Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of NHAcGD2/NHAcGD3 conjugates of bacteriophage MX1 virus-like particles as anticancer vaccines†
Qingyu
Zhao
 a,
Xuefei
Huang
b and
Xuanjun
Wu
a,
Xuefei
Huang
b and
Xuanjun
Wu
 *a
*a
aNational Glycoengineering Research Center and Shandong Key Laboratory of Carbohydrate Chemistry and Glycobiology, NMPA Key Laboratory for Quality Research and Evaluation of Carbohydrate-based Medicine, Shandong University, Qingdao, Shandong 266237, China. E-mail: xuanjun@sdu.edu.cn
bDepartments of Chemistry and Biomedical Engineering, Institute for Quantitative Health Science and Engineering, Michigan State University, East Lansing, Michigan 48824, USA
First published on 19th February 2024
Abstract
The successful development of an anticancer vaccine will be a giant leap forward in cancer prevention and treatment. Herein, the bacteriophage MX1 coat protein virus-like particles (MX1 VLPs) have been conjugated with 9NHAc-GD2 (NHAcGD2) to obtain a MX1-NHAcGD2 conjugate. Intriguingly, vaccinating against this conjugate produced a robust anti-NHAcGD2 IgG response in mice, with an average IgG titer of over 3 million. More interestingly, antibodies induced by the MX1-NHAcGD2 conjugate bound well to IMR-32 neuroblastoma cells and had potent complement-dependent cytotoxic (CDC) effects on IMR-32 cells. Inspired by the superiority of the 9NHAc-GD2 antigen, we also designed another 9NHAc-modified ganglioside antigen, 9NHAc-GD3 (NHAcGD3), to overcome the hydrolytic instability of 9-O-acetylated-GD3. By coupling NHAcGD3 with MX1 VLP, the MX1-NHAcGD3 conjugate was constructed. Strikingly, vaccination of MX1-NHAcGD3 elicited high anti-NHAcGD3 IgG antibodies, which effectively recognized human malignant melanoma SK-MEL-28 cells and had a significant CDC effect against this cell line. This study provides novel MX1-NHAcGD2 and MX1-NHAcGD3 conjugates with broad clinical translational prospects as promising anticancer vaccines.
Cancer is a major human disease with high fatality. Globally, there were 19.3 million new cases of cancer in 2020, including nearly 10 million deaths. The number of cancer cases worldwide is expected to reach 28.4 million by 2040.1 These alarming data urgently require the development of effective cancer treatments. Commonly used cancer treatment methods include chemotherapy,2,3 radiotherapy,4 phototherapy,5–7 and immunotherapy,8–11 of which immunotherapy is attractive because it can call on the immune system to fight cancer cells. As a promising modality of cancer immunotherapy, vaccine immunization can provide long-term protection to the host with few side effects.12–15 Therefore, developing effective and safe anticancer vaccines is crucial.
Antigens are one of the indispensable components of vaccines. As one fantastic class of antigens, tumor-associated carbohydrate antigens (TACAs) include Tn, Tf, STn, Globo-H, GM2, GD2, GD3, etc,16 among which GD2 and GD3 gangliosides are a class of glycosphingolipids with two sialic acid residues linked to lactosylceramides.17 They are widely expressed in most cancers of neuroectodermal or mesodermal origin, including melanoma, neuroblastoma and sarcoma. Nevertheless, GD2 and GD3 are also distributed in normal tissues, leading to adverse side effects. For example, treating patients with high-dose GD2 monoclonal antibodies results in dose-dependent acute and/or chronic toxicity due to high expression of GD2 in peripheral nerves.18 To overcome side effects, more specific target antigens should be explored.
9-O-acetylation is a common natural modification on sialic acid, which impacts many biological phenomena, such as microbial and host interactions.19 Compared with GD2 and GD3 antigens, 9-O-acetyl-GD2 (9OAc-GD2) and 9-O-acetyl-GD3 (9OAc-GD3) are a class of tumor target antigens with higher specificity. 9OAc-GD2 is strongly expressed on the surface of many cancer cells, including neuroblastoma,20 glioblastoma,21 and breast cancer;22 meanwhile, 9OAc-GD3 is overexpressed in melanoma,23 glioblastoma,24 breast cancer,25 and small cell lung cancer.26 Since 9OAc-GD2 and 9OAc-GD3 are rarely expressed in normal tissues, antibodies based on these two antigens or antibodies produced by vaccines are highly specific. However, developing vaccines based on 9-OAc-GD2/9-OAc-GD3 is challenging due to the hydrolytic instability of O-acetylated-GD2/or-GD3. To address this challenge, we recently reported a stable 9NHAc-GD2 (NHAcGD2) antigen that mimics the 9OAc-GD2 antigen.27 It has been coupled to the bacteriophage Qβ virus-like particle (VLP) to produce the Qβ-NHAcGD2 conjugate, which elicited robust anti-NHAcGD2 IgG antibodies.27 The results show that NHAcGD2 is a tumor target antigen with high specificity and indicate the importance of Qβ VLP conjugation in enhancing the anti-NHAcGD2 IgG response.
Building on the above work, we aim to establish more vaccines based on NHAcGD2, such as developing NHAcGD2 conjugates with a new carrier. This is because as the variety of vaccines under development increases, more and more conjugate vaccines share the same carrier; for example, many reported vaccine candidates contain Qβ VLP.27–37 The concomitant use of these vaccines may lead to high levels of pre-existing antibodies against the carrier, possibly suppressing the immune responses against new conjugate vaccines sharing the same carrier moiety.38,39 Therefore, establishing a new carrier is valuable for expanding the arsenal of vaccines.
In this work, we have begun investigating a novel carrier, the coat protein (CP) VLP of Enterobacteria phage MX1 (a strain of Qβ virus), which has never been used in TACA-based vaccines. The pET-28-MX1-CP plasmid was constructed according to the gene ID 1261502. Strikingly, we achieved a high yield (∼80 mg L−1) in preparing MX1 VLPs. Then, we investigated this carrier for NHAcGD2 conjugate vaccine development (Scheme 1a). Intriguingly, MX1-NHAcGD2 elicited high levels of anti-NHAcGD2 IgG responses. Inspired by the superiority of the NHAcGD2 antigen, we envision 9NHAc-GD3 (NHAcGD3) as a promising antigen to mimic 9OAc-GD3. As such, we synthesized the NHAcGD3 antigen and conjugated this new antigen with MX1 VLP to produce MX1-NHAcGD3 conjugate (Scheme 1b). Encouragingly, vaccinating mice with MX1-NHAcGD3 elicited potent anti-NHAcGD3 IgG responses.
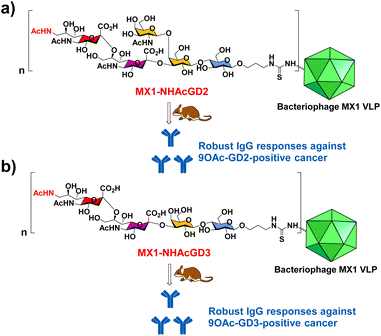 | ||
| Scheme 1 Schematic diagram of the proposed (a) MX1-NHAcGD2 and (b) MX1-NHAcGD3 conjugates for eliciting robust anticancer IgG responses. | ||
Results and discussion
Preparation and characterization of MX1 VLP
The pET28-MX1-CP plasmid was constructed according to the gene ID 1261502 and transformed into BL21 (DE3) competent cells. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was used to induce the expression of MX1 VLP. Afterward, the MX1 VLP-expressing bacteria were collected for ultrasonic disruption. The lysate is then subjected to purification steps, including PEG8000 precipitation, chloroform/n-butanol organic extraction, and sucrose gradient centrifugation. After purification, the Bradford assay was used to quantify total protein content, using bovine serum albumin (BSA) as a standard. Excitingly, the yield of MX1 VLP is ∼80 mg L−1. Previous studies have shown that the yield of Qβ VLP is approximately 30 mg L−1,40,41 which is consistent with the results we have obtained by multiple expressions of Qβ VLP in the past. The very high yield of MX1 VLP facilitates the large-scale preparation of MX1-TACA conjugate vaccines for clinical translation in the future.The purified MX1 VLP was characterized by size-exclusion HPLC (SEC), SDS polyacrylamide gel electrophoresis (SDS-PAGE), MALDI-TOF mass spectrometry (MS), transmission electron microscopy (TEM), and dynamic light scattering (DLS). SEC and SDS-PAGE results showed good purity of the obtained MX1 VLP (Fig. S1 and S2, ESI†). The MALDI-TOF MS result showed that the molecular weight of the MX1 subunit was 14.1 kDa (Fig. S3, ESI†). TEM showed the nanostructure of MX1 with a mean diameter of 26 nm (Fig. 1), which was almost consistent with the DLS result (Fig. S4, ESI†). MX1's VLP structure will confer its ability to present TACA, such as NHAcGD2, in a highly ordered manner,42 thereby eliciting potent anti-TACA IgG antibodies.
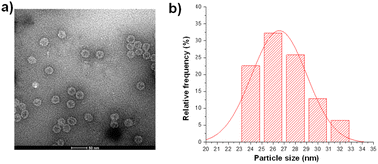 | ||
| Fig. 1 (a) TEM image of the bacteriophage MX1 coat protein virus-like particles (MX1 VLPs). (b) The average diameter of MX1 VLP is 26 nm, as determined by nano-measure software. | ||
Synthesis of NHAcGD2-isothiocyanate (NCS) 1, MX1-NHAcGD2 conjugate 2, CRM197-NHAcGD2 conjugate 3, and MX1-GD2 conjugate 4
The chemoenzymatic synthesis of NHAcGD2-NCS 1 began with LacβProN3 S1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 (Scheme S1, ESI†), which was incubated with Neu5Ac, cytidine-5′-triphosphate (CTP) in Tris–HCl buffer (100 mM, pH 8.5) containing MgCl2, followed by the addition of Neisseria meningitidis CMP-sialic acid synthetase (NmCSS) and Pasteurella multocida α2,3-sialyltransferase (PmST1) to form GM3-N3 S2 in a yield of 92%. Then, S2 was treated with Campylobacter jejuni α2,8-sialyltransferase (CjCstII) and NmCSS in the presence of 9NHAc-Neu5Ac S3,27 CTP and MgCl2, resulting in NHAcGD3-N3 S4 in a yield of 81%. By the addition of UDP-GalNAc and Campylobacter jejuni β1,4 N-acetylgalactosaminyltransferase (CjCgtA) in Tris–HCl buffer (100 mM, pH 7.5) with MgCl2, S4 was converted to NHAcGD2-N3 S5 in a yield of 83%, followed by Pd/C-H2 reduction and isothiocyanate formation producing NHAcGD2-NCS 1. Nuclear magnetic resonance (NMR) spectroscopy and MS were used to characterize the final product and intermediate compounds.
43 (Scheme S1, ESI†), which was incubated with Neu5Ac, cytidine-5′-triphosphate (CTP) in Tris–HCl buffer (100 mM, pH 8.5) containing MgCl2, followed by the addition of Neisseria meningitidis CMP-sialic acid synthetase (NmCSS) and Pasteurella multocida α2,3-sialyltransferase (PmST1) to form GM3-N3 S2 in a yield of 92%. Then, S2 was treated with Campylobacter jejuni α2,8-sialyltransferase (CjCstII) and NmCSS in the presence of 9NHAc-Neu5Ac S3,27 CTP and MgCl2, resulting in NHAcGD3-N3 S4 in a yield of 81%. By the addition of UDP-GalNAc and Campylobacter jejuni β1,4 N-acetylgalactosaminyltransferase (CjCgtA) in Tris–HCl buffer (100 mM, pH 7.5) with MgCl2, S4 was converted to NHAcGD2-N3 S5 in a yield of 83%, followed by Pd/C-H2 reduction and isothiocyanate formation producing NHAcGD2-NCS 1. Nuclear magnetic resonance (NMR) spectroscopy and MS were used to characterize the final product and intermediate compounds.
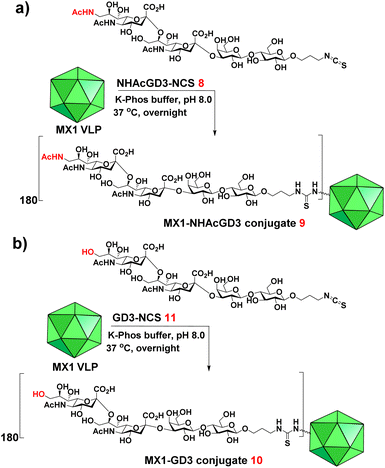
We next exploited the MX1 carrier for the NHAcGD2 conjugate vaccine development. MX1-NHAcGD2 conjugate 2 was synthesized by adding 1 to K-Phos buffer (0.1 M, pH 8.0) with MX1 VLP overnight at 37 °C (Scheme 2a). MALDI-TOF MS analysis of the conjugate 2 showed that each MX1 particle contained an average of 270 NHAcGD2 (Fig. S5, ESI†). In parallel, the CRM197-NHAcGD2 conjugate 3 (Scheme 2b) was fabricated by coupling NHAcGD2-NCS 1 with cross-reactive material CRM197, which has been widely utilized as an anti-microbial vaccine and anticancer vaccine candidate.44–46 It was determined that 3 contained an average of 7 copies of NHAcGD2 per CRM197 (Fig. S6, ESI†). In addition, to verify the effect of 9-NHAc modification of GD2 on vaccine performance, MX1-GD2 conjugate 4 (Scheme 2c) was also made by conjugating GD2-NCS 5 (Scheme S2, ESI†) with MX1 VLP as a control vaccine, which contained 235 GD2 per particle (Fig. S7, ESI†).
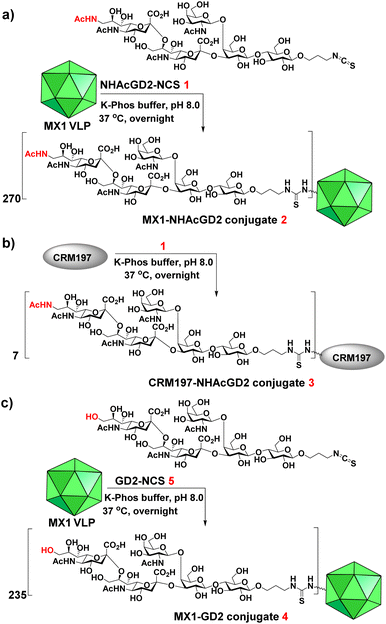 | ||
| Scheme 2 Syntheses of (a) MX1-NHAcGD2 conjugate 2, (b) CRM197-NHAcGD2 conjugate 3 and (c) MX1-GD2 conjugate 4. | ||
Immunological evaluation of MX1-NHAcGD2 conjugate 2, CRM197-NHAcGD2 conjugate 3 and MX1-GD2 conjugate 4
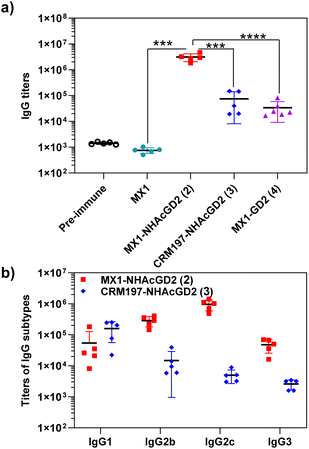
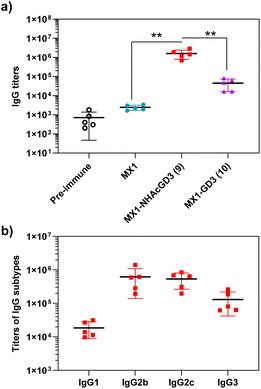
After synthesizing and characterizing conjugates 2–4, we assess their immune properties in C57BL/6 female mice. On days 0, 14, and 28, C57BL/6 female mice were vaccinated with conjugates 2–4 with monophosphoryl-lipid A (MPLA) as adjuvant. Pre- and post-immunization (day 35) sera were collected for immunological testing, including enzyme-linked immunosorbent assay (ELISA), fluorescence-activated cell sorting (FACS), and complement-dependent cytotoxicity (CDC) assay.Firstly, ELISA was used to detect anti-NHAcGD2/GD2 IgG antibody titers produced by the conjugates. To perform ELISA, NHAcGD2-NCS 1 and GD2-NCS 5 were respectively conjugated to bovine serum albumin (BSA) to give BSA-NHAcGD2 conjugate 6 and BSA-GD2 conjugate 7, which were characterized by MALDI-TOF MS (Fig. S8 and S9, ESI†), and then coated on plates for ELISA experiments. It was shown that pre-immunization, MX1-NHAcGD2 conjugate 2, and CRM197-NHAcGD2 conjugate 3-induced average anti-NHAcGD2 IgG titers were 1,425, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 094
094![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 844, and 74
844, and 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472, respectively (Fig. 2a), suggesting that MX1-NHAcGD2 conjugate 2 can elicit a more robust humoral immune response than CRM197-NHAcGD2 conjugate 3. Notably, the MX1-GD2 conjugate 4 induced an average anti-GD2 IgG titer of only 33
472, respectively (Fig. 2a), suggesting that MX1-NHAcGD2 conjugate 2 can elicit a more robust humoral immune response than CRM197-NHAcGD2 conjugate 3. Notably, the MX1-GD2 conjugate 4 induced an average anti-GD2 IgG titer of only 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 768 (Fig. 2a), which was significantly lower than the MX1-NHAcGD2 conjugate 2-induced IgG titer, indicating the importance of the 9NHAc modification of GD2 antigen. In addition, through the analysis of vaccine-induced IgG antibody subtypes, it was found that the IgG subtype produced by CRM197-NHAcGD2 conjugate 3 was predominantly IgG1. In contrast, the IgG produced by MX1-NHAcGD2 conjugate 2 had high levels of various subtypes, including IgG1, IgG2b, IgG2c, and IgG3, with IgG2c being the highest (Fig. 2b). Collectively, MX1-NHAcGD2 conjugate 2 produced high levels of 9-NHAc-GD2 specific IgG antibodies.
768 (Fig. 2a), which was significantly lower than the MX1-NHAcGD2 conjugate 2-induced IgG titer, indicating the importance of the 9NHAc modification of GD2 antigen. In addition, through the analysis of vaccine-induced IgG antibody subtypes, it was found that the IgG subtype produced by CRM197-NHAcGD2 conjugate 3 was predominantly IgG1. In contrast, the IgG produced by MX1-NHAcGD2 conjugate 2 had high levels of various subtypes, including IgG1, IgG2b, IgG2c, and IgG3, with IgG2c being the highest (Fig. 2b). Collectively, MX1-NHAcGD2 conjugate 2 produced high levels of 9-NHAc-GD2 specific IgG antibodies.
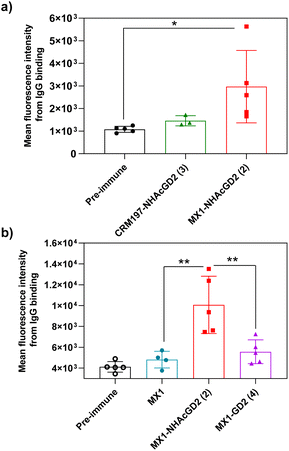
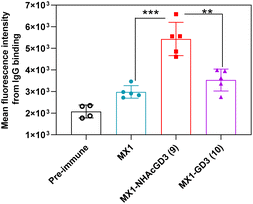
Secondly, FACS was used to detect the ability of serum antibodies to bind to tumors. The serum was co-incubated with tumor cells in FACS buffer. After washing the cells, the FITC-conjugated goat anti-mouse IgG antibody was added for flow cytometry analysis. GD2/9OAc-GD2 was expressed on the surface of EL4 lymphoma cells and human neuroblastoma IMR-32 cells, so these cells were used for FACS studies. The results showed MX1-NHAcGD2 conjugate 2-induced IgG antibody bound to EL4 cells more efficiently than CRM197-NHAcGD2 conjugate 3-generated IgG antibody (Fig. 3a). Notably, MX1 and MX1-GD2 (4) induced antibodies had a weaker binding capacity to IMR-32 cells. In contrast, the MX1-NHAcGD2 conjugate 2-induced IgG has the highest binding ability to IMR-32 cells (Fig. 3b). This result confirms that MX1-NHAcGD2 conjugate 2 produces potent IgG antibodies that bind tightly to tumor cells.
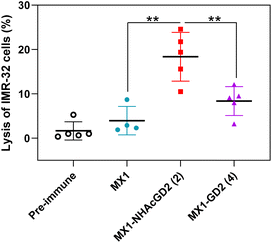
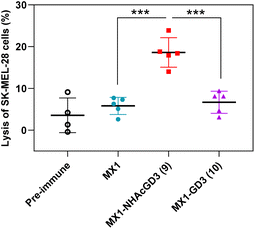
Thirdly, the CDC was used to test the complement's ability to mediate cancer cell killing through serum antibodies. Complement cleaves target cells by binding specific antibodies to the corresponding antigen on the surface of tumor cells, activating the classical complement pathway known as the CDC. Post-immunization serum of MX1, MX1-NHAcGD2 conjugate 2, or MX1-GD2 conjugate 4 was incubated with IMR-32 cells, rabbit complement was then added, and cytotoxicity was calculated using the MTS cell viability assay. The results showed that the proportion of IMR-32 cells lysed upon incubation with MX1-NHAcGD2 conjugate 2 induced IgG was significantly higher than those treated with MX1-GD2 conjugate 4 and MX1-induced IgG (Fig. 4), indicating that the IgG produced by the conjugate 2 had a superior killing effect on IMR-32 cells in the presence of complement.
MX1 VLP is also a superior carrier for NHAcGD3 conjugate vaccine development
9-OAc-GD3 is another important ganglioside antigen that is overexpressed in melanoma,23 glioblastoma,24 breast cancer,25 and small cell lung cancer.26 Inspired by the superiority of the NHAcGD2 antigen, we envision NHAcGD3 as a promising antigen that mimics 9OAc-GD3. To synthesize the 9-NHAc-GD3 antigen for MX1 conjugation, we converted the amine group of NHAcGD3-NH2 S10 to NHAcGD3-NCS 8 with thiophosgene (Scheme S3a, ESI†). 8 was then conjugated with the bacteriophage MX1 VLP in K-Phos buffer (0.1 M, pH 8.0) overnight at 37 °C to give MX1-NHAcGD3 conjugate 9 (Scheme 3a). MALDI-TOF MS analysis of the resulting conjugate showed that the number of NHAcGD3 per MX1 particle was 180 (Fig. S10, ESI†). To benchmark the performance of the 9NHAc modification, we also synthesized MX1-GD3 conjugate 10(Scheme 3b) by conjugating MX1 VLP with GD3-NCS 11 (Scheme S3b, ESI†) in K-Phos buffer (pH 8.0, 0.1 M). MALDI-TOF MS determined the amount of GD3 in MX1-GD3 (10) as 180 copies of GD3 per particle (Fig. S11, ESI†).With conjugate vaccines 9 and 10 in hand, we measured the anti-NHAcGD3/GD3 IgG titers they produced in C57BL/6 mice. NHAcGD3-NCS 8 and GD3-NCS 11 were conjugated to BSA to obtain BSA-NHAcGD3 conjugate 12 and BSA-GD3 conjugate 13, which were characterized with MALDI-TOF MS (Fig. S12 and S13, ESI†). When ELISA was performed, the levels of anti-NHAcGD3 IgG and anti-GD3 IgG were against 12 and 13, respectively. The results showed that the mean anti-NHAcGD3 IgG titers by pre-immunization and those induced by MX1, and MX1-NHAcGD3 conjugate 9 were 711, 1626, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 603
603![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 672, respectively (Fig. 5a). Notably, the mean anti-GD3 IgG titer caused by MX1-GD3 conjugate 10 was only 45
672, respectively (Fig. 5a). Notably, the mean anti-GD3 IgG titer caused by MX1-GD3 conjugate 10 was only 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 054 (Fig. 5a), which was significantly lower than that induced by MX1-NHAcGD3 conjugate 9, indicating the crucial role of 9NHAc modification of GD3 antigen. The vaccine-induced IgG antibody subtype analysis revealed that MX1-NHAcGD3 conjugate 9 produced high levels of various subtypes of IgG, with IgG2b and IgG2c predominating (Fig. 5b). Overall, MX1-NHAcGD3 conjugate 9 induces a high NHAcGD3-specific IgG response.
054 (Fig. 5a), which was significantly lower than that induced by MX1-NHAcGD3 conjugate 9, indicating the crucial role of 9NHAc modification of GD3 antigen. The vaccine-induced IgG antibody subtype analysis revealed that MX1-NHAcGD3 conjugate 9 produced high levels of various subtypes of IgG, with IgG2b and IgG2c predominating (Fig. 5b). Overall, MX1-NHAcGD3 conjugate 9 induces a high NHAcGD3-specific IgG response.
Next, FACS was used to test the ability of antibodies produced by GD3-related conjugate vaccines to bind to tumors. GD3/9OAc-GD3 was expressed on the surface of human melanoma SK-MEL-28 cell line, which was used in FACS studies. The results showed that MX1 and MX1-GD3 conjugate 10-induced IgG antibodies had a weak binding capacity to SK-MEL-28 cells. In contrast, MX1-NHAcGD3 conjugate 9 induced-IgG had strong binding to SK-MEL-28 cells (Fig. 6). This result confirms that MX1-NHAcGD3 conjugate 9 produces potent NHAcGD3-specific IgG antibodies that bind tightly to tumor cells.
Finally, post-immunization serum of MX1, MX1-NHAcGD3 conjugate 9, or MX1-GD3 conjugate 10 was incubated with SK-MEL-28 cells, rabbit complement was added, and cytotoxicity was calculated using the MTS assay. The results showed that the lysis proportion of SK-MEL-28 cells treated with MX1-NHAcGD3 conjugate 9 induced IgG antibodies was significantly higher than those treated with MX1-GD3 conjugate 10 or MX1-induced IgG antibodies (Fig. 7), suggesting that the IgG produced by the conjugate 9 had a superior killing effect on SK-MEL-28 cells in the presence of complement.
Conclusions
In this work, a novel bacteriophage MX1 VLP is reported to construct the MX1-NHAcGD2 conjugate by conjugating the 9NHAc-GD2 (NHAcGD2) antigen with MX1 VLP. The MX1-NHAcGD2 conjugate produces potent NHAcGD2-specific IgG antibodies that bind specifically to IMR-32 neuroblastoma cells, and antibodies mediate good CDC to kill this cell line. In addition, inspired by the NHAcGD2 antigen, the 9NHAc-GD3 (NHAcGD3) antigen used to mimic the 9-OAc-GD3 antigen is investigated for the first time. The conjugate of MX1 to NHAcGD3 induces potent NHAcGD3-specific IgG antibodies, which bound firmly to SK-MEL-28 melanoma cells and mediated significant CDC killing. In summary, the MX1 VLP has excellent potential as a new class of VLP vaccine carrier. The MX1-NHAcGD2 and MX1-NHAcGD3 conjugates can be exciting leads for anticancer vaccines.Author contributions
X. W. conceived the concept of developing MX1-NHAcGD2 and MX1-NHAcGD3 conjugate vaccines and supervised the project. X. W. and Q. Z. contributed to experimental design. Q. Z. performed the experiments. X. W. and Q. Z. wrote the manuscript. All authors revised the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (22007058 and 22377065), the Excellent Youth Fund Project supported by Shandong Provincial Natural Science Foundation (ZR2022YQ17), the Taishan Scholars Program for Young Expert of Shandong Province, and the Qilu Young Scholar Fund of Shandong University. We thank Dr Yawen Zou (Prof. Yi Yang group) at Hunan Agricultural University for giving the PET-28-MX1-CP plasmid. All animal experiments were performed in accordance with the guidelines of the Animal Care and Use Committee of Shandong University and approved by the Animal Ethics Committee of the College of Life Sciences at Shandong University (SYDWLL-2023-044).Notes and references
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, CA Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- H. Dong, X. Huang and X. Wu, Biomater. Sci., 2023, 11, 6160–6166 RSC.
- J. L. Nitiss, Nat. Rev. Cancer, 2009, 9, 338–350 CrossRef CAS.
- D. Schaue and W. H. McBride, Nat. Rev. Clin. Oncol., 2015, 12, 527–540 CrossRef.
- H. Dong, Y. Gao, X. Huang and X. Wu, J. Mater. Chem. B, 2022, 10, 927–934 RSC.
- Q. Chen, C. Wang, Z. Zhan, W. He, Z. Cheng, Y. Li and Z. Liu, Biomaterials, 2014, 35, 8206–8214 CrossRef CAS PubMed.
- J. Wang, C. Yang, Y. Gao and X. Wu, J. Mater. Chem. B, 2023, 11, 6560–6566 RSC.
- A. D. Waldman, J. M. Fritz and M. J. Lenardo, Nat. Rev. Immunol., 2020, 20, 651–668 CrossRef CAS.
- C. Graham, R. Hewitson, A. Pagliuca and R. Benjamin, Clin. Med., 2018, 18, 324–328 CrossRef.
- N. Shimasaki, A. Jain and D. Campana, Nat. Rev. Drug Discovery, 2020, 19, 200–218 CrossRef CAS PubMed.
- F. Marofi, O. F. Abdul-Rasheed, H. S. Rahman, H. S. Budi, A. T. Jalil, A. V. Yumashev, A. Hassanzadeh, M. Yazdanifar, R. Motavalli, M. S. Chartrand, M. Ahmadi, A. Cid-Arreguid and M. Jarahian, Cancer Sci., 2021, 112, 3427–3436 CrossRef CAS PubMed.
- E. Gilboa, Nat. Rev. Cancer, 2004, 4, 401–411 CrossRef CAS PubMed.
- I. Melero, G. Gaudernack, W. Gerritsen, C. Huber, G. Parmiani, S. Scholl, N. Thatcher, J. Wagstaff, C. Zielinski, I. Faulkner and H. Mellstedt, Nat. Rev. Clin. Oncol., 2014, 11, 509–524 CrossRef CAS.
- S. Mohammed, N. Bakshi, N. Chaudri, J. Akhter and M. Akhtar, Adv. Anat. Pathol., 2016, 23, 180–191 CrossRef CAS PubMed.
- M. Saxena, S. H. van der Burg, C. J. M. Melief and N. Bhardwaj, Nat. Rev. Cancer, 2021, 21, 360–378 CrossRef CAS.
- M. M. Wei, Y. S. Wang and X. S. Ye, Med. Res. Rev., 2018, 38, 1003–1026 CrossRef PubMed.
- S. Cavdarli, S. Groux-Degroote and P. Delannoy, Biomolecules, 2019, 9, 311 CrossRef CAS PubMed.
- S. Zhang, C. Cordon-Cardo, H. S. Zhang, V. E. Reuter, S. Adluri, W. B. Hamilton, K. O. Lloyd and P. O. Livingston, Int. J. Cancer, 1997, 73, 42–49 CrossRef CAS PubMed.
- G. N. Rogers, G. Herrler, J. C. Paulson and H. D. Klenk, J. Biol. Chem., 1986, 261, 5947–5951 CrossRef CAS PubMed.
- N. Alvarez-Rueda, A. Desselle, D. Cochonneau, T. Chaumette, B. Clemenceau, S. Leprieur, G. Bougras, S. Supiot, J.-M. Mussini, J. Barbet, J. Saba, F. Paris, J. Aubry and S. Birklé, PLoS One, 2011, 6, e25220 CrossRef CAS PubMed.
- J. Fleurence, D. Cochonneau, S. Fougeray, L. Oliver, F. Geraldo, M. Terme, M. Dorvillius, D. Loussouarn, F. Vallette, F. Paris and S. Birklé, Oncotarget, 2016, 7, 41172–41185 CrossRef PubMed.
- S. Cavdarli, J. H. Dewald, N. Yamakawa, Y. Guérardel, M. Terme, J.-M. Le Doussal, P. Delannoy and S. Groux-Degroote, Glycoconj. J., 2019, 36, 79–90 CrossRef CAS PubMed.
- D. A. Cheresh, R. A. Reisfeld and A. P. Varki, Science, 1984, 225, 844–846 CrossRef CAS PubMed.
- S. M. Birks, J. O. Danquah, L. King, R. Vlasak, D. C. Gorecki and G. J. Pilkington, Neuro-Oncol., 2011, 13, 950–960 CrossRef CAS.
- A. Gocht, G. Rutter and B. Kniep, Histochem. Cell Biol., 1998, 110, 217–229 CrossRef CAS PubMed.
- R. Fuentes, R. Allman and M. D. Mason, Lung cancer, 1997, 18, 21–33 CrossRef CAS.
- X. Wu, J. Ye, A. T. DeLaitsch, Z. Rashidijahanabad, S. Lang, T. Kakeshpour, Y. Zhao, S. Ramadan, P. V. Saavedra, V. Yuzbasiyan-Gurkan, H. Kavunja, H. Cao, J. C. Gildersleeve and X. Huang, Angew. Chem., Int. Ed., 2021, 60, 24179–24188 CrossRef CAS.
- Z. Yin, X. Wu, K. Kaczanowska, S. Sungsuwan, M. Comellas-Aragones, C. Pett, J. Yu, C. Baniel, U. Westerlind, M. G. Finn and X. Huang, ACS Chem. Biol., 2018, 13, 1668–1676 CrossRef CAS.
- Z. Yin, M. Comellas-Aragones, S. Chowdhury, P. Bentley, K. Kaczanowska, L. BenMohamed, J. C. Gildersleeve, M. G. Finn and X. Huang, ACS Chem. Biol., 2013, 8, 1253–1262 CrossRef CAS.
- Z. Yin, S. Dulaney, C. S. McKay, C. Baniel, K. Kaczanowska, S. Ramadan, M. G. Finn and X. Huang, Chembiochem, 2016, 17, 174–180 CrossRef CAS.
- M. O. Mohsen, L. Zha, G. Cabral-Miranda and M. F. Bachmann, Semin. Immunol., 2017, 34, 123–132 CrossRef CAS PubMed.
- D. E. Speiser, K. Schwarz, P. Baumgaertner, V. Manolova, E. Devevre, W. Sterry, P. Walden, A. Zippelius, K. B. Conzett, G. Senti, V. Voelter, J.-P. Cerottini, D. Guggisberg, J. Willers, C. Geldhof, P. Romero, T. Kündig, A. Knuth, R. Dummer, U. Trefzer and M. F. Bachmann, J. Immunother., 2010, 33, 848–858 CrossRef CAS.
- K.-M. Beeh, F. Kanniess, F. Wagner, C. Schilder, I. Naudts, A. Hammann-Haenni, J. Willers, H. Stocker, P. Mueller, M. F. Bachmann and W. A. Renner, J. Allergy Clin. Immunol., 2013, 131, 866–874 CrossRef CAS PubMed.
- X. Wu, Z. Yin, C. McKay, C. Pett, J. Yu, M. Schorlemer, T. Gohl, S. Sungsuwan, S. Ramadan, C. Baniel, A. Allmon, R. Das, U. Westerlind, M. G. Finn and X. Huang, J. Am. Chem. Soc., 2018, 140, 16596–16609 CrossRef CAS.
- Z. Yin, S. Chowdhury, C. McKay, C. Baniel, W. S. Wright, P. Bentley, K. Kaczanowska, J. C. Gildersleeve, M. G. Finn, L. BenMohamed and X. Huang, ACS Chem. Biol., 2015, 10, 2364–2372 CrossRef CAS.
- Z. Rashidijahanabad, S. Ramadan, N. A. O'Brien, A. Nakisa, S. Lang, H. Crawford, J. C. Gildersleeve and X. Huang, Angew. Chem., Int. Ed., 2023, 62, e202309744 CrossRef CAS.
- X. Wu, C. McKay, C. Pett, J. Yu, M. Schorlemer, S. Ramadan, S. Lang, S. Behren, U. Westerlind, M. G. Finn and X. Huang, ACS Chem. Biol., 2019, 14, 2176–2184 CrossRef CAS PubMed.
- R. Dagan, J. Eskola, C. Leclerc and O. Leroy, Infect. Immun., 1998, 66, 2093–2098 CrossRef CAS.
- A. Fattom, Y. H. Cho, C. Chu, S. Fuller, L. Fries and R. Naso, Vaccine, 1999, 17, 126–133 CrossRef CAS.
- S. Sungsuwan, X. Wu, V. Shaw, H. Kavunja, H. McFall-Boegeman, Z. Rashidijahanabad, Z. Tan, S. Lang, S. Tahmasebi Nick, P.-h. Lin, Z. Yin, S. Ramadan, X. Jin and X. Huang, ACS Chem. Biol., 2022, 17, 3047–3058 CrossRef CAS.
- R. Golmohammadi, K. Fridborg, M. Bundule, K. Valegård and L. Liljas, Structure, 1996, 4, 543–554 CrossRef CAS.
- M. F. Bachmann, U. H. Rohrer, T. M. Kündig, K. Bürki, H. Hengartner and R. M. Zinkernagel, Science, 1993, 262, 1448–1451 CrossRef CAS PubMed.
- H. Yu, H. Chokhawala, R. Karpel, H. Yu, B. Wu, J. Zhang, Y. Zhang, Q. Jia and X. Chen, J. Am. Chem. Soc., 2005, 127, 17618–17619 CrossRef CAS.
- R. D. Astronomo and D. R. Burton, Nat. Rev. Drug Discovery, 2010, 9, 308–324 CrossRef CAS.
- C. Song, X.-J. Zheng, H. Guo, Y. Cao, F. Zhang, Q. Li, X.-S. Ye and Y. Zhou, Glycoconj. J., 2019, 36, 399–408 CrossRef CAS PubMed.
- S. J. Danishefsky, Y.-K. Shue, M. N. Chang and C.-H. Wong, Acc. Chem. Res., 2015, 48, 643–652 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra08923a |
| This journal is © The Royal Society of Chemistry 2024 |