Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Serendipitous N,S-difunctionalization of triazoles with trifluoromethyl-β-diketones: access to regioisomeric 1-trifluoroacetyl-3-aryl-5-(2-oxo-2-arylethylthio)-1,2,4-triazoles as DNA-groove binders†
Ranjana Aggarwal *ab,
Prince Kumar
*ab,
Prince Kumar a,
Mona Hooda
a,
Mona Hooda c and
Suresh Kumar
c and
Suresh Kumar a
a
aDepartment of Chemistry, Kurukshetra University, Kurukshetra-136119, Haryana, India
bCouncil of Scientific and Industrial Research-National Institute of Science Communication and Policy Research, New Delhi 110012, India. E-mail: ranjana67in@yahoo.com; ranjanaaggarwal67@gmail.com; Tel: +91-9896740740
cDepartment of Chemistry, Gurugram University, Gurugram-122003, Haryana, India
First published on 23rd February 2024
Abstract
In the present research work, a serendipitous regioselective synthesis of DNA targeting agents, 1-trifluoroacetyl-3-aryl-5-(2-oxo-2-arylethylthio)-1,2,4-triazoles, has been achieved through the one-pot cascade reaction of 3-mercapto[1,2,4]triazoles with trifluoromethyl-β-diktetones in presence of NBS instead of the cyclized thiazolo[3,2-b][1,2,4]triazole. The present protocol offered a unique approach for functionalizing both N-acylation and S-alkylation in a concerted fashion. The structures of the regioisomeric products were thoroughly characterized by heteronuclear 2D NMR experiments. Facile scalability and excellent atom economy through easily available starting reactants are the notable features of the present sustainable protocol. Targeting tumor cell DNA with minor groove-binding small molecules has proven highly effective in the recent past, drawing significant attention for combating tumor-related afflictions. In this context, the synthesized analogs were primarily screened for their ability to bind with the DNA duplex d(CGCGAATTCGCG)2 using molecular modeling tools. Additionally, the most promising compound 14m was deployed as a probe for DNA sensing and interaction mechanisms with calf thymus (ct)DNA through various spectral techniques at a physiologic temperature of 37 °C. It has been found that the compound demonstrated a strong binding affinity (Kb = 1 × 105 M−1) with double-helical DNA, particularly within the minor groove, resulting in the formation of a stable complex through static quenching (Kq = 5.86 ± 0.11 × 1012 M−1 s−1). The fluorescent displacement assay confirmed that the quencher binds to the minor groove of ctDNA, further supported by circular dichroism and viscosity studies.
Introduction
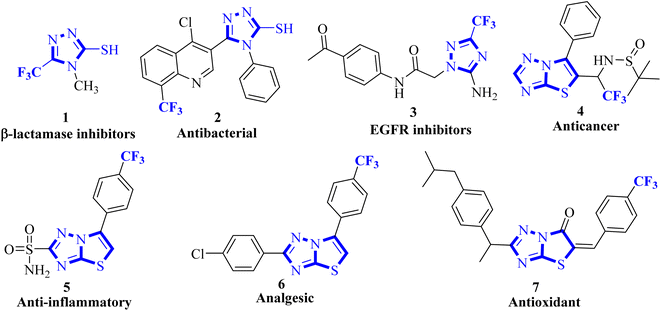
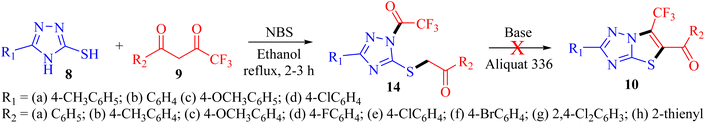
Organofluorine chemistry irrefutably holds immense importance in the realm of scientific exploration, particularly in the pharmaceutical and agrochemical industries, and makes up more than 50% of the best-selling drug molecules approved by the US Food and Drug Administration (FDA).1 Within the category of organofluorine compounds, those containing the trifluoromethyl (CF3) group are particularly noteworthy due to their potent electron-withdrawing properties and considerable hydrophobic surface area. The inclusion of the CF3 group in an organic compound can significantly alter a plethora of physiochemical characteristics, leading to improved effectiveness, specificity, ability to dissolve in fats, ability to be absorbed by the body, resistance to metabolism, electrical charge, ability to pass through membranes, polarity, and durability of C–F bond compared to C–H bond.2,3 Trifluoromethyl group is utilized as a bioisostere for chlorine, methyl, or nitro groups in the development of new medications.4–6 Indeed, numerous successful drugs have capitalized on the benefits of incorporating the trifluoromethyl group into their chemical makeup. It has been well-documented that the linkage of CF3 groups to various heterocyclic molecules renders them highly effective as DNA targeting agents.7–91,2,4-Triazoles belong to an important class of heterocyclic compounds containing three nitrogen atoms in a five-membered aromatic ring. Triazoles are stable for metabolic degradation and show target specificity, as the three nitrogen atoms can act as hydrogen bond acceptors or donors at the active site of the biological receptors and can modulate their activity accordingly.10 Meanwhile, CF3 substituted 3-mercapto[1,2,4]triazole ring system and its fused derivative; thiazolo[3,2-b][1,2,4]triazoles represent an important group of bioactive compounds has captured remarkable attention from researchers owing to their wide array of applications, such as anticancer, anti-inflammatory, analgesic, antifungal, antibacterial, β-lactamase inhibitors, antiviral, and antioxidant agents.11–14 It is well documented that the integration of CF3 with 1,2,4-triazoles plays a substantial role in the drug development program and agrochemical industry.15 Some biologically representative examples of trifluoromethyl-linked mercapto[1,2,4]triazole-based analogs (1-7) are shown in Fig. 1.
Within the realm of pharmacology, DNA is a crucial biological target for the binding of small chemical entities, particularly those utilized in the treatment of cancer.16–18 The research indicated that small organic molecules interact with DNA in two distinct ways; either by intercalating between base pairs or by recognizing the grooves. The major groove offers more opportunities for hydrogen bonding, but its pocket for small molecules is broader and less deep than that of the minor groove. Hence, the minor groove is the preferred site for small ligands, which creates a more hydrophobic environment between the base pairs. In contrast, proteins and peptides tend to favour the major groove for binding.19,20 The minor groove of DNA plays a pivotal role in a variety of molecular interactions and target modifications, rendering it a significant point of interest. Drugs that bind to the minor groove of DNA have garnered considerable attention for their ability to combat tumor-related afflictions.18,21 In recent years, researchers have discovered compounds that bind to DNA and can alter its functions, offering the potential for new forms of chemotherapy. Through extensive biochemical studies, these compounds have been considered as having antibiotic, antiviral, or antitumor properties. Therefore, it is essential to investigate the dynamics of these interactions within the natural environment of living cells and the development of corresponding investigative tools.
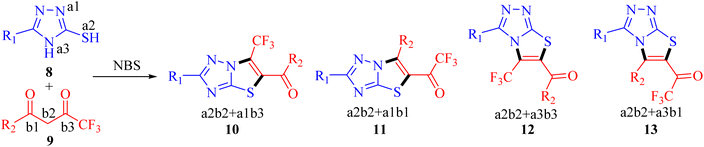
Considering the unique properties of the CF3 group and the significant biological applications of 3-mercapto-1,2,4-triazole derivatives, it was deemed advantageous to incorporate the trifluoromethyl group into the triazole ring. Pertinent to the present research, the development of a general and convenient approach for the assembly of trifluoromethyl decorated 1,2,4-triazole-based derivatives is still highly desirable. In this context, we aimed to synthesize new trifluoromethylated isomers of thiazolo[3,2-b][1,2,4]triazole by reacting 3-mercapto[1,2,4]triazoles 8 with trifluoromethyl-β-diketones 9 in the presence of NBS. However, open-chain analogs were obtained which were investigated for DNA binding properties, and ex vivo studies were performed using various spectroscopic techniques.
Results and discussion
Recently, we have reported visible-light-mediated regioselective synthesis of thiazolo[3,2-b][1,2,4]triazoles by the reaction of 2-bromo-1,3-diketones, with 3-mercapto[1,2,4]triazoles under aqueous conditions in excellent yields.22,23 In the quest for the development of CF3-integrated novel therapeutic agents, we envisaged exploring the reaction of 3-mercapto[1,2,4]triazoles 8 with trifluoromethyl-β-diketones 9 in the presence of NBS to achieve the synthesis of trifluoromethylated thiazolo[3,2-b][1,2,4]triazoles. The reaction, in principle may afford four possible regioisomers; 5-aroyl-6-trifluoromethylthiazolo[3,2-b][1,2,4]triazoles 10, 6-aryl-5-trifluoroacetylthiazolo[3,2-b][1,2,4]triazoles 11, 6-aroyl-5-trifluoroacetylthiazolo[2,3-c][1,2,4]triazole 12 and 5-aryl-6-trifluoroacetylthiazolo[2,3-c][1,2,4]triazole 13, based on their fusion permutation, as illustrated in Scheme 1.To elucidate our work hypothesis, 5-(4-methylphenyl)-3-mercapto[1,2,4]triazole 8a and 4,4,4-trifluoro-1-phenylbutane-1,3-dione 9a in the presence of NBS were selected as the model reaction partners to screen the reaction conditions. The reaction was monitored by TLC (ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) petroleum ether, 30
petroleum ether, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70, v/v as eluent) at regular intervals. The first reaction was carried out under visible-light conditions; similar to those already successfully employed for the regioselective synthesis of non-fluorinated thiazolo[3,2-b]1,2,4 triazoles by our research group,22,23 however, the reaction could not be initiated even after irradiation of 5 h (run 1, Table 1). Thereafter, the reaction was explored in ethanol (EtOH) and a mixture of EtOH and H2O in different ratios under visible light, TLC indicated the partial consumption of reactants and resulted in low yields (runs 2-4, Table 1). Refluxing in EtOH pleasingly afforded the product in 71% yield after 2 h (run 5, Table 1), however, the use of PTSA as an additive did not improve the reaction outcomes (run 6, Table 1). Thereafter, the condensation reaction was screened in different solvents viz. DCM, THF, and DMF under refluxing conditions (run 7-9, Table 1), however, no significant results were observed in terms of yield or reaction time. Shifting the reaction conditions to a solvent-free environment caused a slowdown of the reaction rate and decreased the reaction yield (run 10-11, Table 1). Based on the screening results, the refluxing of substrates in EtOH was selected as the best condition for this transformation. In all the cases, we have notably observed the consistent formation of a single product with the same Rf values.
70, v/v as eluent) at regular intervals. The first reaction was carried out under visible-light conditions; similar to those already successfully employed for the regioselective synthesis of non-fluorinated thiazolo[3,2-b]1,2,4 triazoles by our research group,22,23 however, the reaction could not be initiated even after irradiation of 5 h (run 1, Table 1). Thereafter, the reaction was explored in ethanol (EtOH) and a mixture of EtOH and H2O in different ratios under visible light, TLC indicated the partial consumption of reactants and resulted in low yields (runs 2-4, Table 1). Refluxing in EtOH pleasingly afforded the product in 71% yield after 2 h (run 5, Table 1), however, the use of PTSA as an additive did not improve the reaction outcomes (run 6, Table 1). Thereafter, the condensation reaction was screened in different solvents viz. DCM, THF, and DMF under refluxing conditions (run 7-9, Table 1), however, no significant results were observed in terms of yield or reaction time. Shifting the reaction conditions to a solvent-free environment caused a slowdown of the reaction rate and decreased the reaction yield (run 10-11, Table 1). Based on the screening results, the refluxing of substrates in EtOH was selected as the best condition for this transformation. In all the cases, we have notably observed the consistent formation of a single product with the same Rf values.
| Run | Solvent | Visible light (27 W)/convention heat | Time | Yieldsb (%) |
|---|---|---|---|---|
| a Reaction of 8a (1.0 mmol), 9a (1.0 mmol), NBS (1.0 mmol) in different solvents was processed in the indicated reaction conditions.b Isolated yield; nr: no reaction. | ||||
| 1 | H2O | Visible-light | 5 h | nr |
| 2 | EtOH | Visible-light | 5 h | 25 |
| 3 | EtOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1 H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
Visible-light | 5 h | 20 |
| 4 | EtOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (4 H2O (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
Visible-light | 5 h | 30 |
| 5 | EtOH | Reflux | 2 h | 71 |
| 6 | EtOH/PTSA | Reflux | 2 h | 70 |
| 7 | DCM | rt/reflux | 5 h | nr |
| 8 | THF | Reflux | 5 h | 20 |
| 9 | DMF | Reflux | 5 h | 15 |
| 10 | Solvent-free | 70–80 °C | 5 h | 15 |
| 11 | Solvent-free/PTSA | 70–80 °C | 5 h | 15 |
It is noteworthy to highlight here that inspired by Sherif et al. work on thiazolo[3,2-b][1,2,4]triazoles synthesis in acidified acetic acid (AcOH/H+),24 our study replicated the acidic conditions using 8a and 9a in presence of NBS. However, the outcomes closely mirrored our previous findings,23 revealing the exclusive formation of a single product identified as 6-phenyl-2-(4-methylphenyl)thiazolo[3,2-b][1,2,4]triazole, confirmed through comparison of its melting point and spectral data with literature values.25
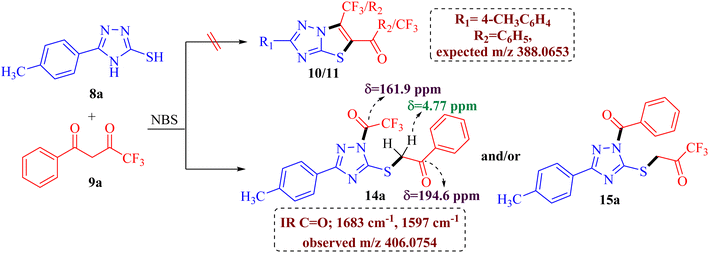
The preliminary examination of the spectral data of the obtained product gestured towards some interesting reaction pathway of 5-(4-methylphenyl)-4H-3-mercapto[1,2,4]triazoles 8a with trifluoromethyl-β-diketone 9a to afford an unexpected product, which does not correlate with the data desired for the expected fluorinated thiazolotriazoles 10, 11, 12 or 13 (Scheme 2), as obtained in our earlier non-fluorinated thiazolotriazole analogs.22,23
The IR spectrum of the obtained product 14a/15a displayed an additional strong C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O absorption band at 1683 cm−1 along with the expected C
O absorption band at 1683 cm−1 along with the expected C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O absorption band at 1597 cm−1 indicating the presence of two carbonyl groups in the product. 13C NMR spectrum also depicted the presence of two carbonyl carbons at δ 194.6 ppm and 161.9 ppm along with the desired aromatic carbons, however, an additional peak for the methylene group at δ 39.2 ppm has been observed in the spectrum. Furthermore, the 1H NMR spectrum also depicted an additional singlet integrating for two protons in the aliphatic region (δ 4.77 ppm) indicating the formation of an unexpected open chain product from the unusual reaction of 8a and 9a. Additionally, the DEPT spectrum depicted the presence of five methines (CH), one methylene (CH2), and one methane (CH3) carbon. To be further certain about the structure; we have recorded the mass spectrum of the obtained product. The mass spectrum depicted an intense peak, m/z value of 406.0754 for [M+1] peak, which is notably higher than the expected value for the fluorinated thiazolotriazoles 10, 11, 12 or 13 value (388.0653). Thus, the preliminary examination of spectra indicated the formation of an open chain analog instead of cyclized thiazolotriazoles, which was identified as 1-trifluoroacetyl-3-(4-tolyl)-5-(2-oxo-2-phenylethylthio)-1,2,4-triazole 14a or 1-benzoyl-3-(4-tolyl)-5-(2-oxo-2-trifluoromethylethylthio)-1,2,4-triazole 15a by the combined analysis of spectral data including IR, 1D NMR and HRMS spectrum.
O absorption band at 1597 cm−1 indicating the presence of two carbonyl groups in the product. 13C NMR spectrum also depicted the presence of two carbonyl carbons at δ 194.6 ppm and 161.9 ppm along with the desired aromatic carbons, however, an additional peak for the methylene group at δ 39.2 ppm has been observed in the spectrum. Furthermore, the 1H NMR spectrum also depicted an additional singlet integrating for two protons in the aliphatic region (δ 4.77 ppm) indicating the formation of an unexpected open chain product from the unusual reaction of 8a and 9a. Additionally, the DEPT spectrum depicted the presence of five methines (CH), one methylene (CH2), and one methane (CH3) carbon. To be further certain about the structure; we have recorded the mass spectrum of the obtained product. The mass spectrum depicted an intense peak, m/z value of 406.0754 for [M+1] peak, which is notably higher than the expected value for the fluorinated thiazolotriazoles 10, 11, 12 or 13 value (388.0653). Thus, the preliminary examination of spectra indicated the formation of an open chain analog instead of cyclized thiazolotriazoles, which was identified as 1-trifluoroacetyl-3-(4-tolyl)-5-(2-oxo-2-phenylethylthio)-1,2,4-triazole 14a or 1-benzoyl-3-(4-tolyl)-5-(2-oxo-2-trifluoromethylethylthio)-1,2,4-triazole 15a by the combined analysis of spectral data including IR, 1D NMR and HRMS spectrum.
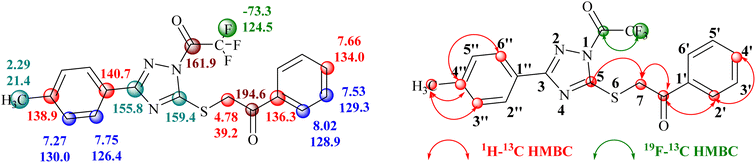
Conclusive evidence for the formation of 1-trifluoroacetyl-3-aryl-5-(2-oxo-2-arylethylthio)-1,2,4-triazole regioisomer 14 was obtained by collective results from the rigorous study of heteronuclear 2D NMR experiments of the compound 14a. The chemical shift values of protons and their corresponding carbons were assigned using (1H–13C) HSQC spectrum. Further, the (1H–13C) HMBC results showed cross-peaks of carbonyl carbon (δ 194.6 ppm) with 2′/6′-H proton (δ 8.02 ppm) of the phenyl ring indicating the presence of carbonyl carbon with aryl ring and the cross peak of carbonyl carbon (δ 194.6 ppm) with CH2 (δ 4.77 ppm) confirmed the presence of CH2–CO-Ar fragment, ruled out the possibility for the formation of regioisomer 15a. Similarly, the cross peak of C-5 (δ 159.4 ppm) of the triazole nucleus with CH2 (δ 4.77 ppm) indicated the presence of 2-oxo-2-arylethylthio group at position 5 of the triazole ring. Other correlations are seen from protons of CH3 (δ 2.29 ppm) with C-4′′ (δ 138.9 ppm), C-3′′/C-5′′ (δ 130.0 ppm). The (19F–13C) HMBC results displayed the correlation of fluorine (δ −73.4 ppm) with carbonyl carbon (δ 161.9 ppm). All expected correlations were validated in the COSY and NOESY spectrum, sufficient to support the substituents pattern around the triazole nucleus and all our data are consistent with structure 14a as the reaction product. The 2D NMR correlation results obtained for compounds 14a and their 1H, 13C, and 19F chemical shift values are presented in Fig. 2.
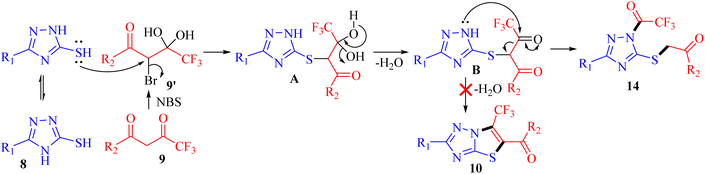
Following an in-depth structural analysis of the unexpected product, we made efforts to induce cyclization of the cleaved product 14 employing Aliquat 336 as a phase transfer catalyst and exploring diverse basic conditions (KOH, K2CO3, C2H5ONa, DABCO, and trimethylamine) to obtain the desired cyclized product 10. Unfortunately, none of these reaction experiments yielded the anticipated outcome (Scheme 3).
To explore the biological potential of the newly synthesized compound, we conducted a comprehensive investigation into the substrate scope and limitations of this process. Variously substituted 3-mercapto[1,2,4]triazole and fluorinated 1,3-diketone compounds were employed, and the results are depicted in Fig. 3. The outcomes indicated that, in all cases, the reaction proceeded smoothly, yielding 14a–o as the final product. Although different substituent groups had a slight effect on the yields, the differences were minimal, and it was challenging to ascertain the specific impact of any individual substituent.
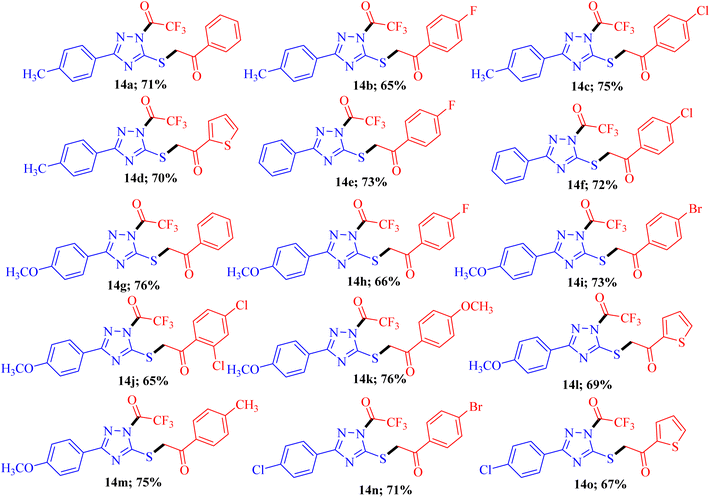 | ||
| Fig. 3 Substrate scopea,b. aReaction conditions: 3-mercapto[1,2,4]triazoles 8a-d (1.0 mmol), diketones 9a-h (1.0 mmol), and NBS (1.0 mmol) reflux in ethanol (10 ml) for 2-3 h; bisolated yields. | ||
Mechanism
The possible mechanism for the synthesis of 1-trifluoroacetyl-3-aryl-5-(2-oxo-2-arylethylthio)-1,2,4-triazoles 14a–o is outlined in Scheme 4. Based on the literature, the bromination of trifluoromethyl-β-diketones 9 with NBS resulted in 2-bromo-4,4,4-trifluoro-3,3-dihydroxybutan-1-ones 9′.26,27 The reaction is initiated by the nucleophilic displacement of bromine of 9′ by sulfur of 3-mercapto[1,2,4]triazole 8 to give an open chain intermediate A. Subsequently, dehydration from diol group to regenerate carbonyl provided the dicarbonyl intermediate B. Nucleophilic attack of amine nitrogen at more electrophilic carbonyl carbon adjacent to the CF3 group followed by C–C bond cleavage provided 1-trifluoroacetyl-3-aryl-5-(2-oxo-2-arylethylthio)-1,2,4-triazoles 14 as the final product instead of cyclization to yield 6-(trifluoromethyl)thiazolo[3,2-b][1,2,4]triazoles 10.DNA binding studies
| Comp. No. | Binding affinity (kcal mol−1) |
|---|---|
| 14a | −8.6 |
| 14b | −8.8 |
| 14c | −8.8 |
| 14d | −8.2 |
| 14e | −8.1 |
| 14f | −8.2 |
| 14g | −7.0 |
| 14h | −8.4 |
| 14i | −8.6 |
| 14j | −8.6 |
| 14k | −8.8 |
| 14l | −8.1 |
| 14m | −9.0 |
| 14n | −8.4 |
| 14o | −8.1 |
| 10a | −6.3 |
| 10b | −6.9 |
| 10c | −7.0 |
| 10e | −6.0 |
| 10g | −6.3 |
| 10h | −6.0 |
| 10j | −6.1 |
| 10k | −6.6 |
| 10l | −6.8 |
| 10o | −6.1 |
| 10n | −7.0 |
| 10p | −6.7 |
| 10m | −7.2 |
| 10q | −6.2 |
| 10r | −6.5 |
Through analysis of docking, it has been discovered that the triazole derivatives bind in the minor groove section of base pairs particularly in the guanine-rich region through various non-covalent interactions, such as hydrophobic interactions (π-alkyl, π-sulfur, etc), conventional hydrogen bonding, halogen interactions, and van der Waals forces. By examining the 2D plots of the best dock complex (DNA-14m), it was determined that both the carbonyl groups of the open-chain functionalized triazole derivative played a crucial role in conventional hydrogen bonding, with additional supportive interactions provided by the sulfur atom, and trifluoromethyl group (Fig. 4).
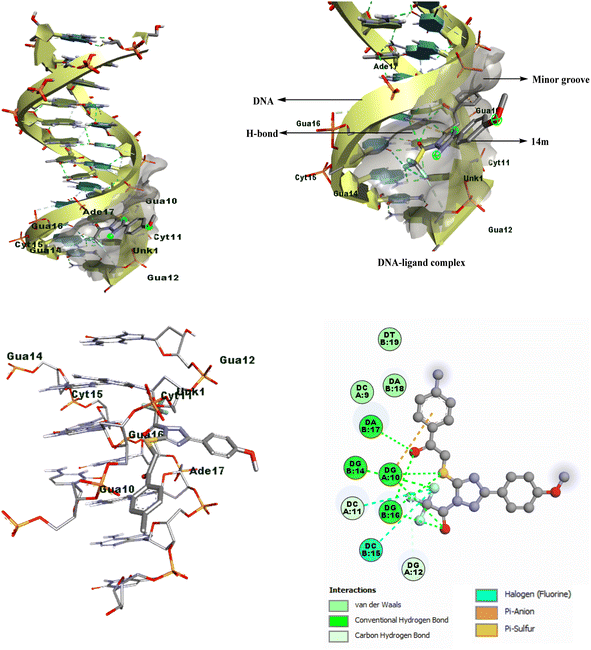 | ||
| Fig. 4 2D/3D binding poses of DNA-14m complex (PDB ID: 1BNA). | ||
It is significant to mention that the corresponding cyclized derivatives 10 had a low docking score (−6.0 to −7.2 kcal mol−1), which may be attributed to the presence of only one carbonyl group, resulting in a decreased likelihood of H-bond formation. Additionally, the cyclized structure limits the flexibility of the molecule, thereby restricting its binding strength.
The docking analysis revealed that among all the compounds, 1-trifluoroacetyl-3-(4-methoxyphenyl)-5-((2-oxo-2-(4-tolyl)ethyl)thio)-1,2,4-triazole 14m had the best docking results. Therefore, it was chosen for further investigation through various spectral techniques to understand its interaction with calf thymus DNA and its mechanisms.
After taking into consideration the aforementioned findings, we proceeded to delve deeper into how the DNA-triazole (14m) complex interacts. We aimed to gain a better understanding of the mode of interactions and binding strength. To this end, we utilized various methods such as fluorescence quenching, CD spectral, and viscosity analysis.
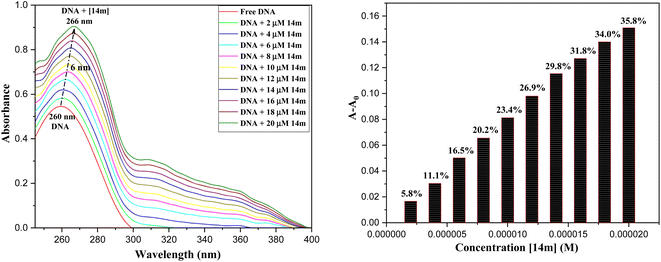
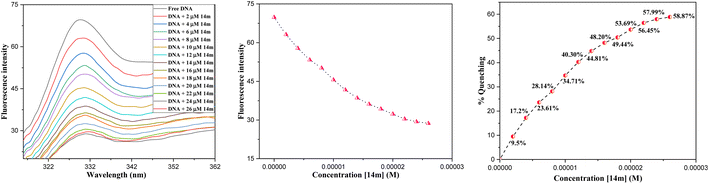
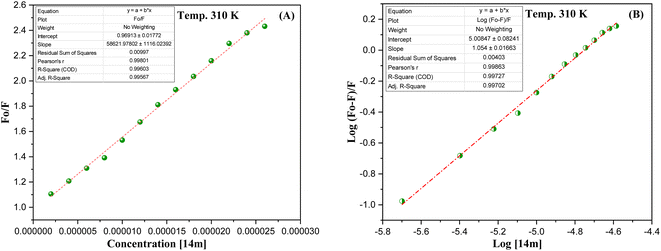
The reduction in fluorescence intensity in the DNA spectrum is usually caused by quenching mechanisms such as static quenching, dynamic quenching, and mixed quenching. Static quenching occurs due to complex forms between biomolecule and quencher in the ground state, while dynamic quenching occurs due to collisions in the excited state. Through the utilization of Stern–Volmer constants (Ksv) and quenching constants (Kq), the efficacy of quenching in the spectrum can be comprehended. To this end, graphs were generated by plotting the ratio of fluorescence blank DNA and DNA+14m (F0/F) against increments of 14m, thus revealing the mechanism (Fig. 7A). The quenching constant was determined by assuming the average lifetime to be 10−8 seconds. The quenching constant (Kq = 5.86 ± 0.11 × 1012 M−1 s−1) was found to be higher than the scattering constant (1010 L mol−1 s−1), indicating that the quencher 14m forms a complex with DNA through static quenching.33
The value of the slope of the graphs determines the Stern–Volmer constant (Ksv) (eqn (1)).
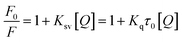 | (1) |
F0 represents the fluorescence intensity of pure DNA, however, F refers to the fluorescence intensity of DNA-14m complex (0–26 μM). [Q] signifies the molar concentration of 14m, τo denotes the average lifetime.
Furthermore, to understand how strongly DNA and 14m are bound together, we have calculated the intrinsic binding constant (Kb) by creating Scatchard graphs (Fig. 7B). The binding constant Kb and number of binding sites n were calculated employing the Scatchard equation (eqn (2)) (Table 3).
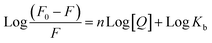 | (2) |
| Ksv × 104 (M−1) | Kq × 1012 (M−1 s−1) | Log Kb | Kb × 105 (M−1) | n | ΔG° (kJ mol−1) |
|---|---|---|---|---|---|
| (5.86 ± 0.11) | (5.86 ± 0.11) | 5.00 ± 0.08 | 1.0 | 1.054 ± 0.01 | −23.738 |
However, a change in Gibbs free energy (ΔG°) helps in the determination of the spontaneity of the binding procedures.
ΔG° = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb Kb
| (3) |
ΔG°, Kb, and R refer to the Gibbs free energy, binding constant at temperature T and gas constant (8.314 J mol−1 K−1) respectively.
The intrinsic binding constant value has been determined to be within the range of 105, signifying the robust and enduring binding between the triazole molecule and DNA through 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry. The results reveal the remarkable and effective bonding properties of the triazole compound with DNA, underscoring its potential as a valuable asset in diverse biological contexts. Additionally, the negative value for Gibbs free energy change ΔG° elegantly supports the feasibility and favorability for the formation of the DNA-14m complex.
1 binding stoichiometry. The results reveal the remarkable and effective bonding properties of the triazole compound with DNA, underscoring its potential as a valuable asset in diverse biological contexts. Additionally, the negative value for Gibbs free energy change ΔG° elegantly supports the feasibility and favorability for the formation of the DNA-14m complex.
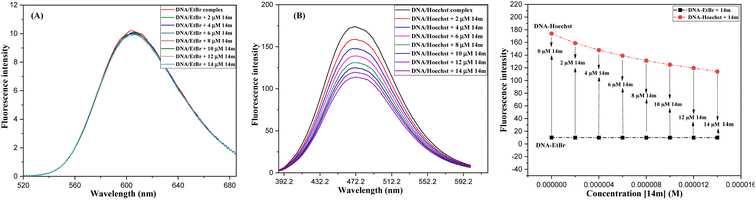
The displacement assay is an important tool for identifying how DNA and a ligand molecule interact. Two well-known dyes, Ethidium bromide (intercalator) and Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 (groove binder), are commonly used to determine whether a ligand molecule intercalates or binds to the grooves of DNA. The study tested the interaction of a triazole derivative (14m) with DNA using two different dyes (EtBr and Hoechst). The results showed that compound 14m did not displace the intercalating dye from the DNA-EtBr complex, indicating it does not interact with DNA through intercalation. However, there was a significant reduction in fluorescence intensity of the DNA-Hoechst complex with increasing concentration of compound 14m at 37 °C, suggesting it interacts with DNA through groove-binding modes (Fig. 8).
258 (groove binder), are commonly used to determine whether a ligand molecule intercalates or binds to the grooves of DNA. The study tested the interaction of a triazole derivative (14m) with DNA using two different dyes (EtBr and Hoechst). The results showed that compound 14m did not displace the intercalating dye from the DNA-EtBr complex, indicating it does not interact with DNA through intercalation. However, there was a significant reduction in fluorescence intensity of the DNA-Hoechst complex with increasing concentration of compound 14m at 37 °C, suggesting it interacts with DNA through groove-binding modes (Fig. 8).
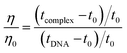 | (4) |
Conclusion
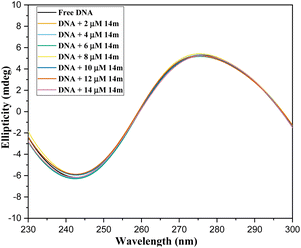
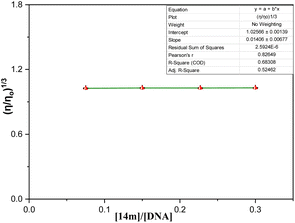
On the whole, a strategically novel one-step protocol for regioselective 1,3 N,S-difunctionalization of triazoles, which allows an expedient construction of CF3-containing architectures through an unprecedented reaction between 3-mercapto[1,2,4]triazoles and trifluoromethyl-β-diketones was disclosed. This methodology for the synthesis of various fluorinated triazoles has great potential for applications in related chemistry. In silico molecular docking analysis evidenced that trifluoromethylated triazole derivatives exhibited superior binding efficacy within the minor groove of the DNA double helix when compared to their cyclic counterparts. Spectroscopic results represented that binding of triazole derivatives to DNA resulted in significant changes in the structure and conformation of DNA in a concentration-dependent manner and acted as groove binder (Kb = 1 × 105 M−1) via a static mode of quenching (Kq = 5.86 ± 0.11 × 1012 M−1 s−1). Moreover, circular dichroism and viscosity measurements supported the DNA groove binding nature of the triazole derivatives.Experimental
Materials and methods
An electrical digital Melting Point Apparatus (MEPA) was used to examine melting points in open capillaries and were not corrected. Analytical TLC was performed using Merck Kieselgel 60 F254 silica gel plates and visualized under UV light (254 nm). IR spectra were recorded on Buck Scientific IR M-500 spectrophotometer in KBr pellets (υmax in cm−1), 1H (400 Hz), and 13C NMR (100 Hz) spectra for the analytical purposes were recorded on a Bruker instrument, using CDCl3 and DMSO as a solvent and the chemical shifts are expressed in parts per million (ppm) and coupling constant J in Hz with TMS as internal standard. High-resolution mass spectra (HRMS) were measured in ESI+ mode at MRC, MNIT, Jaipur. 3-Mercapto-1,2,4 triazoles and 1,3-diketones were prepared as per the literature procedure,23,38–41 however, commercially available NBS (Avra Chemicals, India) were used without any purification.General procedure for the synthesis of 1-trifluoroacetyl-3-aryl-5-(2-oxo-2-arylethylthio)-1,2,4-triazoles 14a-o
Trifluoromethyl-β-diketones 9 (1.0 mmol) and NBS (0.178 g, 1.0 mmol) were homogenized thoroughly in a dry pestle mortar until a thick paste was formed and were subsequently added with an ethanolic solution of 5-aryl-3-mercapto[1,2,4]triazole 8 (1.0 mmol) in the round bottom flask. The reaction mixture was refluxed for 2–2.5 hours. The reaction progress was monitored with TLC using ethyl acetate-petroleum ether (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70, v/v). On completion of the reaction, the solvent was evaporated at reduced pressure on the rotatory evaporator and the residue thus obtained was neutralized with a saturated solution of sodium bicarbonate and extracted with ethyl acetate. The obtained solid was recrystallized with ethanol and dried to afford pure 14 with 65–75% yields. The products were characterized by IR, 1H, 13C NMR, and HRMS spectrometry.
70, v/v). On completion of the reaction, the solvent was evaporated at reduced pressure on the rotatory evaporator and the residue thus obtained was neutralized with a saturated solution of sodium bicarbonate and extracted with ethyl acetate. The obtained solid was recrystallized with ethanol and dried to afford pure 14 with 65–75% yields. The products were characterized by IR, 1H, 13C NMR, and HRMS spectrometry.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1597 (C
O), 1597 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.02–8.00 (m, 2H, 2′,6′-H), 7.75–7.73 (d, 2H, J = 8 Hz, 2′′,6′′-H), 7.65–7.62 (t, 1H, J = 8 Hz, 4′-H), 7.54–7.50 (t, 2H, J = 8 Hz, 3′,5′-H), 7.27–7.22 (m, 2H, 3′′,5′′-H), 4.77 (s, 2H, 7-CH2), 2.29 (s, 3H, 4′′-CH3); 13C NMR (100 MHz, CDCl3) δ (ppm): 194.6, 161.9, 159.4, 155.8, 140.7, 138.9, 136.3, 134.0, 130.1, 129.3, 128.9, 126.4, 124.5, 39.2, 21.4; 19F NMR (376 MHz, CDCl3) δ (ppm): −73.34; HRMS (ESI): 406.0754 [M + H]+.
O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.02–8.00 (m, 2H, 2′,6′-H), 7.75–7.73 (d, 2H, J = 8 Hz, 2′′,6′′-H), 7.65–7.62 (t, 1H, J = 8 Hz, 4′-H), 7.54–7.50 (t, 2H, J = 8 Hz, 3′,5′-H), 7.27–7.22 (m, 2H, 3′′,5′′-H), 4.77 (s, 2H, 7-CH2), 2.29 (s, 3H, 4′′-CH3); 13C NMR (100 MHz, CDCl3) δ (ppm): 194.6, 161.9, 159.4, 155.8, 140.7, 138.9, 136.3, 134.0, 130.1, 129.3, 128.9, 126.4, 124.5, 39.2, 21.4; 19F NMR (376 MHz, CDCl3) δ (ppm): −73.34; HRMS (ESI): 406.0754 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1595 (C
O), 1595 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.12–8.03 (m, 2H, 2′,6′-H), 7.74–7.72 (d, 2H, J = 8 Hz, 2′′,6′′-H), 7.38–7.33 (m, 2H, 3′,5′-H), 7.28–7.26 (d, 2H, J = 8 Hz, 3′′,5′′-H), 4.76 (s, 2H, 7-CH2), 2.30 (s, 3H, 4′′-CH3); 13C NMR (100 MHz, CDCl3) δ (ppm): 193.3, 164.4, 159.3, 155.8, 140.7, 133.0, 132.0–131.9 (d), 130.1, 129.7, 126.4, 126.1, 124.5, 116.4–116.2 (d), 39.1, 21.9; 19F NMR (376 MHz, CDCl3) δ (ppm): −73.32, −105.31; HRMS (ESI): 424.0664 [M + H]+.
O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.12–8.03 (m, 2H, 2′,6′-H), 7.74–7.72 (d, 2H, J = 8 Hz, 2′′,6′′-H), 7.38–7.33 (m, 2H, 3′,5′-H), 7.28–7.26 (d, 2H, J = 8 Hz, 3′′,5′′-H), 4.76 (s, 2H, 7-CH2), 2.30 (s, 3H, 4′′-CH3); 13C NMR (100 MHz, CDCl3) δ (ppm): 193.3, 164.4, 159.3, 155.8, 140.7, 133.0, 132.0–131.9 (d), 130.1, 129.7, 126.4, 126.1, 124.5, 116.4–116.2 (d), 39.1, 21.9; 19F NMR (376 MHz, CDCl3) δ (ppm): −73.32, −105.31; HRMS (ESI): 424.0664 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1600 (C
O), 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.05–8.01 (m, 2H, 2′,6′-H), 7.73–7.71 (m, 2H, 2′′,6′′-H), 7.63–7.57 (m, 2H, 3′,5′-H), 7.28–7.20 (m, 2H, 3′′,5′′-H), 4.75 (s, 2H, 7-CH2), 2.30 (s, 3H, 4′′-CH3); 13C NMR (100 MHz, CDCl3) δ (ppm): 193.8, 161.0, 159.3, 155.8, 140.7, 139.2, 135.0, 130.8, 130.1, 129.43, 129.42, 126.4, 124.4, 39.1, 21.5; 19F NMR (376 MHz, CDCl3) δ (ppm): −73.38; HRMS (ESI): 440.0368 [M + H]+; 442.0365 [M + H+2]+ (3
O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.05–8.01 (m, 2H, 2′,6′-H), 7.73–7.71 (m, 2H, 2′′,6′′-H), 7.63–7.57 (m, 2H, 3′,5′-H), 7.28–7.20 (m, 2H, 3′′,5′′-H), 4.75 (s, 2H, 7-CH2), 2.30 (s, 3H, 4′′-CH3); 13C NMR (100 MHz, CDCl3) δ (ppm): 193.8, 161.0, 159.3, 155.8, 140.7, 139.2, 135.0, 130.8, 130.1, 129.43, 129.42, 126.4, 124.4, 39.1, 21.5; 19F NMR (376 MHz, CDCl3) δ (ppm): −73.38; HRMS (ESI): 440.0368 [M + H]+; 442.0365 [M + H+2]+ (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1600 (C
O), 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.12–8.10 (dd, 1H, J = 4 Hz, J = 1 Hz, 3′-H), 8.03–8.01 (dd, 1H, J = 4 Hz, J = 1 Hz, 5′-H), 7.74–7.72 (m, 2H, 2′′,6′′-H), 7.27–7.22 (m, 3H, 4′,3′′,5′′-H), 4.71 (s, 2H, 7-CH2), 2.29 (s, 3H, 4′′-CH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 187.1, 161.6, 158.8, 155.3, 142.5, 140.2, 135.4, 134.2, 129.6, 128.9, 125.9, 123.9, 38.2, 21.0; 19F NMR (376 MHz, CDCl3) δ (ppm): −80.21; HRMS (ESI): 412.0320 [M + H]+.
O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.12–8.10 (dd, 1H, J = 4 Hz, J = 1 Hz, 3′-H), 8.03–8.01 (dd, 1H, J = 4 Hz, J = 1 Hz, 5′-H), 7.74–7.72 (m, 2H, 2′′,6′′-H), 7.27–7.22 (m, 3H, 4′,3′′,5′′-H), 4.71 (s, 2H, 7-CH2), 2.29 (s, 3H, 4′′-CH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 187.1, 161.6, 158.8, 155.3, 142.5, 140.2, 135.4, 134.2, 129.6, 128.9, 125.9, 123.9, 38.2, 21.0; 19F NMR (376 MHz, CDCl3) δ (ppm): −80.21; HRMS (ESI): 412.0320 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1602 (C
O), 1602 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.14–8.06 (m, 2H, 2′,6′-H), 7.85–7.83 (m, 2H, 2′′,6′′-H), 7.49–7.45 (m, 2H, 3′,5′-H), 7.40–7.33 (m, 3H, 3′′,4′′,5′′-H), 4.77 (s, 2H, 7-CH2); 13C NMR (100 MHz, CDCl3) δ (ppm): 193.3, 161.9, 159.5, 155.7, 133.0, 132.0–131.9 (d), 131.3, 130.9, 129.6, 129.1, 127.2, 126.5, 126.2, 124.4, 116.4–116.2 (d), 39.2; 19F NMR (376 MHz, CDCl3) δ (ppm): −72.88, −104.98; HRMS (ESI): 410.0505 [M + H]+.
O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.14–8.06 (m, 2H, 2′,6′-H), 7.85–7.83 (m, 2H, 2′′,6′′-H), 7.49–7.45 (m, 2H, 3′,5′-H), 7.40–7.33 (m, 3H, 3′′,4′′,5′′-H), 4.77 (s, 2H, 7-CH2); 13C NMR (100 MHz, CDCl3) δ (ppm): 193.3, 161.9, 159.5, 155.7, 133.0, 132.0–131.9 (d), 131.3, 130.9, 129.6, 129.1, 127.2, 126.5, 126.2, 124.4, 116.4–116.2 (d), 39.2; 19F NMR (376 MHz, CDCl3) δ (ppm): −72.88, −104.98; HRMS (ESI): 410.0505 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1602 (C
O), 1602 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.05–8.01 (m, 2H, 2′,6′-H), 7.85–7.82 (m, 2H, 2′′,6′′-H), 7.62–7.58 (m, 2H, 3′,5′-H), 7.49–7.42 (m, 2H, 3′′,5′′-H), 7.39–7.36 (m, 1H, 4′′-H), 4.77 (s, 2H, 7-CH2); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 193.7, 161.8, 159.4, 155.6, 151.8, 138.8, 134.9, 130.7, 129.5, 129.3, 129.1, 127.0, 126.4, 126.1, 39.1; 19F NMR (376 MHz, CDCl3) δ (ppm): −74.21; HRMS (ESI): 426.0200 [M + H]+; 428.0206 [M + H+2]+ (3
O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.05–8.01 (m, 2H, 2′,6′-H), 7.85–7.82 (m, 2H, 2′′,6′′-H), 7.62–7.58 (m, 2H, 3′,5′-H), 7.49–7.42 (m, 2H, 3′′,5′′-H), 7.39–7.36 (m, 1H, 4′′-H), 4.77 (s, 2H, 7-CH2); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 193.7, 161.8, 159.4, 155.6, 151.8, 138.8, 134.9, 130.7, 129.5, 129.3, 129.1, 127.0, 126.4, 126.1, 39.1; 19F NMR (376 MHz, CDCl3) δ (ppm): −74.21; HRMS (ESI): 426.0200 [M + H]+; 428.0206 [M + H+2]+ (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1604 (C
O), 1604 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.03–8.01 (m, 2H, 2′′,6′′-H), 7.86–7.84 (d, 2H, 2′,6′-H, 3J = 8.8), 7.62–7.60 (m, 1H, 4′-H), 7.50–7.46 (m, 2H, 3′′,5′′-H), 6.93–6.91 (d, 2H, 3′,5′-H, 3J = 8.8), 4.66 (s, 2H, 7-CH2), 3.83 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 194.6, 161.3, 159.2, 155.6, 136.2, 134.0, 129.3, 128.9, 128.1, 127.6, 119.7, 115.0, 114.5, 55.8, 38.9; 19F NMR (376 MHz, CDCl3) δ (ppm): −72.54; HRMS (ESI): 422.0712 [M + H]+.
O); 1H NMR (400 MHz, CDCl3) δ (ppm): 8.03–8.01 (m, 2H, 2′′,6′′-H), 7.86–7.84 (d, 2H, 2′,6′-H, 3J = 8.8), 7.62–7.60 (m, 1H, 4′-H), 7.50–7.46 (m, 2H, 3′′,5′′-H), 6.93–6.91 (d, 2H, 3′,5′-H, 3J = 8.8), 4.66 (s, 2H, 7-CH2), 3.83 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 194.6, 161.3, 159.2, 155.6, 136.2, 134.0, 129.3, 128.9, 128.1, 127.6, 119.7, 115.0, 114.5, 55.8, 38.9; 19F NMR (376 MHz, CDCl3) δ (ppm): −72.54; HRMS (ESI): 422.0712 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1602 (C
O), 1602 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 7.99–7.97 (m, 2H, 2′,6′-H), 7.82–7.80 (m, 2H, 2′′,6′′-H), 7.79–7.77 (m, 2H, 3′,5′-H), 7.06–6.98 (m, 2H, 3′′,5′′-H), 4.78 (s, 2H, 7-CH2), 3.81 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 193.5, 160.8, 158.6, 155.1, 134.8, 131.8, 130.4, 127.6, 127.1, 119.2, 114.4, 114.0, 55.3, 38.6; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −78.13, −105.43; HRMS (ESI): 440.0618 [M + H]+.
O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 7.99–7.97 (m, 2H, 2′,6′-H), 7.82–7.80 (m, 2H, 2′′,6′′-H), 7.79–7.77 (m, 2H, 3′,5′-H), 7.06–6.98 (m, 2H, 3′′,5′′-H), 4.78 (s, 2H, 7-CH2), 3.81 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 193.5, 160.8, 158.6, 155.1, 134.8, 131.8, 130.4, 127.6, 127.1, 119.2, 114.4, 114.0, 55.3, 38.6; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −78.13, −105.43; HRMS (ESI): 440.0618 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1605 (C
O), 1605 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 7.99–7.97 (m, 2H, 2′′,6′′-H), 7.86–7.79 (m, 4H, 2′,3′,5′,6′-H), 7.07–7.05 (m, 2H, 3′′,5′′-H), 4.78 (s, 2H, 7-CH2), 3.81 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 193.5, 160.8, 158.5, 155.1, 134.8, 131.8, 130.3, 127.6, 127.2, 119.2, 114.4, 113.9, 55.3, 38.5; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −73.32; HRMS (ESI): 500.9815 [M + H]+.
O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 7.99–7.97 (m, 2H, 2′′,6′′-H), 7.86–7.79 (m, 4H, 2′,3′,5′,6′-H), 7.07–7.05 (m, 2H, 3′′,5′′-H), 4.78 (s, 2H, 7-CH2), 3.81 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 193.5, 160.8, 158.5, 155.1, 134.8, 131.8, 130.3, 127.6, 127.2, 119.2, 114.4, 113.9, 55.3, 38.5; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −73.32; HRMS (ESI): 500.9815 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1600 (C
O), 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 8.18–8.11 (m, 1H, 6′-H), 8.07–8.06 (m, 1H, 3′-H), 7.84–7.77 (m, 2H, 2′′,6′′-H), 7.30–7.28 (m, 1H, 5′), 7.06–7.04 (m, 2H, 3′′,5′′-H), 4.72 (s, 2H, 7-CH2), 3.80 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 187.3, 161.0, 158.8, 157.2, 155.4, 142.5, 135.5, 6134.6, 134.3, 129.0, 127.8, 127.3, 119.2, 114.6, 112.0, 55.4, 38.3; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −74.41; HRMS (ESI): 489.9930 [M + H]+.
O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 8.18–8.11 (m, 1H, 6′-H), 8.07–8.06 (m, 1H, 3′-H), 7.84–7.77 (m, 2H, 2′′,6′′-H), 7.30–7.28 (m, 1H, 5′), 7.06–7.04 (m, 2H, 3′′,5′′-H), 4.72 (s, 2H, 7-CH2), 3.80 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 187.3, 161.0, 158.8, 157.2, 155.4, 142.5, 135.5, 6134.6, 134.3, 129.0, 127.8, 127.3, 119.2, 114.6, 112.0, 55.4, 38.3; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −74.41; HRMS (ESI): 489.9930 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1596 (C
O), 1596 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, CDCl3) δ (ppm): 7.99–7.97 (m, 2H, 2′′,6′′-H), 7.88–7.86 (m, 2H, 2′,6′-H), 6.93–6.89 (m, 4H, 3′′,5′′,3′,5′-H), 4.60 (s, 2H, 7-CH2), 3.86 (s, 3H, OCH3), 3.81 (s, 3H, OCH3); 13C NMR (100 MHz, CDCl3) δ (ppm): 193.0, 164.2, 161.0, 131.1, 128.1, 127.9, 120.8, 114.1, 113.9, 55.5, 55.3, 39.3; 19F NMR (376 MHz, CDCl3) δ (ppm): −73.22; HRMS (ESI): 452.0816 [M + H]+.
O); 1H NMR (400 MHz, CDCl3) δ (ppm): 7.99–7.97 (m, 2H, 2′′,6′′-H), 7.88–7.86 (m, 2H, 2′,6′-H), 6.93–6.89 (m, 4H, 3′′,5′′,3′,5′-H), 4.60 (s, 2H, 7-CH2), 3.86 (s, 3H, OCH3), 3.81 (s, 3H, OCH3); 13C NMR (100 MHz, CDCl3) δ (ppm): 193.0, 164.2, 161.0, 131.1, 128.1, 127.9, 120.8, 114.1, 113.9, 55.5, 55.3, 39.3; 19F NMR (376 MHz, CDCl3) δ (ppm): −73.22; HRMS (ESI): 452.0816 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1602 (C
O), 1602 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 8.16–8.14 (m, 1H, 3′-H), 7.92–7.90 (m, 1H, 5′-H), 7.83–7.81 (m, 2H, 2′′,6′′-H), 7.45–7.37 (m, 1H, 4′), 7.07–7.05 (m, 2H, 3′′,5′′-H), 4.79 (s, 2H, 7-CH2), 3.82 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 192.7, 166.4, 163.9, 160.7, 158.6, 155.1, 132.4, 131.5, 131.4, 127.6, 119.2, 115.9, 115.7, 114.4, 55.3, 38.6; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −72.11; HRMS (ESI): 428.0277 [M + H]+.
O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 8.16–8.14 (m, 1H, 3′-H), 7.92–7.90 (m, 1H, 5′-H), 7.83–7.81 (m, 2H, 2′′,6′′-H), 7.45–7.37 (m, 1H, 4′), 7.07–7.05 (m, 2H, 3′′,5′′-H), 4.79 (s, 2H, 7-CH2), 3.82 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 192.7, 166.4, 163.9, 160.7, 158.6, 155.1, 132.4, 131.5, 131.4, 127.6, 119.2, 115.9, 115.7, 114.4, 55.3, 38.6; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −72.11; HRMS (ESI): 428.0277 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1598 (C
O), 1598 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, CDCl3) δ (ppm): 7.94–7.92 (m, 2H, 2′′,6′′-H), 7.89–7.87 (m, 2H, 3′,5′-H), 7.30–7.28 (m, 2H, 3′′,5′′-H), 7.54–7.45 (m, 2H, 2′,6′-H), 4.61 (s, 2H, 7-CH2), 3.84 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 194.1, 161.3, 159.3, 155.6, 144.5, 133.7, 129.8, 129.0, 128.1, 127.6, 119.7, 115.0, 114.5, 55.8, 39.1, 21.7; 19F NMR (376 MHz, CDCl3) δ (ppm): −79.18; HRMS (ESI): 436.0866 [M + H]+.
O); 1H NMR (400 MHz, CDCl3) δ (ppm): 7.94–7.92 (m, 2H, 2′′,6′′-H), 7.89–7.87 (m, 2H, 3′,5′-H), 7.30–7.28 (m, 2H, 3′′,5′′-H), 7.54–7.45 (m, 2H, 2′,6′-H), 4.61 (s, 2H, 7-CH2), 3.84 (s, 3H, OCH3); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 194.1, 161.3, 159.3, 155.6, 144.5, 133.7, 129.8, 129.0, 128.1, 127.6, 119.7, 115.0, 114.5, 55.8, 39.1, 21.7; 19F NMR (376 MHz, CDCl3) δ (ppm): −79.18; HRMS (ESI): 436.0866 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1606 (C
O), 1606 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 8.08–8.06 (m, 2H, 2′′,6′′-H), 7.90–7.88 (m, 2H, 2′,6′-H), 7.84–7.74 (m, 4H, 3′,5′,3′′,5′′-H), 4.81 (s, 2H, 7-CH2); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 188.0, 165.9, 146.0, 135.4, 132.9, 132.7, 131.9, 131.5, 130.0, 129.6, 129.4, 128.5, 128.4, 128.0, 39.2; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −72.88; HRMS (ESI): 503.9320 [M + H]+.
O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 8.08–8.06 (m, 2H, 2′′,6′′-H), 7.90–7.88 (m, 2H, 2′,6′-H), 7.84–7.74 (m, 4H, 3′,5′,3′′,5′′-H), 4.81 (s, 2H, 7-CH2); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 188.0, 165.9, 146.0, 135.4, 132.9, 132.7, 131.9, 131.5, 130.0, 129.6, 129.4, 128.5, 128.4, 128.0, 39.2; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −72.88; HRMS (ESI): 503.9320 [M + H]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1604 (C
O), 1604 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 8.12–8.10 (m, 1H, 6′-H), 8.04–8.02 (m, 1H, 3′-H), 7.86–7.83 (m, 2H, 2′′,6′′-H), 7.52–7.50 (m, 2H, 3′′,5′′-H), 7.27–7.24 (m, 2H, 5′-H), 4.74 (s, 2H, 7-CH2); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 187.0, 159.3, 154.2, 142.4, 135.8, 135.4, 134.5, 134.2, 129.2, 128.9, 128.8, 127.7, 127.3, 125.5, 38.7; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −73.34; HRMS (ESI): 431.9779 [M + H]+.
O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 8.12–8.10 (m, 1H, 6′-H), 8.04–8.02 (m, 1H, 3′-H), 7.86–7.83 (m, 2H, 2′′,6′′-H), 7.52–7.50 (m, 2H, 3′′,5′′-H), 7.27–7.24 (m, 2H, 5′-H), 4.74 (s, 2H, 7-CH2); 13C NMR (100 MHz, DMSO-d6) δ (ppm): 187.0, 159.3, 154.2, 142.4, 135.8, 135.4, 134.5, 134.2, 129.2, 128.9, 128.8, 127.7, 127.3, 125.5, 38.7; 19F NMR (376 MHz, DMSO-d6) δ (ppm): −73.34; HRMS (ESI): 431.9779 [M + H]+.DNA binding studies
Absorbance values were recorded on a double beam Thermo Scientific's Evolution 300 spectrophotometer and fluorescence measurements were performed on a Hitachi F-4700 quantum north-west 5J20700 fortified with a xenon lamp, using a quartz cuvette of path length 1 cm. The CD measurements of the DNA-14m complex (Far-UV, 200–250 nm) were recorded on a J-815 spectrophotometer (JASCO, Japan) at room temperature using a quartz cuvette with a path length of 10 mm.
Author contributions
R. A. and S. K.; conceptualization, supervision, reviewing and editing. P. K. and M. H.; methodology, investigation, computational calculations, writing and original draft preparation.Conflicts of interest
The authors declare no competing interest.Acknowledgements
We are highly thankful to the Council of Scientific and Industrial Research (CSIR), New Delhi, India for kindly providing financial assistance for JRF & SRF to Prince Kumar (Grant 09/105(0302)/2020-EMR-I) and Mona Hooda (Grant 09/105(0236)/2016-EMR-I).References
- M. Inoue, Y. Sumii and N. Shibata, Contribution of Organofluorine Compounds to Pharmaceuticals, ACS Omega, 2020, 5, 10633–10640, DOI:10.1021/acsomega.0c00830.
- B. Jeffries, Z. Wang, J. Graton, S. D. Holland, T. Brind, R. D. R. Greenwood, J. Y. Le Questel, J. S. Scott, E. Chiarparin and B. Linclau, Reducing the Lipophilicity of Perfluoroalkyl Groups by CF2-F/CF2-Me or CF3/CH3 Exchange, J. Med. Chem., 2018, 61, 10602–10618, DOI:10.1021/acs.jmedchem.8b01222.
- R. Aggarwal, A. Bansal, I. Rozas, E. Diez-Cecilia, A. Kaur, R. Mahajan and J. Sharma, P-Toluenesulfonic acid-catalyzed solvent-free synthesis and biological evaluation of new 1-(4′,6′-dimethylpyrimidin-2′-yl)-5-amino- 4H-3-arylpyrazole derivatives, Med. Chem. Res., 2014, 23, 1454–1464, DOI:10.1007/s00044-013-0751-9.
- Q. Zhou, S. Xu and R. Zhang, Transition-metal-free, visible-light-mediated regioselective C–H trifluoromethylation of imidazo[1,2-a]pyridines, Tetrahedron Lett., 2019, 60, 734–738, DOI:10.1016/j.tetlet.2019.02.003.
- A. S. Nair, A. K. Singh, A. Kumar, S. Kumar, S. Sukumaran, V. P. Koyiparambath, L. K. Pappachen, T. M. Rangarajan, H. Kim and B. Mathew, FDA-Approved Trifluoromethyl Group-Containing Drugs: A Review of 20 Years, Processes, 2022, 10, 2054, DOI:10.3390/pr10102054.
- S. Schiesser, R. J. Cox and W. Czechtizky, The powerful symbiosis between synthetic and medicinal chemistry, Future Med. Chem., 2021, 13, 941–944, DOI:10.4155/fmc-2021-0062.
- R. Aggarwal, S. Kumar, A. Mittal, R. Sadana and V. Dutt, Synthesis, characterization, in vitro DNA photocleavage and cytotoxicity studies of 4-arylazo-1-phenyl-3-(2-thienyl)-5-hydroxy-5-trifluoromethylpyrazolines and regioisomeric 4-arylazo-1-phenyl-5(3)-(2-thienyl)-3(5)-trifluoromethylpyrazoles, J. Fluorine Chem., 2020, 236, 109573, DOI:10.1016/j.jfluchem.2020.109573.
- L. Dai, F. Qin, Y. Xie, B. Zhang, Z. Zhang, S. Liang, F. Chen, X. Huang and H. Wang, Antitumor activity and mechanisms of dual EGFR/DNA-targeting strategy for the treatment of lung cancer with EGFRL858R/T790M mutation, Bioorg. Chem., 2023, 135, 106510, DOI:10.1016/j.bioorg.2023.106510.
- S. Banerjee, E. B. Veale, C. M. Phelan, S. A. Murphy, G. M. Tocci, L. J. Gillespie, D. O. Frimannsson, J. M. Kelly and T. Gunnlaugsson, Recent advances in the development of 1,8-naphthalimide based DNA targeting binders, anticancer and fluorescent cellular imaging agents, Chem. Soc. Rev., 2013, 42, 1601–1618, 10.1039/c2cs35467e.
- A. Abdelli, S. Azzouni, R. Plais, A. Gaucher, M. L. Efrit and D. Prim, Recent advances in the chemistry of 1,2,4-triazoles: Synthesis, reactivity and biological activities, Tetrahedron Lett., 2021, 86, 153518, DOI:10.1016/j.tetlet.2021.153518.
- S. Z. Siddiqui, M. Arfan, M. A. Aziz-ur-Rehman Abbasi, S. A. A. Shah, M. Ashraf, S. Hussain, R. S. Z. Saleem, R. Rafique and K. M. Khan, Discovery of Dual Inhibitors of Acetyl and Butrylcholinesterase and Antiproliferative Activity of 1,2,4-Triazole-3-thiol: Synthesis and In Silico Molecular Study, ChemistrySelect, 2020, 5, 6430–6439, DOI:10.1002/slct.201904905.
- V. M. Patel, N. B. Patel, M. J. Chan-Bacab and G. Rivera, Synthesis, biological evaluation and molecular dynamics studies of 1,2,4-triazole clubbed Mannich bases, Comput. Biol. Chem., 2018, 76, 264–274, DOI:10.1016/j.compbiolchem.2018.07.020.
- H. A. M. El-Sherief, B. G. M. Youssif, S. N. Abbas Bukhari, A. H. Abdelazeem, M. Abdel-Aziz and H. M. Abdel-Rahman, Synthesis, anticancer activity and molecular modeling studies of 1,2,4-triazole derivatives as EGFR inhibitors, Eur. J. Med. Chem., 2018, 156, 774–789, DOI:10.1016/j.ejmech.2018.07.024.
- S. Abbas, S. Zaib, S. Ur Rahman, S. Ali, S. Hameed, M. N. Tahir, K. S. Munawar, F. Shaheen, S. M. Abbas and J. Iqbal, Carbonic Anhydrase Inhibitory Potential of 1,2,4-triazole-3-thione Derivatives of Flurbiprofen, Ibuprofen and 4-tert-butylbenzoic Hydrazide: Design, Synthesis, Characterization, Biochemical Evaluation, Molecular Docking and Dynamic Simulation Studies, Med. Chem., 2018, 15, 298–310, DOI:10.2174/1573406414666181012165156.
- S. A. Shahzad, M. Yar, Z. A. Khan, L. Shahzadi, S. A. R. Naqvi, A. Mahmood, S. Ullah, A. J. Shaikh, T. A. Sherazi, A. T. Bale, J. Kukułowicz and M. Bajda, Identification of 1,2,4-triazoles as new thymidine phosphorylase inhibitors: Future anti-tumor drugs, Bioorg. Chem., 2019, 85, 209–220, DOI:10.1016/j.bioorg.2019.01.005.
- F. J. Groelly, M. Fawkes, R. A. Dagg, A. N. Blackford and M. Tarsounas, Targeting DNA damage response pathways in cancer, Nat. Rev. Cancer, 2023, 23, 78–94, DOI:10.1038/s41568-022-00535-5.
- N. Rezki, F. F. Al-Blewi, S. A. Al-Sodies, A. K. Alnuzha, M. Messali, I. Ali and M. R. Aouad, Synthesis, Characterization, DNA Binding, Anticancer, and Molecular Docking Studies of Novel Imidazolium-Based Ionic Liquids with Fluorinated Phenylacetamide Tethers, ACS Omega, 2020, 5, 4807–4815, DOI:10.1021/acsomega.9b03468.
- V. Sharma, M. Gupta, P. Kumar and A. Sharma, A Comprehensive Review on Fused Heterocyclic as DNA Intercalators: Promising Anticancer Agents, Curr. Pharm. Des., 2020, 27, 15–42, DOI:10.2174/1381612826666201118113311.
- F. Shiri, S. Hadidi, M. Rahimi-Nasrabadi, F. Ahmadi, M. R. Ganjali and H. Ehrlich, Synthesis, characterization and DNA binding studies of a new ibuprofen–platinum(II) complex, J. Biomol. Struct. Dyn., 2020, 38, 1119–1129, DOI:10.1080/07391102.2019.1597769.
- S. Khan, R. Ahmad and I. Naseem, Elucidating the interaction of aminophylline with calf thymus DNA using multispectroscopic and molecular docking approach, J. Biomol. Struct. Dyn., 2021, 39, 970–976, DOI:10.1080/07391102.2020.1722240.
- M. C. Costas-Lago, N. Vila, A. Rahman, P. Besada, I. Rozas, J. Brea, M. I. Loza, E. González-Romero and C. Terán, Novel Pyridazin-3(2H)-one-Based Guanidine Derivatives as Potential DNA Minor Groove Binders with Anticancer Activity, ACS Med. Chem. Lett., 2022, 13, 463–469, DOI:10.1021/acsmedchemlett.1c00633.
- R. Aggarwal, P. Kumar, N. Jain, M. Hooda, S. Kumar, R. Sadana, R. Lwanga, A. Guzman, H. Chugh and R. Chandra, Synthesis and In vitro-In silico Evaluation of Thiazolo-triazole Hybrids as Anticancer Candidates, ChemistrySelect, 2023, 8, e202301368, DOI:10.1002/slct.202301368.
- R. Aggarwal, M. Hooda, P. Kumar and M. C. Torralba, Visible-light-mediated regioselective synthesis of novel thiazolo[3,2-b][1,2,4]triazoles: Advantageous synthetic application of aqueous conditions, Org. Biomol. Chem., 2022, 20, 584–595, 10.1039/d1ob02194j.
- H. El-Sherif, A. Mahmoud, A. Sarhan, Z. Hozien and O. Habib, One pot synthesis of novel thiazolo[3,2-b][1,2,4]triazoles: A useful synthetic application of the acidified acetic acid method, J. Sulfur Chem., 2006, 27, 65–85, DOI:10.1080/17415990500520908.
- G. Ramachandraiah and K. K. Reddy, Synthesis of 2,5-diarylthiazolo[3,2-b]-s-triazoles, Indian J. Chem. - Sect. B Org. Med. Chem., 1985, 24B, 808–810 CAS.
- I. Pravst, M. Zupan and S. Stavber, Solvent-free bromination of 1,3-diketones and β-keto esters with NBS, Green Chem., 2006, 8, 1001–1005, 10.1039/b608446j.
- I. Pravst, M. Zupan and S. Stavber, Halogenation of ketones with N-halosuccinimides under solvent-free reaction conditions, Tetrahedron, 2008, 64, 5191–5199, DOI:10.1016/j.tet.2008.03.048.
- T. A. Wani, N. Alsaif, A. H. Bakheit, S. Zargar, A. A. Al-Mehizia and A. A. Khan, Interaction of an abiraterone with calf thymus DNA: Investigation with spectroscopic technique and modelling studies, Bioorg. Chem., 2020, 100, 103957, DOI:10.1016/j.bioorg.2020.103957.
- R. Arif, P. S. Nayab, Akrema, M. Abid, U. Yadava and Rahisuddin, Investigation of DNA binding and molecular docking propensity of phthalimide derivatives: in vitro antibacterial and antioxidant assay, J. Anal. Sci. Technol., 2019, 10, 19, DOI:10.1186/s40543-019-0177-1.
- M. H. El-Wakil, A. F. El-Yazbi, H. M. A. Ashour, M. A. Khalil, K. A. Ismail and I. M. Labouta, Discovery of a novel DNA binding agent via design and synthesis of new thiazole hybrids and fused 1,2,4-triazines as potential antitumor agents: Computational, spectrometric and in silico studies, Bioorg. Chem., 2019, 90, 103089, DOI:10.1016/j.bioorg.2019.103089.
- R. Aggarwal, M. Hooda, P. Kumar, N. Jain, G. P. Dubey, H. Chugh and R. Chandra, Visible-Light-Prompted Synthesis and Binding Studies of 5,6-Dihydroimidazo[2,1- b]thiazoles with BSA and DNA Using Biophysical and Computational Methods, J. Org. Chem., 2022, 87, 3952–3966, DOI:10.1021/acs.joc.1c02471.
- R. Aggarwal, N. Jain, S. Sharma, P. Kumar, G. P. Dubey, H. Chugh and R. Chandra, Visible-light driven regioselective synthesis, characterization and binding studies of 2-aroyl-3-methyl-6,7-dihydro-5H-thiazolo[3,2-a]pyrimidines with DNA and BSA using biophysical and computational techniques, Sci. Rep., 2021, 11, 22135, DOI:10.1038/s41598-021-01037-4.
- R. Aggarwal, M. Hooda, P. Kumar, S. Kumar, S. Singh and R. Chandra, An expeditious on-water regioselective synthesis of novel arylidene-hydrazinyl-thiazoles as DNA targeting agents, Bioorg. Chem., 2023, 136, 106524, DOI:10.1016/j.bioorg.2023.106524.
- Z. Asadi, Z. Mandegani, M. Asadi, A. H. Pakiari, M. Salarhaji, M. Manassir, H. R. Karbalaei-Heidari, B. Rastegari and M. Sedaghat, Substituted effect on some water-soluble Mn(II) salen complexes: DNA binding, cytotoxicity, molecular docking, DFT studies and theoretical IR & UV studies, Spectrochim. Acta, Part A, 2019, 206, 278–294, DOI:10.1016/j.saa.2018.08.020.
- R. Mehandi, R. Arif, M. Rana, S. Ahmedi, R. Sultana, M. S. Khan, M. Maseet, M. Khanuja, N. Manzoor, Rahisuddin and N. Nishat, Synthesis, characterization, DFT calculation, antifungal, antioxidant, CT-DNA/pBR322 DNA interaction and molecular docking studies of heterocyclic analogs, J. Mol. Struct., 2021, 1245, 131248, DOI:10.1016/j.molstruc.2021.131248.
- A. Kellett, Z. Molphy, C. Slator, V. McKee and N. P. Farrell, Molecular methods for assessment of non-covalent metallodrug-DNA interactions, Chem. Soc. Rev., 2019, 48, 971–988, 10.1039/c8cs00157j.
- M. Dareini, Z. Amiri Tehranizadeh, N. Marjani, R. Taheri, S. Aslani-Firoozabadi, A. Talebi, N. NayebZadeh Eidgahi, M. R. Saberi and J. Chamani, A novel view of the separate and simultaneous binding effects of docetaxel and anastrozole with calf thymus DNA: Experimental and in silico approaches, Spectrochim. Acta, Part A, 2020, 228, 117528, DOI:10.1016/j.saa.2019.117528.
- R. Aggarwal, M. Hooda, N. Jain, D. Sanz, R. M. Claramunt, B. Twamley and I. Rozas, An efficient, one-pot, regioselective synthesis of 2-aryl/hetaryl-4-methyl-5-acylthiazoles under solvent-free conditions, J. Sulfur Chem., 2022, 43, 12–21, DOI:10.1080/17415993.2021.1975119.
- R. Aggarwal, S. Sharma, M. Hooda, D. Sanz, R. M. Claramunt, B. Twamley and I. Rozas, Visible-light mediated regioselective approach towards synthesis of 7-aroyl-6-methyl-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazines, Tetrahedron, 2019, 75, 130728, DOI:10.1016/j.tet.2019.130728.
- R. Aggarwal and M. Hooda, Visible-light promoted serendipitous synthesis of 3,5-diaryl-1,2,4-thiadiazoles via oxidative dimerization of thiobenzamides, J. Sulfur Chem., 2022, 43, 242–251, DOI:10.1080/17415993.2021.2005064.
- R. Aggarwal, G. Singh, D. Sanz, R. M. Claramunt, M. C. Torralba and M. R. Torres, NBS mediated one-pot regioselective synthesis of 2,3-disubstituted imidazo[1,2-a]pyridines and their unambiguous characterization through 2D NMR and X-ray crystallography, Tetrahedron, 2016, 72, 3832–3838, DOI:10.1016/j.tet.2016.04.072.
Footnote |
| † Electronic supplementary information (ESI) available: The Supporting Information consists of additional experimental data, (1H, 13C, HMBC, HSQC NMR, and HRMS spectra) for final compounds. See DOI: https://doi.org/10.1039/d4ra00083h |
| This journal is © The Royal Society of Chemistry 2024 |