Open Access Article
Open Access ArticleMaking wood inspection easier: FTIR spectroscopy and machine learning for Brazilian native commercial wood species identification
Everton Jesusa,
Thiago Francaa,
Camila Calvania,
Miller Lacerdaa,
Daniel Gonçalves bc,
Samuel L. Oliveiraa,
Bruno Marangoni
bc,
Samuel L. Oliveiraa,
Bruno Marangoni a and
Cicero Cena
a and
Cicero Cena *a
*a
aUFMS – Universidade Federal de Mato Grosso do Sul, Optics and Photonic Lab (SISFOTON-UFMS), Campo Grande, MS, Brazil. E-mail: cicero.cena@ufms.br
bUFGD – Universidade Federal da Grande Dourados, Dourados, MS, Brazil
cUEMS – Universidade Estadual de Mato Grosso do Sul, Dourados, MS, Brazil
First published on 1st March 2024
Abstract
The molecular structure of wood is mainly based on cellulose, lignin, and hemicellulose. However, low concentrations of lipids, phenolic compounds, terpenoids, fatty acids, resin acids, and waxes can also be found. In general, their color, smell, texture, quantity, and distribution of pores are used in human sensory analysis to identify native wood species, which may lead to erroneous classification, impairing quality control and inspection of commercialized wood. This study developed a fast and accurate method to discriminate Brazilian native commercial wood species using Fourier Transform Infrared Spectroscopy (FTIR) and machine learning algorithms. It not only solves the limitations of traditional methods but also goes beyond as it allows fast analyses to be obtained at low cost and high accuracy. In this work, we provide the identification of five Brazilian native wood species: Angelim-pedra (Hymenolobium petraeum Ducke), Cambara (Gochnatia polymorpha), Cedrinho (Erisma uncinatum), Champagne (Dipteryx odorata), and Peroba do Norte (Goupia glabra Aubl). The results showed the great potential of FTIR and multivariate analysis for wood sample classification; here, the Linear SVM differentiated the five wood species with an accuracy of 98%. The developed method allows industries, laboratories, companies, and control bodies to identify the nature of the wood product after being extracted and semi-manufactured.
1. Introduction
Wood is a very abundant, renewable, and environmentally friendly material with numerous practical applications. However, accurately identifying wood species is crucial to assessing their value and proper usage. The laboratory identification of wood species is considered complex and difficult to implement on a large scale because it usually involves several time-consuming steps – in which the wood components are isolated or degraded into monomeric fragments to quantify the total fiber content of the plants and analyze the chemical composition of the wood – that can take days to be complete.1–3 This is one of the reasons why the identification of wood species is based on human sensory analysis; the color, smell, texture, quantity, and distribution of pores are some of the factors used to identify each wood species. The absence of an accurate method for classification and traceability, regardless of human training or sensory analysis, leads to erroneous wood classification, impairing quality control and inspection of commercialized wood. The human sensory analysis provides only 65% accuracy, which has motivated the development of studies using artificial intelligence methods to avoid errors and increase classification accuracy.4,5The wood composition is divided into 50% cellulose, 20–25% hemicellulose, 20–30% lignin, and up to 15% other constituents. Cellulose (C6H10O5)n is a linear polysaccharide that constitutes the structural part of the plant cell wall and determines the plant structure, forming the filaments that reinforce the walls of the longitudinal fibers, presenting a higher molecular weight than other wood components.6,7 Lignin (C9H10O2, C10H12O3, C11H14O4) is a random condensed polymer with many aromatic groups and free of carbohydrates (sugars) in its polymeric structure, which has a hydroxyl group (–OH) attached to an aromatic ring.8 Lignin is also responsible for the structural rigidity of lignocellulosic biomass, which generally covers hemicellulose and cellulose.9
Hemicellulose (C6H10O5)n is a group of polysaccharides composed of 5- and 6-carbon ring sugars with an irregular chain backbone with side groups, substituent groups, and sugars along it.10 With hydroxyl groups connected to its main chain and exposure due to its amorphous condition, hemicellulose becomes more susceptible to chemical degradation reactions and is less tolerant to the action of heat.6 The –OH groups and sugar branches of hemicellulose also play an essential role in binding and stabilizing microfibrils, which are highly polar and linked via hydrogen bonds.11
Hemicellulose and Lignin surround the cellulose macromolecules by binding them and providing stiffness and compressive strength to fiber walls.12–15 The wood structure also presents small amounts (0.2 to 1%) of mineral salts, constituting the nourishment of living tissues. Plant species also have resins, oils, and waxes, which are deposited in the cell cavities, producing the characteristic color and smell of the species.7 Wood contains up to 15% of extractives, including lipids, phenolic compounds, terpenoids, fatty acids, resin acids, and waxes.11,14,15
The diverse molecular composition found in wood presents an exciting prospect for creating a rapid and precise technique to detect and categorize wood samples using molecular spectroscopy. In the literature, we can find a few examples that support our hypothesis: (i) three fast-growing tree species from the Amazonian forest (Pashco, Capirona, and Bolaina) were successfully discriminated by using Fourier transform infrared spectroscopy (FTIR) associated with partial least square discrimination analysis (PLS-DA), by using cellulose, lignin, and hemicellulose peaks with accuracy above 91%;16 (ii) FTIR and PLS-DA were also able to differentiate between compression and non-compression wood by analyzing lignin bands from Pinus radiata species;17 (iii) the growing location of wood samples was identified by FTIR spectra – from 24 wood samples (16 hardwood and 8 softwood) – and hierarchical clustering analysis (HCA), followed by principal component analysis (PCA) and linear discriminant analysis (LDA);18 (iv) two Pine woods species and growing location was also determined by using ATR-FTIR (attenuated total reflectance – Fourier transform infrared spectroscopy) and PCA with DA in the lignin and polysaccharide bands range.19 Recently, our group demonstrated the potential use of FTIR associated with PCA and Support Vector Machine (SVM) to classify Eucalyptus species.20
Here, we investigated the potential use of Fourier-transformed infrared spectroscopy (FTIR) and multivariate analysis for accurate identification of five Brazilian native wood species most used commercially in the country: Angelim-pedra (Hymenolobium petraeum Ducke); Cambara (Gochnatia polymorpha); Cedrinho (Erisma uncinatum); Champagne (Dipteryx odorata); Peroba do Norte (Goupia glabra Aubl). Besides the subtle difference between the FTIR spectra of each species, the proper choice of the spectral range showed promising results in building a prediction model based on relatively simple algorithms for sample classification. The developed method can allow industries, laboratories, companies, and control bodies to identify the nature of the product after being extracted and semi-manufactured.
2. Materials and methods
2.1. Sample and data acquisition
Commercial wood samples were obtained from the local wood industry in the midwest region of Brazil (Campo Grande city, Mato Grosso do Sul state). The five wood species analyzed were, Fig. 1: (i) Hymenolobium petraeum Ducke (Angelim-pedra), (ii) Gochnatia polymorpha (Cambará); (iii) Erisma uncinatum (Cedrinho); (iv) Dipteryx odorata (Champagne); and (v) Goupia glabra Aubl (Peroba do Norte).A total of 52 heartwood sawdust samples per species were obtained from 26 different batches. The powder granulometry was uniformized using an analytical sieve with 45 mesh (355 μm) and submitted to natural drying at room temperature (around 30 °C) for several days before the measurements. The sifted sawdust samples' infrared spectrum was obtained in a Fourier transform infrared spectrophotometer (FTIR) – PerkinElmer spectrum 100 model – with an attenuated total reflectance (ATR) accessory. After background collection in air, the sawdust was directly deposited and carefully compressed against the ATR window (ZnSe crystal), and the FTIR spectra were obtained in the 4000 to 600 cm−1 range, with a 4 cm−1 resolution and 10 scans. The average spectra were collected by measuring each sample in duplicate and then used for analysis.
2.2. Data analysis and sample classification
The data analysis was performed using algorithms previously implemented in Python using the Scikit-learn package. First, averaged FTIR raw spectra were subjected to a standard normal variate (SNV) preprocessing step to remove baseline offset and rescale the absorbance values, which is necessary to prevent random experimental variation from interfering with the result.21An unsupervised method initially analyzed the data potential for wood species clustering and discrimination. FTIR-SNV spectra were submitted to the principal component analysis (PCA) responsible for data transformation.22 Here, the data matrix (intensity versus wavenumber) was transformed into a new matrix set by dimension reduction. The data matrix was converted to a score matrix and a loading matrix. The transformation converts correlated variables into uncorrelated variables (principal components), where the first PC has the highest variance, the second PC has the second highest variance, and so on. In this way, PCs can highlight variations and trends in the entire data set, providing a simple way to visualize the data set through the eigenvectors called scores (scores) and the eigenvalues called their weights (loadings). The first analysis used the entire spectral range from 4000 to 600 cm−1. We selected two other spectral ranges, 3000 to 2700 cm−1 and 2000 to 700 cm−1, to improve the clustering and group separation as previously demonstrated in the literature.23,24
The PCA data obtained from the three spectral ranges were used as input data for the supervised analysis step using machine learning algorithms for sample classification. The DA (Discriminant Analysis) algorithm uses the training sample set to determine a boundary (Linear or Quadratic) between different classes in a particular distribution. The SVM (Support Vector Machine) algorithm performs a spatial separation of the training sample using hyperplanes (linear, quadratic, cubic, fine, medium, and coarse). The main difference between LDA and SVM is that LDA assumes a normal probability distribution across samples, the same covariance matrix for all classes, whereas no assumptions are required for SVM. The KNN (Kernel-Nearest Neighbors) algorithm is based on the spatial distribution of points and classifies the validation sample based on the distance to the nearest neighbor's class; it uses the “k” nearest neighbors for classification (fine = 1, medium = 10, and coarse = 100) or using weights for the distances so that the closest has more weight.25,26
Two distinct datasets were constructed to conduct the multivariate analyses. The first was a training set comprising 75% of the total samples. It was used for algorithm training and parameter selection to improve the classification based on accuracy. The accuracy was calculated by leave-one-out cross-validation (LOOCV). In LOOCV, one sample is taken from the data set, and the others are used to build the prediction model. Then, the sample data draw is used to assess the quality of the model. The procedure is repeated until all sample data have been tested. Model quality is measured by accuracy, and the average percentage of correct answers remains in each test.27 After this step, the remaining 25% of the samples, referred to as test sets, were utilized to certify the robustness and reproducibility of the model.
3. Results and discussion
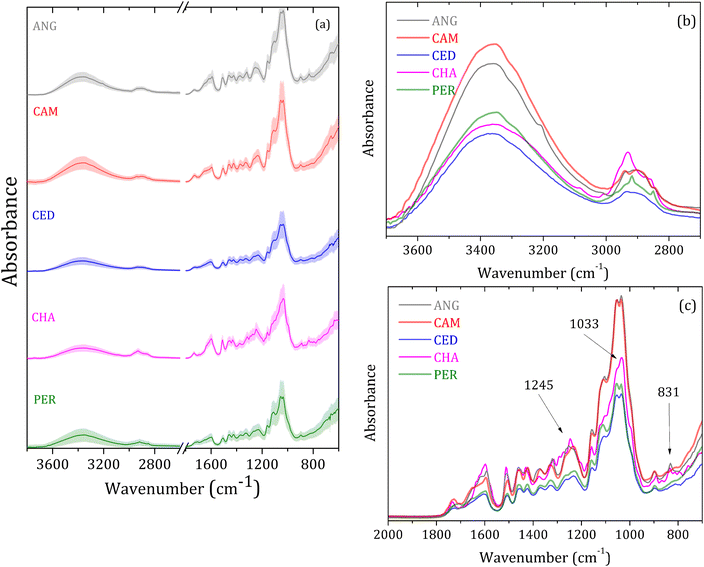
Fig. 2(a) shows the wood species averaged FTIR-SNV spectra in the 4000 to 700 cm−1 range, with respective standard deviations for the groups analyzed: angelim-pedra (ANG), cambará (CAM), cedrinho (CED), Champagne (CHA) e Peroba (PER). The data show a slight deviation in good accordance between the spectra of the same group. The most prominent vibrational bands in the 2000 to 700 cm−1 range are due to the cellulose, hemicellulose, and lignin molecular vibrational modes.28–31 In turn, the samples' wood constituents in small concentrations caused alterations in the vibrational bands, such as a small shoulder, shift, area, or width.The wide vibrational band centered around 3400 cm−1 is assigned to O–H stretching modes, while the small band at around 2900 cm−1 is linked to C–H elongation groups from cellulose.28–31 Fig. 2(b) highlights a distinct variation in band intensity from O–H modes. Additionally, noticeable differences are observed within the wide bands spanning the 3000–2800 cm−1 range, with some exhibiting multiple vibrational peaks.
Fig. 2(c) shows the analyzed groups' spectral differences in the 2000–700 cm−1 range. The 1731 cm−1 band – assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching from carbonyl and acetyl groups from hemicellulose,28,30–32 is more intense for CHA and CAM groups and slightly shifted for higher energy. The broad bands centered at around 1600 cm−1 are very similar for PER and CED, intense for CAM and ANG, and narrow for CHA. The bands in the 1700 to 1450 cm−1 range are assigned to C
O stretching from carbonyl and acetyl groups from hemicellulose,28,30–32 is more intense for CHA and CAM groups and slightly shifted for higher energy. The broad bands centered at around 1600 cm−1 are very similar for PER and CED, intense for CAM and ANG, and narrow for CHA. The bands in the 1700 to 1450 cm−1 range are assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C–O stretching and bending modes in lignin and cellulose.1,28,30,31
C and C–O stretching and bending modes in lignin and cellulose.1,28,30,31
Bands at 1461, 1427, 1371, and 1103 cm−1 are characteristic of C–H vibrations, C–O deformation, bending, or elongation in lignin and carbohydrate.31 The 1427 cm−1 band is associated with aromatic vibrations related to C–H in-plane deformation cellulose. The bands at 1731, 1371, 1245, 1158, 1103, and 1033 cm−1 are attributed to the deformation of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–H, C–O–C, C–O, or stretching vibrations in carbohydrate groups.31 The bands at around 892 and 833 cm−1 are assigned to amorphous material in the cellulose region.28,30
O, C–H, C–O–C, C–O, or stretching vibrations in carbohydrate groups.31 The bands at around 892 and 833 cm−1 are assigned to amorphous material in the cellulose region.28,30
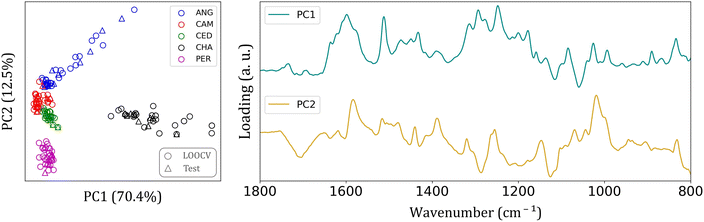
Since many differences were found among the FTIR spectra of each group, it is reasonable to expect an easier clustering formation. Fig. 3 shows the principal component analysis (PCA) results for FTIR-SNV spectra from the five wood species analyzed in the 1800–800 cm−1 interval.
This spectral range provided a more distinct separation among wood species. Fig. 3 also depicts the loading plot of the first and second PCs, which account for 82.9% of the total data variance. The predominant elements within this spectral range are the major constituents of wood (cellulose, hemicellulose, and lignin). This range may also include extractives such as lipids, phenolic compounds, terpenoids, fatty acids, resin acids, carbohydrates, and waxes.14,30–32
The cellulose, lignin, hemicellulose molecules, and small amounts of wood extractives can cause intra and interspecific differences among batches, thereby improving group separation. Each wood species possesses varying levels of extractives, which are influenced by factors such as the specific wood species, the wood age, and the position of the wood within the tree.14 In this study, we analyzed species from the same region, all with a similar age of approximately 25 years. In the loading graph (Fig. 3), a peak can be observed at 1597 cm−1, corresponding to the aromatic skeletal vibration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond in the benzene ring, a characteristic feature of lignin. Additionally, we observe other peaks in the loading graph associated with aromatic skeletal vibrations of C–H bonds in the in-plane deformation of cellulose.
C bond in the benzene ring, a characteristic feature of lignin. Additionally, we observe other peaks in the loading graph associated with aromatic skeletal vibrations of C–H bonds in the in-plane deformation of cellulose.
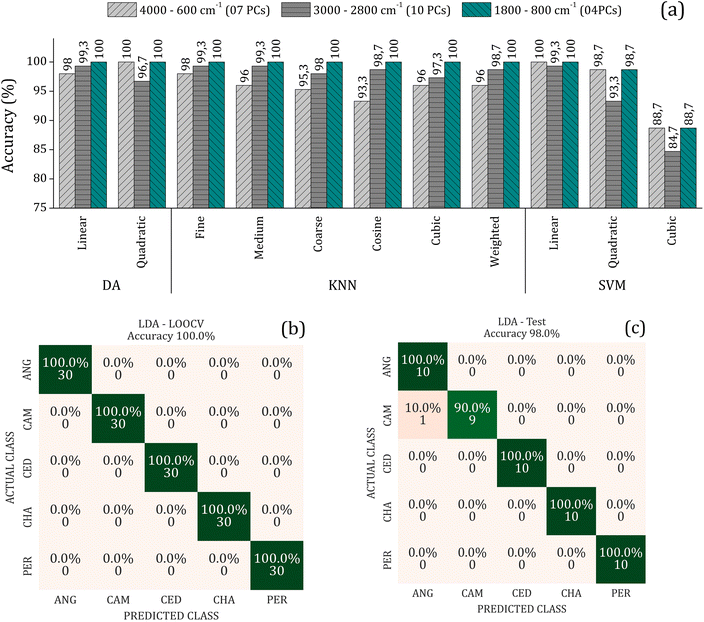
To improve the overall accuracy in sample classification, the Principal Components (PCs) were evaluated by DA, KNN, and SVM models in the leave-one-out cross-validation (LOOCV) using the training set consisting of 75% of the samples. The number of PCs used for each algorithm in the three spectral ranges (4000–600, 3000–2800, and 1800–800 cm−1) were tuned to avoid underfitting and overfitting. Detailed results of these analyses can be found in the supplementary material. Fig. 4(a) presents the overall accuracy for the classification methods utilizing the LOOCV (Leave-One-Out Cross-Validation) and the respective number of PCs used to achieve the best accuracy for each spectral range. The results indicate that the LDA, QDA, Fine KNN, and SVM-linear model has higher accuracy (é maior que ou igual a 98% in the internal validation, regardless of the spectral range).
On the other hand, external validation tests were performed using a sample set (25% of samples) dedicated solely to this purpose. Here, we use the LDA since it uses a minor amount of input data to reach high accuracy in the LOOCV. It is considered a simple and intuitive method for data classification and presents good resistance against overfitting. Fig. 4(b) and (c) shows the confusion matrix depicting the results of the LDA method using 4 PCs in the range of 1800–800 cm−1, with an accuracy of 100% in training (LOOCV) and 98.0% in the external validation achieved. This indicates that the model is reliable and can be generalized with a low possibility of overfitting or underfitting. The confusion matrix is one of the most used approaches to analyze validation results. It provides information about the success rate and the percentages of true negatives (VN), true positives (VP), false positives (FP), and false negatives (FN) predictions. These metrics are essential for assessing the accuracy, sensitivity (actual class rate), and specificity (predicted class rate) of the predictions, allowing the evaluation of the robustness of the protocol.33 The number of wood samples on the main diagonal of the confusion matrix represents the correctly classified ones.
It is worth emphasizing that FTIR spectroscopy was selected as the analytical technique due to its simplicity and ability to perform chemical analysis in a small sample size in a short time. On the other hand, the use of multivariate statistical analysis is highly justified due to its ability to simultaneously study multiple variables (characteristics), such as wood species, and deal with a large data set. By employing multivariate analysis, interrelationships among numerous variables can be analyzed, and the contribution of each variable can be assessed to identify those that can be used in classification protocols. Through this comprehensive analysis, we gain a deeper understanding of the complex data relationships providing valuable insights for further interpretation and decision-making.
This work highlights that the FTIR technique with multivariate analysis and machine learning algorithms could be used for classifying wood species using a large dataset. This approach offers several advantages compared to conventional methods. The combination of FTIR with multivariate analysis enables the examination of multiple variables simultaneously, enhancing the efficiency and completeness of the classification process. Moreover, the application of machine learning algorithms further improves the accuracy and speed of classification. Notably, the reported approach requires smaller sample sizes and shorter analysis times, with FTIR measurements typically taking around 10 min. Furthermore, the findings have practical implications for wood inspection in companies, the wood industry, and commerce. Utilizing the FTIR technique allied with multivariate analysis and machine learning algorithms made it possible to determine the wood species quickly and reliably.
4. Conclusion
Using FTIR with PCA and machine learning classification methods demonstrates its remarkable potential for differentiating wood species. The most favorable outcome was achieved by analyzing the spectral range between 1800–800 cm−1, resulting in improved separation within the studied wood species using the LDA classifier. The LDA classifier exhibited 100% accuracy in internal validation and 98.0% accuracy in external validation. The result, coupled with the straightforward sample preparation, highlights the significant potential of FTIR combined with multivariate analysis for wood species classification. This approach offers a considerable advantage over traditional methods, which typically require 3 to 4 days, while FTIR provides results within 15 min once the samples are ready for characterization.Author contributions
Jesus: data acquisition, data analysis; Franca: data analysis and validation; Lacerda and Calvani: writing and revision; Marangoni: data analysis; Jesus and Goncalves: sample acquisition and writing; Oliveira: writing and revision; Cena: conceptualization, methodology, supervision, writing – review & editing.Conflicts of interest
The authors declare the following financial interests/personal relationships, which may be considered as potential competing interests: Thiago Franca reports that Coordination of Higher Education Personnel Improvement provided financial support.Acknowledgements
Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), code 001. Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPQ), code 403651/2020–5; 302525/2022–0; 440214/2021–1. Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul (FUNDECT), code 007/2019; 360/2022.References
- H. Chen, C. Ferrari, M. Angiuli, J. Yao, C. Raspi and E. Bramanti, Qualitative and Quantitative Analysis of wood samples by Fourier transform infrared spectroscopy and multivariate Analysis, Carbohydr. Polym., 2010, 82, 772–778 CrossRef CAS.
- P. J. Van Soest, Use of detergents in the Analysis of fibrous feeds. II. A rapid method for the determination of the composition of fiber and lignin, J. Assoc. Off. Anal. Chem, 1963, 46, 829–835 CrossRef CAS.
- P. J. Van Soest and R. H. Wine, Use of detergents in the Analysis of fibrous feeds. IV. Determination of plant cell-wall constituents, J. Assoc. Off. Anal. Chem, 1967, 50, 50–55 CrossRef CAS.
- C. Affonso, et al., Deep learning for biological image classification, Expert Systems with Applications, 2017, 85, 114–122 CrossRef.
- N. R. Silva, et al., Automated classification of wood transverse cross-section micro-imagery from 77 commercial Central-African timber species, Ann. For. Sci., 2017, 30 CrossRef.
- M. J. John and S. Thomas, Biofibres and biocomposites, Carbohydr. Polym., 2008, 71, 343–364 CrossRef CAS.
- W. Pfeil and M. Pfeil, Estruturas de Madeira. 6ed, Rio de Janeiro. LTC., 2003 Search PubMed.
- D. Watikins, M. D. Nuruddin, H. Mahesh, A. Tcherbi-Narteh and S. Jeelani, Extraction and characterization of lignina from diferent biomass resources, J. Mater. Res. Technol., 2015, 4, 26–32 CrossRef.
- C. H. Zhou, X. Xia, C. X. Lin, D. S. Tong and J. Beltramini, Catalytic conversion of lignocellulosic biomass to fine chemicals and fuels, Chem. Soc. Rev., 2011, 40, 5588–5617 RSC.
- P. Philipp and M. L. O. D’almeida, Celulose e Papel. Volume I. Tecnologia de Fabricação da Pasta Celulósica, Instituto de Pesquisas Tecnológicas do Estado de São Paulo – Centro Técnico em celulose e papel, São Paulo,2a edição, 1988 Search PubMed.
- N. N. Deshavath, V. D. Veeranki and V. V. Goud, Lignocellulosic feedstocks for the production of bioethanol availability, structure, and composition, J. Sustain. Bioenergy Syst., 2019, 1–19 Search PubMed.
- L. P. Ramos, The chemistry involved in the steam treatment of lignocellulosic materials, Quim. Nova, 2003, 26(6), 863–871 CrossRef CAS.
- F. F. Wangaard, Wood: its Structure and Properties. The Pennsylvania State University, USA, 1979 Search PubMed.
- A. N. Shebani, A. J. van Reenen, A. J. and M. Meincken, The effect wood extractives on the thermal stability of different wood-LLDPE composites, Thermochim. Acta, 2008, 481, 52–56 CrossRef.
- C. M. Popescu, G. Singurel, M.-C. Popescu, C. Vasile, D. S. Argyropoulos and S. Willfor, Vibrational spectroscopy and X-ray diffraction methods to establish the differences between hardwood and softwood, Carbohydr. Polym., 2009, 77, 851–857 CrossRef CAS.
- J. R. Astete, J. Melo and J. J. Davalos, Classification of Amazonian fast-growing tree species and wood chemical determination by FTIR and multivariate analysis (PLS-DA, PLS), Sci. Rep., 2023, 13, 7827 CrossRef PubMed.
- J. P. Mclean, G. Jin, M. Brennan, M. K. Nieuwoudt and P. J. Harris, Using NIR and ATR-FTIR Spectroscopy to Rapidly Detect Compression Wood in Pinus Radiata, Can. J. For. Res., 2014, 820–830 CrossRef CAS.
- V. Sharma, J. Yadav, R. Kumar, D. Tesarova, A. Ekielski and P. K. Mishra, On the rapid and non-destructive approach for wood identification using ATR-FTIR spectroscopy and chemometric methods, Vib. Spectrosc., 2020, 110, 103097 CrossRef CAS.
- M. Traore, J. Kaal and A. M. Cortizas, Differentiation between pine woods according to species and growing location using FTIR-ATR, Wood Sci. Technol., 2018, 52, 487–504 CrossRef CAS PubMed.
- M. Lacerda, T. Franca, C. Calvani, B. Marangoni, P. Teodoro, C. N. S. Campos, F. H. R. Baio, G. B. Azevedo and C. Cena, A simple method for Eucalyptus species discrimination: FTIR spectroscopy and machine learning, Results Chem., 2024, 7, 101233 CrossRef CAS.
- M. Zeaiter and D. Rutledge, Preprocessing Methods. Comprehensive Chemometrics – Chemical and Biochemical Data Analysis, pp. 121–231, 2009 Search PubMed.
- S. Wold, K. Esbensen and P. Geladi, Principal component analysis, Chemom. Intell. Lab. Syst., 1987, 2(i. 1–3), 37–52 CrossRef CAS.
- E. A. Casaril, G. C. Santos, B. S. Marangoni, M. S. Lima, L. H. C. Andrade, W. S. Fernandes, O. M. J. Infran, N. O. Alves, D. G. L. M. Borges, C. Cena and A. G. Oliveira, Intraspecific differentiation of sandflies specimens by optical spectroscopy and multivariate Analysis, J. Biophot., 2020, e202000412 Search PubMed.
- C. I. Olveira, T. Franca, G. Nicolodelli, P. C. Morais, B. Marangoni, G. Bracchetta, D. M. B. P. Milori, Z. C. Alves and C. Cena, Fast and Accurate discrimination of Brachiaria brizantha (A.Rich.) staff seeds by molecular spectroscopy and machine learning, Agric. Sci. Technol., 2021, 443–448 CrossRef.
- T. G. Rios, G. Larios, B. Marangoni, S. L. Oliveira, C. Cena and C. A. D. Ramos, FTIR spectroscopy with machine learning: A new approach to animal DNA polymorphism screening, Spectrochim. Acta - A: Mol. Biomol. Spectrosc., 2021, 261, 120036 CrossRef PubMed.
- T. Franca, D. Goncalves and C. Cena, ATR-FTIR spectroscopy combined with machine learning to classify PVA/PVP blends in low concentrations, Vib. Spectrosc., 2022, 120, 103378 CrossRef CAS.
- T. Wong, Performance evaluation of classification algorithms by k-fold and leave-one-out cross-validation, Pattern Recognit., 2015, 48(9), 2839–2846 CrossRef.
- K. K. Pandey and A. J. Pitman, FTIR studies of the changes in wood chemistry following decay by brown-rot and white-rot fungi, Int. Biodeterior. Biodegrad., 2003, 52, 151–160 CrossRef CAS.
- G. Müller, C. Schöpper, H. Vos, A. Kharazipour and A. Polle, FTIR-ATR spectroscopic analysis of changes in wood properties during particle- and fibreboard production of hard- and softwood trees, Bioresources, 2009, 4, 49–71 Search PubMed.
- M.-C. Popescu, C. M. Popescu, G. Lisa and Y. Sakata, Evaluation of morphological and chemical aspects of different wood species by spectroscopy and thermal methods, J. Mol. Struct., 2011, 988, 65–72 CrossRef CAS.
- M. Poletto, A. J. Zattera and R. M. C. Santana, Structural Differences Between Wood Species: Evidence from Chemical Composition, FTIR Spectroscopy, and Thermogravimetric Analysis, J. Appl. Polym. Sci., 2012, 126, E336–E343 CrossRef.
- M. Schwanninger, J. C. Rodrigues, H. Pereira and B. Hinterstoisser, Effects of short-time vibratory ball milling on the shape of FT-IR spectra of wood and cellulose, Vib. Spectrosc., 2004, 36, 23–40 CrossRef CAS.
- E. C. A. Brito, Paracoccidioidomycosis screening diagnosis by FTIR spectroscopy and multivariate analysis, Photodiagn. Photodyn. Ther., 2022, 39, 102921 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |