavil Olyai and Z. Khodadadi, The index of ideality of correlation: QSAR studies of hepatitis C virus NS3/4A protease inhibitors using SMILES descriptors, SAR QSAR Environ. Res., 2021, 32(6), 495–520 CrossRef CAS PubMed.
- S. Ahmadi, A. P. Toropova and A. A. Toropov, Correlation intensity index: mathematical modeling of cytotoxicity of metal oxide nanoparticles, Nanotoxicology, 2020, 14(8), 1118–1126 CrossRef CAS PubMed.
- A. A. Toropov and A. P. Toropova, Correlation intensity index: building up models for mutagenicity of silver nanoparticles, Sci. Total Environ., 2020, 737, 139720 CrossRef CAS PubMed.
- C. Rojas, P. R. Duchowicz and E. A. Castro, Foodinformatics: Quantitative Structure-Property Relationship Modeling of Volatile Organic Compounds in Peppers, J. Food Sci., 2019, 84(4), 770–781 CrossRef CAS PubMed.
- R. Singh, P. Kumar, M. Devi, S. Lal, A. Kumar, J. Sindhu, A. P. Toropova, A. A. Toropov and D. Singh, Monte Carlo based QSGFEAR: prediction of Gibb's free energy of activation at different temperatures using SMILES based descriptors, New J. Chem., 2022, 46(39), 19062–19072 RSC.
- P. Kumar, A. Kumar and D. Singh, CORAL: development of a hybrid descriptor based QSTR model to predict the toxicity of dioxins and dioxin-like compounds with correlation intensity index and consensus modelling, Environ. Toxicol. Pharmacol., 2022, 93, 103893 CrossRef CAS PubMed.
- A. Kumar, P. Kumar and D. Singh, QSRR modelling for the investigation of gas chromatography retention indices of flavour and fragrance compounds on Carbowax 20 M glass capillary column with the index of ideality of correlation and the consensus modelling, Chemometr. Intelligent Lab. Syst., 2022, 224, 104552 CrossRef CAS.
- M. Duhan, J. Sindhu, P. Kumar, M. Devi, R. Singh, R. Kumar, S. Lal, A. Kumar, S. Kumar and K. Hussain, Quantitative structure activity relationship studies of novel hydrazone derivatives as α-amylase inhibitors with index of ideality of correlation, J. Biomol. Struct. Dyn., 2022, 40(11), 4933–4953 CrossRef CAS PubMed.
- A. P. Toropova, A. A. Toropov, J. B. Veselinović, F. N. Miljković and A. M. Veselinović, QSAR models for HEPT derivates as NNRTI inhibitors based on Monte Carlo method, Eur. J. Med. Chem., 2014, 77, 298–305 CrossRef CAS PubMed.
- S. Ahmadi and A. Akbari, Prediction of the adsorption coefficients of some aromatic compounds on multi-wall carbon nanotubes by the Monte Carlo method, SAR QSAR Environ. Res., 2018, 29(11), 895–909 CrossRef CAS PubMed.
- A. A. Toropov and A. P. Toropova, QSAR as a random event: criteria of predictive potential for a chance model, Struct. Chem., 2019, 30(5), 1677–1683 CrossRef CAS.
- S. Ahmadi, Z. Moradi, A. Kumar and A. Almasirad, SMILES-based QSAR and molecular docking study of xanthone derivatives as α-glucosidase inhibitors, J. Recept. Signal Transduction, 2022, 42(4), 361–372 CrossRef CAS PubMed.
- A. N. Menon, S. Chacko and C. Narayanan, Free and glycosidically bound volatiles of pepper (Piper nigrum L.), J. Essent. Oil Res., 2001, 13(3), 166–169 CrossRef CAS.
- M. Gopalakrishnan, N. Menon, K. Padmakumari, A. Jayalekshmy and C. Narayanan, GC Analysis and Odor Profiles of Four New Indian Genotypes of Piper nigrum L, J. Essent. Oil Res., 1993, 5(3), 247–253 CrossRef CAS.
- A. Martins, L. Salgueiro, R. Vila, F. Tomi, S. Canigueral, J. Casanova, A. P. Da Cunha and T. Adzet, Essential oils from four Piper species, Phytochemistry, 1998, 49(7), 2019–2023 CrossRef CAS.
- A. Orav, I. Stulova, T. Kailas and M. Müürisepp, Effect of storage on the essential oil composition of Piper nigrum L. fruits of different ripening states, J. Agric. Food Chem., 2004, 52(9), 2582–2586 CrossRef CAS PubMed.
- J. K. R. Da Silva, J. R. A. Silva, S. B. Nascimento, S. F. Da Luz, E. N. Meireles, C. N. Alves, A. R. Ramos and J. G. S. Maia, Antifungal activity and computational study of constituents from Piper divaricatum essential oil against Fusarium infection in black pepper, Molecules, 2014, 19(11), 17926–17942 CrossRef PubMed.
- A. N. Menon and K. Padmakumari, Essential oil composition of four major cultivars of black pepper (Piper nigrum L.)—IV, J. Essent. Oil Res., 2005, 17(2), 206–208 CrossRef.
- A. N. Menon and K. Padmakumari, Studies on essential oil composition of cultivars of black pepper (Piper nigrum L.)—V, J. Essent. Oil Res., 2005, 17(2), 153–155 CrossRef.
- A. N. Menon, K. Padmakumari and A. Jayalekshmy, Essential oil composition of four major cultivars of black pepper (Piper nigrum L.) III, J. Essent. Oil Res., 2003, 15(3), 155–157 CrossRef CAS.
- A. N. Menon, K. Padmakumari, A. Jayalekshmy, M. Gopalakrishnan and C. Narayanan, Essential oil composition of four popular Indian cultivars of black pepper (Piper nigrum L.), J. Essent. Oil Res., 2000, 12(4), 431–434 CrossRef CAS.
- D. Sruthi, J. T. Zachariah, N. Leela and K. Jayarajan, Correlation between chemical profiles of black pepper (Piper nigrum L.) var. Panniyur-1 collected from different locations, J. Med. Plants Res., 2013, 7(31), 2349–2357 CrossRef.
- R. Howevertery, R. Seifert, D. Guadagni and L. Ling, Characterization of some volatile constituents of bell peppers, J. Agric. Food Chem., 1969, 17(6), 1322–1327 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra07960k |
| This journal is © The Royal Society of Chemistry 2024 |
